Abstract
Background
Extraintestinal pathogenic Escherichia coli are important pathogens of human and animal hosts. Some human and avian extraintestinal pathogenic E. coli are indistinguishable on the basis of diseases caused, multilocus sequence and phylogenetic typing, carriage of large virulence plasmids and traits known to be associated with extraintestinal pathogenic E. coli virulence.
Results
The gene tkt1 identified by a previous signature-tagged transposon mutagenesis study, was found on a 16-kb genomic island of avian pathogenic Escherichia coli (APEC) O1, the first pathogenic Escherichia coli strain whose genome has been completely sequenced. tkt1 was present in 39.6% (38/96) of pathogenic Escherichia coli strains, while only 6.25% (3/48) of E. coli from the feces of apparently healthy chickens was positive. Further, tkt1 was predominantly present in extraintestinal pathogenic E. coli belonging to the B2 phylogenetic group, as compared to extraintestinal pathogenic E. coli of other phylogenetic groups. The tkt1-containing genomic island is inserted between the metE and ysgA genes of the E. coli K12 genome. Among different extraintestinal pathogenic E. coli of the B2 phylogenetic group, 61.7% of pathogenic Escherichia coli, 80.6% of human uropathogenic E.coli and 94.1% of human neonatal meningitis-causing E. coli, respectively, harbor a complete copy of this island; whereas, only a few avian fecal E. coli strains contained the complete island. Functional analysis showed that Tkt1 confers very little transketolase activity but is involved in peptide nitrogen metabolism.
Conclusion
These results suggest tkt1 and its corresponding genomic island are frequently associated with avian and human ExPEC and are involved in bipeptide metabolism.
Background
Extraintestinal pathogenic Escherichia coli (ExPEC) including uropathogenic E. coli (UPEC), neonatal meningitis E. coli (NMEC), and avian pathogenic E. coli (APEC), cause infection in humans and/or animals [1]. One of the most common diseases caused by ExPEC in animals is systemic colibacillosis due to APEC that often starts as a respiratory tract infection and progresses to septicemia, which is characterized by fibrinous lesions of the internal organs [2]. A variety of factors have been associated with ExPEC virulence including pilus adhesins, the temperature-sensitive hemagglutinin (Tsh), serum resistance traits (e.g., iss and traT), iron acquisition systems (e.g., aerobactin, salmochelin and yersiniabactin), and vacuolating autotransporter toxin (Vat) [2,3]. Chromosomally located virulence genes occur widely among all ExPEC subpathotypes [4,5], but plasmid-linked virulence genes are more common in APEC and NMEC subpathotypes than they are in UPEC [5]. It is also well known that ExPEC strains often contain multiple pathogenicity islands (PAIs), which are horizontally acquired genomic regions of 20 to 200 kb. PAIs are present in pathogenic bacteria but absent from E. coli K12, and carry genes encoding one or more virulence factors.
Since they are horizontally acquired, they differ from the rest of the genome in G+C content and codon usage [6]. The first PAI identified on the APEC chromosome was the VAT-PAI, which contains the vacuolating autotransporter gene, vat, a contributor to APEC virulence. vat has been reported to be present in about half of the APEC, UPEC, and NMEC strains [7]. A selC-associated genomic island of APEC strain BEN2908 was subsequently described. This island is prevalent in ExPEC strains and is involved in carbohydrate uptake and virulence [8]. Two PAIs were characterized in APEC O1. One is the PAI localized in the large plasmid pAPEC-O1-ColBM [9,10], and the other is PAI IAPEC-O1, harboring ireA, the pap operon and the invasion locus tia [11]. The PAI IAPEC-O1-related genes occurred not only in strains belonging to the APEC subpathotype (17.9%) but also in UPEC (10.7%) and NMEC (28.0%).
In a previous study we used signature-tagged transposon mutagenesis (STM) to identify 28 virulence-associated genes in APEC [12]. One of the genes identified, tkt1, encodes a transketolase-like protein whose amino acid sequence shares 68% identity to TktA of a Vibrio cholerae strain [13]. However, it does not show any similarity with the tktA gene of E. coli MG1655 at the nucleotide level. Recent completion of the first APEC genomic sequence (APEC O1) showed that tkt1 is localized on an 'as-yet' uncharacterized genomic island [14]. Here, we sought to better understand the prevalence and function of tkt1 and its associated genomic island in APEC pathogenicity.
Methods
Bacterial strains, plasmids and growth conditions
All bacterial strains and plasmids used in this study are listed in Table 1. APEC O1, an E. coli O1:K1:H7 strain that shares strong similarities with sequenced human ExPEC genomes [14], was used to construct the mutants and as a positive control in virulence and other functional assays. A tktA mutant, BJ502 of an E. coli K12 strain, was used as the control strain in the functional analysis of APEC O1 mutants [15], and E. coli DH5α was employed as a negative control in the virulence assays. A well-characterized collection of APEC, fecal E. coli isolated from the feces of healthy birds (avian fecal E. coli), human UPEC, and human NMEC were used for gene prevalence studies. Strains were grouped phylogenetically using multiplex PCR [16]. Cells were routinely grown at 37°C in Luria Bertani broth (LB) supplemented with an appropriate antibiotic: kanamycin (Km; 50 mg ml-1), chloramphenicol (Cm; 25 mg ml-1), or ampicillin (Amp; 100 mg ml-1), unless otherwise specified.
