Abstract
To search for virulence effector genes of the rice blast fungus, Magnaporthe oryzae, we carried out a large-scale targeted disruption of genes for 78 putative secreted proteins that are expressed during the early stages of infection of M. oryzae. Disruption of the majority of genes did not affect growth, conidiation, or pathogenicity of M. oryzae. One exception was the gene MC69. The mc69 mutant showed a severe reduction in blast symptoms on rice and barley, indicating the importance of MC69 for pathogenicity of M. oryzae. The mc69 mutant did not exhibit changes in saprophytic growth and conidiation. Microscopic analysis of infection behavior in the mc69 mutant revealed that MC69 is dispensable for appressorium formation. However, mc69 mutant failed to develop invasive hyphae after appressorium formation in rice leaf sheath, indicating a critical role of MC69 in interaction with host plants. MC69 encodes a hypothetical 54 amino acids protein with a signal peptide. Live-cell imaging suggested that fluorescently labeled MC69 was not translocated into rice cytoplasm. Site-directed mutagenesis of two conserved cysteine residues (Cys36 and Cys46) in the mature MC69 impaired function of MC69 without affecting its secretion, suggesting the importance of the disulfide bond in MC69 pathogenicity function. Furthermore, deletion of the MC69 orthologous gene reduced pathogenicity of the cucumber anthracnose fungus Colletotrichum orbiculare on both cucumber and Nicotiana benthamiana leaves. We conclude that MC69 is a secreted pathogenicity protein commonly required for infection of two different plant pathogenic fungi, M. oryzae and C. orbiculare pathogenic on monocot and dicot plants, respectively.
Author Summary
Magnaporthe oryzae causes the most devastating fungal disease in rice. M. oryzae secretes a plethora of effector proteins, including several avirulence proteins which are known to be recognized by host resistance proteins activating innate immunity. However, the effectors that are required for virulence activity have not been identified in M. oryzae to date except for an effector protein, Secreted LysM Protein 1 (Slp1) that was recently identified. We performed a large-scale disruption analysis of M. oryzae effector candidates and identified a small protein MC69, which is secreted by the fungus during infection. When MC69 is absent, pathogenicity is severely reduced after penetration into the host cells. Furthermore, deletion of the MC69 orthologous gene in Colletotrichum orbiculare reduced its pathogenicity in the host plants cucumber and Nicotiana benthamiana. Thus, MC69 is conserved in ascomycete fungi and is crucial for establishing compatibility. This is the first report of a single secreted protein that is indispensable for pathogenicity in both monocot and dicot pathogenic fungi. How MC69 contributes to pathogenicity or virulence is unknown but it could be required for the fungus to be a pathogen or might be a classical effector that acts on plant target molecules.
Introduction
Rice blast, caused by an ascomycete fungus Magnaporthe oryzae, is the most severe fungal disease of rice throughout the world [1]. Genetic studies of this pathogen over the last two decades have made the Magnaporthe-rice pathosystem an excellent model for investigating fungus-plant interactions.
Plants are equipped to sense evolutionarily conserved microbial molecular signatures, collectively called Pathogen-Associated Molecular Patterns (PAMPs) or Microbe-Associated Molecular Patterns (MAMPs), and activate PAMP-Triggered Immunity (PTI) [2]–[4]. Pathogens are capable of inhibiting PTI on their host plants by delivering virulence effector proteins into host cells [5]–[9].
In M. oryzae, effector secretion machinery has recently been elucidated [10]–[12]. A Golgi-localized P-type ATPase-encoding gene, MgAPT2 is required for exocytosis during plant infection. Further analysis suggested that MgAPT2 is involved in secretion of a range of extracellular enzymes as well as an AVR effector for the rapid induction of host defense responses in an incompatible reaction in rice cultivar IR-68 [10]. Another study demonstrated that M. oryzae mutants with a defect in an ER chaperone-encoding gene, LHS1, have reduced activities of extracellular enzymes and secretion of AVR-Pita1 [13], [14] blocking Pi-ta R-gene-mediated hypersensitive response. The contribution of LHS1 to protein translocation and secretion of proteins, including effectors, revealed the importance of ER chaperones for successful disease development by rice blast fungus [12]. Live-cell imaging revealed development of the biotrophic interfacial complex (BIC), a structure that accumulates fluorescently labeled effectors secreted by invasive hyphae (IH). The examined BIC-localized secreted proteins were translocated into rice cytoplasm. By contrast, a biotrophy-associated secreted protein BAS4, which uniformly outlines the IH, was not translocated into the host cytoplasm [11]. These results suggest that BIC represents the site of effector translocation in rice blast disease [11].
Several effector protein genes have been cloned and characterized from M. oryzae but all of them were avirulence (AVR) effectors with no virulence function elucidated to date [13]–[20] except for a recently identified virulence effector protein, Slp1 [21]. Slp1 accumulates at the interface between the fungal cell wall and the rice plasma membrane, can bind to chitin, and is able to suppress chitin-induced plant immune responses, including generation of reactive oxygen species and plant defense gene expression [21]. Several effector candidates were identified by using interaction transcriptome in the biotrophic invasion of M. oryzae [22]. In the paper the authors have identified a known effector PWL2 as well as 58 candidate effectors showing >10-fold increase in the expression in the biotrophic invasive hyphae relative to control mycelia using M. oryzae oligoarrays. Four of these candidates were confirmed to be fungal biotrophy-associated secreted proteins [22]. However, virulence function of all the candidates has not been elucidated, and comprehensive gene disruption analyses of the candidates have not been carried out. Therefore, in this study we employed a large-scale disruption analysis of M. oryzae secreted protein genes to search for novel virulence effectors.
Whole-genome draft sequence of M. oryzae was published for the isolate 70-15, a laboratory strain [23]. The genome assembly consists of 37.8 Mb nucleotides encoding 11,109 predicted protein coding genes. We recently retrieved 1,306 putative secreted protein genes from the predicted proteome of 70-15 [20]. From these, a total of 78 genes expressed in the fungus were disrupted and analyzed. We found that disruptants of the 77 genes did not show change in pathogenicity as compared to the wild-type strains. Disruption of only one gene, MC69, showed a severe reduction in pathogenicity. Further analysis showed that MC69 protein is involved in the full pathogenicity of M. oryzae after the penetration stage of infection.
Results
Large-scale disruption analysis of Magnaporthe oryzae secreted protein genes
To search for effector protein genes of Magnaporthe oryzae, we carried out a large-scale targeted gene disruption analysis of the 78 putative secreted protein genes that are expressed during infection (Table 1). Initially we selected 1,306 putative secreted protein genes as described previously [20]. The 78 genes subjected to functional study were selected on the basis of their confirmed expression in the pathogen at the early stages of infection. We focused on secreted protein genes involved in the two stages of infection of M. oryzae. One is the appressorium formation stage and the other is the biotrophic invasion stage. Treatment with cyclic AMP (cAMP) induces appressorium formation on hydrophilic surface [24]. By SAGE (Serial Analysis of Gene Expression) of cAMP-treated conidia on hydrophilic membrane 6 h after the start of treatment [25], we identified several pathogenicity genes, e.g. MPG1, MAS1 and MAC1, already characterized [26]–[28] and thought to be involved in pathogen-host interaction. Therefore, we assumed that a part of effector genes should be expressed during the appressorium formation. To achieve a high efficiency tag-to-gene annotation, we established SuperSAGE method that extracts a 26-bp tag from each cDNA [29]. SuperSAGE of the cAMP-treated M. oryzae strain 70-15 has been done in this study to search for novel effector candidates (Table S1). Furthermore, we also used the SuperSAGE data of invasive hyphae for searching new effectors (Supplemental Data Set 1 in [20]). Indeed, this SuperSAGE analysis revealed that two AVR effector genes, AVR-Pia and AVR-Pii were expressed at the stage of invasive hyphae (Supplemental Data Set 2 in [20]).
Table 1. Gene disruption analysis of 78 putative secreted protein genes.
| Mutant ID | Gene ID | cAMPa | IHb | Mutant ID | Gene ID | cAMPa | IHb |
| HS9 | MGG_04172.6 | + | − | MC28 | MGG_01609.6 | + | + |
| RT11 | MGG_08342.6 | + | − | MC33 | MGG_09378.6 | + | − |
| RT76 | MGG_05716.6 | + | − | MC45 | MGG_00380.6 | − | + |
| HM4 | MGG_04757.6 | + | + | MC47 | MGG_00269.6 | − | + |
| HM5 | MGG_09188.6 | + | − | MC48 | MGG_05989.6 | − | + |
| HM6 | MGG_05798.6 | − | + | MC52 | MGG_10171.6 | − | + |
| HM17 | MGG_09920.6 | + | − | MC55 | MGG_05366.6 | + | − |
| HM18 | MGG_05785.6 | + | + | MC56 | MGG_05608.6 | + | − |
| HM20 | MGG_03356.6 | − | + | MC57 | MGG_10877.5 | + | − |
| HM21 | MGG_00505.6 | + | + | MC58 | MGG_08275.6 | + | + |
| HM22 | MGG_07763.6 | − | + | MC59 | MGG_09246.6 | + | − |
| HM24 | MGG_02245.6 | − | + | MC61 | MGG_09716.6 | + | − |
| HM27 | MGG_10102.6 | − | + | MC62 | MGG_02296.6 | + | − |
| HM30 | MGG_05381.6 | + | − | MC63 | MGG_06069.6 | + | + |
| HM36 | MGG_09460.6 | + | − | MC65 | MGG_05912.6 | + | + |
| HM57 | MGG_00703.6 | + | + | MC69 | MGG_02848.6 | + | − |
| HM63 | MGG_00659.6 | + | − | MC70 | MGG_03347.6 | + | − |
| HM65 | MGG_03245.6 | + | + | MC71 | MGG_12906.6 | + | − |
| HM66 | MGG_02420.6 | − | + | MC72 | MGG_13009.6 | + | − |
| HM68 | MGG_01532.6 | + | + | MC73 | MGG_13275.6 | + | − |
| HM88 | MGG_06951.6 | + | + | MC79 | MGG_08041.6 | + | + |
| HM91 | MGG_00314.6 | + | − | MC81 | MGG_09875.6 | − | + |
| HM93 | MGG_01843.6 | + | + | MC82 | MGG_07560.6 | + | + |
| HM104 | MGG_02987.6 | + | + | MC83 | MGG_00052.6 | + | + |
| HM106 | MGG_03130.6 | + | + | MoCel12A | MGG_00677.6 | − | + |
| HM108 | MGG_07312.6 | + | − | KY5 | MGG_10799.6 | + | + |
| Eco2 | MGG_00269.6 | − | + | KY8 | MGG_03844.6 | − | + |
| HMM14 | MGG_01872.6 | + | + | KY10 | MGG_03870.6 | − | + |
| HMM53 | MGG_06216.6 | + | + | KY22 | MGG_10291.6 | − | + |
| Taka1 | MGG_05232.6 | + | − | KY23 | MGG_10394.6 | − | + |
| Taka2 | MGG_00860.6 | + | − | KY45 | MGG_02898.6 | − | + |
| MC4 | MGG_06840.6 | − | + | KY51 | MGG_01387.6 | − | + |
| MC8 | MGG_07877.6 | + | + | KY55 | MGG_03338.6 | − | + |
| MC11 | MGG_03593.6 | + | + | AI9 | MGG_09742.6 | + | + |
| MC14 | MGG_09465.6 | + | + | AI41 | MGG_07704.6 | + | + |
| MC16 | MGG_00552.6 | + | + | AI43 | MGG_05092.6 | + | + |
| MC19 | MGG_05103.6 | − | + | AI44 | MGG_03316.6 | − | + |
| MC24 | MGG_04952.6 | − | + | AI58 | MGG_07645.6 | + | − |
| MC25 | MGG_03276.6 | − | + | AI59 | MGG_01064.6 | + | + |
Expressed gene in the cAMP-treated M. oryzae on dialysis membrane at 6 hpi in the Table S1.
Expressed gene in the M. oryzae-infected rice leaf sheath at 40 hpi in the Supplemental data set 1 [20].
(+) or (−) indicate the expressed gene or not, respectively in the cAMP-treated or invasive hyphae of M. oryzae.
To investigate the function of the effector candidate genes, we generated disruption mutants for each of the selected 78 genes (Table 1) in M. oryzae by TAG-KO method [30], [31]. To assess the virulence of each mutant, conidial suspension of each mutant was sprayed onto seedlings of a barley cultivar Nigrate, which is susceptible to the wild type M. oryzae. Blast phenotypes of barley infected by KO mutants of all the genes except for MC69 gene were the same as that infected by wild-type strain Ina72. Similarly the 77 mutants did not show reduced virulence in a susceptible rice cultivar Shin No. 2. By contrast, we observed a dramatic reduction in disease symptoms on barley cotyledons and the susceptible rice cultivar, Shin No. 2, inoculated with all of the three independent mc69 mutant lines (Figure 1A). Consequently, we identified the MC69 gene (MGG_02848.6) as required for pathogenicity of M. oryzae after a large-scale targeted gene disruption analysis of the 78 putative secreted protein genes.
