Abstract
Bacterial biofilm has been shown to play a role in delaying wound healing of chronic wounds, a major medical problem that results in significant healthcare burden. A reproducible animal model could be very valuable for studying the mechanism and management of chronic wounds. Our previous work demonstrated that Pseudomonas aeruginosa (PAO1) biofilmchallenge on wounds in diabetic (db/db) mice significantly delayed wound healing. In this wound time course study, we further characterize the bacterial burden, delayed wound healing and certain aspects of the host inflammatory response in the PAO1 biofilm-challenged db/db mouse model. PAO1 biofilms were transferred onto 2 day old wounds created on the dorsal surface of db/db mice. Control wounds without biofilm-challenge healed by 4 weeks, consistent with previous studies; none of the biofilm-challenged wounds healed by 4 weeks; 64% of the biofilm-challenged wounds healed by 6 weeks; and all of the biofilm-challenged wounds healed by 8 weeks. During the wound healing process, P. aeruginosa were gradually cleared from the wounds while the presence of S. aureus (part of the normal mouse skin flora) increased. Scabs from all unhealed wounds contained 107 P. aeruginosa, which was 100 fold higher than the counts isolated from wound beds (i.e. 99% of the P. aeruginosa was in the scab). Histology and genetic analysis showed proliferative epidermis, deficient vascularization and increased inflammatory cytokines. Hypoxia inducible factor (HIF) expression increased 3 fold in 4 week wounds. In summary, our study demonstrates that biofilm-challenged wounds typically heal in approximately 6 weeks, at least 2 weeks longer than non biofilm-challenged normal wounds. These data suggest that this delayed wound healing model enables the in vivo study of bacterial biofilm responses to host defenses and the effects of biofilms on host wound healing pathways. It may also be used to test anti-biofilm strategies the treatment of chronic wounds.
Keywords: Pseudomonas aeruginosa, biofilm, wound infection, keratinocytes, inflammatory response, gene expression
INTRODUCTION
Chronic wounds, such as diabetic foot ulcers, are a major medical problem that results in debilitating disease, extensive healthcare expenditure and lost productivity. Chronic wounds are generally defined as wounds that take longer than 6 weeks to heal [1–3]. Chronic wounds differ from acute wounds as the healing phase is prolonged with bacterial contamination, persistent inflammation and unbalanced cellular defense mechanism [4]. Multiple species of bacteria, including Staphylococcus aureus, followed by coagulase-negative staphylococci, Pseudomonas aeruginosa, Enterococcus spp., and Streptococcus pyogenes [5–7] have been isolated from chronic wounds, even though the wound may not show any clinical signs of localized infection. Multiple bacterial species, usually two to five species, reside concurrently on a single ulcer [7–9]. The chronicity of unhealed wounds is associated with higher proportion of colonization by anaerobic bacteria and greater variety of aerobic species [5]. More recent studies using molecular techniques have shown that microbial communities in chronic wounds are more diverse than indicated by culture-based techniques [10, 11].
Research over the past 20 years has revealed that bacteria in many environments exist as complex surface-attached communities termed biofilms [12]. Biofilms are organized structures made of surface associated bacteria and their extracellular polysaccharides. Biofilm microbes secrete specific toxins, create a hypoxemic microenvironment, and are resistant to antibiotics and the host immune system, all of which may contribute to delayed wound healing [12, 13]. Sixty percent of chronic wound specimens contained microbial biofilm, compared to only 6% of acute wounds [14]. Current topical and systemic antibiotics are minimally effective in the treatment of these microbial communities. In addition, the host’s inflammatory response is ineffective in combating biofilms [15]. In order to systematically study pathologenic mechanisms and test new therapies for chronic wounds, a reliable animal model would be a valuable tool. Researchers have tried several methods to delay wound healing, such as the ischemic rabbit ear model [16, 17], radiation impaired rats [18] and diabetic mice [19, 20]. One method to model diabetic foot ulcer is to induce infection in diabetic mouse wounds. Uninfected wounds in diabetic (db/db) mice steadily progress to complete re-epithelialization [20] although they are significantly delayed compared to normal littermates. On the other hand, bacterial inoculation causes acute infection, enlarged wound size and significant weight loss [21, 22]. It is a challenge when using bacteria to delay wound healing to find the critical balance between delayed wound healing and severe infectious side effects.
We recently developed a biofilm-challenged wound model in the db/db mouse by inoculating the wound with P. aeruginosa biofilm (PAO1) and found that wounds remained unhealed for 28 days as compared to control, non-biofilm-challenged wounds [23]. This biofilm challenged wound is characterized of thick epidermis and dermis, non-vascular wound matrix and delayed re-epithelialization; the scab over the wound bed is comprised of high density of P. aeruginosa biofilms and neutrophils (Figure 1). This model provides a reproducible mouse wound with localized cutaneous infection while avoiding systemic infection. In this study, we further characterize the model to examine the wound healing process, bacteria turnover and host immune response in a time course study from 4 to 8 weeks post-wounding. The result demonstrates that this animal model can be used for both delayed wound healing studies and in vivo studies of microbial biofilm.
Figure 1.
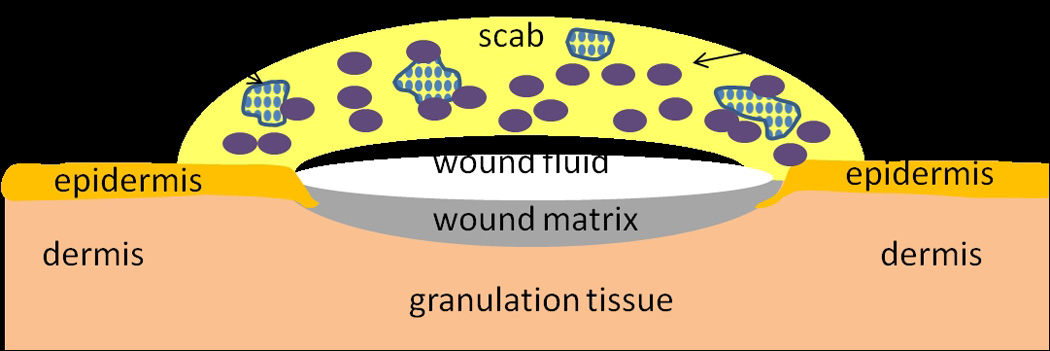
Schematic illustration of biofilm challenged wounds at 28 days post-wounding.
METHODS
Animals and wounding
Forty-two genetically diabetic female mice (db/db; BKS.Cg-Dock7m +/+ Leprdb/J) 10–12 weeks of age were purchased from Jackson Laboratory (Bar Harbor, ME) for the study described below. Mice were housed individually in the University of Washington Department of Comparative Medicine vivarium with ad libitum rodent chow and water. These studies were conducted with University of Washington Internal Animal Care and Use Committee approval in compliance with the NIH guide for the Care and Use of Laboratory Animals, 1996. The mice were anesthetized with an intraperitoneal injection of a mixture of ketamine (0.106mg/g weight) and xylazine (0.0075mg/g weight) (Phoenix Pharmaceuticals, Inc., St. Joseph, MO) in saline. The dorsal skin was shaved, treated with depilatory cream to remove hair, and then cleaned with povidone-iodine solution followed by an alcohol wipe. One circular, full-thickness wound was created on the dorsal skin of each mouse using a 6 mm biopsy punch. Mice were placed on a warming pad (37°C) until they fully recovered from surgery and then recaged.