Table 1.
Bacterial strains and plasmids used in this study
| Strain | Description | Reference |
|---|---|---|
| APEC O1 | O1:K1:H7; fyuA, sitA, chuA, irp2, iroN, ireA, tsh, iucD, fimC, iss, ompA, vat, traT; contains four plasmids, including pAPEC-O1-ColBM | [14] |
| BJ502 | E. coli K12, ΔtktA | [15] |
| DH5α | E. coli K12 | |
| APEC O1-Mtkt1 | APEC O1 derivative, Δtkt1 | this study |
| APEC O1-MtktA | APEC O1 derivative, ΔtktA | this study |
| S17λpir | recA thi pro hsdR- M+ RP4::2-Tc::Mu::Km Tn7 lysogenized with λpir phage | [12] |
| S17pGP tkt1 | S17λpir with plasmid pGP704 tkt1 | this study |
| APEC O1-Ctkt1 | APEC O1 Mtkt1 with plasmid pGP tkt1 inserted into bacterial chromosome | this study |
| APEC O1-P1 | APEC O1 Mtkt1 with plasmid pBAD tkt1 | this study |
| BJ502-P1 | BJ502 with plasmid pBAD24 | this study |
| BJ502-P2 | BJ502 with plasmid pBAD tkt1 | this study |
| BJ502-P3 | BJ502 with plasmid pBAD tktA | this study |
| APEC collection | 452 APEC strains isolated from lesions of birds clinically diagnosed with colibacillosis | [17] |
| Avian fecal E. coli | 106 avian fecal E. coli strains were isolated from the feces of apparently healthy birds | [17] |
| UPEC collection | 200 uropathogenic E. coli strains from from MeritCare Medical Center in Fargo, North Dakota | [18] |
| NMEC collection | 91 human neonatal meningitis-causing E. coli strains from the cerebrospinal fluid of newborns in the Netherlands, isolated from 1989 through 1997 and from Dr. K. S. Kim at John Hopkins. | [19] |
| Plasmids | ||
| pGP704 | Apr, suicide plasmid | [20] |
| pBAD24 | Apr, expression plasmid with arabinose-inducible promoter | [21] |
| pKD46 | Apr; expresses λ red recombinase | [22] |
| pKD3 | cat gene, template plasmid | [22] |
| pGP tkt1 | pGP704 derivative harboring tkt1 gene | this study |
| pBAD tkt1 | pBAD24 derivative, tkt1 gene under the control of PBAD | this study |
| pBAD tktA | pBAD24 derivative, tktA gene under the control of PBAD | this study |
PCR and multiplex PCR
DNA templates were prepared by the rapid boiling-lysis method. Primer pairs used were tkt1- F 5'- cttacggcggtactttcctg-3'and tkt 1-R 5'-gtacgccgcatcctgattat-3'; genomic island left junction primer pair piaL-F 5'-cgacatcatggattcgattg-3'and piaL-R 5'-ggatggtgctggatcgtact-3'; and genomic island right junction primer pair piaR-F 5'-gcgccactcttcttctgttc-3' and piaR-R 5'-tcagctaattgctcggcttt-3' PCR was accomplished under the following reaction conditions: 4 mM magnesium chloride, 0.25 mM deoxynucleotide triphosphates 0.3 uM each primer, and 1 Unit Taq DNA polymerase. Reactions were performed in a Mastercycler EP machine (Eppendorf, Germany) using the following cycling parameters: 94°C for 4 min; 30 cycles of 94°C for 30 sec, 55°C for 30 sec, 72°C for 2 min; and a final cycle of 72°C for 10 min.
Construction of plasmids, mutants and complemented strains
Enzymes used for generation of constructs were purchased from New England Biolabs. The pBAD expression system (Invitrogen) was used for cloning and arabinose-inducible expression of tkt1 and tktA. The coding sequence of tkt1 was amplified by PCR using genomic DNA of APEC O1 as the template. The Advantage™ 2 PCR kit (Clontech, Mountain View, CA) was used in these experiments according to the manufacturer's directions. The primers used for tkt1 gene were the tkt1E-F primer 5'-agctccatggattcacaattactggctaacg-3', which introduces an Ncol site (underlined bases) and the tkt1E-R primer 5'- gcattctagagtcatcctttcaccccttgtgcag-3' which introduces an XbaI site (underlined bases). The primers used for tktA were tktAE-F 5'-agctccatggcctcacgtaaagagcttgcc-3'and tktAE-R 5' gcattctagattgcggcccttctcacaaagcat-3' The complete tkt1 gene and tktA were cloned into the expression vector pBAD24 using the created NcoI and XbaI sites [21] to obtain pBAD tkt1 and pBAD tktA, respectively (Table 1). The APEC O1 mutant strain APEC O1 Mtkt1 with plasmid pBAD tkt1 was designated as APEC O1-P1, and the E. coli K12 mutant strains BJ502 harboring the empty pBAD24, pBAD tkt1 and pBAD tktA plasmids were designated as BJ502 p1, BJ502 p2 and BJ502 p3, respectively.
