Abstract
Tumor-induced osteomalacia (TIO) is one of the paraneoplastic diseases characterized by hypophosphatemia caused by renal phosphate wasting. Because removal of responsible tumors normalizes phosphate metabolism, an unidentified humoral phosphaturic factor is believed to be responsible for this syndrome. To identify the causative factor of TIO, we obtained cDNA clones that were abundantly expressed only in a tumor causing TIO and constructed tumor-specific cDNA contigs. Based on the sequence of one major contig, we cloned 2,270-bp cDNA, which turned out to encode fibroblast growth factor 23 (FGF23). Administration of recombinant FGF23 decreased serum phosphate in mice within 12 h. When Chinese hamster ovary cells stably expressing FGF23 were s.c. implanted into nude mice, hypophosphatemia with increased renal phosphate clearance was observed. In addition, a high level of serum alkaline phosphatase, low 1,25-dihydroxyvitamin D, deformity of bone, and impairment of body weight gain became evident. Histological examination showed marked increase of osteoid and widening of growth plate. Thus, continuous production of FGF23 reproduced clinical, biochemical, and histological features of TIO in vivo. Analyses for recombinant FGF23 products produced by Chinese hamster ovary cells indicated proteolytic cleavage of FGF23 at the RXXR motif. Recent genetic study indicates that missense mutations in this RXXR motif of FGF23 are responsible for autosomal dominant hypophosphatemic rickets, another hypophosphatemic disease with similar features to TIO. We conclude that overproduction of FGF23 causes TIO, whereas mutations in the FGF23 gene result in autosomal dominant hypophosphatemic rickets possibly by preventing proteolytic cleavage and enhancing biological activity of FGF23.
Tumor-induced osteomalacia (TIO) is one of the hypophosphatemic diseases characterized by renal phosphate wasting. Because removal of responsible tumors normalizes phosphate metabolism, an unknown phosphaturic factor sometimes called phosphatonin is believed to be responsible for this paraneoplastic syndrome (1, 2). Although several groups have reported inhibitory activity of renal phosphate transport in conditioned media of tumor cells causing TIO (3–6), the responsible factor for TIO has not been identified. Similar biochemical findings to TIO also are observed in X-linked hypophosphatemic rickets/osteomalacia (XLH), its murine homologue, Hyp, and autosomal dominant hypophosphatemic rickets (ADHR) (7). In addition, several lines of evidence indicate that XLH and Hyp are caused by a humoral mechanism (7–10). Therefore, it is possible that TIO and XLH derive from a common or at least very similar humoral factor(s). Thus, identification of this phosphaturic factor causing TIO is indispensable for understanding normal phosphate metabolism and pathogenesis of several hypophosphatemic diseases. In this report, we describe the cloning of a humoral factor from a TIO tumor and show that this factor has the ability to rapidly induce hypophosphatemia and reproduce clinical, biochemical, and histological features of TIO in vivo.
Methods
Differential cDNA Screening of TIO and Adjacent Normal Bone Tissue.
The clinical course of a patient with TIO has been described (11). We constructed a cDNA library with the Lambda ZAP II vector from the frozen tumor tissue responsible for TIO of this patient and performed a differential cDNA screening using two kinds of probes specific for the tumor and the adjacent normal bone tissue. Total RNAs (0.7 μg each) from both tissues were used to synthesize cDNA by the SMART cDNA synthesis kit (CLONTECH). We prepared tumor-specific and nontumor probes by subtraction between both cDNAs by using the PCR-Select cDNA subtraction system (CLONTECH). The probes were labeled with alkaline phosphatase by the AlkPhos Direct system (Amersham Pharmacia). Plaques generated from the TIO cDNA library were lifted on two nylon membranes (Hybond N+; Amersham Pharmacia). One membrane was hybridized with a tumor-specific probe and the other with a probe specific for the normal bone. Clones specifically hybridized only with a tumor-specific probe were subjected to DNA sequencing.
Expression of Fibroblast Growth Factor 23 (FGF23).
Full-length human FGF23 cDNA was amplified with F1EcoRI primer (5′-CCGGAATTCAGCCACTCAGAGCAGGGCACG-3) containing EcoRI site at the 5′ terminus and LhisNot primer (5′-ATAAGAATGCGGCCGCTCAATGGTGATGGTGATGATGGATGAACTTGGCGAA-3′) containing the sequence encoding the His6 tag and NotI site. The PCR product was digested with EcoRI and NotI and ligated to pcDNA3.1zeo (Invitrogen). A Chinese hamster ovary (CHO) cell line stably producing FGF23 (CHO-FGF23) was cloned by limited dilution.
Northern Blot Analysis.