P. aeruginosa biofilm
Biofilms were prepared by modifying the method developed at the Center for Biofilm Engineering at Montana State University [24]. Six millimeter diameter polycarbonate membrane filters (0.2 µm pore size, GE, Minnetonka, MN) were cut using a 6 mm biopsy punch (Acuderm, Inc., Ft. Lauderdale, FL), and sterilized with exposure to UVC light for 5 minutes on both sides. P. aeruginosa from frozen stock was grown overnight in lysogeny broth (LB) medium at 37°C on a shaker. The PAO1 culture was diluted 1:1000 in sterile phosphate buffer solution (PBS); 2 µl of the diluted solution was placed on each of the 6 mm filters, which were placed on LB agar plates incubated at 37°C for 72 h. Filters with PAO1 were transferred to fresh LB agar plates at 24 and 48 hours. On average, PAO1 organisms grew to levels of 108 colony forming units (CFU) per membrane after 24 hours of incubation.
Biofilm application and animal care
Application of biofilm on mice was performed 48 hours after the initial wounding procedure. The bacteria-inoculated filters were removed from the agar plate after 72 hours of incubation. A single filter was placed on each wound so the surface with bacterial biofilm was in direct contact with the wound. The filter was then removed from the wound, leaving behind the PAO1 biofilm. Thirty-four mice were inoculated with biofilm and 8 control mice did not receive biofilm. Mastisol® (Ferndale Laboratories, Inc., Ferndale, MI) liquid adhesive was applied to the skin surrounding the wound and allowed to dry for 2 min. Wounds were then covered with a 2 cm diameter transparent and semi-occlusive dressing (Tegaderm®, 3M, St. Paul, MN). All the dressings were removed at day 14. Mice were observed twice a week for the first 28 days, then at 42 or 56 days post-wounding. To evaluate overall health of the mice, weight was monitored throughout the experiment. Mice losing more than 30% weight were euthanized.
Wound harvesting /Blood glucose
At harvest days 28, 42 or 56 after wounding, mice were euthanized with a 0.25 ml intraperitoneal injection of pentobarbital sodium (390 mg/ml) and phenytoin sodium (50 mg/ml) (Schering-Plough Animal Health, Union, NJ). A One Touch® Blood Glucose Meter (LifeScan Inc., Milpitas, CA) was used to measure blood glucose levels. The upper limit of blood glucometer was 600mg/dL. Blood samples (around 0.3 ml) drawn from 2 mice in each group were spread on LB agar plate for blood culture.
For histological analysis, wounds including a 0.5 cm margin of surrounding skin were removed, and were bisected. Half of each wound was placed in sterile PBS for bacterial CFU counting, and the other half was embedded in O.C.T. (Sakura Finetek Inc., Torrance, CA) for cryosectioning and immunohistochemistry. Additional samples were collected for molecular analysis at 4, 6 and 8 weeks post-wounding. Tissue samples included: wound bed, adjacent skin to wounds, and normal skin at least 2 cm away from the wounds. All the samples for molecular analysis were removed by 6 mm biopsy punch and immediately stored at 1 ml RNALater (Ambion, Austin, TX).
Macrophotography and image analysis
Macrophotographs of the mouse wounds and a metric ruler were photographed using a Nikon D1 digital camera equipped with a Micro Nikkor macro lens and dual electronic flash (Nikon, Tokyo, Japan). Polarizing filters were fitted over both the lens and flash. Cross-polarization of these filters were necessary to remove spectral reflections from the Tegaderm® and wound surface. Wound size was measured by image calibration using the metric ruler and subsequent calculation of wound area using image analysis (Photoshop with IP tools Plug-in, Reindeer Graphics, Asheville, NC).
Colony formation unit (CFU) counting
Scabs and excised wounds were placed into separate tubes with 1 ml PBS. Tissue samples were then homogenized at 26,000rpm for 30 seconds (Polytran® PT 3100 Benchtop Large-Scale Homogenizer, Capitol Scientific Inc., Austin, TX). The resulting solutions were serially diluted and plated on LB agar and incubated overnight at 37°C. CFUs were determined by standard colony counting method.
Histology
Wound samples from 8 db/db control mice and 34 db/db biofilm-challenged mice were evaluated for histologic study. Six micron O.C.T. frozen sections were stained with hematoxylineosin (H&E) or Gram staining. H&E stained tissue was evaluated for re-epithelialization, granulation tissue, and inflammatory response. Gram stained sections were used to evaluate the presence of bacteria. To determine the percentage of wound closure, the distance between two epithelial tongues in the center of the wound was divided by the distance between original wound edges. The original wound edge was determined as the point where there was a clear margin in which subcutaneous fat tissue, mature collagen or mature hair follicles were absent.
Immunohistochemistry (IHC)
Six-micron sections of O.C.T.-embedded implant specimens were immunolabeled using routine immunoperoxidase methods as previously described [25]. Primary antibodies used were: pooled Pankeratin (Panker) ( rabbit, 1:1000, Dako, Carpenteria, CA) and Keratin 14 (K14) (rabbit, 1:1000, Covance, Princeton, NJ), platelet/endothelial cell adhesion molecule-1 (PECAM-1 or CD-31) (rat, 1:400, Research Diagnostics, Concord, MA), P3C8 – laminin 332 (rat, 1:10, gift from Dr. William Carter), and Ki67 (rabbit, 1:1000, Abcam Inc., Cambridge, MA). Secondary antibodies used were biotinylated goat anti-rat (1:1600, Jackson ImmunoResearch, West Grove, PA) or biotinylated goat anti-rabbit (1:300, Vector Laboratories, Burlingame, CA). Following secondary antibody labeling, sections were incubated with strept-avidin-biotin complex [Vectastain Elite ABC kit (peroxidase), Vector Laboratories, Burlingame, CA], with 0.12% 3,3’-diaminobenzidine used as chromogen. Glycergel (Dako, Carpenteria, CA) was used as mounting medium. Tissue sections were viewed using a Nikon Microphot-SA microscope. Images were captured using a Photometrics Sensys digital camera controlled by IP Lab software (Scanalytics, Fairfax, VA) or a Spot Flex digital color camera (Diagnostic Instruments, Sterling Heights, MI) for brightfield and differential interference contrast (DIC) images of H&E and IHC stained tissues. Photoshop® (Adobe Systems Inc., San Jose, CA) was used for image color adjustment, image analysis and figure preparation.
Polymerase chain reaction
Samples were stored in RNALater at 4°C overnight and then stored at -20°C before processing. RNA was extracted using an Aurum™ Total RNA Fatty and Fibrous Tissue Kit (Bio-Rad, Hercules, CA). RNA quality was evaluated using a 2100 Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA was reverse transcribed into cDNA using the iScript™ cDNA synthesis Kit (Bio-Rad). Real time PCR was performed using SsoFast™ EvaGreen Supermix (Bio-Rad) on an Eppendorf Mastercycler® ep realplex Thermal Cyclers (Eppendorf, Hamburg, Germany). The following primers were purchased from Qiagen: GAPDH QT01658692, transforming growth factor β (TGFβ1 QT00145250), interleukin 6 (IL-6 QT00098875), vascular endothelial growth factor α (VEGFα QT00160769), hypoxia inducible factor 1α (HIF-1α QT01039542), tumor necrosis factor (TNF QT00104006), matrix metalloproteinase 10 (MMP10 QT0011552), interleukin 1β (IL-1β QT01048355), heat shock protein 90α (HSP90α QT00246967).
Data analysis
Mouse weight, wound closure and histology data were pooled from two mouse experiments. First experiment included 6 control and 12 challenged mice; second experiment included 2 control and 22 challenged mice. Bacterial counting and genetic data were collected from second experiment. Data were analyzed using Excel 2007 (Microsoft Inc., Redmond, WA). Values were expressed as means ± standard error. Significance level was set at α=0.05. Differences in wound sizes, glucose levels and gene expression levels between groups were determined using general linear model ANOVA and one sided student t-tests.
