Abstract
Background
NLRC5 is a member of the CARD domain containing, nucleotide-binding oligomerization (NOD)-like receptor (NLR) family, which recognizes pathogen-associated molecular patterns (PAMPs) and initiates an innate immune response leading to inflammation and/or cell death. However, the specific role of NLRC5 as a modulator of the inflammatory immune response remains controversial. It has been reported to be a mediator of type I IFNs, NF-kB, and MHC class I gene. But no study on NLRC5 function has been reported to date in chickens. In the current study, we investigated the role of NLRC5 in the regulation of IFNA, IFNB, IL-6, and MHC class I in the chicken HD11 macrophage cell line, by using RNAi technology. HD11 cells were transfected with one of five siRNAs (s1, s2, s3, negative-siRNA, or a mixture of s1, s2, s3-siRNAs). After 24 hours, cells were exposed to LPS or poly (I:C) or a vehicle control. Gene expression of NLRC5, IFNA, IFNB, IL-6, and MHC class I at 2, 4, 6, and 8 hours post stimulation (hps) was quantified by qPCR.
Results
The expression of NLRC5, IFNA, IFNB, and IL-6 genes in negative irrelevant transfection controls was up-regulated at 2 hps after LPS treatment compared to the vehicle controls. S3-siRNA effectively knocked down NLRC5 expression at 4 hps, and the expression of IFNA and IFNB (but not IL-6 and MHC class I) was also down-regulated at 4 hps in s3-siRNA transfected cells, compared to negative irrelevant transfection controls. Stimulation by LPS appeared to relatively restore the decrease in NLRC5, IFNA, and IFNB expression, but the difference is not significant.
Conclusions
Functional characterization of chicken NLRC5 in an in vitro system demonstrated its importance in regulating intracellular molecules involved in inflammatory response. The knockdown of NLRC5 expression negatively mediates gene expression of IFNA and IFNB in the chicken HD11 cell line; therefore, NLRC5 likely has a role in positive regulation of IFNA and IFNB expression. No direct relationship was found between NLRC5 knockdown and IL-6 and MHC class I expression. Future studies will further clarify the roles of NLRC5 and other NLRs in infectious diseases of chickens and may increase the efficacy of antiviral vaccine design.
Background
The host innate immune system recognizes various pathogen-associated molecular patterns (PAMPs) and danger-associated molecular patterns (DAMPs) through pattern-recognition receptors (PRRs) and triggers the inflammatory response to defense against microorganisms invasion [1]. There are three classes of PRRs in vertebrates, Toll-like receptors (TLRs), nucleotide-binding oligomerization (NOD)-like receptors (NLRs), and the retinoid acid-inducible gene-I (RIG-I)-like receptors (RLRs) [1-6]. These molecules showed different subcellular localization, for example, most TLRs identify extracellular PAMPs, whereas NLRs and RLRs sense intracellular PAMPs [7,8]. Recently, the NLRs have gained attention because of their involvement in mediating innate immune responses to microbial invasion and controlling innate immune pathways. To date, at least 22 members of the NLR family have been identified in humans [9], and some of them have been well characterized. For example, two members of the NLRC (NLR containing a caspase-recruitment domain (CARD)) family, NLRC1 and NLRC2, recognize bacteria-derived molecules and result in activation of downstream signaling pathways, including NF-kB and mitogen-activated protein kinase (MAPK)s, to induce production of inflammatory cytokines [10,11]. NLRP1 and NLRP3 (NLR containing a pyrin domain) play key roles in activating caspase-1 inflammasomes as a response to PAMPs and DAMPs, leading to maturation and secretion of pro-inflammatory cytokines interleukin (IL)-1B and IL-18 [12]. In addition, NLRC1, NLRC2, NLRP1, NLRP3, and MHC class II transactivator (CIITA) are all associated with susceptibility to chronic inflammatory diseases [13-20].