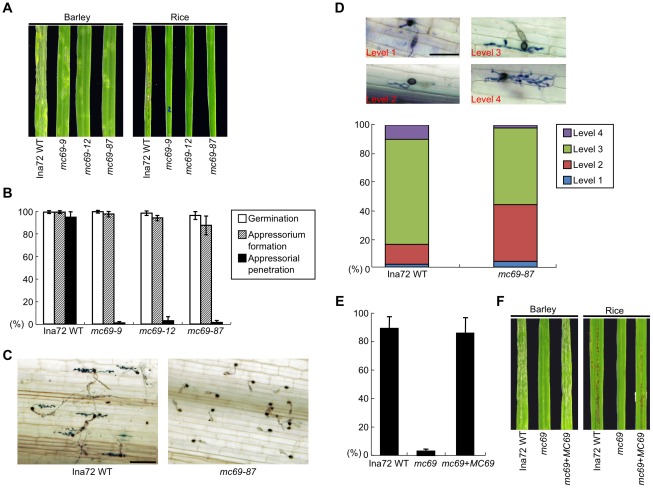
Figure 1. MC69 is involved in appressorial penetration and pathogenicity of M. oryzae.
(A) MC69 is required for pathogenicity of M. oryzae strain Ina72. Conidial suspension of the wild-type strain Ina72 (Ina72 WT) and the mc69 mutants (mc69-9, -12, -87) were inoculated on barley (cv. Nigrate) and rice (cv. Shin No. 2) leaves, and incubated for 4 and 7 days, respectively. (B) Germination, appressorium formation and appressorial penetration of Ina72 WT and the mc69 mutants. The ratio of germination was calculated as the mean percentage of conidia germinated after 24 h on rice (cv. Shin No. 2) leaf sheath cells. The mean percentage of appressorium formation on rice leaf sheath cells among the germinated conidia is presented. Three replicates of ∼50 conidia were counted for each observation. The mean percentage of appressorial penetration by the mc69 mutants 32 h after inoculation is presented. Standard errors are indicated by the vertical bars. (C) Appressorial penetration assays on rice leaf sheath cells. Conidia from Ina72 WT and mc69-87 germinated and formed melanized appressoria. All appressoria formed by Ina72 WT penetrated and produced infectious hyphae but no appressoria formed by mc69-87 produced infectious hyphae in most of area. Photographs were taken 32 h after inoculation. Scale bar = 40 µm. (D) Invasive growth rating of rice leaf sheath cells 32 h after inoculating with Ina72 WT and mc69-87. The levels for invasive growth rating are given above. For details of the invasive growth levels and rating see Materials and Methods. Scale bar = 20 µm. (E), (F) Complementation of mc69 mutant with the wild type allele of MC69. (E) Appressorial penetration by Ina72 WT, mc69-87 (mc69) and the MC69 re-introduced strain (mc69+MC69). Mean percentage of appressorial penetration is recorded 32 h after inoculation in rice leaf sheath cells. Four replicates of ∼50 appressoria were counted for each observation. (F) Blast symptoms caused by Ina72 WT, mc69 and mc69+MC69 on barley (cv. Nigrate) and on rice (cv. Shin No 2) 3 and 4 days after inoculation, respectively.
In summary, we found that targeted disruption of MC69 affected pathogenicity of M. oryzae and disruption of the other 77 genes had no effect on its pathogenicity.
MC69 is required for appressorial penetration and pathogenicity of M. oryzae
To investigate the physiological and molecular function of MC69 in detail, we generated MC69 disruptants in M. oryzae strain Ina72 by targeted gene disruption as described above. Colony growth, color and the production of conidia were the same as the wild-type strain (Figure S2A and B). We observed a remarkable reduction in disease symptoms on barley and rice inoculated with the mc69 mutants compared to those inoculated with the wild type strain 4 and 7 days after inoculation suggesting an important role of MC69 in fungal pathogenicity (Figure 1A). Subsequently, we performed a detailed phenotypic analysis of the mc69 mutants. The mc69 mutants exhibited a defect in appressorium-mediated penetration in rice leaf sheath cells but neither in conidial germination nor appressorium formation (Figure 1B and C). We studied 200 appressoria of each mc69 mutant, 97∼99% with failed penetration (no visible hyphae) and 1∼2% with post-penetration blockage. Therefore, we conclude that MC69 is required for appressorial penetration and pathogenicity of M. oryzae. Although most of appressoria formed by the mc69 mutants could neither penetrate nor produce infectious hyphae in the inoculated rice leaf sheath cells, we further analyzed invasive growth at 50 appressorial penetration sites by rating the hyphal growth from the level 1 (low) to 4 (high; see Materials and Methods, Figure 1D). In Ina72 WT, 84% of penetration sites showed invasive growth levels 3 or 4, by contrast in the mc69 mutant infectious growth within the inner epidermal tissue was relatively limited (levels 2 and 3) 32 hours after inoculation, suggesting that the loss of MC69 also affects infectious growth to some extent at the post-penetration stage (Figure 1D). To test whether the observed phenotypes of the mc69 mutants were solely caused by disruption of MC69, an intact copy of MC69 was introduced into the mc69 mutant mc69-87 (Figure S1B) for complementation. The MC69-reintegrated strain showed normal appressorial penetration rate and the strain developed blast disease symptoms on barley and rice leaves with a similar extent to the wild type (Figure 1E and F). These results demonstrate that disruption of MC69 gene caused defects in appressorial penetration and development of blast symptoms by M. oryzae.
MC69 is expressed in conidia and all stages of infection, secreted but not translocated into rice cells
MC69 was found in the SuperSAGE list of the cAMP-treated conidia (Table S1). MC69-EST was found in MGOS databases for mycelium, conidia, germinated conidia and appressoria [32]. RL-SAGE tags of MC69 were also found in the fungus grown on a minimum medium for three days [33]. These data suggest that MC69 is constitutively expressed in M. oryzae.
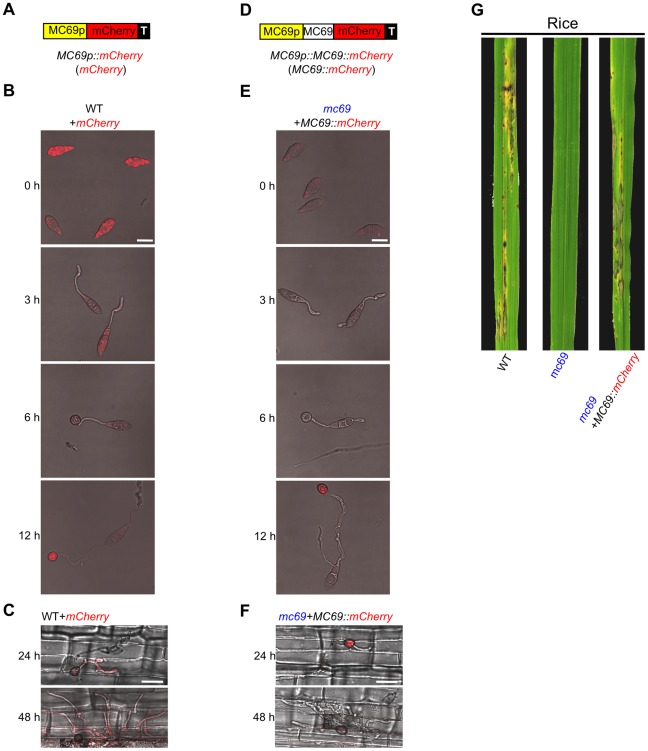
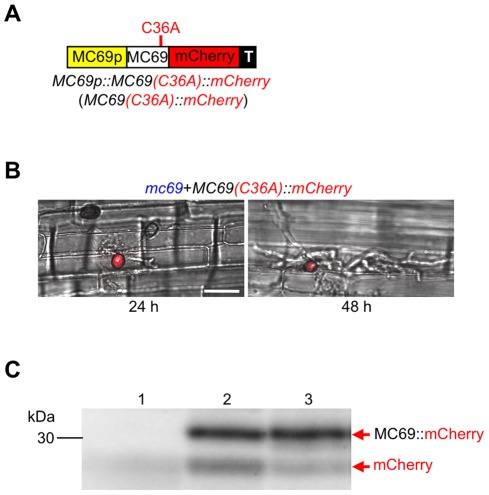
To investigate the expression pattern of MC69 in detail, we produced an M. oryzae strain Ina72 harboring a vector containing the MC69 promoter fused with a reporter protein gene mCherry (MC69p::mCherry; Figure 2A) generating WT+mCherry. The mCherry fluorescence was observed in all morphological stages with enhanced fluorescence in conidia before germination (0 h) and matured appressoria (12 h after incubation) on glass coverslips under confocal laser-scanning microscope (Figure 2B). To determine the mode of expression and spatial localization of the MC69 protein, a construct MC69p::MC69::mCherry was prepared (Figure 2D) and used for transformation of the mc69 mutant generating mc69+MC69::mCherry, and the mCherry fluorescence was then observed on glass coverslips (Figure 2E). The transgenic M. oryzae mutant mc69 expressing MC69::mCherry restored pathogenicity (Figure 2G), showing that the fusion protein MC69::mCherry is functional for infectivity. The mCherry fluorescence was detected in all the developmental stages like WT+mCherry, but the intensity of fluorescence in the strain mc69+MC69::mCherry was weaker than that of the strain WT+mCherry presumably because of secretion and diffusion of MC69::mCherry fusion protein (Figure 2B and E). To observe the fluorescence of WT+mCherry and mc69+MC69::mCherry in the infected tissues, we inoculated these conidial suspensions to rice leaf sheath. The mCherry fluorescence was detected in the invaded hyphae 24 and 48 hours after inoculation with WT+mCherry, but not detected in that inoculation with mc69+MC69::mCherry (Figure 2C and F). These results suggest that the MC69 gene is expressed throughout infection: conidia, infection-related morphogenesis and subsequent growth stage. The fluorescence from MC69::mCherry fusion proteins was not detected in the invaded hyphae in planta presumably because they have been secreted.
Figure 2. Microscopic analysis suggests that MC69 promoter is constitutively active and MC69::mCherry fusion protein is secreted.
(A) and (D) Schematic diagrams of mCherry and MC69::mCherry fusion protein expression constructs. (B) and (E) Conidia from WT+mCherry and mc69+MC69::mCherry were harvested, and appressorium development was observed for 12 h on glass coverslips. Merged DIC and mCherry (red) images were taken. Scale bars = 10 µm. (C) and (F) Merged DIC and mCherry images of the rice leaf sheath cells infected with mCherry- and MC69::mCherry-expressing transformants 24 h and 48 h after inoculation. Scale bars = 20 µm. (G) Blast symptoms caused by Ina72 WT, mc69 and mc69+MC69::Cherrry on rice (cv. Shin No. 2) 7 days after inoculation.
To obtain direct evidence that the MC69 protein is actually produced in the invasive hyphae, we made mc69 mutant harboring a construct MC69p::MC69::HA (mc69+MC69::HA) or a MC69p::MC69::3xFLAG (mc69+MC69::3xFLAG) to perform immunodetection of MC69::HA or MC69::3xFLAG proteins in planta, respectively. Both of mc69+MC69::HA and mc69+MC69::3xFLAG restored appressorial penetration and invasive growth in rice leaf sheath (Figure S3A and C), showing that the fusion proteins MC69::HA and MC69::3xFLAG are functional for pathogenicity. To clarify whether the MC69::HA and MC69::3xFLAG are expressed or not, mc69+MC69::HA- and mc69+MC69::3xFLAG-infected rice leaf sheath extracts were analyzed by SDS-PAGE gel blot analysis. We extracted total protein at 24 and 48 hours after leaf sheath inoculation. Note that at 24 hours after inoculation, most conidia develop appressoria but hyphal invasion is still limited, whereas at 48 hours extensive hyphal growth develops. Both of HA- and 3xFLAG-tagged MC69 were detected only faintly 24 hours after inoculation, but these proteins were abundant 48 hours after inoculation (Figure S3B and D) indicating that MC69 protein was indeed produced in invasive hyphae. Furthermore, we tried to express an AVR effector gene, AVR-Pia [18], [20] from the MC69 promoter to see whether AVR-Pia avirulence function is supported by MC69 promoter in rice plant harboring Pia R-gene. We hypothesized that only when MC69 promoter allows expression of AVR-Pia in invasive hyphae, a sufficient amount of AVR-Pia protein would be translocated inside rice cells to be recognized by Pia, NBS-LRR-type cytoplasmic R-proteins [34]. We performed transformation of the M. oryzae isolate Ina86-137 (that lacks AVR-Pia and can infect rice cultivars possessing Pia R-gene) [20] with a construct MC69p::AVR-Pia. We used the wild-type stain (Ina86-137 WT) and a transformant harboring an intact copy of AVR-Pia (+AVR-Piap::AVR-Pia) [20] as negative and positive controls, respectively. In contrast with the Ina86-137 WT, +AVR-Piap::AVR-Pia and the transformants harboring MC69p::AVR-Pia (+MC69p::AVR-Pia-1 and -2) failed to cause disease in the rice cultivar Sasanishiki possessing Pia (Figure S4A). Both Ina72-WT and the transformants successfully infected rice cultivar Shin No. 2 that lacks Pia, suggesting that their inability in infecting cv Sasanishiki was caused by Pia-AVR-Pia interactions and that the MC69 promoter is expressed during invasive growth. Active transcription of the AVR-Pia in the transformants was confirmed by RT-PCR (Figure S4B) [20].