RESULTS
Mouse weight
At day 0 the db/db mice ranged in weight from 37.8 to 48.2 g. Average weight of the control group was 44.3 g, and of the mice with PAO1 biofilm-challenged wounds (referred as PAO1 group below) was 42.8 g. The average weight of the PAO1 group was statistically significantly lower than the control group at 1 week (weight difference 2.1g, p=0.048) and 2 week (2.4g difference, p=0.041) post-wounding (Figure 2a). However, the percentage weight loss showed no statistical difference. By week 4, the control group lost 7.7% body weight and the PAO1 group lost 9.2%. By week 8, the PAO1 group lost 12.3% body weight (Figure 2b). None of the animals had to be euthanized because of weight loss (our threshold for euthanasia was a 30% loss). Blood glucose levels, another factor that may impact wound healing, were also comparable between control and PAO1 groups. The average baseline glucose level was >568 mg/dL. At week 4, the glucose level in the control group was >600 mg/dL. The glucose levels were >596, >591, and >564 mg/dL in the PAO1 group at week 4, 6 and 8, respectively.
Figure 2.
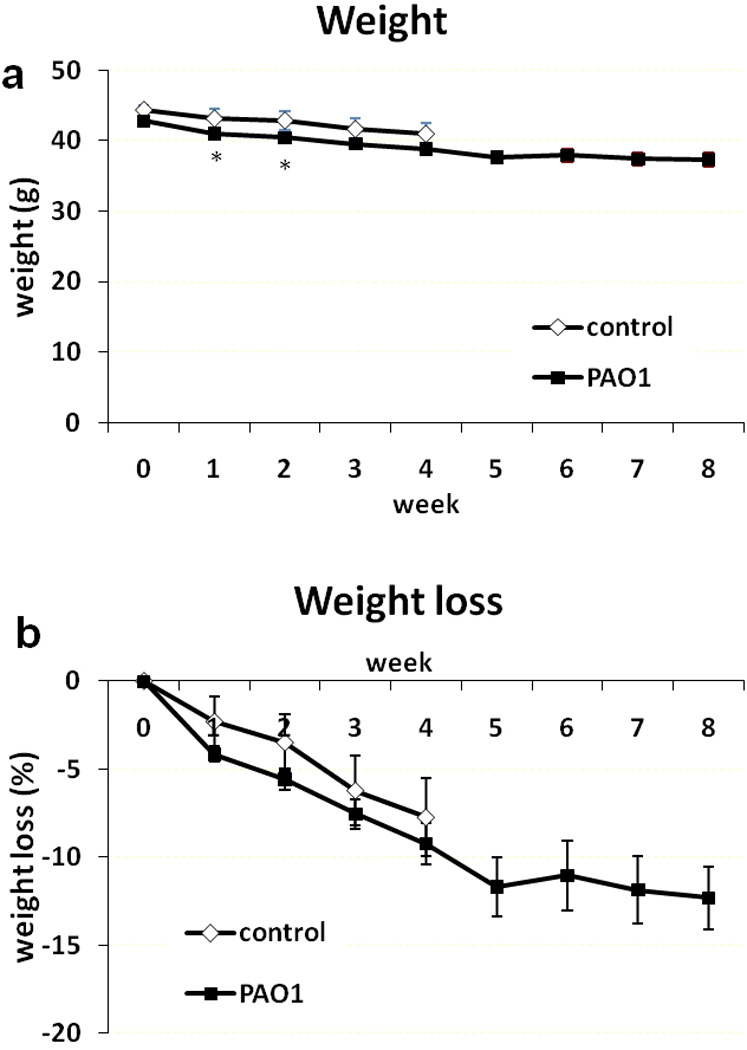
Weight measurements as a function of time. Actual weight in grams (a) and percentage weight loss (b) in control (◊, n=8) and PAO1 (■, n=34) groups from 0 to 8 weeks post-wounding. Data are presented as mean±SD. *P<0.05.
Gross observations of wounds
The experimental wounds created by the 6 mm biopsy punch were consistent in size and shape without significant bleeding. Representative photographs of the morphological changes in wounds over time are shown in Figure 3. Thick scabs (crusts of dried blood, and exudate over wound during healing) formed on unhealed wounds between 3 to 6 weeks post-wounding. The scabs were then lost and wounds proceeded to close. New hair grew around the PAO1 wounds.
Figure 3.
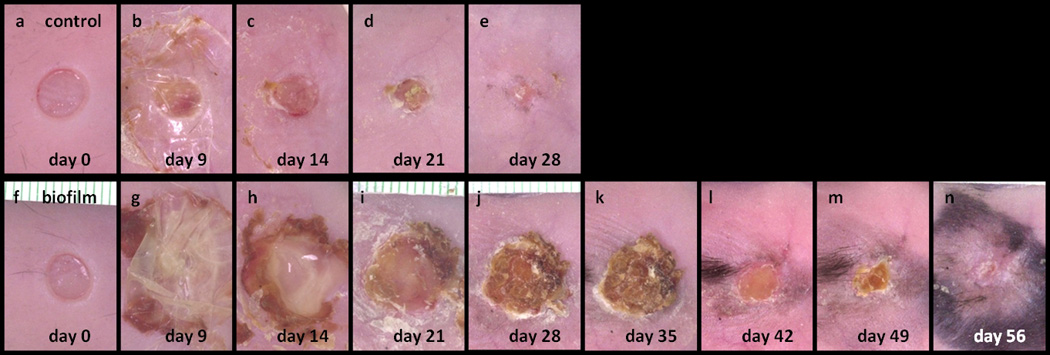
Macro photographs of wounds taken from 0 to 56 days post-wounding of a control mouse (a–e) and a PAO1 biofilm challenged mouse (f–n).
Wound closure
Seven of 8 control wounds (88%) healed with full re-epithelialization by week 4 (Figure 4a). In contrast, none of 13 wounds infected with P. aeruginosa (0%) healed at week 4. Instead, healing was significantly delayed; 7 of 11 wounds (64%) healed by week 6, and all 10 wounds (100%) healed by week 8. The newly formed epidermis and dermis in control group were significantly thicker than the adjacent normal skin, with higher cellular density (Figure 4). The center of the open wound was filled with a mixture of non-vascular debris and infiltrated polymorphonuclear leukocytes (neutrophils). The clusters of neutrophils formed microabcesses. The average wound diameter as a measurement of closure in all groups is summarized in Table 1.
Figure 4.