Recently, NLRC5, a newly identified member of NLR family, with the most evolutionary relationship to NLRC1 and NLRC2 [21,22], has been implicated in regulation of the innate immune response [23]. NLRC5 is highly expressed in lymphocytic and macrophage/monocytic cell lineages [24] and immune tissues [9]. NLRC5 can be potently induced by interferon gamma (IFNG) and modestly induced by the microbial-derived molecules, lipopolysaccharide (LPS) and polyinosinic:polycytidylic acid (poly (I:C)) [9]. The specific role of NLRC5 in acting as a regulator of proinflammatory pathways is controversial [24]. NLRC5 is suggested to positively regulate the interferon (IFN) pathway in human cervical carcinoma HeLaS3 cells and human acute monocytic cell line, THP-1 [25]. Overexpression of NLRC5 leads to the activation of IFN-specific response elements (ISRE) in HeLa cells [25]. siRNA-mediated knockdown of endogenous NLRC5 decreased type I IFN pathway-dependent responses mediated by Sendai virus and poly (I:C) in THP-1 cells [26]. In contrast, overexpression of NLRC5 was shown to repress the activation of NF-kB-, type I IFN-, and AP-1-dependent signaling pathways in human embryonic kidney cell line HEK293T, and the knockdown of NLRC5 increased secretion of proinflammatory cytokines in mouse leukaemic monocyte macrophage cell line, RAW264.7 [9]. Moreover, Cui et al. demonstrated that NLRC5 negatively modulated NF-kB and type I IFN signaling pathways and its absence resulted in elevated expression levels of Tumor necrosis factor (TNF) A, IL-6, and IL-1B [27]. In addition, opposite effects have also been described for the role of NLRC5 as a transcriptional regulator of MHC class I expression. It binds to the MHC class I promoter and up-regulates MHC class I expression in lymphoid and epithelial cell lines. NLRC5 was also proven to be required in the pathway of efficient induction of MHC class I by IFNG stimulation [28]. In contrast, knockdown of NLRC5 was shown to increase cell surface expression of MHC class I in RAW 264.7 cells, which indicated a negative role of NLRC5 in regulating MHC class I expression [9]. Therefore, the effects of NLRC5 in regulating immune-related components and inflammatory responsive pathways are very complex, and likely either cell type- or species-specific [24].
To date, no study on the functional role of NLRC5 in chickens has been reported. In the current study, we characterized the functions of NLRC5 gene in the regulation of IFNA, IFNB, IL-6, and MHC class I by utilizing RNA-interference technology and stimulation with LPS from Salmonella typhimurium or poly (I:C) in the chicken HD11 macrophage cell line. These specific ligands were selected to complement and expand upon our previously published studies [29-31]. The expression levels, over time, for five genes were determined by qPCR to elucidate the association of NLRC5 with IFNA, IFNB, IL-6, and MHC class I. The present study reports the initial characterization of chicken NLRC5 and its roles in the regulation of innate immune response to bacterial components. Our findings are novel and of significant scientific value for a better understanding of host response to infectious diseases in chickens.
Results and discussion
Expression of NLRC5, IFNA, IFNB, and IL-6 in negative irrelevant siRNA transfection controls was up-regulated at 2 hours post LPS stimulation
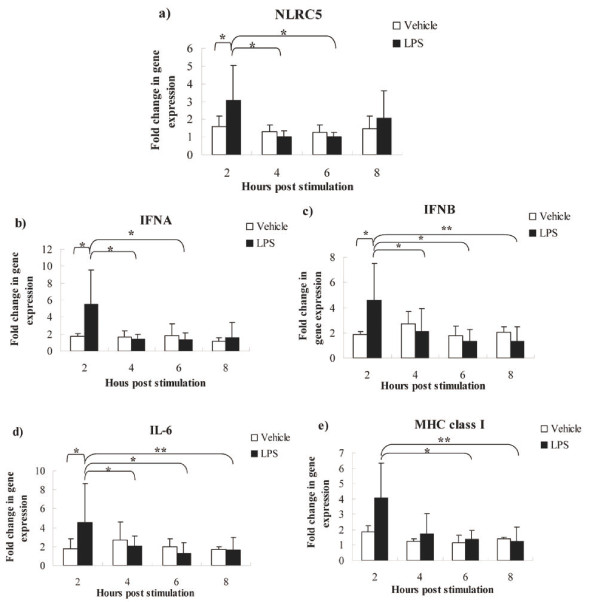
The expression of NLRC5, IFNA, IFNB, and IL-6 was significantly up-regulated, and MHC class I showed an increasing trend, in LPS-treated HD11 cells compared to vehicle controls at 2 hours post stimulation (hps) (Figure 1). The expression of NLRC5 and IFNA after LPS treatment at 2 hps was higher than 4 and 6 hps (Figure 1a,b). IFNB and IL-6 genes expression at 2 hours post LPS stimulation was higher than 4, 6, and 8 hps (Figure 1c,d). MHC class I gene expression was also higher at 2 hours post LPS treatment than these at 6 and 8 hours (Figure 1e). However, poly (I:C) had no effect on gene expression, based upon lack of significant difference between the poly (I:C) treated group and non-poly (I:C)-treated controls at any time points in negative siRNA transfected cells (data not shown), which may be because of miscalculation in the dilution of the stock resulting in the low concentration of poly (I:C) used.
Figure 1.
Gene expression of NLRC5, IFNA, IFNB, IL-6, and MHC class I after LPS treatment in negative siRNA transfection controls. HD11 macrophages were stimulated for 2, 4, 6 and 8 hours with 1 μg/ml LPS. Data are shown as the fold change in mRNA expression between LPS treatment and vehicle-treatment for each gene. a: NLRC5; b: IFNA; c: IFNB; d: IL-6; e: MHC class I; RNA samples were assayed in triplicates by qPCR. P values are flagged with asterisks when lower than 0.05(*) or 0.01 (**).
