Abstract
Tumor resistance to antitubulin drugs resulting from Pgp drug-efflux activity, increased expression of the βIII tubulin isotype, and alterations in the drug-binding sites are major obstacles in cancer therapy. Consequently, novel antitubulin drugs that overcome these challenges are of substantial interest. Here, we study a novel chemotype named furan metotica that localizes to the colchicine-binding site in β-tubulin, inhibits tubulin polymerization, and is not antagonized by Pgp. To elucidate the structure-activity properties of this chiral chemotype, the enantiomers of its most potent member were separated and their absolute configurations determined by X-ray crystallography. Both isomers were active and inhibited all 60 primary cancer cell lines tested at the US National Cancer Institute. They also efficiently killed drug-resistant cancer cells that overexpressed the Pgp drug-efflux pump 106-fold. In vitro, the R-isomer inhibited tubulin polymerization at least 4-fold more potently than the S-isomer, while in human cells the difference was 30-fold. Molecular modeling showed that the two isomers bind to β-tubulin in distinct manners: the R-isomer binds in a colchicine-like mode and the S-isomer in a podophyllotoxin-like fashion. In addition, the dynamic binding trajectory and occupancy state of the R-isomer were energetically more favorable then those of the S-isomer, explaining the observed differences in biological activities. The ability of a racemic drug to assume the binding modes of two prototypical colchicine-site binders represents a novel mechanistic basis for antitubulin activity and paves the way towards a comprehensive design of novel anticancer agents.
Keywords: Antitubulin, Pgp, MDR1, colchicine, drug-ligand modeling, structure-activity relationship
Introduction
Since most solid tumors are characterized by uncontrolled proliferation, a large number of anticancer drugs target cell division (1). As a dividing cell partitions its replicated chromosomes along the mitotic spindle, antiproliferation drugs include compounds that target spindle microtubule (MT) formation or activity. MT-stabilizing agents such as taxanes and ixabepilone, or MT-destabilizing drugs, such as vinca alkaloids and estramustine, are effective against a broad spectrum of tumors (2). However, the resistance of tumors to antitubulin drugs is a substantial problem. This is due, in particular, to a pathological expression of the ABC transporter P-glycoprotein (Pgp), encoded by MDR1, which engages in drug efflux (3). Additional factors contributing to antitubulin drug resistance may include mutations in the tubulin subunits, changes in the tubulin isotype composition of MTs, enhanced levels of βIII tubulin expression, altered expression or binding of MT-regulatory proteins including Tau, mutations in or reduced levels of γ-actin, and/or a reduced apoptotic response (2). To deal with resistance, new drugs against alternative mitotic targets are being developed, and, concomitantly, antitubulin therapeutics are being designed to evade detection by Pgp (2, 4).
We previously identified five compounds comprising a new chemotype named furan metotica (6-furan-2-yl-3-methyl-4-oxo-4,5,6,7-tetrahydro-1H-indole-2-carboxylic acid derivatives). The compounds were originally found in a screen for compounds that inhibit the binding of the kinetochore Ndc80 complex to taxol-stabilized MTs in vitro. The compounds proved to prevent Ndc80 complex binding by acting at the MT level as they inhibited tubulin polymerization and depolymerized mitotic spindles when added to HeLa cells. This observation characterized them as MT-destabilizers. Competition experiments next showed that they acted at the colchicine site in β-tubulin (5).
Here, we investigate the furan metotica chemotype by focusing on its most active member. We describe the first chiral separation of its enantiomers and report the biochemical, functional and computational dissection of how they interact with tubulin. Importantly, both enantiomers killed all primary cancer cell lines tested, including those with pathological Pgp levels. Of note, the R-enantiomer was 25- to 35-fold more potent than the S-isomer. In vitro biochemical measurements of antitubulin activity confirmed that the R-enantiomer was more potent than the S-isomer. The discrepancy in isomer activity led us to search for the molecular basis of this difference. We did this by determining how both enantiomers localized to and interacted with the colchicine site. Molecular modeling showed that the colchicine site is stereoselective for the enantiomers as the isomers mimicked the binding mode of two different tubulin inhibitors, colchicine and podophyllotoxin. Docking experiments further pointed to the R-isomer entering β-tubulin via its furan ring, whereas the S-isomer entered by adapting its flexible ester side chain. In addition, the localization trajectory and docked pose of the colchicine-mimetic R-isomer were energetically more favorable than those of the podophyllotoxin-mimicking S-isomer, explaining their differences in biological activity. The unique mechanistic behavior of our stereoisomeric chemotype promises to advance the rational design of a novel class of antitubulin drugs that act at the colchicine-binding site and are not antagonized by Pgp. The analyses implemented here will also prove useful to study the activity and structure-based behavior of isomers of racemic drugs in general.