Deletion of tkt1 was achieved using the method of Datsenko and Wanner [22]. The Cm resistance cassette in pKD3, flanked by 5' and 3' sequences of tkt1, was amplified from genomic DNA of strain APEC O1 using primers tkt1M-F (5'-ttagcgggctggtttcagcccgccagacagagagagctgaagtgtgtaggctggagctgcttcga-3') and tkt1M-R (5'-tcaaggggtaaaaggtcatcctttcaccccttgtgcaggtcatatgaatatcctccttag-3') and was introduced into APEC O1 by homologous recombination using λ Red recombinase. Successful Δtkt1::Cm mutation was confirmed by PCR, using primers flanking the tkt1 region. The Δtkt1::Cm derivative of APEC O1 was designated APEC O1 Mtkt1. The mutant strain APEC O1 MtktA (Table 1), a ΔtktA::Cm derivative of APEC O1, was generated using primer pair tktAM-F 5'-aagggccgcatttgcggcccttctcacaaagcatcttaccgagtgtaggctggagctgcttcga-3' and tktAM-R 5'-cgttaagggcgtgcccttcatcatccgatctggagtcaaacatatgaatatcctccttag-3'. The Δtkt1 mutant strain APEC O1 Mtkt1, was complemented by single-copy integration of the plasmid pGPtkt1. The tkt1 operon, including the 300-bp upstream DNA sequence, was amplified by PCR using primers tkt1C-F 5'-tgacagatctgggctatgcagcgatttactac-3' and tkt1C-R 5'-cagttctagatgtgcaggtttagctgttcagt-3'. Plasmid pGPtkt1 was constructed by cloning this BglII-XbaI (underlined bases) fragment into the same sites of suicide vector pGP704 [10,20]. PGPtkt1 was conjugated from strain S17- pGPtkt1 to strain APEC O1 Mtkt1. A strain that was resistant to Amp and found to contain a full-length copy of the tkt1 gene, as confirmed by PCR, was designated APEC O1 Ctkt1 (Table 1).
Phenotype microarray
The Phenotype Microarray (PM) assay was performed essentially according to published methods using Biolog PM plates (Biolog Inc., CA). APEC O1 and APEC O1Δtkt1 were grown overnight at 37°C in BUG_B agar (Biolog Inc., CA). Cells were washed with IF-0 GN Base inoculating fluid (Biolog Inc., CA), and then resuspended in IF-10 GN Base inoculating fluid (Biolog Inc., CA) at a density corresponding to 85% transmittance (approximately 0.185 OD600 nm). The suspensions were then inoculated into microplate PM1-8 for the metabolism test (Biolog Inc., CA) at a volume of 100 μl per well. Cell growth was monitored by measuring the respiration-dependent color change of tetrazolium violet in each well. The PM assay was performed twice.
Results
tkt1 is strongly associated with APEC and ExPEC of the B2 phylogenetic group
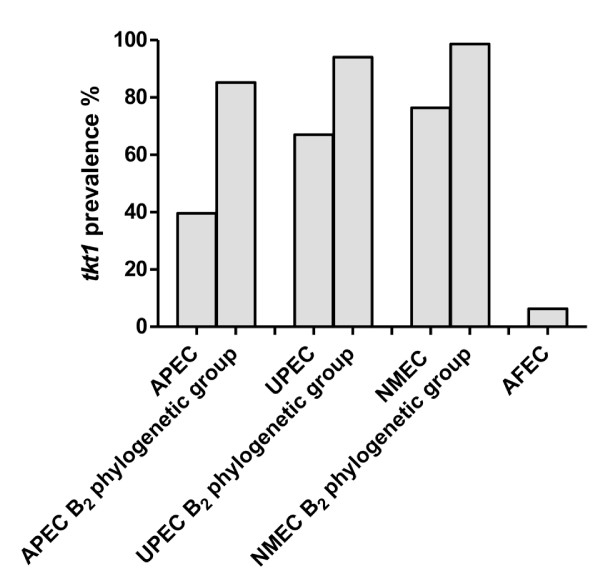
tkt1 was initially identified as an APEC-specific gene by genomic subtraction [23]. Here, we examined its prevalence in a collection of APE and avian fecal E. coli. A pair of primers designated tkt1F and tkt1R (Table 1) were used to test 96 APEC and 48 avian fecal E. coli strains by PCR. Thirty-eight strains from the APEC group (39.6%) were positive for tkt1; while only three strains from avian fecal E. coli group were positive (6.25%). Thus, tkt1 is significantly more likely to be present in pathogenic strains (P < 0.001). Interestingly, 12 out of 14 (85.7%) APEC strains from phylogenetic group B2 were tkt1 positive; while the prevalence of tkt1 in APEC strains from any other phylogenetic group was much lower (group A, 16.1%; group B1, 12.5% and group D, 47.4%). Since only 14 strains of phylogenetic group B2 were used, a number inadequate for statistical analysis, an additional 47 APEC strains of phylogenetic B2 group were selected from our collection and examined. A total of 52 out of 61 APEC (85.2%) from phylogenetic group B2 was found to be positive for tkt1 (Figure 1), demonstrating that tkt1 is significantly (P < 0.01) associated with APEC strains belonging to phylogenetic group B2.