LPS and poly (I:C), as agonists of TLR4 and TLR3, respectively, have been extensively used to mimic bacterial and viral infection to induce inflammation in host innate immune response studies. In mammals, LPS is recognized by LPS-binding protein (LBP)3 and transferred to another LPS binding protein CD14, which delivers the LPS to a complex of myeloid differentiation protein-2 (MD-2) and TLR4 [32]. The TLR4/MD-2/CD14 complex transduces the LPS signal through the recruitment of various adaptor molecules at the Toll/IL-1R (TIR) domain of the TLR4 receptor, and triggers downstream signaling events, such as activation of nuclear factors, NF-kB and AP-1, and subsequent production of proinflammatory cytokines and interferons [33]. Two predominant intracellular pathways, the MyD88-dependent and independent pathways are activated by TLR agonists to induce inflammatory responses [34]. The MyD88-dependent pathway utilizes MyD88 and TIRAP and results in activation of NF-kB and production of the responsive immunoregulatory molecules, such as IL-1, IL-2, IL-6, TNF, and IFNG [35,36]. MyD88-independent pathway, also called the TRAM/TRIF-dependent pathway, induces phosphorylation of IFN regulatory factor (IRF)3 and the subsequent production of type I IFNs as well as a delayed NF-kB response [37-39]. TLR4 is an unique member of TLR family, which activates both the MyD88-dependent and the independent pathways [40,41]. However, TLR3, the sensor of poly (I:C), exclusively triggers the MyD88-independent pathway, leading to production of inflammatory cytokines such as TNF, IL-6, and IFNB [40-42].
Keestra et al. recently reported that a functional LPS-induced MyD88-independent pathway is absent in chicken, based upon their observation of no change of IFNB gene expression after LPS treatment (5 μg/ml of Salmonella Enteritidis (SE) or Salmonella Gallinarum (SG)-LPS) in chicken cells [43]. Interestingly, conflicting results were obtained in the current study, as well as in research recently reported by Esnault et al. [44]. Use of different sources and doses of LPS may partly explain the conflicting results among the studies. In the current study, gene expression of IFNA, IFNB, and IL-6 was up-regulated after Salmonella typhimurium (ST)-LPS treatment (1 μg/ml) at 2 hps. These results indicate that LPS stimulation induced elevated expression of type I IFN genes (IFNA and IFNB) and also an NF-kB-reponsive gene (IL-6). Similar results were reported by Esnault et al. [44], who found that the expression of IFNA, IFNB and IL-8 was strongly up-regulated in chicken epithelial cell line (CLEC213) after E. coli (EC)-LPS (10 μg/ml) stimulation between 4 hps and 24 hps. Collectively, these studies suggest that specific gene expression patterns after LPS stimulation are dependent upon the distinct LPS sources and dosage.
Additionally, in the present study, LPS treatment up-regulated the expression of the NLRC5 gene in chicken HD11 macrophage cells at 2 hps. Although LPS and poly (I:C) are considered to be moderate regulators of NLRC5, the reported effects seem to be varied and dependent upon the cell type tested. For example, in mouse splenic B lymphocytes, LPS induced a moderate increase in NLRC5 expression compared with IFNG, which is a strong regulator of NLRC5 [9]. But in murine bone marrow-derived macrophages (BMDMs), LPS simulation did not alter NLRC5 mRNA expression, whereas, poly (I:C) treatment increased its expression [9]. In RAW 264.7 cells, LPS stimulation up-regulated NLRC5 at mRNA and protein levels, peaked at 6 hours post treatment, while only a weak increase of NLRC5 was observed after poly (I:C) stimulation [27].