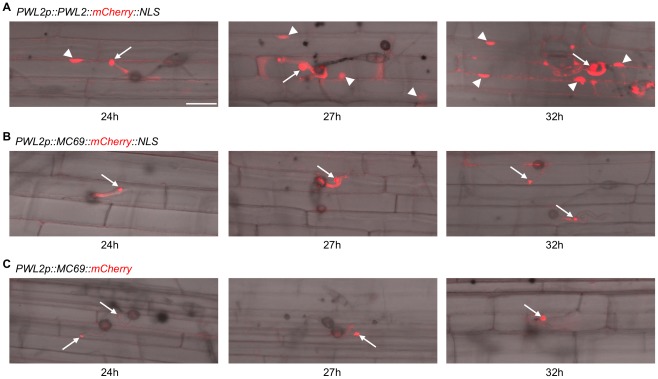
Khang et al. (2010) reported that secreted fluorescent effectors preferentially accumulate in biotrophic interfacial complexes (BICs) at the invasive hyphae-rice cell interface [11]. By fusing nuclear localization signal (NLS) to the fluorescent effectors to facilitate visualization of translocation, they also showed that the two BIC-localized secreted proteins, PWL2 and BAS1 were translocated into rice cytoplasm [11]. To test translocation of MC69::mCherry in rice cells, we added a modified small NLS from simian virus large T-antigen [35] at the C terminus of the MC69::mCherry fusion downstream of the PWL2 promoter (PWL2p::MC69::mCherry::NLS) and transformed M. oryzae strain Sasa2 with the construct. A transformant M. oryzae harboring PWL2p::PWL2::mCherry::NLS was used as positive control. PWL2::mCherry::NLS exhibited significant fluorescence in BIC and in nuclei of invaded host cells at successful infection sites 24, 27 and 32 hours after inoculation (Figure 3A) whereas MC69::mCherry::NLS did not show fluorescence in nuclei of the invaded rice cells, but showed weaker fluorescence in BIC than that of PWL2::mCherry::NLS (Figure 3B). To eliminate the possibility that the NLS influences BIC localization, a transformant strain harboring PWL2p::MC69::mCherry was inoculated to rice leaf sheath. The result showed that MC69::mCherry was also detected in the BIC (Figure 3C). In addition, we observed mCherry fluorescence with different pinhole settings to compare the signals in the BIC among the three strains 27 hours after inoculation. The result showed that BIC accumulation signals of MC69::mCherry::NLS and MC69::mCherry were significantly weaker than that of PWL2::mCherry::NLS (Figure S5). These results suggest that the MC69 does not translocate into the infected rice cells, but localizes in BIC, however the accumulation level of MC69 in BIC is significantly lower than that of PWL2.
Figure 3. MC69::mCherry is not translocated into the rice cytoplasm.
Merged DIC and mCherry images of rice leaf sheath cells infected by M. oryzae Sasa2 strain harboring (A) PWL2p::PWL2::mCherry::NLS, (B) PWL2p::MC69::mCherry::NLS, and (C) PWL2p::MC69::mCherry 24, 27 and 32 h after inoculation as observed by confocal laser scanning microscopy. Arrows indicate BICs and triangles indicate rice nuclei. Pinhole setting is 240 µm for all panels. Scale bar = 20 µm.
Two cysteine residues are essential for MC69 function
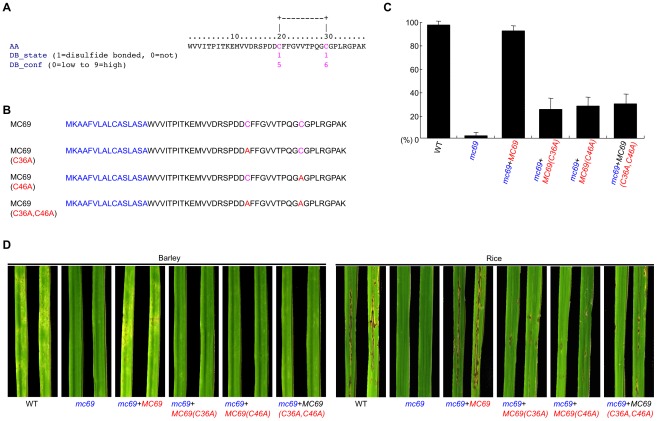
MC69 homologs were found in other filamentous fungi Colletotrichum orbiculare (AB669186), Glomerella graminicola (EFQ29542), Verticillium albo-atrum (EEY15898), V. dahliae (EGY20943), Neurospora crassa (XP_965292), N. tetrasperma (EGO52621), Myceliophthora thermophila (XP_003659994), Podospora anserina (XP_00190740), Grosmannia clavigera (EFX05010), Fusarium oxysporum (EGU75378), Gibberella zeae (XP_388669), Trichoderma atroviride (EHK44387), T. virens (EHK23962), Metarhizium acridum (EFY93067), M. anisopliae (EFY97094) and Cordyceps militaris (EGX95034) (Figure S6 and S7). However, these amino acid sequences did not contain known domains/motifs that would allow the prediction of their function. Nevertheless, MC69 homologs contain two conserved cysteine residues in the mature protein region C-terminal to the signal peptide (Figure S6). A software DISULFIND (http://disulfind.dsi.unifi.it/; [36]) predicted that the two cysteine residues in mature MC69 can form a disulfide bond (Figure 4A). To test whether these cysteines are necessary for MC69 function, mutant alleles of MC69 were generated in which each or both of C36 and C46 were replaced with alanine (Figure 4B). Mutant alleles with one amino acid replacement (MC69(C36A); MC69(C46A)) or two replacements (MC69(C36A,C46A)) were expressed in the mc69 mutant (mc69+MC69(C36A), mc69+MC69(C46A) or mc69+MC69(C36A,C46A)). In all cases, appressorial penetration rate and blast symptoms on barley and rice were slightly restored, but still significantly reduced as compared to the wild type (Figure 4C and D). In addition, we further analyzed invasive growth rating of the 50 appressorial penetration sites. Infectious growth of mc69+MC69(C36A), mc69+MC69(C46A) and mc69+MC69(C36A,C46A) within the inner epidermal tissue was slightly restored as compared to the mc69 mutant (Figure S8). These results indicate that C36 and C46, presumably involved in disulfide bond, are necessary for MC69 to exert its pathogenicity in M. oryzae.
Figure 4. Two cysteine residues are important for MC69 virulence activity.
(A) DISULFIND (http://disulfind.dsi.unifi.it/) output of the mature form of MC69 protein. (B) Amino acid sequences of disulfide mutants of MC69. The predicted signal peptide is indicated in blue. (C) Appressorial penetration by M. oryzae wild-type strain Ina72 (WT) and transformants with various versions of MC69. Mean percentage of invasion in rice (cv. Shin No. 2) leaf sheath cells 30 h after inoculation is presented. Four replicates of ∼50 appressoria were counted for each observation. (D) Blast symptoms caused by WT, the mc69 mutant (mc69), the MC69-, MC69(C36A)-, MC69(C46A)- and the MC69(C36A,C46A)-re-introduced strains [mc69+MC69, mc69+MC69(C36A), mc69+MC69(C46A) and mc69+MC69(C36A,C46A)] on barley (cv. Nigrate) and on rice (cv. Shin No. 2) 4 and 7 days after inoculation, respectively.
To see whether C36 and C46 are important for MC69 secretion/localization, spatial localization of the MC69(C36A) protein was tested by transforming mc69 mutant with a construct MC69p::MC69(C36A)::mCherry, resulting in mc69+MC69(C36A)::mCherry (Figure 5A). We inoculated conidial suspension of the strain to rice leaf sheath to observe the mCherry fluorescence in the infected tissue. The mCherry fluorescence was detected in appressoria but not in the invaded hyphae 24 and 48 hours after inoculation (Figure 5B). The result suggests that the MC69(C36A)::mCherry protein was secreted into the plant and diffused below the detection limit like MC69::mCherry (Figure 2F and 5B). To clarify whether the MC69::mCherry and MC69(C36A)::mCherry are secreted or not, extracellular proteins secreted by Magnaporthe after liquid culture were analyzed by SDS-PAGE gel blot analysis. We used the wild-type strain expressing mCherry under MC69 promoter (WT+mCherry; Figure 2B and C) as negative control. Western blot analysis (Figure 5C) revealed the presence of mCherry-tagged MC69 and mCherry-tagged MC69(C36A) in the culture medium. Faint signals of cleaved mCherry were observed as well for the transformants mc69+MC69::mCherry and mc69+MC69(C36A)::mCherry. The molecular weight (MW) of the fusion proteins was around 30 kDa, in line with the predicted MW of mature MC69::mCherry. These data strongly suggest that both of MC69 and MC69(C36A) are secreted to the medium, and C36 is not important for MC69 secretion. It could be possible that mutation of the Cys residues may impact pathogenicity by reducing the stability of the protein after secretion in planta.
Figure 5. Secretion of MC69(C36A)::mCherry and MC69::mCherry fusion proteins.
(A) Schematic diagram of MC69(36A)::mCherry fusion protein expression construct. (B) Merged DIC and mCherry images of rice leaf sheath cells infected by the MC69(C36A)::mCherry-expressing transformants 24 h and 48 h after inoculation. Scale bar = 20 µm. (C) Western blot probed with an anti-DsRed antibody. Samples were loaded as follows: lane 1, culture filtrate from mCherry-expressing strain; lane 2, culture filtrate from MC69::mCherry-expressing strain; lane 3, culture filtrate from MC69(C36A)::mCherry-expressing strain.
MC69 is commonly required for pathogenicity in M. oryzae
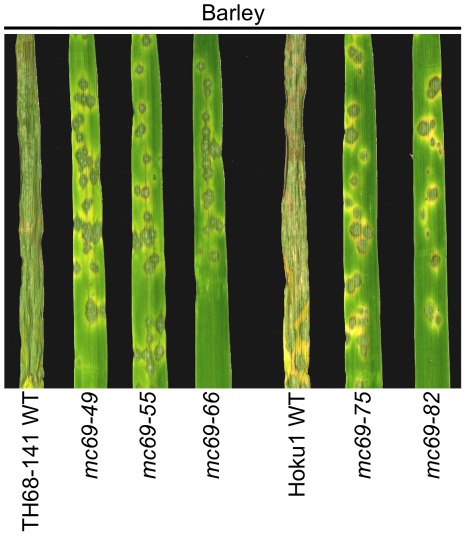
To investigate whether pathogenicity function of MC69 is conserved in M. oryzae, we produced mc69 disruptants in other two Japanese field isolates of TH68-141 and Hoku1, in addition to the isolate Ina72. We used the MC69 knockout vector used for Ina72 to generate mc69 disruptants of both TH68-141 and Hoku1 isolates. Generated mc69 disruptants of the two isolates showed a reduced pathogenicity on barley leaves as compared to the wild type strains (Figure 6), indicating the importance of MC69 in virulence of TH68-141 and Hoku1.
Figure 6. MC69 is required for pathogenicity of other two different Japanese field isolates TH68-141 and Hoku1.
Conidial suspension of wild-type strain TH68-141 (TH68-141 WT), the three independent mc69 mutants (mc69-49, mc69-55 and mc69-66), wild-type strain Hoku1 (Hoku1 WT) and the two independent mc69 mutants (mc69-75 and mc69-82) were inoculated on barley (cv. Nigrate) leaves and incubated for 5 days.
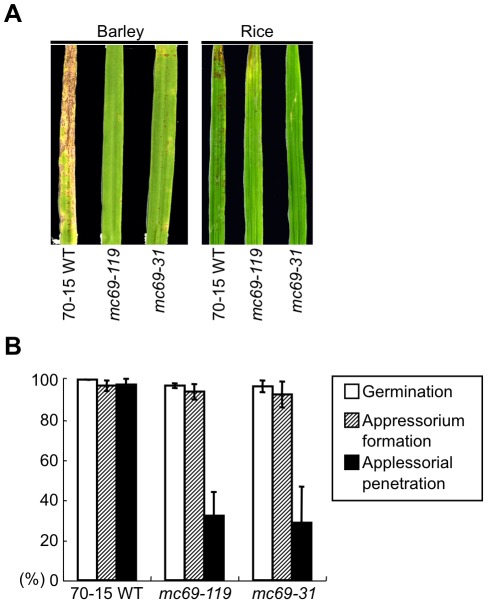
The whole genome sequence of 70-15, a well-studied laboratory strain of M. oryzae, was published [23]. We found that 70-15 showed poor virulence as compared to the Japanese strains in the previous study. It caused intermediate responses in all of the 13 tested rice cultivars: infection caused reddish lesions of various sizes, but they did not further develop into typical susceptible brown spindle-shaped necrotic lesions [20]. To investigate whether MC69 is required for pathogenicity in 70-15, MC69 gene disruption analysis was performed in the 70-15 background (Figure S1). Two independent MC69-KO lines (mc69-119 and mc69-31) and wild-type 70-15 were sprayed onto barley cotyledons and rice leaves. The barley and rice infected by mc69 mutants showed much weaker symptoms as compared to the 70-15-infected plants (Figure 7A), indicating the importance of MC69 in pathogenicity of 70-15. Appressorial penetration rates of the mutants in rice leaf sheath cells were significantly lower than that of 70-15 but the rates of germination and appressorium formation were same with the wild type (Figure 7B). In addition, we further analyzed invasive growth rating of 50 appressorial penetration sites. Infectious growth of the mc69 mutants within the inner epidermal tissue was restricted as compared to the wild type (Figure S9). However, the colony growth and conidiation of the mutants on oatmeal agar media were similar to the wild type (Figure S2C and D). Thus, the mc69 mutants of 70-15 have a defect in appressorial penetration and development of blast symptoms, which is similar to the phenotype of the mc69 disruptants of Ina72. These results suggest that MC69 is commonly required for appressorial penetration and subsequent colonization in various M. oryzae strains.