Three kinds of human multiple tissue blots (CLONTECH) derived from 23 tissues were used. Those tissues were heart, brain, placenta, lung, liver, skeletal muscle, kidney, pancreas, spleen, thymus, prostate, testis, ovary, small intestine, colon, peripheral blood leukocyte, stomach, thyroid, spinal cord, lymph node, trachea, adrenal gland, and bone marrow. The amplified FGF23 cDNA fragment using F1 (5′-AGCCACTCAGAGCAGGGCAC-3′) and L1 (5′-CACGTTCAAGGGGTCCCGCT-3′) primers was labeled and used for hybridization. Total RNAs were isolated from whole kidney with Isogen (Nippon Gene, Toyama, Japan) and subjected to Northern blotting for renal 25-hydroxyvitamin D 1α-hydroxylase (1α-OHase). A 1,090-bp cDNA fragment for mouse 1α-OHase was amplified by using 1AF (5′-CAGACAGAGACATCCGTGTAG-3′) and 1AR (5′-CCACATGGTCCAGGTTCAGTC-3′) primers, labeled and used for hybridization. The filters were washed to the stringency of 0.1× SSC with 0.1% of SDS at 50°C for 40 min. A BAS imaging system (Fuji) was used for identification of signals.
Reverse Transcriptase–PCR.
Each 0.3 ng of cDNA of various human tissues (MTC human panel, CLONTECH) was amplified with F1 and L1 primers for FGF23. The PCR products were analyzed by Southern blotting using the 32P-labeled FGF23 cDNA fragment. The filter was washed to the stringency of 0.1× SSC with 0.5% of SDS at 65°C for 30 min.
Western Blot Analysis.
Twenty microliters of conditioned medium from CHO-FGF23 cells was resolved by 10–20% gradient SDS/PAGE under reduced condition and electroblotted onto a poly(vinylidene difluoride) membrane. The membrane was incubated with anti-His (C-term)-horseradish peroxidase antibody (Invitrogen), and signals were detected by the ECL system (Amersham Pharmacia).
Amino Acid Sequence.
The recombinant proteins separated by SDS/PAGE were electroblotted to a poly(vinylidene difluoride) membrane and stained with Coomassie brilliant blue. The band corresponding to each recombinant protein was dissected and subjected to automated Edman degradation in gas phase protein sequencer model 492A (Perkin–Elmer).
Recombinant Protein Preparation.
Ten liters of conditioned medium from CHO-FGF23 cells was filtrated through 0.2-μm membrane (SuporCap, Pall Gelman Laboratory, Ann Arbor, MI) at 4°C. The filtrate was applied to SP-Sepharose FF (Amersham Pharmacia), and the column was washed with 50 mM sodium phosphate buffer, pH 6.7. The retained proteins were eluted with a linear gradient of NaCl ranging from 0 to 0.7 M in a buffer of 50 mM sodium phosphate, pH 6.7. The mature full-length polypeptide was collected at ≈0.3 M NaCl. Purified mature recombinant FGF23 was concentrated into a buffer consisting of 5 mM Hepes and 0.1 M NaCl, pH 6.9.
Animals and Experimental Designs.
Approximately 4 μg of recombinant FGF23 was i.p. administered to 6-week-old male BALB/c mice (SLC, Hamamatsu, Japan). To examine the long-term effects of FGF23, ≈1 × 107 CHO-FGF23 or wild-type CHO cells in 0.1 ml of PBS were s.c. implanted into both sides of backs of 6-week-old male BALB/c athymic nude mice (SLC). The control group was injected with 0.1 ml of PBS as well. Tumor volume was calculated according to the formula of 1/2 × (long diameter) × (short diameter)2. The calculated result by this formula was proved to correlate well with the volume measured by water displacement (r = 0.98) (12). To collect urine samples, mice were bred in metabolic cages for 24 h. Blood samples were taken under anesthesia with diethyl ether. All animals received a commercial rodent diet (CE-2; CLEA Japan, Osaka) containing 1.1% phosphate and 1.0% calcium. Diets and tap water were provided ad libitum throughout the experiments. All experiments were reviewed and approved by the institutional animal care and use committee at the Pharmaceutical Research Laboratory, Kirin Brewery and were conducted in the specific pathogen-free area where no infectious problem has been reported.
Measurement of 1,25-Dihydroxyvitamin D [1,25(OH)2D] in Serum.
Serum 1,25(OH)2D was measured by RIA (Immunodiagnostic Systems, Boldon, U.K.). Because 500 μl of serum was required for one measurement, 250 μl of serum from individual animals in the same groups (n = 5) was pooled and subjected to duplicate measurements. The detection limit was 5 pmol/liter.
Roentgenography.
To take soft roentgenographs, we used an x-ray imaging system (μFX-1000; Fuji). Exposure of imaging plates was conducted at 25 kV, 0.1 mA. Exposure period was 10 sec for the whole body and 5 sec for isolated femurs. Exposed plates were analyzed by a BAS imaging system.
Histological Examination.