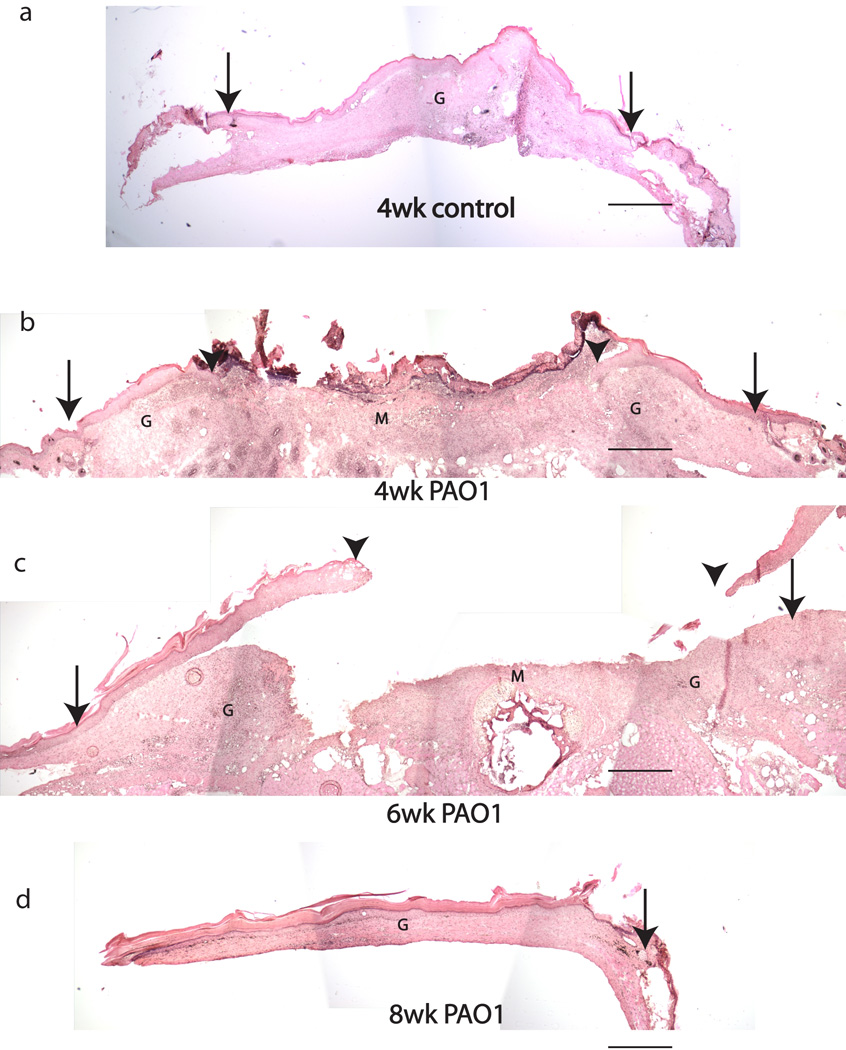
H&E stained sections of 4 week control wound (a) and PAO1 group wounds at 4, 6 and 8 weeks post-wounding (b–d). Original wound edge (arrow), migrating epithelial tongue (arrow head), granulation tissue (G) and wound matrix (M.
Table 1.
| Group | healed | unhealed | % wound healing | |
|---|---|---|---|---|
| control | 4 wk | 7 | 1 | 97 |
| PAO1 | 4 wk | 0 | 13 | 56 |
| 6 wk | 7 | 4 | 85 | |
| 8 wk | 10 | 0 | 100 |
Bacterial colony counts
The number of viable bacteria in scabs and wounds were quantified by plating wound and scab homogenates, and enumerating colonies after 2 days of growth. Population of different bacterial species were differentiated by colony color, shape and size, then confirmed by biochemical methods (Figure 5). Control wounds had an average of 6×103 CFU S. aureus In the PAO1 group, at 4 weeks post-wounding, both scabs and wounds contained only P. aeruginosa, with the majority of bacteria (99%) located in the scabs. At week 6, healed PAO1 wounds had similar amounts of S. aureus as in the control group. Unhealed wounds at 6 weeks were covered by scabs. A mixture of S. aureus and P. aeruginosa colonies were identified from these wound beds. In contrast, the scabs contained an average of 3.5×106 CFU P. aeruginosa. At 8 weeks post-wounding, all wounds were healed with moderate presence of S. aureus. Blood cultures from both control and biofilm challenged mice were all negative.
Figure 5.
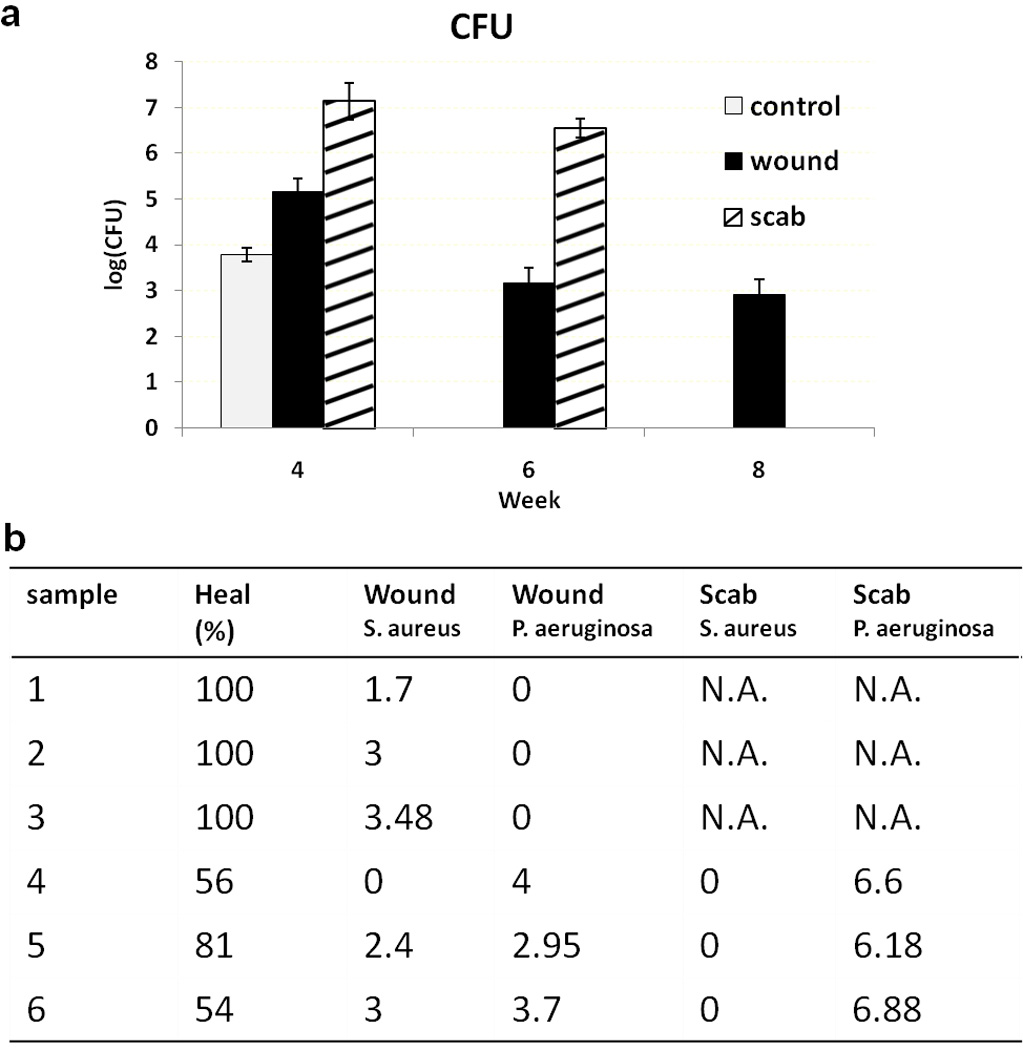
CFU of control wounds, PAO1 wounds and PAO1 scabs at 4, 6 and 8 weeks post-wounding (a). Percent wound healing, log(CFU) of bacterial species in wounds and scabs from 6 PAO1 mice at 6 weeks post-wounding are shown in (b).
Immunohistochemistry
Immunohistologic staining revealed cellular changes in the wounds. Keratinocytes in the migrating epithelial tongue were identified by immunolabeling with a pooled panker/K14 antibody. The new wound epidermis was much thicker than normal skin (Figure 6a). The basement membrane, which separates the epidermis from dermis, was immunolocalized with an anti-laminin 332 antibody and showed a typical linear structure (Figure 6b–c). In several PAO1 wounds, the epidermis separated from the dermis at the dermal epidermal junction along the epidermal tongue. The gap between the epidermis and dermis was filled with numerous cells (Figure 6d insert), which may represent local abscesses. PECAM immunoreactive endothelial cells were seen in granulation tissue under the re-epithelialized area (Figure 6e), but not in the wound matrix, a mixture of fibrin clot, platelet, collagen and inflammatory cells. (Figure 6f). In healed wounds, PECAM immunoreactive cells were observed throughout the dermis. In peripheral unwounded skin, Ki67 immunoreactive proliferating basal keratinocytes can be seen (Figure 6g) which was comparable to unwound normal skin (Figure 6h). In the re-epithelialized area, more Ki67 positive cells were seen in basal and differentiated keratinocytes (Figure 6i).