S3-siRNA knocked down NLRC5 expression, and IFNA and IFNB were down-regulated in s3-siRNA transfected cells
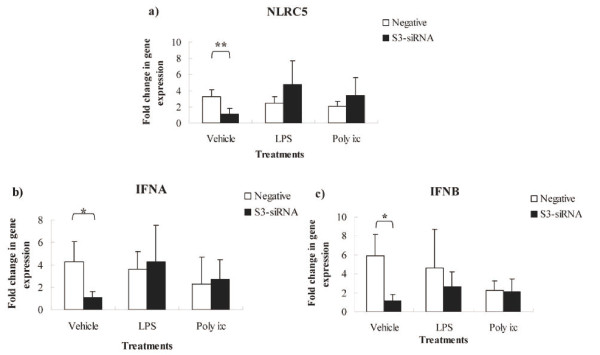
To further investigate the roles of NLRC5 in regulating expression of type I IFNs and NF-kB responsive genes in chickens, we utilized RNA interference to knock down NLRC5 expression, and then treated cells with LPS or poly (I:C), as well as a vehicle control. The gene expression of type I IFNs (IFNA and IFNB), an NF-kB responsive gene (IL-6), and also an antigen presentation molecule (MHC class I) was quantified by qPCR. Of three siRNAs designed, only s3-siRNA down-regulated the expression of NLRC5 compared to negative irrelevant siRNA. NLRC5 expression was reduced by 65% at 4 hps (Figure 2a) with a transfection efficiency 75%. Interestingly, IFNA and IFNB exhibited down-regulated expression patterns consistent with that of NLRC5 in s3-siRNA transfected cells (Figure 2b,c). Although gene expression of NLRC5, IFNA, and IFNB in the s3-siRNA LPS treatment group is not significantly higher than that in s3-siRNA vehicle control group (P > 0.05), their gene expression has numerically higher values in the s3-siRNA LPS group (Figure 2), and it appeared that the gene expression decrease is relatively alleviated after LPS treatment. These results indicated that knockdown of NLRC5 negatively modulated gene expression of IFNA and IFNB in chicken HD11 cells. There was no significant change in IL-6 and MHC class I gene expression in NLRC5 knocked-down cells.
Figure 2.
Gene expression of NLRC5, IFNA, and IFNB in s3-siRNA transfected HD11 macrophages cells after vehicle-treatment, LPS (1 μg/ml), and poly (I:C) (0.1 ng/ml) treatment at 4 hps. Data are shown as the fold change in expression for each gene between s3-siRNA and negative siRNA transfections, among vehicle-treatment, LPS treatment, and poly (I:C) treatment. a: NLRC5; b: IFNA c: IFNB. RNA assays were carried out in triplicates by qPCR. P values are flagged with asterisks when lower than 0.05 (*), 0.01 (**).
NLRC5 is a highly conserved member of the NLR family and has been reported to be involved in type I IFN and NF-kB signaling pathways, as well as regulating MHC class I [9,22-24,26-28]. However, the specific role of NLRC5 in regulating inflammatory immune response remains unsolved [24,45]. Cui et al. showed that NLRC5 is a negative modulator of NF-kB and the type I IFN pathways. NLRC5 potently inhibited the NF-kB pathway through blocking phosphorylation of two subunits of IKK complex, IKKA and IKKB. It also repressed RLR-mediated type I IFN response by interaction with RIG-I and MDA5 [27]. Similar results have been reported that NLRC5 overexpression resulted in down-regulation of NF-kB and ISRE in the type I IFN-dependent pathway in HEK293T cells [9]. NLRC5 knockdown in RAW264.7 cells induced secretion of proinflammatory cytokines, such as TNF, IL-6, and IL-1B [9]. In contrast, Kuenzel et al. reported that overexpression of NLRC5 in epithelial cells induced greater mRNA levels of IFNA, PRKRIR, and OAS1 [25]. siRNA-mediated knockdown of NLRC5 significantly impaired induction of IFNA in human foreskin fibroblasts cells (HEF) after cytomegalovirus (CMV) infection [25], which indicates NLRC5 functions as a positive regulator of type I IFN. The same conclusion was proposed by Neerincx et al. [26], who showed that NLRC5 knockdown reduced the secretion of IFNB in THP-1 cells infected with Sendai virus, which predominately induces type I IFNs [46]. However, in the same study, overexpression of NLRC5 failed to induce NF-kB, IFNB, IRF3, IRF7, and ISRE in HEK293T cells, which revealed that impact of NLRC5 on downstream pathways might depend on the specific cell type examined. Cell-specificity of response was also supported by Kumar et al. [23], who reported that there was no difference in expression of IFNB, IL-6, and IFNA between wild type (WT) and NLRC5-deficient mice after infection with RNA viruses, DNA virus, and bacteria in macrophages and dendritic cells. Expression levels of TNFA, IL-6, and CCL5 in GM-CSF-induced bone marrow dendritic cells (GMDCs) after LPS treatment were not different between NLRC5-deficient and WT mouse. The authors suggested that NLRC5 may be not necessary for induction of inflammatory cytokines under physiologic conditions after viral and bacterial infections [23]. The roles of NLRC5 in inflammatory responsive pathways appear to depend on the cell types or specific species [24]. In the present study, expression of IFNA and IFNB was down-regulated in NLRC5 knocked-down HD11 cells. These results suggest that NLRC5 positively regulates type I IFNs in the chicken HD11 macrophage cell line. In addition, NLRC5 has been shown to regulate MHC class I gene expression, but reports on the direction of gene regulation are controversial. NLRC5 was reported to positively regulate the expression of MHC class I gene through binding to the promoter region of MHC class I in lymphoid and epithelial cell lines [28]. However, a conflicting effect was observed in RAW 264.7 cells, where knockdown of NLRC5 increased expression of MHC class I on cell surface [9]. In the present study, we detected expression of MHC class I, but there was no significant difference between NLRC5 knocked-down HD11 cells and controls, regardless of LPS or poly (I:C) treatment. These results indicate that NLRC5 is not a major regulator of MHC class I in chicken macrophages.