Freshly isolated tibiae were fixed and subjected to the Villanueva bone stain. The tissues were embedded in methyl methacrylate resin (Wako Pure Chemical, Osaka), and undecalcified sections with 4-μm thickness were prepared with the microtome (Supercut RM2065; Leica, Solms, Germany). Villanueva-Goldner counterstain for these sections was conducted.
In Vitro Transport Assay and Measurement of cAMP Production.
Opossum kidney (OK) cells were obtained from the American Type Culture Collection (CRL-1840). Phosphate, glucose, and alanine transport were measured separately at 0.1 mM solute concentration with [32P] dibasic potassium phosphate, [14C] d-glucose, or [3H] l-alanine in the uptake solution containing 137 mM NaCl, 5.4 mM KCl, 2.8 mM CaCl2, 1.2 mM MgSO4, and 10 mM Hepes, pH 7.4 (13). Na-independent uptake was measured with Na-free uptake solution in which NaCl was replaced by choline chloride. To examine cAMP production, OK cells were incubated with FGF23 for 10 min in the presence of 1 mM 3-isobuthyl-1-methylxanthine. cAMP in cells was extracted and measured by EIA (Amersham Pharmacia).
Statistical Analyses.
All measurements were expressed as mean ± SEM. Statistical significance was evaluated by either Student's t test or one-way ANOVA followed by Dunnett's test for comparison of multiple means. A P value less than 0.05 was considered to be significant.
Results
Cloning of cDNAs Specifically Expressed in a Tumor Causing TIO.
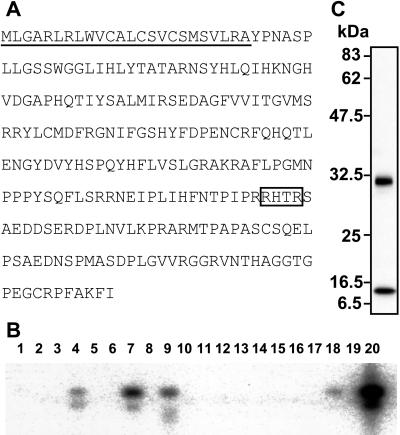
To isolate a putative phosphaturic factor, differential cDNA screening was carried out. Of ≈320,000 clones, 456 clones dominantly expressed in the tumor were selected and sequenced. All sequences obtained were compared with each other and analyzed against the GenBank database. Consequently, each clone was classified into contigs, which were aligned by the number of consisting clones to reflect the frequency of appearance in the screening. The most frequently detected contig was dentine matrix protein-1 (DMP-1) followed by heat shock protein-90, osteopontin, and a novel sequence contig named OST311 that was composed of nine different cDNA clones (Table 1). We also obtained a contig termed OST190 that has been recently identified as a novel molecule, matrix extracellular phosphoglycoprotein (MEPE), produced by TIO tumors (14). The longest cDNA clone for OST311 contained 2,270-bp nucleic acids and had an ORF encoding 251-aa residues with putative molecular mass of 28 kDa (Fig. 1A). The sequence coding OST311 protein derived from three exons spanning more than 10 kbp on 12p13.3 where the responsible gene for ADHR exists (15). Deduced amino acid sequence shows that OST311 protein has a signal peptide sequence at its N terminus and has a homology to FGF family members, especially in the middle portion of the protein. OST311 protein also has a unique C-terminal portion characterized by the presence of several potential phosphorylation sites by casein kinase. Recent reports indicated that OST311 is identical with FGF23 (16, 17).
Table 1.
The tumor-specific contigs by frequency of appearance
| Contig ID | Appearance frequency | Gene name |
|---|---|---|
| OST 131 | 240 | DMP-1 |
| OST 1 | 47 | Heat shock protein-90 |
| OST 2 | 24 | Osteopontin |
| OST 311 | 9 | Unknown |
| OST 190 | 6 | Unknown |
| OST 584 | 6 | Fibronectin-1 |
| OST 666 | 5 | Translationally controlled tumor protein |
| OST 133 | 5 | β-2 microglobulin |
| OST 1001 | 4 | CD44 antigen |
| OST 562 | 3 | Annexin II |
The most frequently cloned 10 contigs in the differential screening are listed. The sequence data obtained in this study were sorted into these contigs. The gene names were identified by comparing with the sequence data in Genbank on November 10, 1999. The nucleotide sequences of the contigs that did not hit any data in Genbank by the blast homology search at that time were described as unknown. OST 311 and OST 190 later turned out to be FGF23 and MEPE, respectively.
Figure 1.
(A) Amino acid sequence of human FGF23. The signal peptide sequence is underlined. The consensus proteolytic cleavage site is boxed. (B) Expression profile of FGF23 in adult normal tissues analyzed by reverse transcriptase–PCR followed by Southern blotting. Template cDNAs are as follows: lanes 1, bone marrow; 2, brain; 3, colon; 4, heart; 5, kidney; 6, leukocyte; 7, liver; 8, lung; 9, lymph nude; 10, muscle; 11, ovary; 12, pancreas; 13, placenta; 14, prostate; 15, small intestine; 16, spleen; 17, testis; 18, thymus; 19, tonsil; 20, TIO tumor. (C) Western blotting with anti-His (c-term) antibody that recognizes the carboxyl-terminal tag sequence of recombinant FGF23 proteins secreted into media by CHO-FGF23 cells.