Materials and Methods
Resolution and characterization of (−)-A8 and (+)-A8
4-Dimethylaminopyridine (5 µmol/L) and di-tert-butyl dicarbonate (Boc2O, 0.45 mmol/L) were added to 0.40 mmol/L of (±)-A8 (2-(ethylthio)ethyl-6-(furan-2-yl)-3-methyl-4-oxo-4,5,6,7,tetrahydro-1H-indole-2-carboxylate; Chembridge) in CH3CN (10 mL) and cooled to 0°C. The mixture was stirred (room temperature, 6 h) and the volatiles removed under reduced pressure. The mixture was purified by silica gel flash chromatography (cyclohexane/ethyl acetate 4:1) to give (±)-Boc-A8 (N-tert-butyl carbamate-A8) as a white solid (155 mg, 85%); Rf = 0.6 (cyclohexane/ethyl acetate 7:3). [M+H]+ = 448.1. The 1H NMR spectrum of (±)-Boc-A8 is shown in Supplementary Fig. S1. (−)-Boc-A8 and (+)-Boc-A8: colorless needles from 2-propanol; melting point (m.p., uncorrected value): 73°C; [α]D20= −36.7 (c = 0.5, CHCl3); and + 36.7 (c = 0.5, CHCl3), respectively. HPLC retention times were 20.9 and 40.5 min, respectively. Next, (−)-Boc-A8 or (+)-Boc-A8 (0.11 mmol/L) were treated with a 30% CH2Cl2 solution of trifluoroacetic acid (1 mL) at 0°C. The solution was stirred (room temperature, 4 h), and a saturated aqueous solution of NaHCO3 (10 mL) and CH2Cl2 (10 mL) were added. The organic layer was separated, dried over anhydrous Na2SO4 and evaporated under reduced pressure. The crude material was purified by silica gel chromatography and gave (−)-A8 or (+)-A8 as white powders (30 mg, 77%; Rf = 0.35) (cyclohexane/ethyl acetate 7:3). (−)-A8 and (+)-A8: colorless crystals from 2-propanol (m.p.: 169°C); [α]D20= −36.8 (c = 1.05, CHCl3) and [α]D20= + 36.6 (c = 1.00, CHCl3), respectively.
X-Ray crystallography
Crystals of (+)-A8 were generated in acetonitrile/chloroform (6:1) by slow solvent evaporation at 4°C (18 d). Diffraction data were collected from a specimen with dimensions 0.750×0.050×0.025 mm3 at room temperature using Mo Ka radiation (λ = 0.71073 Å) on a three-circle Bruker Smart Apex diffractometer equipped with a CCD ApexII area detector. Measured intensities were corrected for absorption by using SADABS (6). The crystal structure was solved by direct methods and was refined against F2 using SHELX (7). Hydrogen atoms were placed in calculated positions. Due to its small dimensions and disorder, the crystal produced a significant number of weak reflections, leading to a large weighted wR2 factor (0.2055, as derived from 4,501 independent data points within 2ϑ= 45 deg). Actually, the ethyl groups of the thioether moieties were disordered in both the symmetry-independent A and B molecules, with refined occupancy factors as large as 40.9% and 59.1%. However, this did not prevent us from reliably determining the crystal structure and the absolute configuration of the compound. The Cambridge Crystallographic Data Centre (CCDC 820759) contains the crystallographic data for this paper (8).
Tubulin binding and activity assays
[3H]colchicine binding to tubulin was measured by the DEAE-cellulose filter method (9). Inhibition of tubulin polymerization was assayed in GTP-containing buffer comprising 0.625 to 20 µmol/L compound, 1.9 mg/mL porcine brain tubulin and 10 µmol/L fluorescent reporter (Cytoskeleton BK011P). Reporter incorporation (increase in fluorescence) during MT polymerization (37°C) was tracked photospectrometrically for 1 h (Tecan Infinite F200). The inhibition (%) of polymerization by a compound was expressed relative to that measured in the positive (5 mmol/L CaCl2 plus1% DMSO) and negative control (1% DMSO) samples. IC50 values (compound concentration showing 50% activity) were determined by nonlinear regression analysis of the area underneath the activity curve (Graphpad Prism 3.03).