Figure 1.
Prevalence of tkt1 in ExPEC strains.
Several recent studies have shown that most human ExPEC strains belong to the B2 phylogenetic group [4,24], and analysis of the genomic sequences of UPEC strains CFT073 and 536 revealed that they contained tkt1. Such results suggest that tkt1 might also be prevalent among human ExPEC. To verify this hypothesis, 94 UPEC strains and 89 NMEC strains were examined by PCR for the presence of tkt1. As expected, 67% of UPEC and 76.4% of NMEC strains were positive for tkt1 gene. As was the case with APEC, the majority of UPEC (94%) and NMEC (98.6%) belonging to phylogenetic group B2 were positive for tkt1. Therefore, tkt1 gene has been significantly associated with ExPEC strains from human and avian hosts, especially with strains of phylogenetic group B2.
tkt1 is located on a novel genomic island prevalent in ExPEC of the B2 phylogenetic group
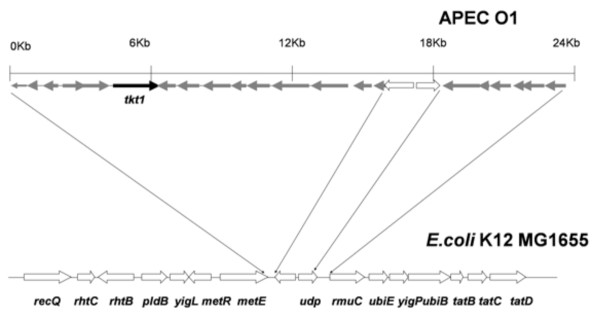
Study of the genomic sequences of several ExPEC showed that tkt1 is located in a 16-kbp genomic island inserted between metE and ysgA [14,25,26]. The overall G+C content of this island is 48.57%, whereas the average G+C content of the E. coli K-12 genome is 50.8%. This discrepancy in G+C content suggests that this particular stretch of DNA does not belong to the E. coli backbone and is foreign. The entire genomic island contains 15 ORFs, including tkt1, with the function of most of them 'as yet' unknown. Products encoded by certain ORFs have been assigned hypothetical functions, including a putative permease, putative glucose-specific IIBC component of a PTS system, carbonate kinase-like protein, and putative transcriptional regulators. Besides this genomic island, there is another small genomic islet of about 5 Kb located between the udp and rmuC genes. This small islet contains 6 ORFs with unknown functions (Figure 2).
Figure 2.
Genetic organization of the 16 Kb tkt1 genomic island and its flanking regions within the APEC O1 genome, drawn to scale. The ORFs present in this genomic island are listed in the Table 2. There is an islet containing 6 ORFs between the udp and rmuC genes.
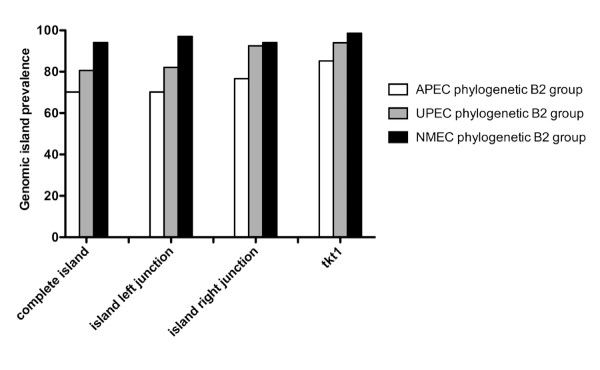
A multiplex PCR panel was developed to determine the presence of the tkt1-containing genomic island in ExPEC of the B2 phylogenetic group. Three pairs of primers were designed to amplify the left and right junctions, as well as the tkt1 gene in 61 APEC, 67 UPEC and 68 NMEC belonging to phylogenetic group B2. The results suggest that 70.2% of APEC, 80.6% of UPEC and 94.1% of NMEC strains from B2 phylogenetic group carry a complete copy of this genomic island (Figure 3). Thus, these data demonstrate that this genomic island is significantly associated with ExPEC strains belonging to the B2 phylogenetic group.
Figure 3.
The prevalence of tkt1 genomic island in phylogenetic group B2 of ExPEC strains.