Figure 7. MC69 is necessary for appressorial penetration and pathogenicity of a laboratory strain 70-15.
(A) MC69 is required for pathogenicity of M. oryzae strain 70-15. Conidial suspension of the wild-type strain 70-15 (70-15 WT) and the mc69 mutants (mc69-119, -31) were inoculated on barley (cv. Nigrate) and rice (cv. Shin No. 2) leaves, and incubated for 7 days. (B) Germination, appressorium formation and appressorial penetration of 70-15 WT and the mc69 mutants. The ratio of germination was calculated as the mean percentage of conidia germinated after 32 h on rice (cv. Shin No. 2) leaf sheath cells. The mean percentage of appressorium formation on rice leaf sheath cells among the germinated conidia is presented. Three replicates of ∼50 conidia were counted for each observation. The mean percentage of appressorial penetration by the mc69 mutants is presented 32 h after inoculation. Standard errors are indicated by the vertical bars.
An ortholog of MC69 is required for pathogenicity of Colletotrichum orbiculare
The importance of MC69 in M. oryzae raised a possibility that MC69 orthologs are also involved in pathogenicity of other fungal pathogens. To assess this point, we investigated whether MC69 ortholog is involved in pathogenicity of the cucumber anthracnose fungus C. orbiculare (Figure S6). A gene homologous to MC69 was isolated from C. orbiculare in this study. The isolated gene, designated CoMC69, comprises 220 bp interrupted by an intron and encodes a predicted protein of 54 amino acids (Figure 8A). Intron/exon organization in MC69 orthologs in filamentous fungi indicated that most of them have one intron only followed by an exon (140–156 bp) except for the genes in T. virens and Gibberella zeae (Figure S10). First exons in all genes encode a common region containing two conserved cysteine residues in the mature proteins (Figure S6).
Figure 8. CoMC69 is involved in fungal pathogenicity of C. orbiculare.
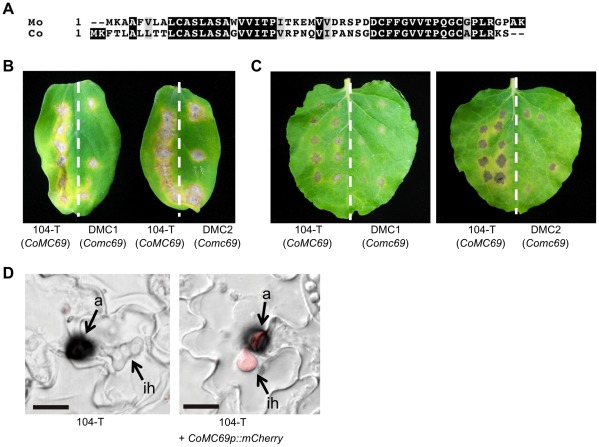
(A) Sequence alignment of MC69 between M. oryzae (Mo) and C. orbiculare (Co). Amino acid sequences were aligned using Clustal W program [52]. Identical amino acids are indicated as white letters on a black background. Similar residues are shown by gray background. Gaps introduced for alignment are indicated by hyphens. (B) Pathogenicity test of the Comc69 mutants on cucumber. Conidial suspensions were inoculated on detached cotyledons of cucumber (Cucumis sativa). On the left half of the cotyledons, the wild-type strain 104-T was inoculated as positive control. On the right half, the Comc69 strains (DMC1 and DMC2) were inoculated. Inoculated cotyledons were incubated for 7 days. (C) Pathogenicity test of the Comc69 mutants on N. benthamiana. On the left half of the detached leaves of N. benthamiana, the strain 104-T was inoculated as positive control. On the right half, the Comc69 strains (DMC1 and DMC2) were inoculated. Inoculated leaves were incubated for 7 days. (D) mCherry-based reporter assay for expression of the CoMC69 gene. Conidia from the C. orbiculare strain carrying the CoMC69 promoter-mCherry fusion gene (CoMC69p::mCherry) was inoculated onto the lower surfaces of cucumber cotyledons, and the inoculated plant was incubated for 4 days. a, appressorium; ih, intracellular hypha. Scale bars = 10 µm.
To investigate whether CoMC69 is involved in the pathogenicity of C. orbiculare, we produced CoMC69 disruption mutants. The plasmid pCBGDMC69 was designed to replace the CoMC69 gene in the wild-type strain 104-T through double crossover homologous recombination (Figure S11A and B).
The colony morphology and conidiogenesis of Comc69 mutants grown on PDA medium were similar to that of 104-T (Figure S11C and data not shown). We next investigated their pathogenicity on host cucumber leaves. Conidial suspensions from the Comc69 mutants were spotted on detached cucumber leaves and incubated for 7 days. The Comc69 mutants exhibited clear reduction in lesion development in comparison with the wild-type strain 104-T (Figure 8B). C. orbiculare 104-T is able to infect Nicotiana benthamiana, which is not closely related to cucumber [37]. The Comc69 mutants also exhibited reduced pathogenicity on N. benthamiana (Figure 8C). These results indicate that CoMC69 is required for pathogenicity of C. orbiculare, suggesting conserved roles of the MC69 proteins in pathogenicity of both M. oryzae and C. orbiculare. To investigate the gene expression of CoMC69 in plant infection of C. orbiculare, we generated C. orbiculare strains carrying a reporter plasmid containing the 1.4 kb 5′ upstream region of CoMC69 fused with mCherry. As a result, we found the mCherry fluorescence in appressoria and primary intracellular hyphae of the transgenic C. orbiculare, indicating the expression of CoMC69 in the plant infection stage of C. orbiculare (Figure 8D).
Discussion
In this study, we show that MC69, a novel secreted protein of Magnaporthe oryzae, is essential for successful appressorial penetration and blast symptom development in rice and barley cultivars. The MC69 gene (MGG_02848.6) resides on chromosome VII of the M. oryzae genome. The MC69 protein comprises 54 amino acids and is predicted to harbor a putative N-terminal secretion signal peptide (Figure 4B). MC69 seems to be a solitary gene without any paralogs in the genome. It lacks known sequence motifs associated with enzymatic function. Although MC69 homologs were found in other filamentous fungi (Figure S6), their functions are also not known. Expression of MC69 was observed in mycelia, conidia and all stages of infection (Figure 2). The mc69 disruptants were unable to invade plant cells to establish compatible interaction with the host plant (Figure 1, 6 and 7). However in mc69 mutants, other phases of infection-related development such as conidial germination and appressorium formation were unaffected (Figure 1B and 7B).
MC69 of M. oryzae, which is predicted to comprise 38 amino acids after cleavage of the signal peptide, contained no known functional domains. Therefore it is unlikely that MC69 has an enzymatic function. The two cysteine residues (C36 and C46) conserved among the MC69 homologs may be involved in disulfide bridge formation and were shown to be important for pathogenicity function of MC69 (Figure 4 and S6). Pep1 is a novel effector protein from the corn smut fungus Ustilago maydis that is essential during penetration. Disruption mutants of pep1 are not affected in saprophytic growth and develop normal infection structures, but are arrested during the penetration of epidermal cells of maize leaves. In addition, two of the four cysteine residues in Pep1 were shown to be essential for the virulence function [38]. The authors consider that the importance of two of the four cysteine residues for secretion of Pep1 to make a compact structure with disulfide bridge structure of Pep1 [38]. To address this possibility we observed localization of MC69::mCherry and MC69(C36A)::mCherry in M. oryzae. We were unable to detect red fluorescence in infectious hyphae and the fusion proteins were detected in the culture filtrate of both strains (Figure 2 and 5). These results indicated that the substitution of cysteine residues of MC69 did not affect secretion, but affected pathogenicity of M. oryzae.
On the other hand, disruption of a total of 77 secreted protein genes in M. oryzae did not affect its pathogenicity within our experimental condition. Since there is no systematic bias in our selection of secreted protein genes for disruption, we extrapolate that 77/78 = 99% of secreted protein genes do not show clear reduction in pathogenicity even after knockout. Several secreted avirulence (AVR) effector genes have been isolated from M. oryzae, including PWL effectors [16], [19], AVR-Pita [13], [14], AVR1-CO39 [15], AVR-Piz-t [17], AVR-Pia, AVR-Pii and AVR-Pik/km/kp [18], [20], but the virulence functions of the genes are still unknown. In fact, the AVR-Pita effector is dispensable for virulence on rice [14], [39]. According to our results and available information on M. oryzae AVR effectors, we hypothesize two mutually non-exclusive possibilities: (1) virulence contribution of most of effectors is too small to be detected by conventional assays; (2) effectors have redundant activities and more than one effector participate in the same virulence pathway.
A recent report of M. oryzae indicates that the fungus overcomes the first line of defense (PAMP-Triggered Immunity) by secreting an effector protein, Slp1 during invasion of new rice cells [21]. There are several reports for secreted effectors of other fungal pathogens. Pep1 of U. maydis was described above. Several hydrophobins or repellent genes that encode secreted proteins of U. maydis were examined for their roles in virulence. Single knock-outs of these genes did not affect virulence, but a double knockout of the repellent-encoding gene Rsp1 and Hum3 (a gene encoding a protein containing both, a hydrophobin domain and a repellent region) were arrested at an early stage of penetration. This indicates that Rsp1 and Hum3 are effectors with a partly redundant virulence function during the early stages of infection [40]. We speculate a similar situation may occur in M. oryzae. It would be a good way to focus on effector candidates exhibiting higher similarities and knockout or silence multiple genes simultaneously, to identify the multiple effectors that act redundantly.
We showed that MC69 was required for pathogenicity of the additional three strains of M. oryzae in addition to the strain Ina72, indicating conserved roles of MC69 in M. oryzae. To investigate whether the pathogenicity function of the MC69 ortholog in the well-studied dicot fungal pathogen, we isolated an ortholog, CoMC69 from the cucumber anthracnose fungus, Colletotrichum orbiculare. Notably, in C. orbiculare the deletion of CoMC69 reduced pathogenicity on the hosts cucumber and Nicotiana benthamiana leaves (Figure 8).
Phylogenetic analyses were performed with M. oryzae MC69 (MoMC69), with 16 homologs from other phytopahogenic (C. orbiculare, Glomerella graminicola, Verticillium albo-atrum, V. dahliae, Grossmannia clavigera, Fusarium oxysporum and Gibberella zeae), entomopathogenic (Metarhizium acridum and M. anisopliae), caterpillar killer (Cordyceps militaris), fungal parasite (Trichoderma atroviride and T. virens) or saprophytic (Neurospora crassa, N. tetrasperma, Myceliophthora thermophila and Podospora anserina) fungi (Figure S7). MoMC69 and CoMC69 are closely related to the Verticillium wilt pathogens V. albo-atrum, V. dahliae and the cereal plants anthracnose fungus Glomerella graminicola. A conserved motif containing the two cysteine residues showed high homology among all MC69 homologs (Figure S6). Thus, it will be interesting to determine whether genes orthologous to MoMC69 also contribute to the pathogenicity of various plant, fungus, entomo or caterpillar pathogenic fungi. However, MC69 orthologs also occur in saprophytes suggesting that a possibility that the primary function of the protein is in relation to the structure or function of the fungus itself, and that the function must be intact for the fungus to succeed as a pathogen.