Expression of FGF23.
Expression of FGF23 could not be detected in all 23 human tissues by Northern blot analysis using about 2 μg of poly(A)+ RNA even after 24 h exposure to the imaging plate (data not shown). Abundant expression of FGF23 was observed in the tumor tissue responsible for TIO by reverse transcriptase–PCR followed by Southern blot analysis (Fig. 1B). Weak expression of FGF23 also was detected in liver, lymph node, thymus, and heart. The expression of FGF23 could not be observed in other tissues, including kidney, even by this method. These results indicate that a few normal tissues can express a small amount of FGF23, but a tumor responsible for TIO abundantly expresses FGF23.
Preparation and Analysis of Recombinant FGF23.
To investigate the biological activity of FGF23, we prepared recombinant CHO-FGF23 cells. Western blotting detecting the C terminus of recombinant proteins revealed that conditioned media contained two recombinant products with molecular masses of ≈30 and 10 kDa (Fig. 1C). Amino acid sequencing showed the larger product was a mature protein lacking signal sequence, which is composed of the first 24 aa residues of FGF23. The smaller product had 180Ser residue at its N terminus. The preceding amino acid sequence of 180Ser, 176Arg-177His-178Thr-179Arg, agrees with the consensus proteolytic cleavage sequence of RXXR (18). The proteolytic processing also was supported by the detection of about a 16-kDa N-terminal fragment using a polyclonal antibody raised against the synthetic peptide corresponding to amino acids 48Arg to 67Gln (data not shown).
Biological Activity of FGF23.
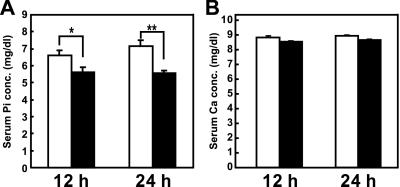
Recombinant mature FGF23 was injected into mice three times with intervals of 5 h, and blood samples were collected at 12 and 24 h after the initial injection. Administration of mature FGF23 reduced serum phosphate significantly at both times without affecting serum calcium (Fig. 2). To investigate the mechanism of this hypophosphatemia, 24-h urine samples were collected. Serum was obtained just after the urine collection. In this experiment, treatment with FGF23 reproduced hypophosphatemia and increased renal phosphate excretion (P < 0.001) (Table 2). These hypophosphatemic and phosphaturic effects were reproduced in two other experiments (data not shown). These data indicate that reduced renal phosphate reabsorption by FGF23 at least in part contributed to the development of hypophosphatemia in those animals. Serum and urinary levels of calcium and other solutes did not change with FGF23 (Table 2). Urinary excretion of measurable 11 kinds of amino acids was determined. Because the absolute value of each amino acid was variable, we calculated the ratio of amino acid excretion in the FGF23 group to that in the control group. As shown in Table 2, FGF23 did not increase renal excretion of amino acids. Furthermore, we did not observe increased urinary cAMP levels in FGF23-treated mice. Thus, FGF23 specifically reduced renal reabsorption and decreased the serum level of phosphate.
Figure 2.
Effects of recombinant FGF23. The changes of serum phosphate (A) and calcium (B) concentrations in mice treated with purified recombinant protein (n = 6; filled columns) or vehicle (n = 6; open columns). FGF23 was i.p. injected three times with intervals of 5 h. Blood samples were collected at 12 and 24 h after the first injection. Each column represents the mean ± SEM. *, P < 0.05; **, P < 0.001 by Student's t test.
Table 2.