Figure 6.
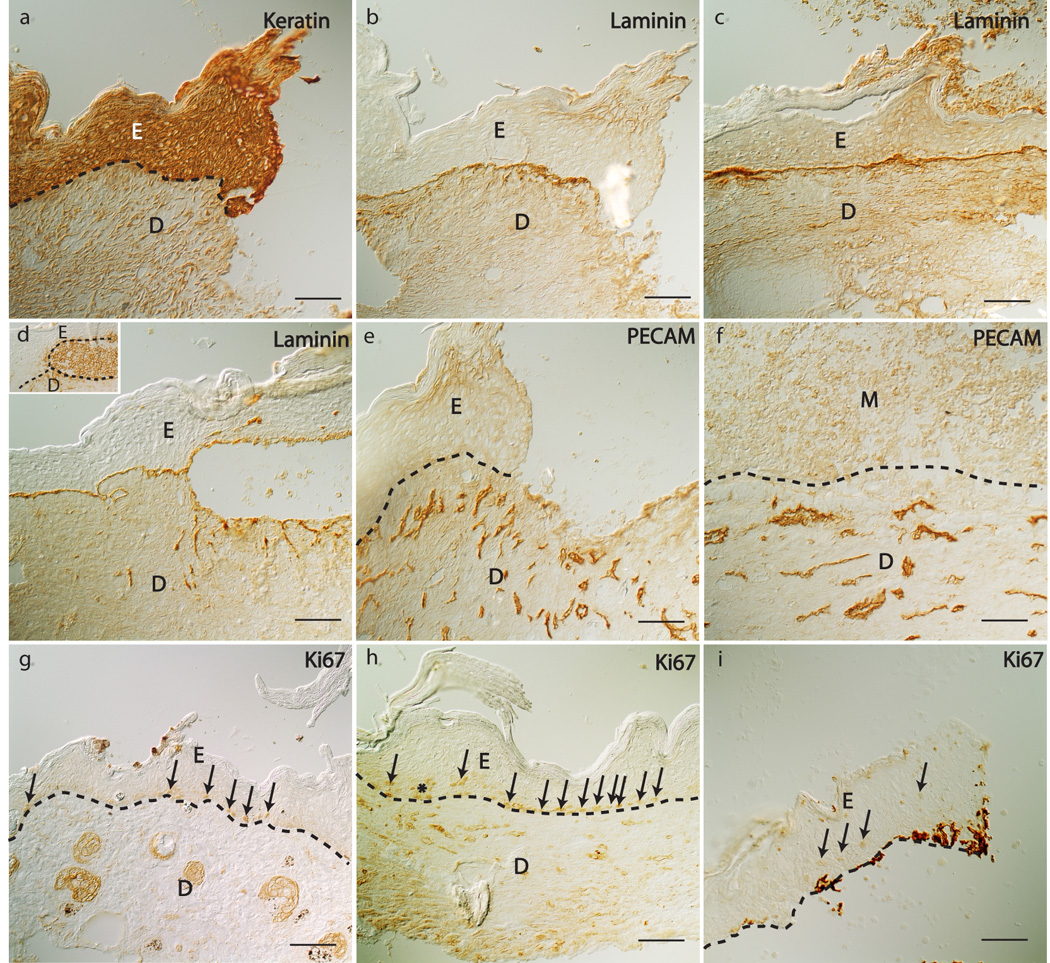
Keratin, laminin 332, PECAM-1, and Ki67 immunostaining of wound tissue sections 4 weeks post-wounding. Keratin in PAO1 wound (a), cleaved laminin 332 in PAO1 wound (b) and control wounds (c, d), PECAM in PAO1 wound (e, f), and Ki67 in peri-wound tissue of control (g), control wound (h) and PAO1 wound (i). Insert in Figure (d) is a PECAM stained section showing accumulation of cells between the epidermis and dermis in healing edge of a PAO1 wound. Epidermis (E), dermis (D), basement membrane (dashed lines), granulation tissue (G), wound matrix (M), Ki67 immunostained cells (arrows indicate single cells, and * indicate a cluster of cells). Scale bar = 100µm.
Gene expression
We also analyzed gene expression in the skin of normal db/db mice, 4 week control wounds, 4 week PAO1-infected wounds, and peri-wound tissue at 4, 6 and 8 week in the PAO1 groups. IL-1β, IL-6 and MMP-10 increased more than 10 fold in the 4 week unhealed PAO1 wound compared normal skin (p<0.05) (Figure 7). HIF-α level was 3 fold higher in the 4 week PAO1 wounds compared normal skin, but had no statistical difference. TNFα, VEGF, TGFβ1 and HSP90α expression levels were not different among all groups (Figure 8).
Figure 7.
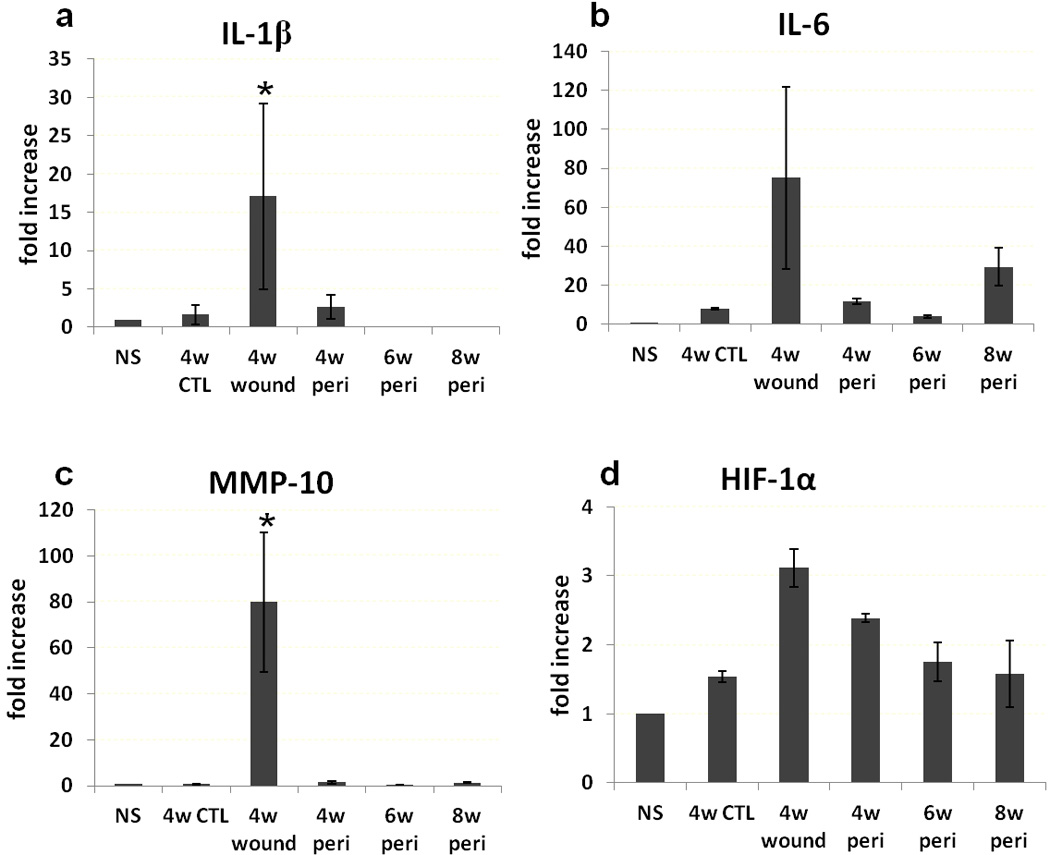
Gene expression. IL-1β (a), IL-6 (b), MMP-10 (c) and HIF-1α (d) expression in normal skin away from the wounds (NS), 4 week control wounds (4w CTL), 4 week PAO1 wounds (4w wound) and peri-wound PAO1 tissue at 4, 6 and 8 weeks (4w peri, 6w peri and 8w peri, respectively). *p<0.05 compared to other groups.