Tkt1 could not complement TktA in E. coli K12
Recently, genome sequencing of APEC O1 revealed that tkt1 gene encodes a transketolase-like protein whose amino acid sequence shares 68% identity to TktA of a V. cholerae strain [13], although tkt1 does not show any similarity to tktA of E. coli MG1655 at the nucleotide level. To explore the function of Tkt1, mutants with single deletions of tkt1 and tktA were constructed in the APEC O1 strain using the method of Datsenko and Wanner [22], and their growth was compared to each other and the wild type in M9 plates with L-arabinose as the sole carbon source. The results showed that both mutants of APEC O1 were able to grow in M9 with the tktA mutant growing slightly slower than the tkt1 mutant. However, the control strain E. coli K12 BJ502, which has a mutation in the tktA, failed to grow in M9 plates with L-arabinose (Figure 4) [15]. These results suggested that, APEC O1 has another gene that is capable of complementing the tktA mutation. To ascertain if it were tkt1, a Tkt1 over-expression plasmid pBAD tkt1 and a TktA over-expression plasmid pBAD tktA, in which the tkt1 or the tktA gene is under the control of the PBAD promoter, were constructed. Induction of TktA expression could recover growth of BJ502-P3 on M9 plates with L-arabinose as the sole carbon source, while Tkt1 expression could not recover growth of BJ502-P2 (Figure 4). These results suggested that Tkt1 has very little transketolase activity, if any.
Figure 4.
Tkt1 could not complement TktA in E. coli K12. 1, APEC O1; 2, APEC O1 Mtkt1; 3, APEC O1 MtktA; 4, BJ502; 5, BJ502-P1; 6, BJ502-P2 and 7 BJ502-P3.
Tkt1 is involved in peptide nitrogen metabolism
Transketolase TktA is involved in carbon metabolism, and Tkt1 shows a high similarity (68%) to transketolase TktA. To determine if this transketolase-like protein is involved in metabolism, we performed the PM assay under a total of 760 culture conditions (carbon sources, nitrogen sources, phosphorus and sulfur sources, nutrient supplements, and peptide nitrogen sources). Growth of wild-type APEC O1 and its tkt1 isogenic mutant was measured using the PM assay system. The time course of cell growth was monitored by measuring the cell density-dependent increase in respiration. No difference between the tkt1 mutant and its wild type in the utilization of carbon sources was detected nor were differences in the use of nitrogen, phosphorus and sulfur sources or nutrient supplements observed. Interestingly, the tkt1 mutant showed defects in the use of Pro-Ala or Phe-Ala as a peptide nitrogen source. These defective phenotypes were reproducible, and induction of Tkt1 expression in APEC O1-P1 resulted in the use of both peptides as nitrogen sources reverting the lost phenotype. Complementation assay was also done by using Biolog plates and 0.2% arabinose was added to induce expression of Tkt1.
Discussion
Human and avian ExPEC are both important pathogens that cause widely prevalent and/or highly significant extraintestinal diseases. The gene tkt1, encoding a transketolase-like protein and sharing 68% amino acid identity with TktA of a V. cholerae strain [13], was firstly identified as a virulence-associated gene from APEC strains by genomic subtractive hybridization [23]. This gene was also thought to be involved in APEC virulence from the results of a previous STM study [12]. Unlike tktA or tktB, which are unequivocally present in both avian fecal E. coli and APEC, tkt1 was predominantly present among APEC (39.6%) but absent from most of the intestinal E. coli (6.25%) examined [27], suggesting that this gene may play a significant role in the pathogenesis of avian colibacillosis. APEC and human ExPEC have recently been shown to possess remarkable similarities in their disease-causing abilities, serogroups associated with disease, and virulence genotypes and traits, such as the presence of adhesins, iron acquisition systems, toxins, protectins and invasins that enable them to grow and cause disease in the extraintestinal host environments [5,28]. The inference of a close genetic relationship between APEC and human ExPEC strains was further substantiated by the distribution of tkt1. About 67% of UPEC and 76.4% of NMEC strains examined in this study harbor tkt1. Like many other virulence genes of ExPEC, tkt1 is also phylogenetically distributed. Of the ExPEC belonging to B2 phylogenetic group, 85.2% APEC, 94.0% of UPEC and 98.6% of NMEC were positive for tkt1. E. coli from phylogenetic group B2 have already been experimentally and epidemiologically associated with extraintestinal infections [29,30]. These results also suggest that tkt1 may play a role in the pathogenesis of human ExPEC as well as APEC.
Genomic sequencing of APEC O1 revealed more than 40 genomic islands; several of them are theoretically involved in virulence [9]. Common features of most, if not all PAIs, include that they encode one or more virulence factors; range in size from 10 to 200 kb; and are likely introduced into the genome via horizontal transfer, resulting in G-C ratios and codon usage that may deviate from the organism's typical pattern. Often PAIs are flanked by small direct repeats and are associated with the 3' end of tRNA genes. PAIS may be phage-derived, but some are thought to originate from plasmids. They may contain mobility elements, such as integrons, transposons, and insertion sequences, and if they move, are likely carried on plasmids, conjugative transposons, or phages, whose loss may spontaneously convert a virulent into an avirulent organism [6]. Similarly, the genomic island encoding tkt1 is 16 Kb in size and present in the APEC O1 genome but absent from the sequenced genome of the E. coli K12 strain MG1655. Moreover, the overall G+C content of this island is 48.57%, whereas the average G+C content of the E. coli K-12 genome is 50.8%. This discrepancy in G+C content further suggests that this particular stretch of DNA does not belong to the E. coli K12 backbone and is foreign-derived. Also, the genomic island encoding tkt1 is localized in close proximity to tRNA genes. Unlike classical PAIs, no flanking direct repeats or mobility elements such as integrases or transposases were found in this genomic island.