Effects of recombinant FGF23 on serum and urinary biochemical parameters
| Vehicle | FGF23 | Probability | |
|---|---|---|---|
| Serum concentration | |||
| Pi (mg/dl) | 9.55 ± 0.21 | 7.85 ± 0.26 | P < 0.001 |
| Ca (mg/dl) | 9.86 ± 0.14 | 9.68 ± 0.10 | |
| Glucose (mg/dl) | 178.8 ± 5.4 | 181.4 ± 8.5 | |
| Na (meq/liter) | 152.0 ± 0.9 | 151.2 ± 0.6 | |
| K (meq/liter) | 5.33 ± 0.18 | 5.30 ± 0.18 | |
| Cl (meq/liter) | 111.9 ± 0.6 | 112.7 ± 0.4 | |
| Renal excretion | |||
| Pi (dl/mg) | 1.38 ± 0.06 | 1.77 ± 0.05 | P < 0.001 |
| Ca (× 10−2 dl/mg) | 1.50 ± 0.05 | 1.55 ± 0.07 | |
| Glucose (× 10−3 dl/mg) | 2.45 ± 0.14 | 2.85 ± 0.24 | |
| Na (× 10−2 dl/mg) | 4.61 ± 0.30 | 4.81 ± 0.16 | |
| K (dl/mg) | 2.37 ± 0.21 | 2.23 ± 0.10 | |
| Cl (× 10−2 dl/mg) | 9.17 ± 0.39 | 9.42 ± 0.31 | |
| Amino acid (T/C ratio) | 1.0 ± 0 | 1.04 ± 0.04 | |
| cAMP (nmol/mg) | 83.3 ± 10.7 | 60.2 ± 7.1 |
Purified recombinant FGF23 (n = 12) or vehicle (n = 12) was injected three times as shown in Methods. All biochemical markers except for amino acids were measured in each mouse. Renal excretion was evaluated by the following formula, urinary concentration divided by serum concentration and urinary creatinine. Using pooled urine, concentrations of 11 kinds of amino acids (Thr, Asn, Glu, Gln, Pro, Gly, Ala, Phe, His, Lys, and Arg) were determined by HPLC. The ratio of each amino acid excretion in FGF23-treated mice to that of control mice (T/C ratio) was calculated. The values for Gly, Ala, and Lys, which are commonly excreted in aminoacidurias, were 1.08, 0.99 and 0.99, respectively. The average of the ratios from 11 amino acids is shown. Results represent the mean ± SEM. P < 0.001 by Student's t test.
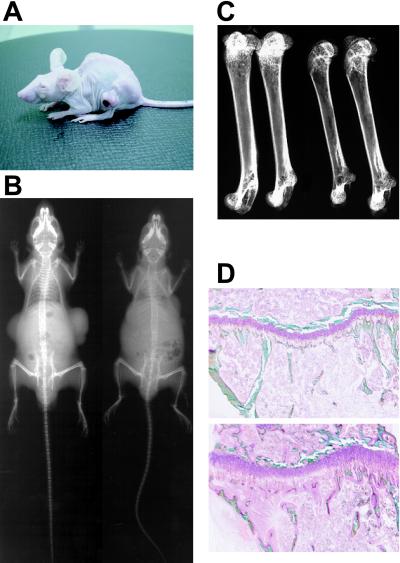
To further investigate the biological function of FGF23 and its role in the development of TIO, we used the tumor-bearing nude mouse system. To examine the time course of biochemical and phenotypic changes, six mice in each group were killed sequentially at indicated time points. Mice with CHO-FGF23 cells showed hypophosphatemia as expected from the experiments of recombinant protein injection. This hypophosphatemia was accompanied by inappropriately increased renal phosphate clearance after 6 days of implantation (Fig. 3 A and B). In addition, serum alkaline phosphatase activity increased in CHO-FGF23 mice as in patients with TIO (Fig. 3C). Furthermore, inappropriately reduced level of 1,25(OH)2D in the presence of hypophosphatemia is reported in TIO patients (1, 2). In agreement with these observations, the serum level of 1,25(OH)2D in CHO-FGF23 mice was reduced after 6 days of tumor cell implantation compared with that in control and wild-type CHO mice (49.1 ± 2.6 pmol/liter, 273.7 ± 10.9 pmol/liter, and 184.6 ± 14.2 pmol/liter, respectively). To confirm the abnormal vitamin D metabolism, we examined the expression level of renal 1α-OHase by Northern blot analysis using kidneys isolated on day 3 after implantation. The decreased expression of 1α-OHase in CHO-FGF23 mice was observed (Fig. 4). Mice with CHO-FGF23 cells showed growth retardation after 2 weeks, although the amount of food intake of CHO-FGF23 mice on day 45 (5.85 ± 0.62 g) was comparable to those of vehicle-treated (5.99 ± 0.63 g) and control CHO-implanted mice (6.23 ± 0.59 g). Fig. 5A shows the typical appearance of CHO-FGF23 mice, which is characterized by leanness and a round back. All mice in the CHO-FGF23 group, but no mice in other groups, showed such a phenotype at 45 days after the implantation. Soft x-ray images showed that bone mineral content of a mouse bearing CHO-FGF23 cells was markedly reduced compared with a mouse bearing CHO cells (Fig. 5B). Distortion of rib cage also was observed in a mouse bearing CHO-FGF23 cells. Bone mineral content in epiphysis and metaphysis of femurs from mice bearing CHO-FGF23 was remarkably reduced compared with that of control mice (Fig. 5C). The ratio of ash weight to dry weight of femurs in CHO-FGF23 mice was significantly decreased compared with that of wild-type CHO mice (0.534 ± 0.007 vs. 0.608 ± 0.004, n = 5, P < 0.001). Fig. 5D shows the histological appearance of tibial proximal metaphysis. There was a prominent increase of osteoid and widening of growth plate in a mouse with CHO-FGF23 cells. Osteomalacia and muscle weakness are typical phenotypes of TIO. In addition, growth retardation and skeletal deformities are reported in younger patients (1). Thus, we could reproduce biochemical, clinical, and histological features of TIO by implanting CHO-FGF23 cells into nude mice. These biochemical and phenotypic changes by CHO-FGF23 cells were confirmed in other independent experiments.