Figure 8.
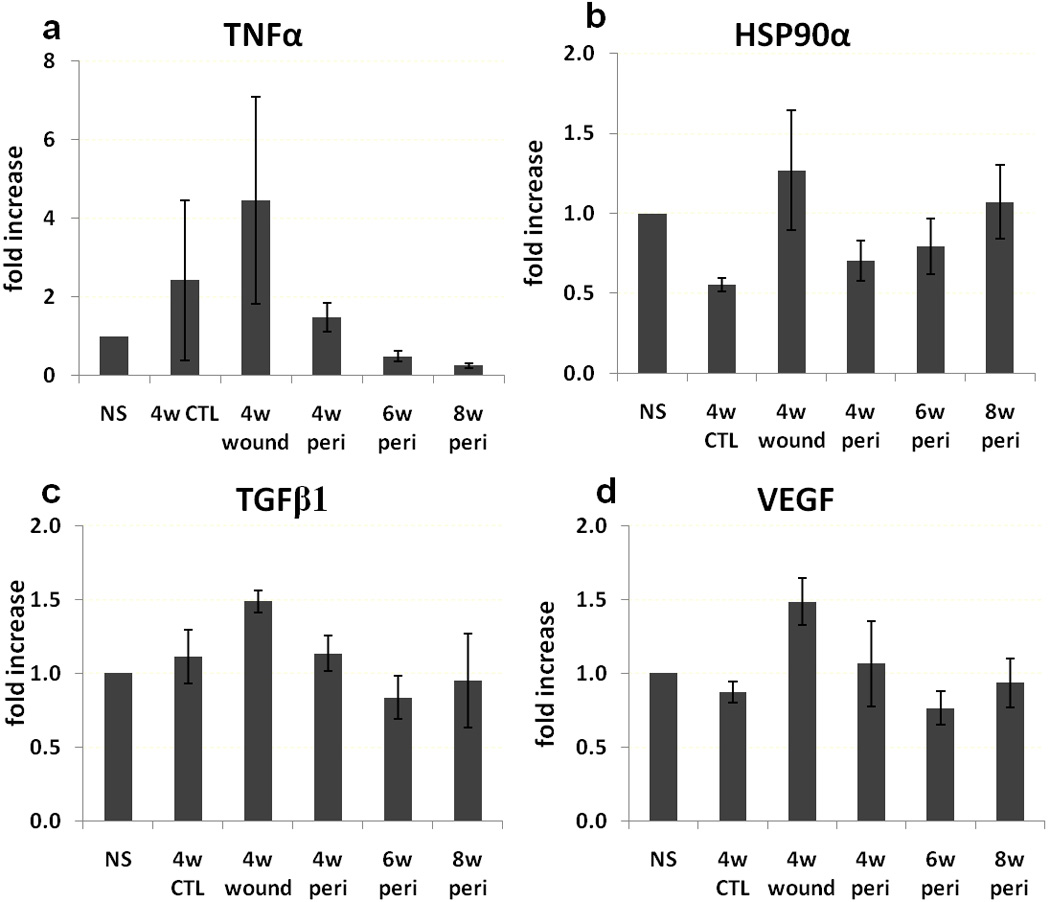
Gene expression. TNFα (a), HSP90α (b), TGFβ (c) and VEGF (d) expression in normal skin away from the wounds (NS), 4 week control wound (4w CTL), 4 week PAO1 wound (4w wound) and peri-wound PAO1 tissue at 4, 6 and 8 weeks (4w peri, 6w peri and 8w peri, respectively).
DISCUSSION
This study demonstrated that P. aeruginosa biofilm infection reproducibly delayed wound healing in diabetic mice by an average of 2 weeks. Typically, non-bacterially challenged wounds healed before 4 weeks; while biofilm-challenged wounds remained unhealed. Unhealed wounds showed similar histological features to chronic wounds in human, including hyperproliferative epithelium [26] and dermis, limited vascular growth in the center of wounds and infiltration of inflammatory cells.
Since the rate of healing of non-bacterially-challenged mice was consistent with numerous previous studies conducted in our laboratory, we were able to use fewer animals for a control group. Occasionally some of the non-challenged control wounds did not heal within 4 weeks. Culture results showed that these wounds had higher numbers of S. aureus, suggesting bacterial colonization is associated with delayed wound healing. More careful handling of the animals in BSL2 level facility can lower this contamination. The biofilm-challenged wounds healed within a range of 6 to 8 weeks. The variability in healing time of the PAO1 group was thought to be due to differences in bacterial response to host defenses, overall health and nutrition of the host and individual differences in host response to biofilm challenge. Similar to the situation in patients, wounds on young, healthy patients heal at a predicted rate, while the rate of chronic wound healing varies widely, from months to years. This is due to the variance of many other factors affecting wound healing in patients, such as aging, original wound size, nutrition, venous status and other comorbidities. Although this animal model for chronic wounds does not completely eliminate variables, using a genetically identical diabetic mouse strain provides a more controlled in vivo model for study.
We confirmed our previous observation that 99% of the P. aeruginosa aggregated in scabs, suggesting physical removal of scabs may benefit chronic wound healing. One of the most effective methods in treatment of biofilm plaque in teeth is physical scaling. Although biofilms in infectious kidney stones, endocarditis or cystic fibrosis airways are not easily debrided, biofilms in open wounds can be reduced if the caregivers can perform adequate debridement. If debridement cannot be adequately performed, a nonsurgical technique must be sought. This model may be helpful for finding an alternative means of reducing biofilm in wounds.
Our previous study showed that bacteria clustered in the matrix, forming organized biofilms. In this study, we found that during wound healing, the amount of P. aeruginosa decreased over time while the amount of S. aureus increased in the wound bed as a function of healing time. When wounds were healed, regardless of whether they were challenged with P. aeruginosa, the dominant colonization was by S. aureus which has been shown in a prior study to be the most abundant organism on normal mouse skin [27]. This observation suggests that clearance of P. aeruginosa is a critical component of wound healing. P. aeruginosa is a major biofilm-forming pathogen that is associated with many diseases, and one of the most commonly isolated and abundant organisms in chronic wounds [13, 28]. P. aeruginosa infected wounds are more severe and slower-healing than those not containing P. aeruginosa [8, 29, 30]. The effects of pseudomonas biofilm may not require direct invasion in the host epithelium. Similarly in this study, P. aeruginosa biofilms delayed wound healing even though the majority of the bacteria were not located in the wound bed. The mice also did not show signs of systemic infection and blood cultures were negative.
In our model, biofilms in the scabs were separated from wound beds by a gap filled with wound fluid. We hypothesize that biofilm may impede wound healing from a distance through depletion of oxygen or releasing diffusible biochemical factors. It has been previously shown that bacteria living in a biofilms can influence epithelial migration while separated from the keratinocytes by a Millipore filter [31]. This suggests that humeral soluble factors produced by the biofilm may adversely influence normal epithelial function. In cystic fibrosis patients the bacterial density can reach very high levels in the sputum overlying the epithelium without invasive dissemination into lung epithelium. Some researchers have proposed that the presence of P. aeruginosa in biofilms, and the lack of the concomitant elimination by host innate immune system, contribute to unhealed wounds [32]. This model will be useful for the further study of host response to P. aeruginosa, as well as the in vivo adaptation of P. aeruginosa to host defenses. It is also noteworthy that this model may be used for other bacteria such as S. aureus or even polymicrobial biofilms.