However, such mobility elements may have been lost during the evolutionary process. Horizontal transfer of genes by genomic islands or PAIs is a common phenomenon in extracellular bacterial pathogens. The acquisition of genes in this way allows bacteria to adapt to a new or changing environment thus contributing to the fitness and/or virulence of the recipient organism.
PAIs have been described in several well-known ExPEC strains, including E. coli strains 536, CFT073, J96, UTI189, RS218 and APEC O1. Indeed, comparative analysis of the APEC O1 genome and other ExPEC genomes revealed that APEC and human ExPEC share more than 28 pathogenicity (genomic) islands [9,25,26,31]. Among them, the genomic island encoding tkt1 was notable in that it was found among all sequenced ExPEC genomes. The multiplex PCR results of this study further demonstrated that a complete copy of this genomic island is significantly associated with both avian and human ExPEC strains of phylogenetic group B2. These observations suggest that the tkt1 genomic island may contribute to the virulence/fitness of both avian and human ExPEC.
Though Tkt1 shares 68% amino acid identity with TktA of a V. cholerae strain [13], it does not show any homology at the nucleotide level with tktA of E. coli MG1655. In E. coli K12, tktA encodes the transketolase A, which is responsible for the major enzymatic activity of transketolase in E. coli. Transketolase is a link between glycolysis and the pentose phosphate pathway and is involved in the catabolism of pentose sugars, formation of D-ribose 5-phosphate, and provision of D-erythrose 4-phosphate which is a precursor of aromatic amino acids, aromatic vitamins and pyridoxine [32]. A previous study showed that the E. coli K12 mutant BJ502 that carries a mutation in tktA was unable to use L-arabinose or D-Xylose as the sole carbon source and required aromatic acids for growth on a minimal medium. The functional analysis in this study demonstrated that over-expression of Tkt1 in E. coli K12 mutant strain BJ502 could not recover its growth in M9 medium with L-arabinose as the sole carbon source; while over-expression of TktA could. These results suggest that tkt1 could not complement the tktA mutation in E. coli K12 and Tkt1 confers very little transketolase activity, if any.
Most studies of bacterial pathogenesis have focused on classical virulence factors such as toxins, adhesins, iron uptake systems and factors that confer resistance to innate and adaptive immune mechanisms. However, the role of metabolism in virulence and fitness of bacterial pathogens is becoming better appreciated. Recently, a selC-associated genomic island of APEC strain BEN2908 was found to be involved in carbohydrate uptake and virulence [8]. Also in the same APEC strain, a carbohydrate metabolic operon (frz) that is highly associated with ExPEC promotes fitness under stressful conditions and invasion of eukaryotic cells [33]. Our STM results showed that one tkt1 STM-mutant was out-competed by the wild type from two to a thousand fold in lung, heart, liver, kidney and spleen of 5-week-old chickens. The functional analysis using phenotype microarray revealed that a tkt1 mutant has defects in use of Pro-Ala or Phe-Ala as a nitrogen source. These results strongly suggest that tkt1 is involved in bipeptide metabolism and contributes to fitness and virulence of APEC. Interestingly, dipeptide transport proteins, DppA and OppA, were identified to be up-regulated when UPEC strain CFT073 was cultured in human urine compared to CFT073 cultured in LB depleted with iron [34]. The greatest challenges confronted by bacterial pathogens are environmental changes, including the rapid changes they encounter in nutrient availability [35]. In the course of evolution, pathogenic organisms have developed several mechanisms to sense and utilize available nutrient sources associated with particular niches or to favor the most efficiently metabolizable nutrient sources when exposed to a range of choices [36]. Thus, genes involved in metabolism, which are required for bacterial growth in specific infection sites, contribute to fitness and virulence. On the other hand, the efficiency of metabolism of a nutrient source or the presence of a specific nutrient source might serve as a signal to switch 'on' or 'off' specific virulence genes in particular infection niches [36].
Conclusions
A previously identified virulence-associated gene tkt1 was further characterized in this study. The results demonstrated that this gene is strongly associated with ExPEC strains of phylogenetic group B2 from human and avian origin and is localized in a genomic island. Function analyses showed that Tkt1 has very little transketolase activity and seems to be involved in peptide nitrogen metabolism.
Authors' contributions
The project was designed by GL, LN, LW. Experiments were performed by GL, SK,KT, YW, CL under supervision of GL and LN. The paper was co-drafted by LG and LN. All authors approved the final version of the manuscript.
Table 2.