Figure 3.
Biochemical changes in mice transplanted with CHO cells producing FGF23. Time course of changes of serum phosphate level (A), renal phosphate excretion index (B), and serum alkaline phosphatase activity (C) in nude mice bearing CHO-FGF23 cells (n = 6; ▴) were compared with those in mice with wild-type CHO cells (n = 6; ■) and control nude mice without CHO cells (n = 6; ○). Blood and 24-h urine samples were collected on days 1, 6, 20, and 45 after the implantation. Renal phosphate excretion index was urinary phosphate divided by serum phosphate and urinary creatinine. Data are means ± SEM at each time point. Significant difference from control group by one-way ANOVA followed by Dunnett's multiple comparison test; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure 4.
Expression of renal 1α- OHase in mice implanted with CHO cells or CHO-FGF23 cells. Vehicle-treated mice (n = 3), control CHO-implanted mice, and CHO-FGF23-implanted mice were killed on day 3 after implantation. Total RNAs (20 μg each) were used for the determination of expression level of 1α-OHase by Northern blot analysis. Expression of glyceraldehyde-3-phosphate dehydrogenase (G3PDH) also was determined by using the same blot.
Figure 5.
(A) Typical appearance of a mouse bearing CHO-FGF23 cell tumors. (B) Whole skeletal soft roentgenogram of a mouse bearing wild-type CHO cell tumors (Left) and a mouse bearing CHO-FGF23 cell tumors (Right) on day 45 after implantation. (C) Soft roentgenogram of femurs isolated from control mice (Left) and mice bearing CHO-FGF23 cell tumors (Right). (D) Histological appearance of proximal metaphysis of tibia from a control mouse (Upper) and a mouse bearing CHO-FGF23 (Lower) on day 45. The undecalcified tissue sections were subjected to the Villanueva bone stain and the Villanueva-Goldner stain to discriminate mineralized bone tissues (green) from osteoid tissues (pink).
In Vitro Activity of FGF23.
Because several reports showed the inhibitory activity on phosphate transport in conditioned media of tumor cells causing TIO (3–6), we evaluated in vitro activity of recombinant FGF23 in OK cells. However, although phosphonoformic acid, a specific inhibitor for Na-dependent phosphate transport, clearly inhibited phosphate transport, FGF23 did not inhibit transport of phosphate (Table 3). Transport of glucose and alanine did not change by FGF23, either. In addition, FGF23 did not increase cAMP production in OK cells (Table 3).
Table 3.
Effects of FGF23 on Na-dependent transport of phosphate, glucose, and alanine, and cAMP production in OK cell cultures
| Treatment | Na-dependent
transport, nmol/mg per 6 min
|
cAMP, nmol/mg | ||
|---|---|---|---|---|
| Phosphate | Glucose | Alanine | ||
| Control | 2.09 ± 0.08 | 0.81 ± 0.03 | 3.33 ± 0.08 | 12.3 ± 1.0 |
| PFA (10 mM) | 0.62 ± 0.05** | — | — | — |
| FGF23 (4 ng/ml) | 2.08 ± 0.06 | 0.83 ± 0.05 | 3.40 ± 0.05 | — |
| FGF23 (40 ng/ml) | 2.05 ± 0.07 | 0.79 ± 0.03 | 3.33 ± 0.09 | — |
| FGF23 (400 ng/ml) | 2.04 ± 0.04 | 0.78 ± 0.03 | 3.32 ± 0.10 | 13.5 ± 3.1 |
OK cells were exposed to FGF23 for 20 h and then subjected to the transport assay (n = 4). Phosphonoformic acid (PFA), a specific inhibitor for Na-dependent phosphate transport, was added to an assay buffer. Cytosolic cAMP was extracted after the incubation with FGF23 in the presence of 3-isobuthyl-1-methylxanthine for 10 min, and measured by EIA (n = 4). Results are expressed as means ± SEM. **, P < 0.001 by Student's t test. Dash indicates not determined.
Discussion
Causative factors of TIO have been sought for a long time. Using molecular biological screening as described, DMP-1, MEPE, and OST311, which turned out to be identical to FGF23, were picked up as candidates. To evaluate the biological activity as a causative factor of TIO, we produced recombinant cells that secrete these candidate gene products. Then we examined the in vivo effects of these gene products so that we could identify the causative factor for TIO even if it does not work on renal tubular cells directly. We first evaluated the most frequently cloned DMP-1 by the tumor-bearing system shown here. However, mice with CHO cells secreting DMP-1 showed neither hypophosphatemia nor increased renal phosphate clearance. Mice with CHO cells secreting MEPE did not show hypophosphatemia either (data not shown). The abundant expression of MEPE was demonstrated in TIO tumors (14), and DMP-1 is a candidate gene for dentinogenesis imperfecta (19). Both DMP-1 and MEPE are members of matrix proteins that may have some role in calcification. However, our results show that these proteins are not causative factors of TIO. We could observe hypophosphatemia only with CHO cells secreting FGF23.