We generated transgenic rice overexpressing MC69 to examine susceptibility to M. oryzae wild-type strain or the mc69 mutant infection. However, overexpression of MC69 in rice neither enhanced the pathogenicity of M. oryzae wild-type strain nor complemented the pathogenicity deficiency of the mc69 mutant (data not shown). We hypothesize three possibilities why overexpression of MC69 did not affect M. oryzae wild-type strain and the mc69 mutant infection. One possibility is that the localization of MC69 in the infection sites of M. oryzae in rice cells is important. We produced M. oryzae transformant harboring PWL2p::MC69::mCherry. After inoculation of the strain to the rice leaf sheath, mCherry fluorescence was detected in biotrophic interfacial complex (BIC) (Figure 3C). However, MC69::mCherry fusion protein expressed by MC69 promoter was not detected in BIC (Figure 2F). It might be because MC69 promoter activity was weaker than PWL2 promoter activity or PWL2 promoter leads MC69::mCherry to BIC accumulation but MC69 promoter did not. To test these possibilities, mc69 mutant expressing MC69::EGFP fusion protein downstream of MC69 promoter has been produced because EGFP fluorescence was relatively stronger than mCherry fluorescence. When the transformant was inoculated to the rice leaf sheath, MC69::EGFP was shown to be accumulated to the BIC (data not shown). These results indicate that BIC localization of MC69 is important for virulence of M. oryzae. Therefore, ectopic overexpression of MC69 in rice neither enhanced pathogenicity of wild-type strain nor complemented deficiency of mc69 mutant of M. oryzae in trans because of the MC69 protein would not be localized in BIC. The second possibility is that post translational modification of MC69 protein in M. oryzae might be different from that in planta even though the secreted MC69::mChrerry protein shows an expected molecular size (Figure 5C). The third possibility is that MC69 affects the physiology of the fungus but does not directly affect the physiology of the plant so that expression of MC69 in rice did not complement the defect in mc69 mutant of M. oryzae.
Khang et al. (2010) demonstrated that BIC-localized secreted proteins PWL2 and BAS1 were translocated into the rice cytoplasm but a secreted protein BAS4, which uniformly outlines the invasive hyphae, was not [11]. Interestingly, when M. oryzae transformants secreted fluorescent MC69 fusion protein during epidermal cell invasion, the fluorescent protein was observed in BICs (Figure 3). To investigate whether this feature is specific to MC69 or not, we generated M. oryzae strains expressing fluorescence-labeled version of four putative secreted proteins (HMM14, MC55, GAS1 or GAS2) with NLS. HMM14 and MC55 are the secreted protein genes studied here (Table 1) and GAS1 and GAS2 are secreted protein genes involved in virulence of M. oryzae [28]. The generated strains were inoculated to rice leaf sheaths, and localization of each protein was investigated. The result showed that all four proteins were localized to the BICs, and both GAS1 and GAS2 were then translocated into the rice cytoplasm, which is similar to PWL2. By contrast, HMM14 and MC55 were not translocated into the rice cytoplasm like MC69 (data not shown). However, fluorescent signals of BIC accumulations of MC69::mCherry::NLS, MC69::mCherry, HMM14::mCherry::NLS and MC55::mCherry::NLS were significantly weaker than that of PWL2::mCherry::NLS, GAS1::mCherry::NLS and GAS2::mCherry::NLS (Figure 3, S5 and data not shown). These finding indicate that BIC accumulation level of secreted proteins might be important for translocation to the infected rice cells.
A virulence effector Slp1 sequesters chitin oligosaccharides to prevent PAMP-triggered immunity in rice, thereby facilitating rapid spread of the fungus within host tissue [21]. Slp1 contains two putative LysM domains, which have previously been shown to bind carbohydrates [41]. An effector known as Ecp6 that also contains LysM domains was identified from a fungal pathogen Cladosporium fulvum that causes leaf mold of tomato [42]. Another effector AVR4 of C. fuluvum binds to chitin present in fungal cell walls and that, through this binding AVR4 can protect these cell walls against hydrolysis by plant chitinases [43]. Growth of the Δpep1 mutants of U. maydis are arrested during penetration of the epidermal cell and elicit a strong plant defense response such as formation of large papillae, induction of strong cell wall autofluorescence, H2O2 accumulation and defense related gene expression [38]. We tried to elucidate the roles of MC69 by addressing differences in H2O2 accumulation and expression of defense related genes in rice infected by mc69 mutant and wild type M. oryzae. However, results showed no difference between mc69 and wild type so that we have no evidence that MC69 suppresses plant defense responses at the moment (data not shown). Taken together, we demonstrated that MC69 has a pathogenicity function (required for the fungus to be a pathogen), but its function has yet to be elucidated.
Materials and Methods
Fungal strains, medium and transformation
All isolates of M. oryzae used in this study are stored at the Iwate Biotechnology Research Center. Fungal strains used were the wild-type strains 70-15, Ina72, TH68-141, Hoku1, Sasa2 and Ina86-137 [20]. To obtain protoplasts, hyphae of M. oryzae strains were incubated for 3 days in 200 mL of YG medium (0.5% yeast extract and 2% of glucose, w/v). Protoplast preparation and transformation were performed as described previously [44]. Hygromycin- or bialaphos-resistant transformants were selected on plates with 300 µg ml−1 of hygromycin B (Wako Pure Chemicals, Osaka, Japan) or 250 µg ml−1 of bialaphos (Wako Pure Chemicals). C. orbiculare (Berk. & Mont.) Arx (syn. C. lagenarium [Pass.] Ellis & Halst.) strain 104-T (MAFF240422) was used as the wild-type strain. All C. orbiculare strains were maintained on 3.9% (w/v) PDA (Difco Laboratories, Detroit, MI) at 24°C. Preparation of protoplasts and transformation of C. orbiculare were performed according to a method described previously [45].
SuperSAGE of cAMP-treated M. oryzae strain 70-15
Mycelia of M. oryzae were grown on oatmeal agar medium (30 g l−1 oatmeal, 5 g l−1 sucrose and 16 g l−1 agar). To enhance conidia formation, the fungus was first grown on oatmeal agar medium for 9 days at 25°C, and then exposed to Black Light Blue light (Toshiba FS20S/BLB 20W; Toshiba, Tokyo, Japan) for 4 days at 22°C, after aerial hyphae of the colonies had been washed away with sterilized distilled water. Conidia of M. oryzae were suspended in 50 mM cAMP to a final density of 1×106 conidia ml−1. This suspension was then poured onto dialysis membranes (2.5 ml of suspension/25 cm2 membrane surface; Spectra/Por, cutoff 1,000 Da; Spectrum Medical Industries, Terminal Annez, LA) and incubated at 25°C in dark [25]. Total RNA was extracted from germinating conidia incubated for 6 h on dialysis membranes, as described below. 32 sheets of membranes containing the germinating conidia were crushed and homogenized in liquid nitrogen with mortar and pestle. The homogenate was transferred to a centrifuge tube containing 40 ml of TRI Reagent (SIGMA-ALDRICH, St. Louis, MO), homogenized by vigorous shaking and incubated at room temperature for 5 min. Then 8 ml of chloroform was added, homogenized by vigorous shaking for 15 sec and incubated at room temperature for 3 min. After centrifugation at 1000× g for 15 min at 4°C, the upper aqueous phase was transferred to a new centrifuge tube, and the total RNA was precipitated by the addition of 20 ml of isopropanol after incubation at room temperature for 10 min. The pellet was rinsed with 70% ethanol. SuperSAGE library was made from total RNA as described [46], [47]. Di-tag fragments were sequenced by the 454 FLX sequencer (454 Life Sciences). Each 26-bp tag sequence was used for BLASTN search against M. oryzae 70-15 genome sequence. A total of 23,491 tags to comprising 26-bp sequence were recovered. Number of tags for each of putative secreted protein genes of M. oryzae is given in Table S1.
Plasmid construction
To construct the gene-disruption vector pGPSMC69-44, a 8.7-kb fragment containing the MC69 gene amplified with the primers MC69S1 (5′- TTATGACGGGAGCACAGGCACAGCACAC-3′) and MC69AS1 (5′- TGGCCGACGTTGTGCTCTTTCAGTTCCT-3′) was cloned into pCR-XL-TOPO to generate pXLMC69 using the TOPO XL PCR Cloning Kit (Invitrogen, Carlsbad, CA). MC69 was mutated using an adaptation of the TAG-KO method using pGPS-HYG-CAM [30], [31]. The pXLMC69 containing MC69 was used as the target. An insertion was formed within the coding region of MC69 (at 11 amino acids) in pXLMC69, which resulted in pGPSMC69-44 (Figure S1).
For complementation assay of an mc69 mutant with MC69, a 5.7-kb fragment containing MC69 was amplified with the primers NMU1 (5′-ATAAGAATGCGGCCGCTGATTCTCAATGCCCTCTGTCCTTT-3′; the NotI site is underlined) and MC69AS1. The PCR product was digested with NotI and XbaI (exists in the middle of the PCR product after 1.1-kb far from the poly A signal recognition site of MC69) to generate 3.1-kb fragment containing MC69, and ligated to the same restriction sites of which carries the bialaphos-resistant (bar) gene [48], creating pCB1531-MC69.
To substitute a cysteine residue at 36 amino acids in MC69 by alanine, single point mutation was introduced in plasmid pCB1531-MC69 using a primer BMC36AU4 (5′- CAGGTCACCAGACGACGCCTTCTTTGG -3′; mutation site is underlined and the BstEII site is indicated in italics). A 1.7-kb fragment containing a half 3′-terminal part of the MC69 ORF and terminator was amplified with the primers BMC36AU4 and M13F (5′- CGCCAGGGTTTTCCCAGTCACGA-3′). The PCR product was digested with BstEII and XbaI, and exchanged to the BstEII/XbaI fragment of pCB1531-MC69, generating pCB1531-MC69(C36A) (Figure 4B). To substitute a cysteine residue at 46 amino acids in MC69 by alanine, single point mutation was introduced in plasmid pCB1531-MC69 using a primer MC46AU5 (5′-CGTCACGCCGCAAGGCGCCGGGTATGTTCTGGG-3′; mutation site is underlined). A 1.7-kb fragment was amplified with the primers MC46AU5 and M13F, and the PCR product was used as a template for another PCR with the primers BMC46AU4 (5′- CAGGTCACCAGACGACTGCTTCTTTGGTGTCGTCACGCCGCAAGGCGCCGG-3′; mutation site is underlined and the BstEII site is indicated in italics) and M13F. The PCR product was digested with BstEII and XbaI, and exchanged to the BstEII/XbaI fragment of pCB1531-MC69, generating pCB1531-MC69(C46A) (Figure 4B). To substitute two cysteine residues at 36 and 46 amino acids in MC69 by alanine, double point mutations were introduced in plasmid pCB1531-MC69 using a primer BMC36&46AU4 (5′- CAGGTCACCAGACGACGCCTTCTTTGGTGTCGTCACGCCGCAAGGCGCCGG-3′; mutation sites are underlined and the BstEII site is indicated in italics). A 1.7-kb fragment was amplified with the primers MC46AU5 and M13F, and the PCR product was used as a template for another PCR with the primers BMC36&46AU4 andM13F. The PCR product was digested with BstEII and XbaI, and exchanged to the BstEII/XbaI fragment of pCB1531-MC69, generating pCB1531-MC69(C36A,C46A) (Figure 4B).
For construction of the MC69-EGFP gene fusion vector pCB1531-MC69-EGFP, a 1.7-kb fragment containing MC69 gene was amplified with the primers NMU1 and XMG5L1 (5′-GCTCTAGA CCACCACCACCACCTTTGGCAGGTCCGCGAAGAGGG-3′; XbaI site is indicated in italics) which was designed with five glycine codons (underlined) as a spacer peptide between MC69 and EGFP. The PCR product encoding MC69-Gly5 was digested with NotI and XbaI, and exchanged to the NotI/XbaI fragment of the Tef promoter in pBAGFP [49], generating pCB1531-MC69-EGFP. For construction of the MC69-mCherry gene fusion vector pCB1531-MC69-mCherry, a 0.7-kb mCherry cDNA fragment was amplified with the primers XmU1 (5′- GCTCTAGACATGGTGAGCAAGGGCGAGG-3′; XbaI site is underlined) and BmL1 (5′- CGGGATCCTACTTGTACAGCTCGTCCAT-3′; BamHI site is underlined) using pmCherry (Clontech, Mountain View, CA) as a template. The PCR product was digested with XbaI and BamHI, and exchanged to the XbaI/BamHI fragment of EGFP cDNA in pCB1531-MC69-EGFP, generating pCB1531-MC69-mCherry (Figure 2D), a 1.6-kb fragment of MC69 promoter (MC69p) and MC69(C36A) ORF was amplified with the primers NMU1 and XMG5L1. The PCR product was digested with NotI and XbaI, and exchanged to the NotI/XbaI fragment of MC69p-MC69 in pCB1531-MC69-mCherry, generating pCB1531-MC69(C36A)-mCherry (Figure 5A). A 1.4-kb fragment of MC69p was amplified with the primers NMU1 and XMpL2 (5′- GCTCAGACCTTCGTAGGCCTGGAACGAGACGCTTCC-3′; XbaI site is underlined). The PCR product was digested with NotI and XbaI, and exchanged to the NotI/XbaI fragment of MC69p-MC69 in pCB1531-MC69-mCherry, generating pCB1531-MC69p-mCherry (Figure 2A).