We used a culture-based method to quantify the bacterial shift on the wounds. Although many studies have shown that conventional culture does not identify whole microbiota [27], S. aureus and P. aeruginosa are still the two major bacteria indentified in clinical chronic wounds by molecular methods [33]. We previously showed that staphylococci were the predominant species on wild type mice housed under specific pathogen free (SPF) condition [27]. A recent study examined the longitudinal shift of colonizing microbiota on db/db skin in response to skin injury [34]. By sequencing 16S rRNA genes, S. aureus was found to be the main species and composed approximately 50% of the total microbiota. Molecular methods to identify all of the bacteria present in the wounds will be important for future studies.
Normal wound healing is usually divided into three phases: 1) inflammation, 2) epithelialization and granulation tissue formation, and 3) matrix formation and remodeling. Chronic wounds are characterized by a prolonged inflammatory response with unbalanced cellular and molecular components [4]. In this study, we evaluated selective cytokines and growth factors involved in the delayed wound healing. Proinflammatory cytokines, including IL-1 and IL-6, are upregulated in both acute and chronic wounds. They are produced by neutrophils, macrophages and monocytes, and induce proliferation or migration of keratinocytes. The upregulation of these cytokines in PAO1 wounds confirms that the inflammatory phase in the db/db wounds is prolonged, impacting healing. TNFα is also an important cytokine that increases during wound healing. The effect of TNFα on wound healing depends on its level in tissue and synergy with other factors. We observed a trend of increased TNFα in PAO1 challenged wounds, but the difference was not statistically significant. TNFα and IL-1 have been shown to have paracrine expression, and increase expression of interstitial collagenases, gelatinases and MMPs. High levels of MMP degrade extracellular matrix, inhibits cell migration and collagen deposition, and break down other growth factors, all of which contribute to delayed wound healing. Therefore, our data support the hypothesis that delayed healing caused by infection is associated with prolonged inflammation.
Oxygen is another essential component of wound healing. Lower oxygen pressure (<30 mmHg) at the wound site is associated with delayed wound healing [35]. In diabetic patients, microvascular disease is a common complication and a significant factor in the pathogenesis of chronic foot ulcers. The presence of biofilm may aggravate underlying tissue hypoxia. Measurement of in vitro biofilm shows significant decrease of oxygen 20–30 µm from the biofilm surface (unpublished observation). HIF-1α is a transcription factor that responds to the hypoxic environment by regulating the cell cycle through targeting both coding and noncoding genes [36]. A recent study found that up-regulated HIF-1α expression is associated with inhibition of keratinocyte proliferation in mice [37]. We didn’t found statistically significant change in HIF-1 α, probably due to small sample size. Future studies with larger sample size will be helpful to evaluate the presence of tissue hypoxia in these wounds.
TGFβ and VEGF regulate epithelialization, connective tissue formation and angiogenesis. Their levels vary in wounds. Usually they are increased in acute wounds and decreased in chronic wounds [4]. Researchers have tried to promote wound healing by introducing these growth factors to chronic wounds. However, the results of clinical trials have showed modest benefits at best. No growth factor, except PDGF, has been approved by the FDA for chronic wound treatment. In our animal wound model, there was no difference of TGFβ and VEGF among normal skin, healed wounds and open wounds, suggesting that the 4 week open wounds in this model were in an intermediate status between acute and chronic wounds.
In conclusion, this model showed that P. aeruginosa biofilm-challenged wounds in diabetic mice reliably healed 2–4 weeks later than control non-challenged wounds. These delayed wounds showed 99% of the biofilm bacteria living in the scabs covering the wounds. Histological and gene expression profile were similar to chronic wounds in humans. The results suggest that bacterial infection delayed wound healing, and this process may be mediated by prolonged inflammatory response. This model can be used to identify changes in host healing pathways in response to bacteria as well as adaptive mechanisms of bacterial biofilm to host defenses and can also used to evaluate clinical anti-biofilm strategies for treating chronic wound.
ACKNOWLEDGMENTS
This study was funded by National Institute of Health Grant P20 GM078445 and the George F. Odland Endowed Research Fund and the generous support by Dr. Marvin and Judy Young, Dr. John and Darcy Halloran, Dr. Peter Odland and Dr. Peter Byers. Dr. Pradeep K. Singh was supported by grants from NIH, the Cystic Fibrosis Foundation, and the Burroughs Wellcome Fund. We appreciate the technical support provided by Elizabeth Bauerle for biofilm development and analysis, and generous donation of antibodies from Dr. William Carter.
REFERENCES
- 1.Fonder MA, Lazarus GS, Cowan DA, Aronson-Cook B, Kohli AR, Mamelak AJ. Treating the chronic wound: A practical approach to the care of nonhealing wounds and wound care dressings. J Am Acad Dermatol. 2008;58(2):185–206. doi: 10.1016/j.jaad.2007.08.048. [DOI] [PubMed] [Google Scholar]
- 2.Fowler E. Chronic wounds: an overview. In: Krasner D, editor. Chronic wound care: a clinical source book for healthcare professionals. King of Prussia, PA: Health Management Publications, Inc; 1990. pp. 12–18. [Google Scholar]
- 3.Singh A, Halder S, Menon GR, Chumber S, Misra MC, Sharma LK, Srivastava A. Meta-analysis of randomized controlled trials on hydrocolloid occlusive dressing versus conventional gauze dressing in the healing of chronic wounds. Asian J Surg. 2004;27(4):326–332. doi: 10.1016/S1015-9584(09)60061-0. [DOI] [PubMed] [Google Scholar]
- 4.Tomic-Canic M, Agren MS, Alvares OM. Epidermal Repair and Chronic Wounds. In: Rovie DT, Maibach H, editors. The Epidermis in Wound Healing. New York: CRS Press; 2004. p. 26. [Google Scholar]
- 5.Bowler PG, Davies BJ. The microbiology of infected and noninfected leg ulcers. Int J Dermatol. 1999;38(8):573–578. doi: 10.1046/j.1365-4362.1999.00738.x. [DOI] [PubMed] [Google Scholar]
- 6.Hansson C, Hoborn J, Moller A, Swanbeck G. The microbial flora in venous leg ulcers without clinical signs of infection. Repeated culture using a validated standardised microbiological technique. Acta Dermato-Venereologica. 1995;75(1):24–30. doi: 10.2340/00015555752430. [DOI] [PubMed] [Google Scholar]