To prove that FGF23 induces hypophosphatemia, we purified recombinant FGF23 protein and administered it into normal mice (Fig. 2). Furthermore, we examined the time course of changes of biochemical markers in tumor-bearing nude mice (Fig. 3). Administration of recombinant protein induced a reproducible and significant decrease of serum phosphate with increased renal phosphate excretion. Long-term exposure to FGF23 induced severer hypophosphatemia, osteomalacia, and inappropriately low 1,25(OH)2D level with decreased 1α-OHase expression. Although the magnitude of the decrease of serum phosphate by recombinant FGF23 was smaller than that in nude mice bearing CHO-FGF23 cells, this disparity may be caused by the difference in serum concentration of FGF23 or the exposed period to FGF23 in these systems.
It is possible that FGF23 has a stimulatory effect on tumor growth and large tumor burden indirectly modified phosphate and bone metabolism in mice with CHO-FGF23. However, the tumor burden was not statistically different between CHO-FGF23 mice and wild-type CHO mice at 45 days after the implantation (0.74 ± 0.15 vs. 0.99 ± 0.41). In addition, neither enhanced growth of CHO-FGF23 cells nor growth stimulation of CHO cells by recombinant FGF23 was observed (data not shown). Furthermore, serum phosphate was clearly decreased in mice with CHO-FGF23 cells at 6 days after the implantation when tumors were barely detectable. These results indicate that tumor burden cannot account for the abnormal phosphate and bone metabolism in mice with CHO-FGF23.
To clarify the mechanism of actions of FGF23 on phosphate metabolism, we examined the effects of recombinant FGF23 on sodium-dependent phosphate transport in OK cells. In contrast to previous reports (3–6), we did not observe a clear inhibitory activity of FGF23. There are several possibilities that can explain these results. For example, FGF23 may not act on renal cells directly, but affects renal phosphate reabsorption indirectly in vivo. It is also possible that FGF23 needs some modification in vivo to exert its effect on phosphate metabolism. Because actual target cells of FGF23 are unknown at the moment, these possibilities should be examined in future experiments.
It is interesting to note that the responsible gene for ADHR has been recently identified and named FGF23 (17). The missense mutations at 176Arg and 179Arg are responsible for ADHR. These amino acids are in the consensus proteolytic cleavage sequence of RXXR. Lack of expression of FGF23 in kidney (Fig. 1B) indicates that ADHR also is caused by a humoral mechanism. Because recombinant FGF23 produced by CHO cells was partially cleaved between 179Arg and 180Ser (Fig. 2A), it is possible that mutations at 176Arg and 179Arg prevent proteolytic cleavage. Taking into account our present data, mutations of the FGF23 gene in ADHR are considered to result in the enhancement of biological activities of FGF23 as a humoral factor.
The involvement of an unknown humoral factor(s) also is proposed for XLH and its murine homologue, Hyp (4, 5). Even though XLH/Hyp is caused by mutations in the putative endopeptidase, a phosphate-regulating gene with homologies to endopeptidases on the X-chromosome (PHEX/Phex) (20), the mechanism by which mutations of PHEX/Phex cause phosphaturia is unclear. It has been postulated that PHEX/Phex protein inactivates a putative phosphaturic factor, phosphatonin, by its proteolytic activity and maintains serum phosphate level (2). It is necessary to investigate whether FGF23 interacts with PHEX protein. Further studies are also necessary to clarify the relationship between FGF23 and a putative humoral factor causing XLH/Hyp. Because both the overproduction and the missense mutations of FGF23 cause hypophosphatemia with renal phosphate wasting, we conclude that FGF23 is at least one of the causative factors of TIO and is an important regulator of phosphate and bone metabolism. A recent report showing overexpression of FGF23 in several tumors causing TIO also supports this notion (21). Elucidation of the detailed actions of FGF23 is indispensable for understanding normal phosphate metabolism and will lead to the development of novel diagnostic and therapeutic approaches for several diseases with abnormal phosphate metabolism.
Acknowledgments
We thank Prof. T. J. Martin for valuable comments on the manuscript and Dr. Shinichiro Kato for helpful discussions. This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science, and Technology, the Ministry of Health, Labor, and Welfare, Japan.