Conclusion
This is the first study reporting the role of NLRC5 in regulating type I IFNs (IFNA and IFNB), an NF-kB responsive gene (IL-6), and antigen presenting pathway gene (MHC class I) in the chicken. The expression of both IFNA and IFNB was down-regulated in NLRC5 knocked-down cells, and their expression was relatively restored by LPS treatment (P > 0.05). The consistent expression patterns between NLRC5 and IFNA and IFNB indicates that NLRC5 is a positive modulator in type I IFN pathway in chicken. In addition, we found that ST-LPS treatment could induce IFNB expression in the chicken HD11 macrophage cell line, although a functional LPS-specific MyD88-independent pathway is reportedly absent in chickens. Stimulation with LPS from different sources and doses may be responsible for differing reports on induction of IFNB expression.
Methods
Cell culture, siRNA transfection, and stimulation of cells with LPS and poly (I:C)
The chicken HD11 macrophage cell line [47], was cultured at 37°C and 5% CO2 concentration in RPMI1640 medium (Sigma-Aldrich Co.) supplemented with 10% heat-inactivated fetal calf serum, 10 mM HEPES, 0.1 mM non-essential amino acids, 2 mM glutamine, 1 mM sodium pyruvate, 100 U/ml penicillin, 100 μg/ml streptomycin, and 5 × 10-5 M 2-mercaptoethanol (pH 7.3). Three siRNAs, named s1, s2, s3-siRNA, were designed by using BLOCK-iT™ RNAi Designer (Invitrogen, Carlsbad, CA). s1-siRNA: 5'-CAUGGACGUGUCAUCAGCUUCUAAA-3', s2-siRNA: 5'-GGACGUUUAUCAUGUUGCUAGCUGA-3', s3-siRNA: 5'-CAUAACACUGCAGUCCUGAGGUUUA-3'. Five siRNAs (s1-siRNA, s2-siRNA, s3-siRNA, mixture of s1, s2, s3-siRNAs, and negative-siRNA) at 100 pM were used to transfect cells by following the manufacturer's instructions for use of Lipofectamine™ RNAiMAX (invitrogen). Transfection efficiency was evaluated by using a positive control, BLOCK-iT™ Alexa Fluor® Red Fluorescent after 6 hours of transfection under a fluorescent microscope. Twenty-four hours post transfection, the HD11 cells were exposed to ST-LPS (1 μg/ml) (Sigma-Aldrich Co.) or poly (I:C) (Invivogen, Carlsbad, CA) (0.1 ng/ml), or vehicle as a control. At 2, 4, 6, and 8 hours post simulation, the cells were collected for RNA extraction. Three biological triplicates were used in each group.
RNA isolation and quantitative reverse transcriptase-PCR
RNA was isolated by using RNAqueous® Kit (Ambion, Austin, TX) followed by DNA treatment using DNA-free™ Kit (Applied Biosystems, Foster City, CA). The expression of NLRC5, IFNA, IFNB, IL-6, and MHC class I was quantified by qRT-PCR using QuantiTect SYBR Green RT-PCR (Qiagen, Waltham, MA). The primers for 28 s, IL-6, and MHC class I have been previously reported [48,49]. The primers specific for the other genes were as follows: NLRC5 (F-5'- TGAGCTACACGTCAGGAAGGA-3', R-5'-GCTCTGCAGAATGGACACAA-3'); IFNA (F-5'- GACAGCCAACGCCAAAGC-3', R-5'-GTCGCTGCTGTCCAAGCATT-3'); IFNB (F-5'- CTGGATTGACCGCACACGCCA-3', R-5'- GGGAGCGCGTGCCTTGGTTTA-3'); Each reaction was run in triplicate and in a final volume of 25 ul with 50 ng/μl or 75 ng/ul total RNA, 12.5 ul QuantiTect SYBR Green master mix, 0.25 ul QuantiTect RT mix, forward and reverse primers, and RNase-free water. Samples were randomly assigned to PCR plates. The slopes for genes were determined with 10-fold serial dilutions. Adjusted cycle threshold (C(t)) values were calculated by following equation: 40 - [C (t) sample mean + (C(t) 28s median -C(t) 28s mean) * (gene slope/28s slope)]. Data were analyzed with the JMP software (SAS Institute, Cary, NC). The main fixed effects were siRNA, treatment, time, and interactions of these effects. Plate was included as a random effect. Multiple comparisons of least squares (LS) means for siRNA, treatment, and time effects were determined by the Tukey-Kramer Honestly Significant Differences test (JMP, SAS Institute, 2005). This test was selected because it allows multiple comparisons among the different treatments (five siRNAs and three treatments including LPS, poly (I:C), and non-treatment controls) without dividing the whole data set, which results in a more robust analysis. Differences were considered to be statistically significant when the P value was less than 0.05. Results were described as fold-change determined by 2-ΔΔCt method.