A 0.6-kb fragment of PWL2 promoter was amplified the primers Ppwl2-5′ (5′-GAGGAGAAGCGGCCGCGTTAACAACGCGGTGTAAAGATTC-3′; NotI site is underlined) and Ppwl2-3′ (5′-GAGAGGAGAAGGATCCACTAGTTCTAGATTTGAAAGTTTTTAATTTTAAAAAGAGATTTTCCGAG-3′; BamHI-SpeI-XbaI sites are underlined). The PCR product was digested with NotI and BamHI, and exchanged to the NotI/BamHI fragment of Tefp-EGFP in pBAGFP [49], generating pCB-Ppwl2. mCherry cDNA fragment was amplified with primers mCherry-N (5′-GAGAGGAGAAGGATCCAGATCTCTCGAGACCATGGTGAGCAAGGGCGAGGAG-3′; BamHI-BglII-XhoI sites are underlined) and mCherry-C (5′-GAGAGGAGAAGAATTCGCTAGCGTCGACCTTGTACAGCTCGTCCATG-3′; EcoRI-NheI-SalI sites are underlined). The PCR product was digested with BamHI and EcoRI, and introduced into pCB-Ppwl2, to produce pCB-Ppwl2-mCherry. pCB-Ppwl2-mCherry was digested with SalI, fill in with Klenow fragment and performed self-ligation, generating pCB-Ppwl2-mCherry-stop. A modified SV40 NLS-coding double stranded fragment [35] was produced annealing with the oligos mSV40NLS (5′-TCGACGGTCCAGGTGGAGCTGGACCAGGTAGAAAGAGGCCACCAAAGAAAAAGAGAAAGGTAGATTATGGAGCTTAAG-3′; SalI protruding end is underlined) and c-mSV40NLS (5′-AATTCTTAAGCTCCATAATCTACCTTTCTCTTTTTCTTTGGTGGCCTCTTTCTACCTGGTCCAGCTCCACCTGGACCG-3′; EcoRI protruding end is underlined). The annealed product was introduced into pCB-Ppwl2-mCherry, to produce pCB-Ppwl2-mCherry-NLS. From 1 µg of total RNA of cAMP-treated M. oryzae strain 70-15, single-stranded cDNA was synthesized by using oligo(dT) primer and ReverTra Ace reverse transcriptase (Toyobo, Osaka, Japan). A 0.4-kb PWL2 cDNA fragment was amplified from the total cDNA as a template with the primers PWL2-N (5′-GAGAGGAGAATCTAGAAAAATGAAATGCAACAACATCATCCTC-3′; XbaI site is underlined) and PWL2-C (5′-GAGAGGAGAAGGATCCCATAATATTGCAGCCCTCTTCTC-3′; BamHI site is underlined). The PCR product was digested with XbaI and BamHI, and introduced into pCB-Ppwl2-mCherry-NLS, generating pCB-Ppwl2-PWL2-mCherry-NLS (Figure 3A). A 0.2-kb MC69 cDNA fragment was amplified from the total cDNA with the primers XMU2 (5′-GCTCTAGAAAATAAAAATGAAGGCCGCT-3′; XbaI site is underlined) and XML1 (5′-CCGCTCGAGTTTGGCAGGTCCGCGAAGAGGGCCGC-3′, XhoI site is underlined). The PCR prodct was digested with XbaI and XhoI, and introduced into pCB-Ppwl2-mCherry-NLS and pCB-Ppwl2-mCherry-stop, to produce pCB-Ppwl2-MC69-mCherry-NLS and pCB-Ppwl2-MC69-mCherry, respectively (Figure 3B and C).
To make the MC69-HA gene fusion vector pCB1531-MC69-HA, HA-tagged full cDNA of MC69 (MC69HA) was amplified from the total cDNA with the primers XMU2 and BMHAL1 (5′- CGggatccTCAAGCATAATCTGGAACATCGTATGGATA ACCACCTTTGGCAGGTCCGCGAAGAGGGCCGC-3′; BamHI site and HA tag sequence are indicated in lower cases and italics, respectively) which was designed with two glycine codons (underlined) as a spacer peptide between MC69 and HA tag. The PCR product was digested with XbaI and BamHI, and exchanged mCherry gene at the same sites of pCB1531-MC69p-mCherry, generating pCB1531-MC69-HA (Figure S3). MC69-3xFLAG gene fusion construct pUC57-MC69-3xFLAG was custum-synthesized (GenScript, Piscataway, NJ). MC69-3xFLAG was amplified from pUC57-MC69-3xFLAG with the primers SMU2 (5′-GACTAGTGAAAATAAAAATGAAGGCCGCTTTCGTTCTCGC-3′; SpeI site is underlined) and BFL1 (5′- CGGGATCCTCACCCATCATGATCCTTGTAATCG-3′; BamHI site is underlined). The PCR product was digested with SpeI and BamHI, and exchanged mCherry gene at XbaI and BamH sites of pCB1531-MC69p-mCherry, generating pCB1531-MC69-3xFLAG (Figure S3).
For construction of the MC69p::AVR-Pia epression vector pCB1531-MC69p-AVR-Pia, a 0.3-kb fragment containing AVR-Pia gene was amplified from pCB1004-pex22 [20] with the primers XP22U2 (5′-GCTCTAGACAAAATGCATTTTTCGACAATTTTC-3′; XbaI site is underlined) and BP22L2 (5′-CGGGATCCTAGTAAGGCTCGGCAGCAAGCC-3′; BamHI site is underlined). The PCR product was digested with XbaI and BamHI, and exchanged mCherry gene at the same sites of pCB1531-MC69p-mCherry, generating pCB1531-MC69p-AVR-Pia (Figure S4).
CoMC69 was isolated from genome of C. orbiculare 104-T by PCR using degenerate primers designed in amino acid sequences of MC69 homologs in fungal pathogens including C. graminicola. To construct the gene replacement vector pGDCOMC69, the 3.0-kb fragment containing the 5′ flanking region of CoMC69 was amplified by PCR with the primers COMC5S (5′-ATAAGAATGCGGCCGCCCAGTGCTTTGTCATGTTGC-3′; NotI site is underlined) and COMC5AS (5′-CCCAAGCTTCGCTGGTTGCGAAGAATGCG-3′; HindIII site is underlined). The amplified fragment was digested with NotI and HindIII, and introduced into pCB1636 [48], which contained the hph gene, to produce plasmid pCB5MC69. The 3-kb fragment that contained the 3′ flanking region of CoMC69 was amplified by PCR with the primers COMC3S (5′-GAAGGGCCCCCGGTCACCACGCATGTGTGATACG-3′; ApaI site is underlined) and COMC3AS (5′-GGGGTACCACGTGTGCACTCTTAAGGAG-3′; KpnI site is underlined). The amplified fragment was digested with ApaI and KpnI, and introduced into pCB5MC69 to produce pGDCOMC69 (Figure S11A). To generate the reporter construct pBATCoMC69pro-mCherry, the 1.4 kb 5′ upstream region of CoMC69 and mCherry were amplified using PCR with the two primer sets, (i) CoMC69pro-NotI-f (5′-ATAAGAATGCGGCCGCGTCTTTCGTCTTTTCGGTCT-3′; NotI site is underlined) and CoMC69pro-BamHI-r(c) (5′-CGGGATCCCGTGTCGATGTATTTGTTGTG-3′; BamHI site is underlined), and (ii) mCherry-BamHI-f (5′-GCGGATCCATGGTGAGCAAGGGCGAGGAGGATAAC-3′; BamHI site is underlined) and mCherry-EcoRI-r(c) (5′-CCGGAATTCTTACTTGTACAGCTCGTCCATGCC-3′; EcoRI site is underlined), respectively. The amplified fragments were introduced into each corresponding site of pBAT [49], resulting in pBAT-CoMC69pro-mCherry.
Pathogenicity assays
Barley leaf and rice leaf inoculation were performed as follows: conidial suspension (1×105 conidia ml−1) containing Tween 20 (0.01% in final concentration) was sprayed onto susceptible barley cotyledons (cv. Nigrate) and rice seedlings (cv. Shin No. 2 or cv. Sasanishiki) of the fourth leaf stage. Inoculated plants were placed in a dew chamber at 27°C for 24 h in the dark, and then transferred to the growth chamber with a photoperiod of 16 h. A rice leaf sheath inoculation test was performed according to the method described previously [50]. To investigate the function of appressorium-mediated penetration of the inner epidermal tissue of rice leaf sheath, penetration hyphae were stained with lactophenol-trypan blue and destained in saturated chloral hydrate as described previously [51]. Invasive growth rating of the 50 appressorial penetration sites in rice leaf sheath cells were scored 32 h after inoculation. Invasive growth were classified into 4 levels: Level 1, invasive hypha length is shorter than 10 µm with no branch; Level 2, invasive hyphae length is 10–20 µm with 0–2 branches; Level 3, invasive hyphae length is longer than 20 µm and/or with more than 2 branches within one cell; Level 4, invasive hyphae are spread more than one cell (Figure 1D). To test fungal pathogenicity of C. orbiculare, conidial suspensions of tested C. orbiculare strains (approximately 5×105 conidia/ml) were spotted onto detached leaves of cucumber or N. benthamiana.
Confocal laser-scanning microscopy
Germinated conidia and appressoria were observed on glass coverslips, and invaded hyphae were observed in epidermal cells of rice leaf sheath. mCherry fluorescence was observed using an Olympus FluoView FV1000-D confocal laser-scanning microscope (Olympus, Tokyo, Japan) equipped with a Multi argon laser, a HeNe G laser, a 40× UPlanSApo (0.9 numerical aperture) and a 60× UPlanFLN (0.9 numerical aperture) objective lens. To assess fluorescent signal in the reporter strains of C. orbiculare, conidia of the reporter strain were inoculated on the lower surfaces of cucumber cotyledons. Detection of mCherry fluorescence was performed using an Olympus FluoView FV500 confocal laser-scanning microscope (Olympus) with a Nikon 60× PlanApo (1.4 numerical aperture) oil-immersion objective (Nikon, Tokyo, Japan). Samples were mounted in water under cover slips and excited with the He/Ne laser. We used diachronic mirror DM488/543/633, SDM630 beam splitter, and emission filter BA560-600.
Preparation of M. oryzae-infected rice leaf sheath extract and Western blot analysis
Conidial suspension (1×105 conidia ml−1) was injected into rice (cv. Shin No. 2) leaf sheath and placed in a dew chamber at 25°C for 32 h in the dark. The infected leaf sheaths were ground in liquid nitrogen, thawed in X µl of extraction buffer (250 mM Tris-HCl pH 7.5, 2.5 mM EDTA, 0.1% ascorbic acid (w/v), 1 mM PMSF, 0.01% PI cocktail (v/v) (SIGMA-ALDRICH), 0.1% Triton X-100 (v/v)) for X mg sample, vortex for 10 min at 4°C, and centrifuged at 15,000× g for 20 min at 4°C in a microcentrifuge. The crude extracts (15 µl per lane) were separated on a 10–20% precast e-PAGEL (ATTO, Tokyo, Japan) and the proteins were transferred on to Immobilon Transfer Membranes (Millipore, Billerica, MA). The blots were blocked in 2% ECL Advance Blocking Agent (GE Healthcare, Buckinghamhire, UK) in TTBS (10 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.1% Tween 20 (v/v)) for 1 h at room temperature with gentle agitation. For immunodetection, blots were probed with anti-HA (3F10)-HRP (Roche, Mannheim, Germany) or anti-FLAG M2-HRP (SIGMA-ALDRICH) in a 1∶10,000 dillution in TTBS for 2 h. After washing the membrane for 10 min three times, the reactions were detected using an ECL Advance Western blotting detection reagents (GE Healthcare) and a Luminescent Image Analyzer LAS-4000 (Fujifilm, Tokyo, Japan).
Preparation of culture filtrate and Western blot analysis
M. oryzae strains were cultured in 20 ml YG medium at 25°C at 120 rpm for 48 h. The culture was filtrated with Miracloth (Merck, Darmstadt, Germany), concentrated and desalted by ultrafiltration with Amicon Ultra-15 (10K) (Millipore). The culture filtrates (20 µg of protein per lane) were separated on a 12.5% SDS-PAGE gel and the proteins were transferred on to Immobilon Transfer Membranes (Millipore). The blots were blocked in 5% nonfat dry milk in TTBS for 1 h at room temperature with gentle agitation. For immunodetection, blots were probed with Living Colors DsRed Polyclonal Antibody (Clontech) in a 1∶1,000 dilution in TTBS for 2 h. After washing the membrane with TTBS for 10 min three times, Anti-Rabbit IgG HRP conjugate (Promega W401B) (Promega, Madison, WI) in a 1∶10,000 dilution in TTBS was used as secondary antibody and incubated for 1 h at room temperature with gentle agitation. After washing the membrane for 10 min three times, the reactions were detected using an ECL Western blotting detection reagents (GE Healthcare) and a Luminescent Image Analyzer LAS-4000 (Fujifilm).
Accession numbers
Sequence data of MC69 and the homologs from this article can be found in the GenBank/EMBL data libraries accession number MGG_02848.6 (Mo), AB669186 (Co), EFQ29542 (Gg), EEY15898 (Va), EGY20943 (Vd), XP_965292 (Nc), EGO52621 (Nt), XP_003659994 (Mt), XP_00190740 (Pa), EFX05010 (Gc), EGU75378 (Fo), XP_388669 (Gz), EHK44387 (Ta), EHK23962 (Tv), EFY93067 (Mac), EFY97094 (Man), and EGX95034 (Cm).