- 7.Gjodsbol K, Christensen JJ, Karlsmark T, Jorgensen B, Klein BM, Krogfelt KA. Multiple bacterial species reside in chronic wounds: a longitudinal study. Int Wound J. 2006;3(3):225–231. doi: 10.1111/j.1742-481X.2006.00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirketerp-Moller K, Jensen PO, Fazli M, Madsen KG, Pedersen J, Moser C, Tolker-Nielsen T, Hoiby N, Givskov M, Bjarnsholt T. Distribution, organization, and ecology of bacteria in chronic wounds. J Clin Microbiol. 2008;46(8):2717–2722. doi: 10.1128/JCM.00501-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, Rhoads D. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP) PLoS One. 2008;3(10):e3326. doi: 10.1371/journal.pone.0003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price LB, Liu CM, Melendez JH, Frankel YM, Engelthaler D, Aziz M, Bowers J, Rattray R, Ravel J, Kingsley C, Keim PS, Lazarus GS, Zenilman JM. Community analysis of chronic wound bacteria using 16S rRNA gene-based pyrosequencing: impact of diabetes and antibiotics on chronic wound microbiota. PLoS One. 2009;4(7):e6462. doi: 10.1371/journal.pone.0006462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, Wolcott RD. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008;8:43. doi: 10.1186/1471-2180-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 13.James GA, Swogger E, Wolcott R, Pulcini E, Secor P, Sestrich J, Costerton JW, Stewart PS. Biofilms in chronic wounds. Wound Repair Regen. 2008;16(1):37–44. doi: 10.1111/j.1524-475X.2007.00321.x. [DOI] [PubMed] [Google Scholar]
- 14.Hurlow J, Bowler PG. Clinical experience with wound biofilm and management: a case series. Ostomy/Wound Manage. 2009;55(4):38–49. [PubMed] [Google Scholar]
- 15.Kelly NM, Kluftinger JL, Pasloske BL, Paranchych W, Hancock RE. Pseudomonas aeruginosa pili as ligands for nonopsonic phagocytosis by fibronectin-stimulated macrophages. Infect Immun. 1989;57(12):3841–3845. doi: 10.1128/iai.57.12.3841-3845.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahn ST, Mustoe TA. Effects of ischemia on ulcer wound healing: a new model in the rabbit ear. Ann Plast Surg. 1990;24(1):17–23. doi: 10.1097/00000637-199001000-00004. [DOI] [PubMed] [Google Scholar]
- 17.Gurjala AN, Geringer MR, Seth AK, Hong SJ, Smeltzer MS, Galiano RD, Leung KP, Mustoe TA. Development of a novel, highly quantitative in vivo model for the study of biofilm-impaired cutaneous wound healing. Wound Repair Regen. 2011;19(3):400–410. doi: 10.1111/j.1524-475X.2011.00690.x. [DOI] [PubMed] [Google Scholar]
- 18.Gu Q, Wang D, Gao Y, Zhou J, Peng R, Cui Y, Xia G, Qing Q, Yang H, Liu J, Zhao M. Expression of MMP1 in surgical and radiation-impaired wound healing and its effects on the healing process. J Environ Pathol Toxicol Oncol. 2002;21(1):71–78. [PubMed] [Google Scholar]
- 19.Greenhalgh DG, Sprugel KH, Murray MJ, Ross R. PDGF and FGF stimulate wound healing in the genetically diabetic mouse. Am J Pathol. 1990;136(6):1235–1246. [PMC free article] [PubMed] [Google Scholar]
- 20.Gibran NS, Jang YC, Isik FF, Greenhalgh DG, Muffley LA, Underwood RA, Usui ML, Larsen J, Smith DG, Bunnett N, Ansel JC, Olerud JE. Diminished neuropeptide levels contribute to the impaired cutaneous healing response associated with diabetes mellitus. J Surg Res. 2002;108(1):122–128. doi: 10.1006/jsre.2002.6525. [DOI] [PubMed] [Google Scholar]
- 21.Brown RL, Greenhalgh DG. Mouse models to study wound closure and topical treatment of infected wounds in healing-impaired and normal healing hosts. Wound Repair Regen. 1997;5(2):198–204. doi: 10.1046/j.1524-475X.1997.50213.x. [DOI] [PubMed] [Google Scholar]
- 22.Shi CM, Nakao H, Yamazaki M, Tsuboi R, Ogawa H. Mixture of sugar and povidoneiodine stimulates healing of MRSA-infected skin ulcers on db/db mice. Arch Dermatol Res. 2007;299(9):449–456. doi: 10.1007/s00403-007-0776-3. [DOI] [PubMed] [Google Scholar]
- 23.Zhao G, Hochwalt PC, Usui ML, Underwood RA, Singh PK, James GA, Stewart PS, Fleckman P, Olerud JE. Delayed wound healing in diabetic (db/db) mice with Pseudomonas aeruginosa biofilm challenge: a model for the study of chronic wounds. Wound Repair Regen. 2010;18(5):467–477. doi: 10.1111/j.1524-475X.2010.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderl JN, Franklin MJ, Stewart PS. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother. 2000;44(7):1818–1824. doi: 10.1128/aac.44.7.1818-1824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukano Y, Knowles NG, Usui ML, Underwood RA, Hauch KD, Marshall AJ, Ratner BD, Giachelli C, Carter WG, Fleckman P, Olerud JE. Characterization of an in vitro model for evaluating the interface between skin and percutaneous biomaterials. Wound Repair Regen. 2006;14(4):484–491. doi: 10.1111/j.1743-6109.2006.00138.x. [DOI] [PubMed] [Google Scholar]
- 26.Usui ML, Mansbridge JN, Carter WG, Fujita M, Olerud JE. Keratinocyte migration, proliferation, and differentiation in chronic ulcers from patients with diabetes and normal wounds. J Histochem Cytochem. 2008;56(7):687–696. doi: 10.1369/jhc.2008.951194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tavakkol Z, Samuelson D, Delancey Pulcini E, Underwood RA, Usui ML, Costerton JW, James GA, Olerud JE, Fleckman P. Resident bacterial flora in the skin of C57BL/6 mice housed under SPF conditions. J Am Assoc Lab Anim Sci. 2010;49(5):588–591. [PMC free article] [PubMed] [Google Scholar]
- 28.Fazli M, Bjarnsholt T, Kirketerp-Moller K, Jorgensen B, Andersen AS, Krogfelt KA, Givskov M, Tolker-Nielsen T. Nonrandom distribution of pseudomonas aeruginosa and staphylococcus aureus in chronic wounds. J Clin Microbiol. 2009;47(12):4084–4089. doi: 10.1128/JCM.01395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2003;100(24):14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eriksson G, Eklund AE, Kallings LO. The clinical significance of bacterial growth in venous leg ulcers. Scand J Infectious Dis. 1984;16(2):175–180. doi: 10.3109/00365548409087138. [DOI] [PubMed] [Google Scholar]
- 31.Kirker KR, Secor PR, James GA, Fleckman P, Olerud JE, Stewart PS. Loss of viability and induction of apoptosis in human keratinocytes exposed to Staphylococcus aureus biofilms in vitro. Wound Repair Regen. 2009;17(5):690–699. doi: 10.1111/j.1524-475X.2009.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bjarnsholt T, Kirketerp-Moller K, Jensen PO, Madsen KG, Phipps R, Krogfelt K, Hoiby N, Givskov M. Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regen. 2008;16(1):2–10. doi: 10.1111/j.1524-475X.2007.00283.x. [DOI] [PubMed] [Google Scholar]
- 33.Thomsen TR, Aasholm MS, Rudkjobing VB, Saunders AM, Bjarnsholt T, Givskov M, Kirketerp-Moller K, Nielsen PH. The bacteriology of chronic venous leg ulcer examined by culture-independent molecular methods. Wound Repair Regen. 2010;18(1):38–49. doi: 10.1111/j.1524-475X.2009.00561.x. [DOI] [PubMed] [Google Scholar]
- 34.Grice EA, Snitkin ES, Yockey LJ, Bermudez DM, Liechty KW, Segre JA. Longitudinal shift in diabetic wound microbiota correlates with prolonged skin defense response. Proc Natl Acad Sci U S A. 2010;107(33):14799–14804. doi: 10.1073/pnas.1004204107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathieu D, Mani R. A review of the clinical significance of tissue hypoxia measurements in lower extremity wound management. Int J Low Extrem Wounds. 2007;6(4):273–283. doi: 10.1177/1534734607310299. [DOI] [PubMed] [Google Scholar]
- 36.Biswas S, Roy S, Banerjee J, Hussain SR, Khanna S, Meenakshisundaram G, Kuppusamy P, Friedman A, Sen CK. Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proc Natl Acad Sci U S A. 2010;107(15):6976–6981. doi: 10.1073/pnas.1001653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scortegagna M, Martin RJ, Kladney RD, Neumann RG, Arbeit JM. Hypoxia-inducible factor-1alpha suppresses squamous carcinogenic progression and epithelial-mesenchymal transition. Cancer Res. 2009;69(6):2638–2646. doi: 10.1158/0008-5472.CAN-08-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]