Abbreviations
- TIO
tumor-induced osteomalacia
- FGF
fibroblast growth factor
- XLH
X-linked hypophosphatemic rickets/osteomalacia
- ADHR
autosomal dominant hypophosphatemic rickets
- CHO
Chinese hamster ovary
- 1α-OHase
1α-hydroxylase
- 1,25(OH)2D
1,25-dihydroxyvitamin D
- OK
opossum kidney
- DMP-1
dentine matrix protein-1
- MEPE
matrix extracellular phosphoglycoprotein
Footnotes
References
- 1.Drezner M K. In: Primer on Metabolic Bone Diseases and Disorders of Mineral Metabolism. Favus M J, editor. Philadelphia: Lippincott; 1999. pp. 331–337. [Google Scholar]
- 2.Kumar R. Bone. 2000;27:333–338. doi: 10.1016/s8756-3282(00)00334-3. [DOI] [PubMed] [Google Scholar]
- 3.Cai Q, Hodgson S F, Kao P C, Lennon V A, Klee G G, Zinsmiester A R, Kumar R. N Engl J Med. 1994;330:1645–1649. doi: 10.1056/NEJM199406093302304. [DOI] [PubMed] [Google Scholar]
- 4.Wilkins G E, Granleese S, Hegele R G, Holden J, Anderson D W, Bondy G P. J Clin Endocrinol Metab. 1995;80:1628–1634. doi: 10.1210/jcem.80.5.7745010. [DOI] [PubMed] [Google Scholar]
- 5.Nelson A E, Namkung H J, Patava J, Wilkinson M R, Chang A C, Reddel R R, Robinson B G, Mason R S. Mol Cell Endocrinol. 1996;124:17–23. doi: 10.1016/s0303-7207(96)03928-7. [DOI] [PubMed] [Google Scholar]
- 6.Rowe P S, Ong A C, Cockerill F J, Goulding J N, Hewison M. Bone. 1996;18:159–169. doi: 10.1016/8756-3282(95)00458-0. [DOI] [PubMed] [Google Scholar]
- 7.Econs M J. Bone. 1999;25:131–135. doi: 10.1016/s8756-3282(99)00108-8. [DOI] [PubMed] [Google Scholar]
- 8.Morgan J M, Hawley W L, Chenoweth A I, Retan W J, Diethelm A G. Arch Intern Med. 1974;134:549–552. [PubMed] [Google Scholar]
- 9.Meyer R A, Jr, Meyer M H, Gray R W. J Bone Miner Res. 1989;4:493–500. doi: 10.1002/jbmr.5650040407. [DOI] [PubMed] [Google Scholar]
- 10.Nesbitt T, Coffman T M, Griffiths R, Drezner M K. J Clin Invest. 1992;89:1453–1459. doi: 10.1172/JCI115735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukumoto S, Takeuchi Y, Nagano A, Fujita T. Bone. 1999;25:375–377. doi: 10.1016/s8756-3282(99)00170-2. [DOI] [PubMed] [Google Scholar]
- 12.Euhus D M, Hudd C, Laregina M C, Johnson F E. J Surg Oncol. 1986;31:229–234. doi: 10.1002/jso.2930310402. [DOI] [PubMed] [Google Scholar]
- 13.Biber J, Malmstrom K, Reshkin S, Murer H. Methods Enzymol. 1991;191:494–505. doi: 10.1016/0076-6879(90)91032-2. [DOI] [PubMed] [Google Scholar]
- 14.Rowe P S N, De Zoysa P A, Dong R, Wang H R, White K E, Econs M J, Oudet C L. Genomics. 2000;67:54–68. doi: 10.1006/geno.2000.6235. [DOI] [PubMed] [Google Scholar]
- 15.Econs M J, McEnery P T, Lennon F, Speer M C. J Clin Invest. 1997;100:2653–2657. doi: 10.1172/JCI119809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamashita T, Yoshioka M, Itoh N. Biochem Biophys Res Commun. 2000;277:494–498. doi: 10.1006/bbrc.2000.3696. [DOI] [PubMed] [Google Scholar]
- 17.White K E, Evans W E, O'Riordan J L H, Speer M C, Econs M J, Lorenz B, Grabowski M, Meitinger T, Strom T M. Nat Genet. 2000;26:345–348. [Google Scholar]
- 18.van de Loo J W, Creemers J W, Bright N A, Young B D, Roebroek A J, Van de Ven W J. J Biol Chem. 1997;272:27116–27123. doi: 10.1074/jbc.272.43.27116. [DOI] [PubMed] [Google Scholar]
- 19.MacDougall M, Gu T T, Simmons D. Connect Tissue Res. 1996;35:267–272. doi: 10.3109/03008209609029200. [DOI] [PubMed] [Google Scholar]
- 20.The Hyp Consortium. Nat Genet. 1995;11:130–136. doi: 10.1038/ng1095-130. [DOI] [PubMed] [Google Scholar]
- 21.White K E, Jonsson K B, Carn G, Hampson G, Spector T D, Mannstadt M, Lorenz-depiereux B, Miyauchi A, Yang I M, Ljunggren O, et al. J Clin Endocrinol Metab. 2001;86:497–500. doi: 10.1210/jcem.86.2.7408. [DOI] [PubMed] [Google Scholar]