Abbreviations
AP-1: Activator protein; CCL: Chemokine (C-C motif) ligand; GM-CSF: Granulocyte-macrophage colony-stimulating-factor; IKK: Inhibitor of kappa B (IkB) kinase complex; MDA: Melanoma differentiation-associated protein; MHC: Major histocompatibility complex; MyD88: myeloid differentiation primary response gene 88; NF-kB: Nuclear factor kappa-light-chain-enhancer of activated B cells; OAS1: 2'-5'-oligoadenylate synthetase 1; PRKRIR: protein-kinase, interferon-inducible double stranded RNA dependent inhibitor, repressor P58 repressor; TIRAP: TIR domain containing adaptor protein; TRAM: TRIF-related adapter molecule; TRIF: TIR-domain-containing adapter-inducing IFNB.
Authors' contributions
LL carried out the experiments and data analysis for qPCR, and drafted the manuscript. CC designed the study and participated in the experiment, data interpretation, and revision of the manuscript. GBC performed a pre-experiment. JDH participated in qPCR experiments. SJL participated in the design of the study, data interpretation and revision of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Ling Lian, Email: lianlinglara@126.com.
Ceren Ciraci, Email: cerenciraci@gmail.com.
Guobin Chang, Email: passioncgb@163.com.
Jingdong Hu, Email: hjd1968@163.com.
Susan J Lamont, Email: sjlamont@iastate.edu.
Acknowledgements
This research was supported by Animal Health Funds distributed from the United States Department of Agriculture to Iowa State University. We thank the State Scholarship Fund of China Scholarship Council for support of Ling Lian as a joint training PhD student in Iowa State University, the National Natural Science Fund of China (31172199) and the International Cooperation Program for Excellent Lecturers of 2009 from Jiangsu Provincial Education Department and Yangzhou University for support of Guobin Chang's visiting scholar period at Iowa State University, and the International Cooperation Program for Excellent Lecturers of 2009 from Shandong Provincial Education Department, China, for support of Jingdong Hu's visiting scholar period at Iowa State University.
References
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6(9):644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- Inohara C, McDonald C, Nunez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449(7164):819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442(7098):39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- Ting JP, Kastner DL, Hoffman HM. CATERPILLERs, pyrin and hereditary immunological disorders. Nat Rev Immunol. 2006;6(3):183–195. doi: 10.1038/nri1788. [DOI] [PubMed] [Google Scholar]
- Fritz JH, Girardin SE. How Toll-like receptors and Nod-like receptors contribute to innate immunity in mammals. J Endotoxin Res. 2005;11(6):390–394. doi: 10.1179/096805105X76850. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. MDA5/RIG-I and virus recognition. Curr Opin Immunol. 2008;20(1):17–22. doi: 10.1016/j.coi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Benko S, Magalhaes JG, Philpott DJ, Girardin SE. NLRC5 limits the activation of inflammatory pathways. J Immunol. 2010;185(3):1681–1691. doi: 10.4049/jimmunol.0903900. [DOI] [PubMed] [Google Scholar]
- Chen G, Shaw MH, Kim YG, Nunez G. NOD-like receptors: role in innate immunity and inflammatory disease. Annu Rev Pathol. 2009;4:365–398. doi: 10.1146/annurev.pathol.4.110807.092239. [DOI] [PubMed] [Google Scholar]
- Park JH, Kim YG, McDonald C, Kanneganti TD, Hasegawa M, Body-Malapel M, Inohara N, Nunez G. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J Immunol. 2007;178(4):2380–2386. doi: 10.4049/jimmunol.178.4.2380. [DOI] [PubMed] [Google Scholar]
- Tschopp J, Martinon F, Burns K. NALPs: a novel protein family involved in inflammation. Nat Rev Mol Cell Biol. 2003;4(2):95–104. doi: 10.1038/nrm1019. [DOI] [PubMed] [Google Scholar]
- Feldmann J, Prieur AM, Quartier P, Berquin P, Certain S, Cortis E, Teillac-Hamel D, Fischer A, de Saint Basile G. Chronic infantile neurological cutaneous and articular syndrome is caused by mutations in CIAS1, a gene highly expressed in polymorphonuclear cells and chondrocytes. Am J Hum Genet. 2002;71(1):198–203. doi: 10.1086/341357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampe J, Cuthbert A, Croucher PJ, Mirza MM, Mascheretti S, Fisher S, Frenzel H, King K, Hasselmeyer A, MacPherson AJ, Bridger S, van Deventer S, Forbes A, Nikolaus S, Lennard-Jones JE, Foelsch UR, Krawczak M, Lewis C, Schreiber S, Mathew CG. Association between insertion mutation in NOD2 gene and Crohn's disease in German and British populations. Lancet. 2001;357(9272):1925–1928. doi: 10.1016/S0140-6736(00)05063-7. [DOI] [PubMed] [Google Scholar]