Supporting Information
Targeted gene disruption of MC69. (A) MC69 locus and the disruption vector pGPSMC69-44. pGPSMC69-44 contains the GPS-HYG-CAM cassette (the hygromicin resistant gene HYG and the chloramphenicol resistant gene CAM) flanked by border sequences from MC69. (B) Genomic PCR analysis of wild-type Ina72 (lane 1, 10 and 19), three independent mc69 mutants (lane 2∼4, 11∼13 and 20∼22), two independent MC69 re-introduced strains (lane 5, 6, 14, 15, 23 and 24), wild-type 70-15 (lane 7, 16 and 25), two independent mc69 mutants (lane 8, 9, 17, 18, 26 and 27). The transformants were analyzed by PCR with primers indicated in A (MC69F/MC69R, lane 1∼9), with HYG-specific primers (lane 10∼18) or with bialaphos-resistant gene specific primers (lane 19∼27).
(TIF)
Colony growth and conidiation of the mc69 mutants. (A,C) Colony color and arial hyphae production of mc69 mutants were normal. Photos were taken 7 days after incubation of wild type (Ina72), mc69 mutants (mc69-9, mc69-12 and mc69-87), wild type (70-15) and mc69 mutants (mc69-119 and mc69-31) on oatmeal agar. (B,D) Growth and conidiation of Ina72 (bar 1), mc69-9 (bar 2), mc69-12 (bar 3), mc69-87 (bar 4), 70-15 (bar 5), mc69-119 (bar 6) and mc69-31 (bar 7). Mean values of colony diameter (cm) were measured 7 days of growth on oatmeal agar. Mean values are calculated from 3 replicates. Conidiogenesis was assessed in 3 replicate experiments. Means are expressed as numbers of conidia ×104 of conidial suspension/cm2 of culture.
(TIF)
MC69 protein is produced in the invasive hyphae. (A,C) In planta growth of MC69::HA- and MC69::3xFLAG-expressing transformants (mc69+MC69::HA and mc69+MC69::3xFLAG) at the post-invasion stage. Invasive mycelia inside the rice (cv. Shin No. 2) leaf sheath cells were photographed 48 h after incubation. Scale bar = 20 µm. (B,D) Western blots probed with an anti-HA and an anti-FLAG antibodies. Protein extracts of rice leaf sheaths 24 h and 48 h after inoculation with Ina72 wild type (WT), mc69+MC69::HA and mc69+MC69::3xFLAG were loaded.
(TIF)
AVR-Pia avirulence function is retained under the MC69 promoter. (A) The isolate Ina86-137 does not have AVR-Pia function and thus can cause disease on Sasanishiki harboring the R gene Pia. Ina86-137 strains transformed with AVR-Piap::AVR-Pia (+AVR-Piap::AVR-Pia) [20] or MC69p::AVR-Pia (+MC69p::AVR-Pia-1, -2) became imcompatible with Sasanishiki. Both Ina86-137 wild type, Ina86-137 containing AVR-Piap::AVR-Pia, or MC69p::AVR-Pia were able to cause disease on a rice cultivar Shin No. 2 lacking Pia, suggesting that the effect of transformation with AVR-Piap::AVR-Pia and MC69p::AVR-Pia is Pia dependent. (B) Confirmation of active AVR-Pia transgene by RT-PCR in M. oryzae transformants during infection. RT-PCR analysis of Ina86-137 WT (lane 1), +AVR-Piap::AVR-Pia (lane 2), +MC69p::AVR-Pia-1 and -2 (lane 3 and 4) with AVR-Pia- or Mg-Actin-specific primers [20].
(TIF)
MC69::mCherry confers BIC localization with weaker fluorescence than that of PWL2::mCherry. Merged DIC and mCherry images of rice leaf sheath cells infected by M. oryzae Sasa2 strain harboring (A) PWL2p::PWL2::mCherry::NLS, (B) PWL2p::MC69::mCherry::NLS, and (C) PWL2p::MC69::mCherry 27 h after inoculation as observed by confocal laser scanning microscopy. Arrows indicate BICs and triangles indicate rice nuclei. Pinhole settings are 80 µm for left panels and 240 µm for right panels. Scale bar = 20 µm.
(TIF)
Predicted amino acid sequence alignment of MC69 with homologs from other filamentous fungi. Amino acid sequences of MC69 (Mo), MC69 homologs of Colletotrichum orbiculare (Co), Glomerella graminicola (Gg), Verticillium albo-atrum (Va), V. dahliae (Vd), Neurospora crassa (Nc), N. tetrasperma (Nt), Myceliophthora thermophila (Mt), Podospora anserina (Pa), Grosmannia clavigera (Gc), Fusarium oxysporum (Fo), Gibberella zeae (Gz), Trichoderma atroviride (Ta), T. virens (Tv), Metarhizium acridum (Mac), M. anisopliae (Man) and Cordyceps militaris (Cm) were aligned using the Clustal W program [52]. Identical amino acids are indicated as white letters on a black background. Similar residues are shown on gray backgrounds. Gaps introduced for alignment are indicated by dashes. The predicted signal peptide and two conserved cysteine residues (C36 and C46) are indicated on top.
(TIF)
Phylogenetic tree of M. oryzae MC69 protein sequence and 16 homologs from other fungi. Phylogenetic analyses were performed with M. oryzae MC69 (Mo), with 16 homologs are shown in Figure S6 legend.
(TIF)
Invasive growth rating of rice leaf sheath cells 32 h after inoculating with Ina72 WT, mc69, mc69+MC69, mc69+MC69(C36A), mc69+MC69(C46A) and mc69+MC69(C36A,C46A). For details of the invasive growth levels and rating see Materials and Methods.
(TIF)
Invasive growth rating of rice leaf sheath cells 32 h after inoculating with 70-15 WT and mc69-31. For details of the invasive growth levels and rating see Materials and Methods.
(TIF)
Intron/exon organization in M. oryzae MC69 gene and 16 orthologous genes from other fungi. Abbreviations of fungus names are shown in Figure S6 legend.
(TIF)
Gene disruption of CoMC69 in C. orbiculare. (A) CoMC69 locus and the gene disruption vector pGDCOMC69. By homologous recombination through double crossing over, the CoMC69 gene was replaced by a hygromycin resistance gene cassette (HYG). (B) Genomic PCR analysis of the Comc69 mutants of C. orbiculare. Genomic DNAs were isolated from the wild-type strain 104-T and the comc69 strains (DMC1 and DMC2). The 0.3 kb product containing the entire CoMC69 gene was amplified from the genome DNA of 104-T with the two primers, indicated by arrows, COMC69F (5′-CGAAAGCAAGGCAGCTATTC-3′) and COMC69R (5′-CTCAGAGGACTACAGACATG-3′). In contrast, the 1.6 kb product was amplified from the genome DNA of both comc69 strains, which is consistent with gene replacement shown in (A). Lane 1, λ Hind III marker; lane 2, 104-T; lane 3, DMC1; lane 4, DMC2. (C) Colony phenotype of the C. orbiculare mc69 mutants. The wild-type strain 104-T and Comc69 mutants (DMC1 and DMC2) were grown on PDA for 12 days.
(TIF)
SuperSAGE result of cAMP-treated Magnaporthe oryzae strain 70-15.
(XLS)
Acknowledgments
We thank Shunichi Kosugi for producing pCB-Ppwl2-mCherry constructs.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by the ‘Program for Promotion of Basic Research Activities for Inovative Biosciences (PROBRAIN)’ (Japan) and JSPS grants no. 18310136 and 20380027, and ‘The Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation PMI-0010)’; and by JSPS grant no. 22780040 to H. Saitoh. We thank PROBRAIN for two research grants to R. Terauchi and Y. Takano. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Valent B. Rice blast as a model system for plant pathology. Phytopathology. 1990;80:33–36. [Google Scholar]
- 2.Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nat Immunol. 2005;6:973–978. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- 3.Bittel P, Robatzek S. Microbe-associated molecular patterns (MAMPs) probe plant immunity. Curr Opin Plant Biol. 2007;10:335–341. doi: 10.1016/j.pbi.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 4.Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 5.Abramovitch RB, Janjusevic R, Stebbins CE, Martin GB. Type III effector AvrPtoB requires intrinsic E3 ubiqutin ligase activity to suppress plant cell death and immunity. Proc Natl Acad Sci U S A. 2006;103:2851–2856. doi: 10.1073/pnas.0507892103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boller T, He SY. Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science. 2009;324:742–744. doi: 10.1126/science.1171647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui H, Wang Y, Xue L, Chu J, Yan C, et al. Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe. 2010;7:164–175. doi: 10.1016/j.chom.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grant SR, Fisher EJ, Chang JH, Mole BM, Dangl JL. Substerfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu Rev Microbiol. 2006;60:425–449. doi: 10.1146/annurev.micro.60.080805.142251. [DOI] [PubMed] [Google Scholar]
- 9.Zhou J-M, Chai J. Plant pathogenic bacterial type III effectors subdue host responses. Curr Opin Microbiol. 2008;11:179–185. doi: 10.1016/j.mib.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Gilbert MJ, Thornton CR, Wakley GE, Talbot NJ. A P-type ATPase required for rice blast disease and induction of host resistance. Nature. 2006;440:535–539. doi: 10.1038/nature04567. [DOI] [PubMed] [Google Scholar]
- 11.Khang CH, Berruyer R, Giraldo MC, Kankanala P, Park S-Y, et al. Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell. 2010;22:1388–1403. doi: 10.1105/tpc.109.069666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi M, Chi M-H, Khang CH, Park S-Y, Kang S, et al. The ER chaperone LHS1 is involved in asexual development and rice infection by the blast fungus Magnaporthe oryzae. Plant Cell. 2009;21:681–695. doi: 10.1105/tpc.107.055988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khang CH, Park S-Y, Lee Y-H, Valent B, Kang S. Genome organization and evolution of the AVR-Pita avirulence gene family in the Magnaporthe grisea species complex. Mol Plant Microbe Interact. 2008;21:658–670. doi: 10.1094/MPMI-21-5-0658. [DOI] [PubMed] [Google Scholar]
- 14.Orbach MJ, Farrall L, Sweigard JA, Chumley FG, Valent B. A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell. 2000;12:2019–2032. doi: 10.1105/tpc.12.11.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farman ML, Leong SA. Chromosome walking to the AVR1-CO39 avirulence gene of Magnaporthe grisea: discrepancy between the physical and genetic maps. Genetics. 1998;150:1049–1058. doi: 10.1093/genetics/150.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang S, Sweigard JA, Valent B. The PWL host specificity gene family in the blast fungus Magnaporthe grisea. Mol Plant Microbe Interact. 1995;8:939–948. doi: 10.1094/mpmi-8-0939. [DOI] [PubMed] [Google Scholar]
- 17.Li W, Wang B, Wu J, Lu G, Hu Y, et al. The Magnaorthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein thet troggers the immunity in rice mediated by the blast resistance gene Piz-t. Mol Plant Microbe Interact. 2009;22:411–420. doi: 10.1094/MPMI-22-4-0411. [DOI] [PubMed] [Google Scholar]
- 18.Miki S, Matsui K, Kito H, Otsuka K, Ashizawa T, et al. Molecular cloning and characterization of the AVR-Pia locus from a Japanese field isolate of Magnaporthe oryzae. Mol Plant Pathol. 2009;10:361–374. doi: 10.1111/j.1364-3703.2009.00534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sweigard JA, Carroll AM, Kang S, Farrall L, Chumley FG, et al. Identification, cloning, and chracterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell. 1995;7:1221–1233. doi: 10.1105/tpc.7.8.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida K, Saitoh H, Fujisawa S, Kanzaki H, Matsumura H, et al. Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell. 2009;21:1573–1591. doi: 10.1105/tpc.109.066324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mentlak TA, Kombrink A, Shinya T, Ryder LS, Otomo I, et al. Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. Plant Cell. 2012;24:322–335. doi: 10.1105/tpc.111.092957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosquera G, Giraldo MC, Khang CH, Coughlan S, Valent B. Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1-4 as biotrophy-associated secreted proteins in rice blast disease. Plant Cell. 2009;21:1273–1290. doi: 10.1105/tpc.107.055228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005;434:980–986. doi: 10.1038/nature03449. [DOI] [PubMed] [Google Scholar]
- 24.Lee Y-H, Dean RA. cAMP regulates infection structure formation in the plant pathogenic fungus Magnaporthe grisea. Plant Cell. 1993;5:693–700. doi: 10.1105/tpc.5.6.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irie T, Matsumura H, Terauchi R, Saitoh H. Serial analysis of gene expression (SAGE) of Magnaporthe grisea: genes involved in appressorium formation. Mol Genet Genomics. 2003;270:181–189. doi: 10.1007/s00438-003-0911-6. [DOI] [PubMed] [Google Scholar]