Viability analysis of tumor cell lines
Growth inhibition analysis of the NCI60 cell lines by R- and S-A8 was performed by the Developmental Therapeutics Program at the NCI/NIH as described (10). To study growth inhibition of cells characterized by pathological Pgp levels, we purchased the isogenic human uterus sarcoma MES-SA and MES-SA/DX5 cell lines from the European Collection of Cell Cultures (ECACC). The latter cell line is characterized by a 106-fold overexpression of MDR1. The cells were authenticated by genome profiling (StemElite ID System, Promega) and confirmed to be free of Mycoplasmas by PCR (11). Both cell lines were seeded onto 96-well plates at 3,000 cells/well in McCoy's GlutaMAX medium (GIBCO) supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 U/mL streptomycin. The cells were incubated at 37°C for 24 h, the compounds then added (0.5 nmol/L to 0.05 mmol/L), and cell viability measured after 72 h of incubation (CellTiter-Glo Luminescent Cell Viability Assay (Promega). GI50 values (compound concentration that inhibited cell growth by 50%) were calculated as the measure of activity (Graphpad Prism 3.03).
Modeling the binding poses of active enantiomers
The binding models were generated using Discovery Studio 2.5 (Accelrys, Inc) and analyzed with HINT (EduSoft, LLC) (12). Simulations employed a CHARMM force field with CFF91 potentials and a distance-dependent dielectric of 1 and nonbonded interactions limited to 13 Å. The 1SA0 crystal structure of αβ-tubulin in complex with DAMA-colchicine (13) was selected as a template for docking studies, and the protein structure was prepared for modeling as previously described (14). It was observed that the ligand-based conformations of the R-isomer assumed a colchicine-like shape in the fold of its key pharmacophoric features and that the S-isomer assumed a podophyllotoxin-like fold. Based on these observations, the R-isomer was manually docked into the 1SA0 colchicine-binding site such that its conformation mapped the 3D conformation of DAMA-colchicine from the 1SA0 structure. The S-isomer was similarly docked to map the 3D conformation of podophyllotoxin from the related 1SA1 crystal structure (13). The docked poses of the R- and S-isomers were subsequently refined. Refinement involved iterative cycles of minimization and analysis of the atom-atom interaction quality with HINT. In the initial stage, the bound ligand conformation was minimized with the tubulin structure fixed in Cartesian space. Unfavorable atom-atom interactions were identified and relieved using manual changes to the torsional bond angles or relative orientation of the bound ligands, or changes to the torsional bond angles of amino acid side chains, followed by energy minimization in which only the tubulin polypeptide backbone was fixed in Cartesian space. Minimization and manual changes were repeated using the HINT score of the protein-ligand interactions, which indicate the favorability of interactions. To achieve the final binding models, the tubulin-ligand complexes were energy minimized without constraints. Extracting the bound ligand and the nearest neighbor residues, the binding energies were calculated using the standard settings in DiscoveryStudio, and found to be −35.6 and −22.5 kcal/mol for the R- and S-isomer, respectively. As a reference, similar calculations for bound combretastatin A-4, colchicine, and podophyllotoxin gave binding energies of −39.4, −37.1, and −34.9 kcal/mol, respectively. For combretastatin A-4, its binding energy was calculated from its docked pose in the binding site modeled for the R- and S-isomer. For colchicine and podophyllotoxin, the binding energies were calculated using the 1SA0 and 1SA1 crystal structures as templates, respectively, in which the bound ligand and nearest neighbor residues were extracted, and in situ minimization was employed.