- Hoffman HM, Mueller JL, Broide DH, Wanderer AA, Kolodner RD. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat Genet. 2001;29(3):301–305. doi: 10.1038/ng756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411(6837):599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- Jin Y, Mailloux CM, Gowan K, Riccardi SL, LaBerge G, Bennett DC, Fain PR, Spritz RA. NALP1 in vitiligo-associated multiple autoimmune disease. N Engl J Med. 2007;356(12):1216–1225. doi: 10.1056/NEJMoa061592. [DOI] [PubMed] [Google Scholar]
- Miceli-Richard C, Lesage S, Rybojad M, Prieur AM, Manouvrier-Hanu S, Hafner R, Chamaillard M, Zouali H, Thomas G, Hugot JP. CARD15 mutations in Blau syndrome. Nat Genet. 2001;29(1):19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nunez G, Cho JH. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411(6837):603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- Villani AC, Lemire M, Fortin G, Louis E, Silverberg MS, Collette C, Baba N, Libioulle C, Belaiche J, Bitton A, Gaudet D, Cohen A, Langelier D, Fortin PR, Wither JE, Sarfati M, Rutgeerts P, Rioux JD, Vermeire S, Hudson TJ, Franchimont D. Common variants in the NLRP3 region contribute to Crohn's disease susceptibility. Nat Genet. 2009;41(1):71–76. doi: 10.1038/ng.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. Evolutionary relationships of vertebrate NACHT domain-containing proteins. Immunogenetics. 2006;58(10):785–791. doi: 10.1007/s00251-006-0148-8. [DOI] [PubMed] [Google Scholar]
- Proell M, Riedl SJ, Fritz JH, Rojas AM, Schwarzenbacher R. The Nod-like receptor (NLR) family: a tale of similarities and differences. PLoS One. 2008;3(4):e2119. doi: 10.1371/journal.pone.0002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, Pandey S, Zou J, Kumagai Y, Takahashi K, Akira S, Kawai T. NLRC5 deficiency does not influence cytokine induction by virus and bacteria infections. J Immunol. 2011;186(2):994–1000. doi: 10.4049/jimmunol.1002094. [DOI] [PubMed] [Google Scholar]
- Davis BK, Roberts RA, Huang MT, Willingham SB, Conti BJ, Brickey WJ, Barker BR, Kwan M, Taxman DJ, Accavitti-Loper MA, Duncan JA, Ting JP. Cutting edge: NLRC5-dependent activation of the inflammasome. J Immunol. 2010;186(3):1333–1337. doi: 10.4049/jimmunol.1003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuenzel S, Till A, Winkler M, Hasler R, Lipinski S, Jung S, Grotzinger J, Fickenscher H, Schreiber S, Rosenstiel P. The nucleotide-binding oligomerization domain-like receptor NLRC5 is involved in IFN-dependent antiviral immune responses. J Immunol. 2010;184(4):1990–2000. doi: 10.4049/jimmunol.0900557. [DOI] [PubMed] [Google Scholar]
- Neerincx A, Lautz K, Menning M, Kremmer E, Zigrino P, Hosel M, Buning H, Schwarzenbacher R, Kufer TA. A role for the human nucleotide-binding domain, leucine-rich repeat-containing family member NLRC5 in antiviral responses. J Biol Chem. 2010;285(34):26223–26232. doi: 10.1074/jbc.M110.109736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Zhu L, Xia X, Wang HY, Legras X, Hong J, Ji J, Shen P, Zheng S, Chen ZJ, Wang RF. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell. 2010;141(3):483–496. doi: 10.1016/j.cell.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner TB, Li A, Biswas A, Lee KH, Liu YJ, Bayir E, Iliopoulos D, van den Elsen PJ, Kobayashi KS. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc Natl Acad Sci USA. 2010;107(31):13794–13799. doi: 10.1073/pnas.1008684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman JH, Lillehoj HS, Lamont SJ. Reduced nitric oxide production and iNOS mRNA expression in IFN-gamma-stimulated chicken macrophages transfected with iNOS siRNAs. Vet Immunol Immunopathol. 2008;125(3-4):375–380. doi: 10.1016/j.vetimm.2008.05.015. [DOI] [PubMed] [Google Scholar]