- 26.Choi W, Dean RA. The adenylate cyclase gene MAC1 of Magnaporthe grisea controls appressorium formation and other aspects of growth and development. Plant Cell. 1997;9:1973–1983. doi: 10.1105/tpc.9.11.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell. 1993;5:1575–1590. doi: 10.1105/tpc.5.11.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue C, Park G, Choi W, Zheng Li, Dean RA, et al. Two novel fungal virulence genes specifically expressed in appressoria of the rice blast fungus. Plant Cell. 2002;14:2107–2119. doi: 10.1105/tpc.003426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsumura H, Reich S, Ito A, Saitoh H, Kamoun S, et al. Gene expression analysis of plant host-pathogen interactions by SuperSAGE. Proc Natl Acad Sci U S A. 2003;100:15718–15723. doi: 10.1073/pnas.2536670100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamer L, Adachi K, Montenegro-Chamorro MV, Tanzer MW, Mahanty SK, et al. Gene discovery and gene function assignment in filamentous fungi. Proc Natl Acad Sci U S A. 2001;98:5110–5115. doi: 10.1073/pnas.091094198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saitoh H, Fujisawa S, Ito A, Mitsuoka C, Berberich T, et al. SPM1 encoding a vacuole-localized protease is required for infection-related autophagy of the rice blast fungus Magnaporthe oryzae. FEMS Microbiol Lett. 2009;300:115–121. doi: 10.1111/j.1574-6968.2009.01769.x. [DOI] [PubMed] [Google Scholar]
- 32.Soderlund C, Haller K, Pampanwar V, Ebbole D, Farman M, et al. MGOS: a resource for studying Magnaporthe grisea and Oryza sativa interactions. Mol Plant Microbe Interact. 2006;19:1055–1061. doi: 10.1094/MPMI-19-1055. [DOI] [PubMed] [Google Scholar]
- 33.Takur S, Jha S, Roy-Barman S, Chattoo B. Genomic resources of Magnaporthe oryzae (GROMO): a comprehensive and integrated database on rice blast fungus. BMC Genomics. 2009;10:316–323. doi: 10.1186/1471-2164-10-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okuyama Y, Kanzaki H, Abe A, Yoshida K, Tamiru M, et al. A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR genes. Plant J. 2011;66:467–479. doi: 10.1111/j.1365-313X.2011.04502.x. [DOI] [PubMed] [Google Scholar]
- 35.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Nuclear export signal defined using a localization-based yeast selection system. Traffic. 2008;8:2053–2062. doi: 10.1111/j.1600-0854.2008.00825.x. [DOI] [PubMed] [Google Scholar]
- 36.Ceroni A, Passerini A, Vullo A, Frasconi P. DISULFIND: a disulfide bonding state and cysteine connectivity prediction server. Nucleic Acids Res. 2006;34:W177–W181. doi: 10.1093/nar/gkl266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takano Y, Takayanagi N, Hori H, Ikeuchi Y, Suzuki T, et al. A gene involved in modifying transfer RNA is required for fungal pathogenicity and stress tolerance of Colletotrichum lagenarium. Mol Microbiol. 2006;60:81–92. doi: 10.1111/j.1365-2958.2006.05080.x. [DOI] [PubMed] [Google Scholar]
- 38.Doehlman G, van der Linde K, Amann D, Schwammbach D, Hof A, et al. Pep1, a secreted effector protein of Ustilago maydis, is required for successful invasion of plant cells. PLoS Path. 2009;5:e1000290. doi: 10.1371/journal.ppat.1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jia Y, McAdams SA, Brayan GT, Hershey HP, Valent B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 2000;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Müller O, Schreier PH, Uhrig JF. Identification and characterization of secreted and pathogenesis-related proteins in Ustilago maydis. Mol Genet Genomics. 2008;279:27–39. doi: 10.1007/s00438-007-0291-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buist G, Steen A, Kok J, Kuipers OP. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol. 2008;68:838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- 42.de Jonge R, van Esse HP, Kombrink A, Shinya T, Desaki Y, et al. Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science. 2010;329:953–955. doi: 10.1126/science.1190859. [DOI] [PubMed] [Google Scholar]
- 43.Van den Burg HA, Harrison SJ, Joosten MH, Vervoort J, de Wit PJ. Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol Plant Microbe In. 2006;19:1420–1430. doi: 10.1094/MPMI-19-1420. [DOI] [PubMed] [Google Scholar]
- 44.Takano Y, Komeda K, Kojima K, Okuno T. Proper regulation of cyclic AMP-dependent protein kinase is required for growth, conidiation, and appressorium function in the anthracnose fungus Colletotrichum lagenarium. Mol Plant Microbe Interact. 2001;14:1149–1157. doi: 10.1094/MPMI.2001.14.10.1149. [DOI] [PubMed] [Google Scholar]
- 45.Yamauchi J, Takayanagi N, Komeda K, Takano Y, Okuno T. cAMP-PKA signaling regulates multiple steps of fungal infection cooperatively with Cmk1 MAP kinase in Colletotrichum lagenarium. Mol Plant Microbe Interact. 2004;12:1355–1365. doi: 10.1094/MPMI.2004.17.12.1355. [DOI] [PubMed] [Google Scholar]
- 46.Matsumura H, Reich S, Ito A, Saitoh H, Kamoun S, et al. Gene expression analysis of plant host-pathogen interactions by SuperSAGE. Proc Natl Acad Sci USA. 2003;100:15718–15723. doi: 10.1073/pnas.2536670100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terauchi R, Matsumura H, Kruger DH, Kahl G. SuperSAGE: The most advanced transcriptome technology for functional genomics. In: Kahl G, Meksem K, editors. The Handbook of Plant Functional Genomics. Weinhaim, Germany: Wiley-VCH; 2008. pp. 37–54. [Google Scholar]
- 48.Sweigard J, Chumley F, Carroll A, Farrall L, Valent B. A series of vectors for fungal transformation. Fungal Genet Newsl. 1997;44:52–55. [Google Scholar]
- 49.Kimura A, Takano Y, Furusawa I, Okuno T. Peroxisomal metabolic function is required for appressorium mediated plant infection by Colletotrichum lagenarium. Plant Cell. 2001;13:1945–1957. doi: 10.1105/TPC.010084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Namai T, Nukina M, Togashi J. Effects of two toxins and a derivative of one toxin produced by rice blast fungus on its infection to inner epidermal tissue of rice leaf sheath. Ann Phytopathol Soc Jpn. 1996;62:114–118. [Google Scholar]
- 51.Koch E, Slusarenko A. Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell. 1990;2:437–445. doi: 10.1105/tpc.2.5.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Targeted gene disruption of MC69. (A) MC69 locus and the disruption vector pGPSMC69-44. pGPSMC69-44 contains the GPS-HYG-CAM cassette (the hygromicin resistant gene HYG and the chloramphenicol resistant gene CAM) flanked by border sequences from MC69. (B) Genomic PCR analysis of wild-type Ina72 (lane 1, 10 and 19), three independent mc69 mutants (lane 2∼4, 11∼13 and 20∼22), two independent MC69 re-introduced strains (lane 5, 6, 14, 15, 23 and 24), wild-type 70-15 (lane 7, 16 and 25), two independent mc69 mutants (lane 8, 9, 17, 18, 26 and 27). The transformants were analyzed by PCR with primers indicated in A (MC69F/MC69R, lane 1∼9), with HYG-specific primers (lane 10∼18) or with bialaphos-resistant gene specific primers (lane 19∼27).
(TIF)
Colony growth and conidiation of the mc69 mutants. (A,C) Colony color and arial hyphae production of mc69 mutants were normal. Photos were taken 7 days after incubation of wild type (Ina72), mc69 mutants (mc69-9, mc69-12 and mc69-87), wild type (70-15) and mc69 mutants (mc69-119 and mc69-31) on oatmeal agar. (B,D) Growth and conidiation of Ina72 (bar 1), mc69-9 (bar 2), mc69-12 (bar 3), mc69-87 (bar 4), 70-15 (bar 5), mc69-119 (bar 6) and mc69-31 (bar 7). Mean values of colony diameter (cm) were measured 7 days of growth on oatmeal agar. Mean values are calculated from 3 replicates. Conidiogenesis was assessed in 3 replicate experiments. Means are expressed as numbers of conidia ×104 of conidial suspension/cm2 of culture.
(TIF)
MC69 protein is produced in the invasive hyphae. (A,C) In planta growth of MC69::HA- and MC69::3xFLAG-expressing transformants (mc69+MC69::HA and mc69+MC69::3xFLAG) at the post-invasion stage. Invasive mycelia inside the rice (cv. Shin No. 2) leaf sheath cells were photographed 48 h after incubation. Scale bar = 20 µm. (B,D) Western blots probed with an anti-HA and an anti-FLAG antibodies. Protein extracts of rice leaf sheaths 24 h and 48 h after inoculation with Ina72 wild type (WT), mc69+MC69::HA and mc69+MC69::3xFLAG were loaded.
(TIF)
AVR-Pia avirulence function is retained under the MC69 promoter. (A) The isolate Ina86-137 does not have AVR-Pia function and thus can cause disease on Sasanishiki harboring the R gene Pia. Ina86-137 strains transformed with AVR-Piap::AVR-Pia (+AVR-Piap::AVR-Pia) [20] or MC69p::AVR-Pia (+MC69p::AVR-Pia-1, -2) became imcompatible with Sasanishiki. Both Ina86-137 wild type, Ina86-137 containing AVR-Piap::AVR-Pia, or MC69p::AVR-Pia were able to cause disease on a rice cultivar Shin No. 2 lacking Pia, suggesting that the effect of transformation with AVR-Piap::AVR-Pia and MC69p::AVR-Pia is Pia dependent. (B) Confirmation of active AVR-Pia transgene by RT-PCR in M. oryzae transformants during infection. RT-PCR analysis of Ina86-137 WT (lane 1), +AVR-Piap::AVR-Pia (lane 2), +MC69p::AVR-Pia-1 and -2 (lane 3 and 4) with AVR-Pia- or Mg-Actin-specific primers [20].
(TIF)
MC69::mCherry confers BIC localization with weaker fluorescence than that of PWL2::mCherry. Merged DIC and mCherry images of rice leaf sheath cells infected by M. oryzae Sasa2 strain harboring (A) PWL2p::PWL2::mCherry::NLS, (B) PWL2p::MC69::mCherry::NLS, and (C) PWL2p::MC69::mCherry 27 h after inoculation as observed by confocal laser scanning microscopy. Arrows indicate BICs and triangles indicate rice nuclei. Pinhole settings are 80 µm for left panels and 240 µm for right panels. Scale bar = 20 µm.
(TIF)
Predicted amino acid sequence alignment of MC69 with homologs from other filamentous fungi. Amino acid sequences of MC69 (Mo), MC69 homologs of Colletotrichum orbiculare (Co), Glomerella graminicola (Gg), Verticillium albo-atrum (Va), V. dahliae (Vd), Neurospora crassa (Nc), N. tetrasperma (Nt), Myceliophthora thermophila (Mt), Podospora anserina (Pa), Grosmannia clavigera (Gc), Fusarium oxysporum (Fo), Gibberella zeae (Gz), Trichoderma atroviride (Ta), T. virens (Tv), Metarhizium acridum (Mac), M. anisopliae (Man) and Cordyceps militaris (Cm) were aligned using the Clustal W program [52]. Identical amino acids are indicated as white letters on a black background. Similar residues are shown on gray backgrounds. Gaps introduced for alignment are indicated by dashes. The predicted signal peptide and two conserved cysteine residues (C36 and C46) are indicated on top.
(TIF)
Phylogenetic tree of M. oryzae MC69 protein sequence and 16 homologs from other fungi. Phylogenetic analyses were performed with M. oryzae MC69 (Mo), with 16 homologs are shown in Figure S6 legend.
(TIF)
Invasive growth rating of rice leaf sheath cells 32 h after inoculating with Ina72 WT, mc69, mc69+MC69, mc69+MC69(C36A), mc69+MC69(C46A) and mc69+MC69(C36A,C46A). For details of the invasive growth levels and rating see Materials and Methods.
(TIF)
Invasive growth rating of rice leaf sheath cells 32 h after inoculating with 70-15 WT and mc69-31. For details of the invasive growth levels and rating see Materials and Methods.
(TIF)
Intron/exon organization in M. oryzae MC69 gene and 16 orthologous genes from other fungi. Abbreviations of fungus names are shown in Figure S6 legend.
(TIF)
Gene disruption of CoMC69 in C. orbiculare. (A) CoMC69 locus and the gene disruption vector pGDCOMC69. By homologous recombination through double crossing over, the CoMC69 gene was replaced by a hygromycin resistance gene cassette (HYG). (B) Genomic PCR analysis of the Comc69 mutants of C. orbiculare. Genomic DNAs were isolated from the wild-type strain 104-T and the comc69 strains (DMC1 and DMC2). The 0.3 kb product containing the entire CoMC69 gene was amplified from the genome DNA of 104-T with the two primers, indicated by arrows, COMC69F (5′-CGAAAGCAAGGCAGCTATTC-3′) and COMC69R (5′-CTCAGAGGACTACAGACATG-3′). In contrast, the 1.6 kb product was amplified from the genome DNA of both comc69 strains, which is consistent with gene replacement shown in (A). Lane 1, λ Hind III marker; lane 2, 104-T; lane 3, DMC1; lane 4, DMC2. (C) Colony phenotype of the C. orbiculare mc69 mutants. The wild-type strain 104-T and Comc69 mutants (DMC1 and DMC2) were grown on PDA for 12 days.
(TIF)
SuperSAGE result of cAMP-treated Magnaporthe oryzae strain 70-15.
(XLS)