ORFs present within the tkt1 genomic island
| ORF No. | ORF name Location of ORF Function | G+C content | |
|---|---|---|---|
| APECO1_2646 | 4312693..4312950 hypothetical protein | NC_008563 | |
| APECO1_2645 | 4312947..4313438 hypothetical protein | NC_008563 | |
| APECO1_2644 | 4313787..4314080 hypothetical protein | NC_008563 | |
| APECO1_2643 | 4314532..4315122 putative sugar isomerase | NC_008563 | |
| APECO1_2642 | 4315164..4316672 PTS system, glucose-specific IIBC component | NC_008563 | |
| APECO1_2641 | tkt1 | 4316723..4318720 Putative transketolase | NC_008563 |
| APECO1_2640 | 4318750..4319595 putative transcriptional regulatory | NC_008563 | |
| APECO1_2639 | 4319796..4320701 putative transcriptional regulatory | NC_008563 | |
| APECO1_2638 | 4320779..4322002 putative permease | NC_008563 | |
| APECO1_2637 | 4322028..4322417 hypothetical protein | NC_008563 | |
| APECO1_2636 | 4322434..4323390 catalyzes the reversible synthesis of carbamate | NC_008563 | |
| and ATP from carbamoyl phosphate and ADP | |||
| APECO1_2635 | yahG | 4323383..4324858 hypothetical protein | NC_008563 |
| APECO1_2634 | yahF | 4324804..4326363 hypothetical protein | NC_008563 |
| APECO1_2633 | yahE | 4326458..4327318 hypothetical protein | NC_008563 |
| APECO1_2632 | 4327324..4327992 putative isochorismatase hydrolase | NC_008563 | |
Contributor Information
Ganwu Li, Email: liganwu@iastate.edu.
Subhashinie Kariyawasam, Email: sxk91@psu.edu.
Kelly A Tivendale, Email: kellyat@unimelb.edu.au.
Yvonne Wannemuehler, Email: ywanne@iastate.edu.
Christa Ewers, Email: ewers.christa@vetmed.fu-berlin.de.
Lothar H Wieler, Email: wieler.lothar@vetmed.fu-berlin.de.
Catherine M Logue, Email: cmlogue@iastate.edu.
Lisa K Nolan, Email: lknolan@iastate.edu.
Acknowledgements
This work was supported by USDA NRICGP Microbial Functional Genomics Program (20083560418805).
References
- Russo TA, Johnson JR. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J Infect Dis. 2000;181(5):1753–1754. doi: 10.1086/315418. [DOI] [PubMed] [Google Scholar]
- Dziva F, Stevens MP. Colibacillosis in poultry: unravelling the molecular basis of virulence of avian pathogenic Escherichia coli in their natural hosts. Avian Pathol. 2008;37(4):355–366. doi: 10.1080/03079450802216652. [DOI] [PubMed] [Google Scholar]
- Li G, Tivendale KA, Liu P, Feng Y, Wannemuehler Y, Cai W, Mangiamele P, Johnson TJ, Constantinidou C, Penn CW. et al. Transcriptome analysis of avian pathogenic Escherichia coli O1 in chicken serum reveals adaptive responses to systemic infection. Infect Immun. 2011;79(5):1951–1960. doi: 10.1128/IAI.01230-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers C, Li G, Wilking H, Kiessling S, Alt K, Antao EM, Laturnus C, Diehl I, Glodde S, Homeier T. et al. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int J Med Microbiol. 2007;297(3):163–176. doi: 10.1016/j.ijmm.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Johnson TJ, Wannemuehler Y, Johnson SJ, Stell AL, Doetkott C, Johnson JR, Kim KS, Spanjaard L, Nolan LK. Comparison of extraintestinal pathogenic Escherichia coli strains from human and avian sources reveals a mixed subset representing potential zoonotic pathogens. Appl Environ Microbiol. 2008;74(22):7043–7050. doi: 10.1128/AEM.01395-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper JB, Hacker J, editor. The concept of pathogenicity islands. Washington, D.C: ASM Press; 1999. [Google Scholar]
- Parreira VR, Gyles CL. A novel pathogenicity island integrated adjacent to the thrW tRNA gene of avian pathogenic Escherichia coli encodes a vacuolating autotransporter toxin. Infect Immun. 2003;71(9):5087–5096. doi: 10.1128/IAI.71.9.5087-5096.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouikha I, Germon P, Bree A, Gilot P, Moulin-Schouleur M, Schouler C. A selC-associated genomic island of the extraintestinal avian pathogenic Escherichia coli strain BEN2908 is involved in carbohydrate uptake and virulence. J Bacteriol. 2006;188(3):977–987. doi: 10.1128/JB.188.3.977-987.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TJ, Johnson SJ, Nolan LK. Complete DNA sequence of a ColBM plasmid from avian pathogenic Escherichia coli suggests that it evolved from closely related ColV virulence plasmids. J Bacteriol. 2006;188(16):5975–5983. doi: 10.1128/JB.00204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Feng Y, Kariyawasam S, Tivendale KA, Wannemuehler Y, Zhou F, Logue CM, Miller CL, Nolan LK. AatA is a novel autotransporter and virulence factor of avian pathogenic Escherichia coli. Infect Immun. 2010;78(3):898–906. doi: 10.1128/IAI.00513-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariyawasam S, Johnson TJ, Nolan LK. The pap operon of avian pathogenic Escherichia coli strain O1:K1 is located on a novel pathogenicity island. Infect Immun. 2006;74(1):744–749. doi: 10.1128/IAI.74.1.744-749.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Laturnus C, Ewers C, Wieler LH. Identification of genes required for avian Escherichia coli septicemia by signature-tagged mutagenesis. Infect Immun. 2005;73(5):2818–2827. doi: 10.1128/IAI.73.5.2818-2827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L. et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406(6795):477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TJ, Kariyawasam S, Wannemuehler Y, Mangiamele P, Johnson SJ, Doetkott C, Skyberg JA, Lynne AM, Johnson JR, Nolan LK. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J Bacteriol. 2007;189(8):3228–3236. doi: 10.1128/JB.01726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephson BL, Fraenkel DG. Transketolase mutants of Escherichia coli. J Bacteriol. 1969;100(3):1289–1295. doi: 10.1128/jb.100.3.1289-1295.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont O, Cordevant C, Bonacorsi S, Marecat A, Lange M, Bingen E. Automated ribotyping provides rapid phylogenetic subgroup affiliation of clinical extraintestinal pathogenic Escherichia coli strains. J Clin Microbiol. 2001;39(12):4549–4553. doi: 10.1128/JCM.39.12.4549-4553.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TJ, Wannemuehler Y, Doetkott C, Johnson SJ, Rosenberger SC, Nolan LK. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J Clin Microbiol. 2008;46(12):3987–3996. doi: 10.1128/JCM.00816-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron EZ. Host specificity of septicemic Escherichia coli: human and avian pathogens. Curr Opin Microbiol. 2006;9(1):28–32. doi: 10.1016/j.mib.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Johnson JR, Oswald E, O'Bryan TT, Kuskowski MA, Spanjaard L. Phylogenetic distribution of virulence-associated genes among Escherichia coli isolates associated with neonatal bacterial meningitis in the Netherlands. J Infect Dis. 2002;185(6):774–784. doi: 10.1086/339343. [DOI] [PubMed] [Google Scholar]
- Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170(6):2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177(14):4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouler C, Koffmann F, Amory C, Leroy-Setrin S, Moulin-Schouleur M. Genomic subtraction for the identification of putative new virulence factors of an avian pathogenic Escherichia coli strain of O2 serogroup. Microbiology. 2004;150(Pt 9):2973–2984. doi: 10.1099/mic.0.27261-0. [DOI] [PubMed] [Google Scholar]
- Moulin-Schouleur M, Reperant M, Laurent S, Bree A, Mignon-Grasteau S, Germon P, Rasschaert D, Schouler C. Extraintestinal pathogenic Escherichia coli strains of avian and human origin: link between phylogenetic relationships and common virulence patterns. J Clin Microbiol. 2007;45(10):3366–3376. doi: 10.1128/JCM.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzuszkiewicz E, Bruggemann H, Liesegang H, Emmerth M, Olschlager T, Nagy G, Albermann K, Wagner C, Buchrieser C, Emody L. et al. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc Natl Acad Sci USA. 2006;103(34):12879–12884. doi: 10.1073/pnas.0603038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch RA, Burland V, Plunkett G, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J. et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 2002;99(26):17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers C, Antao EM, Diehl I, Philipp HC, Wieler LH. Intestine and environment of the chicken as reservoirs for extraintestinal pathogenic Escherichia coli strains with zoonotic potential. Appl Environ Microbiol. 2009;75(1):184–192. doi: 10.1128/AEM.01324-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin-Schouleur M, Schouler C, Tailliez P, Kao MR, Bree A, Germon P, Oswald E, Mainil J, Blanco M, Blanco J. Common virulence factors and genetic relationships between O18:K1:H7 Escherichia coli isolates of human and avian origin. J Clin Microbiol. 2006;44(10):3484–3492. doi: 10.1128/JCM.00548-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun. 1999;67(2):546–553. doi: 10.1128/iai.67.2.546-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181(1):261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- Swenson DL, Bukanov NO, Berg DE, Welch RA. Two pathogenicity islands in uropathogenic Escherichia coli J96: cosmid cloning and sample sequencing. Infect Immun. 1996;64(9):3736–3743. doi: 10.1128/iai.64.9.3736-3743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Winkler ME. An Escherichia coli K-12 tktA tktB mutant deficient in transketolase activity requires pyridoxine (vitamin B6) as well as the aromatic amino acids and vitamins for growth. J Bacteriol. 1994;176(19):6134–6138. doi: 10.1128/jb.176.19.6134-6138.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquet G, Porcheron G, Barra C, Reperant M, Chanteloup NK, Schouler C, Gilot P. A metabolic operon in extraintestinal pathogenic Escherichia coli promotes fitness under stressful conditions and invasion of eukaryotic cells. J Bacteriol. 2009;191(13):4427–4440. doi: 10.1128/JB.00103-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alteri CJ, Smith SN, Mobley HL. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog. 2009;5(5):e1000448. doi: 10.1371/journal.ppat.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville GA, Proctor RA. At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol Mol Biol Rev. 2009;73(2):233–248. doi: 10.1128/MMBR.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncet S, Milohanic E, Maze A, Abdallah JN, Ake F, Larribe M, Deghmane AE, Taha MK, Dozot M, De Bolle X. et al. Correlations between Carbon Metabolism and Virulence in Bacteria. Contrib Microbiol. 2009;16:88–102. doi: 10.1159/000219374. [DOI] [PubMed] [Google Scholar]