- Ciraci C, Lamont SJ. Avian-specific TLRs and downstream effector responses to CpG-induction in chicken macrophages. Dev Comp Immunol. 2010;35(3):392–398. doi: 10.1016/j.dci.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Ciraci C, Tuggle CK, Wannemuehler MJ, Nettleton D, Lamont SJ. Unique genome-wide transcriptome profiles of chicken macrophages exposed to Salmonella-derived endotoxin. BMC Genomics. 2010;11:545. doi: 10.1186/1471-2164-11-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson-McDermott EM, O'Neill LA. Signal transduction by the lipopolysaccharide receptor, Toll-like receptor-4. Immunology. 2004;113(2):153–162. doi: 10.1111/j.1365-2567.2004.01976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Genovese KJ, Nisbet DJ, Kogut MH. Involvement of phosphatidylinositol-phospholipase C in immune response to Salmonella lipopolysacharide in chicken macrophage cells (HD11) Int Immunopharmacol. 2006;6(12):1780–1787. doi: 10.1016/j.intimp.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Bagchi A, Herrup EA, Warren HS, Trigilio J, Shin HS, Valentine C, Hellman J. MyD88-dependent and MyD88-independent pathways in synergy, priming, and tolerance between TLR agonists. J Immunol. 2007;178(2):1164–1171. doi: 10.4049/jimmunol.178.2.1164. [DOI] [PubMed] [Google Scholar]
- Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4(7):499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- Leshchinsky TV, Klasing KC. Profile of chicken cytokines induced by lipopolysaccharide is modulated by dietary alpha-tocopheryl acetate. Poult Sci. 2003;82(8):1266–1273. doi: 10.1093/ps/82.8.1266. [DOI] [PubMed] [Google Scholar]
- Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat Immunol. 2008;9(4):361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimura N, Saitoh S, Matsumoto F, Akashi-Takamura S, Miyake K. Roles for LPS-dependent interaction and relocation of TLR4 and TRAM in TRIF-signaling. Biochem Biophys Res Commun. 2008;368(1):94–99. doi: 10.1016/j.bbrc.2008.01.061. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Hemmi H, Uematsu S, Hoshino K, Kaisho T, Takeuchi O, Takeda K, Akira S. TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signaling pathway. Nat Immunol. 2003;4(11):1144–1150. doi: 10.1038/ni986. [DOI] [PubMed] [Google Scholar]
- Hoebe K, Du X, Georgel P, Janssen E, Tabeta K, Kim SO, Goode J, Lin P, Mann N, Mudd S, Crozat K, Sovath S, Han J, Beutler B. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 2003;424(6950):743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 1999;11(1):115–122. doi: 10.1016/S1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat Immunol. 2003;4(2):161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- Keestra AM, van Putten JP. Unique properties of the chicken TLR4/MD-2 complex: selective lipopolysaccharide activation of the MyD88-dependent pathway. J Immunol. 2008;181(6):4354–4362. doi: 10.4049/jimmunol.181.6.4354. [DOI] [PubMed] [Google Scholar]
- Esnault E, Bonsergent C, Larcher T, Bed'hom B, Vautherot JF, Delaleu B, Guigand L, Soubieux D, Marc D, Quere P. A novel chicken lung epithelial cell line: characterization and response to low pathogenicity avian influenza virus. Virus Res. 2011;159(1):32–42. doi: 10.1016/j.virusres.2011.04.022. [DOI] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Henao-Mejia J, Flavell RA. Regulation of the antimicrobial response by NLR proteins. Immunity. 2011;34(5):665–679. doi: 10.1016/j.immuni.2011.05.007. [DOI] [PubMed] [Google Scholar]
- Hua J, Liao MJ, Rashidbaigi A. Cytokines induced by Sendai virus in human peripheral blood leukocytes. J Leukoc Biol. 1996;60(1):125–128. doi: 10.1002/jlb.60.1.125. [DOI] [PubMed] [Google Scholar]
- Beug H, von Kirchbach A, Doderlain G, Conscience JF, Graf T. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell. 1979;18:375–390. doi: 10.1016/0092-8674(79)90057-6. [DOI] [PubMed] [Google Scholar]
- Abdul-Careem MF, Hunter BD, Sarson AJ, Parvizi P, Haghighi HR, Read L, Heidari M, Sharif S. Host responses are induced in feathers of chickens infected with Marek's disease virus. Virology. 2008;370(2):323–332. doi: 10.1016/j.virol.2007.09.013. [DOI] [PubMed] [Google Scholar]
- Kaiser P, Underwood G, Davison F. Differential cytokine responses following Marek's disease virus infection of chickens differing in resistance to Marek's disease. J Virol. 2003;77(1):762–768. doi: 10.1128/JVI.77.1.762-768.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]