Results
Resolution and configuration analysis of (−)- and (+)-A8 enantiomers
The enantiomers of a chiral antitubulin compound may have distinct molecular shapes, and therefore, may express different affinities for tubulin. To investigate the potential role of chiral selectivity towards antitubulin activity, we separated the enantiomers of our most active chemotype compound, named A8 (2-(ethylthio)ethyl 6-(furan-2-yl)-3-methyl-4-oxo-4,5,6,7,tetrahydro-1H-indole-2-carboxylate; Fig. 1A). To separate its isomers, we explored a number of differently functionalized cellulose- and amylose-based phases in combination with various eluent solutions and flow rates (15–17). However, no condition led to the separation of the enantiomers. We speculated that this was due to the NH group of the pyrrole ring forming a strong hydrogen bond with the stationary phase, thereby abolishing the subtle differences in interaction energies between the enantiomers and the stationary phase that otherwise would have allowed for their separation. Hence, the NH groups of (±)-A8 were neutralized by conversion into N-tert-butyl carbamates (Boc) (Fig. 1A). The derivative compounds were then subjected to semi-preparative HPLC, and excellent enantiomer separation was obtained with tris-[3,5-dimethylphenyl]carbamoyl amylose (α = 2.09; Rs = 6.5), allowing us to collect a substantial amount of the isomers in enantiomeric excess (>99%) (Fig. 1B). Next, the Boc group was removed from both enantiomers, thereby yielding the (−)-A8 and (+)-A8 isomers (Fig. 1A), which were characterized by optical rotations of [α]D20 of −36.8 and +36.6, respectively. To establish their absolute configurations, we focused on the (+)-A8 enantiomer. We succeeded in generating needle-like monoclinic crystals in a 6/1 (v:v) acetonitrile/chloroform mixture combined with slow evaporation (18 days) at 4°C (Fig. 2A). X-ray data collection gave the following results: space group P21 (n. 4); a = 12.956(2), b = 5.0942(9), c = 27.033(5) Å, β = 96.935(8) deg; V = 1771.1(5) Å3; Z = 4, Z' = 2, Dcalc = 1.303 g·cm−3; µ = 0.204 mm−1; F(000) = 736. The final conventional agreement factor R1 was 0.0707 (derived from 2,437 unique reflections with F > 4σ(F)). The final ΔF synthesis extremes were as low as +0.28 and –0.29 e·Å−3. The presence of anomalous scatterers (sulfur atoms) in the unit cells allowed us to establish with great confidence that the absolute configuration of the (+)-A8 isomer at its C13 stereocenter is S (refined Flack parameter: 0.0(2); (18)) (Figs. 2B–D, Supplementary Tables S1,S2).
Figure 1.
Separation of the A8 enantiomers. A, Route used to convert the NH group of the pyrrole ring of racemic (±)-A8 into N-tert-butyl carbamates (Boc), resulting in (±)-Boc-A8. Its isomers were separated by HPLC (hexane/isopropanol 9:1; 20 mL/min) using a chiral tris-[3,5-dimethylphenyl]carbamoyl amylose stationary phase (KROMASIL 5-AmyCoat, 21.2×250 mm, AkzoNobel). The Boc group was then removed to give (−,R)-A8 and (+,S)-A8. B, HPLC chromatogram showing the separation (−)-Boc-A8 and (+)-Boc-A8. Retention times were 20.9 and 40.5 min, respectively.
Figure 2.
Crystallographic analysis of (+)-A8. A, the (+)-A8 crystal specimen selected for the single-crystal X-ray diffraction analysis. B, packing scheme of (+)-A8 viewed down the b-axis. The H⋯O intermolecular contacts (Supplementary Information Table S2) are highlighted as dotted lines. Other hydrogen atoms were omitted for clarity. The asymmetric unit of S-A8 consists of two symmetry-independent homochiral molecules, designated A and B, that differ from each other by the relative orientation of the ester backbone with respect to the plane of the adjacent fused ring system. Specifically, the torsion angle (τ) between the condensed rings and the ester moiety [τ(O2-C5-C6-N1)] was as large as −5(1) deg in molecule A and +178.5(8) deg in molecule B. Consequently, the O2 and N1 atoms are cis-oriented in A and trans-oriented in B. Another significant difference lies in the orientation of the indole derivative and the furan ring, τ(C14-C13-C15-O4) being +168.7(8) deg in A and +44(1) deg in B (Supplementary Table S1). As to the crystal packing, the two symmetry-independent molecules A and B form an alternating motif along the a-axis, each layer being connected by short N–H⋯O contacts (Supporting Table S2). C, X-ray structure of the A conformer within the asymmetric unit of (+,S)-A8. D, X-ray structure of the B conformer within the asymmetric unit of (+,S)-A8. The labels of H atoms bonded to disordered ethyl groups were omitted for clarity. Thermal ellipsoids were drawn at the 25% probability level (Diamond v3.2f.)
Biochemical analysis of A8 enantiomer activity
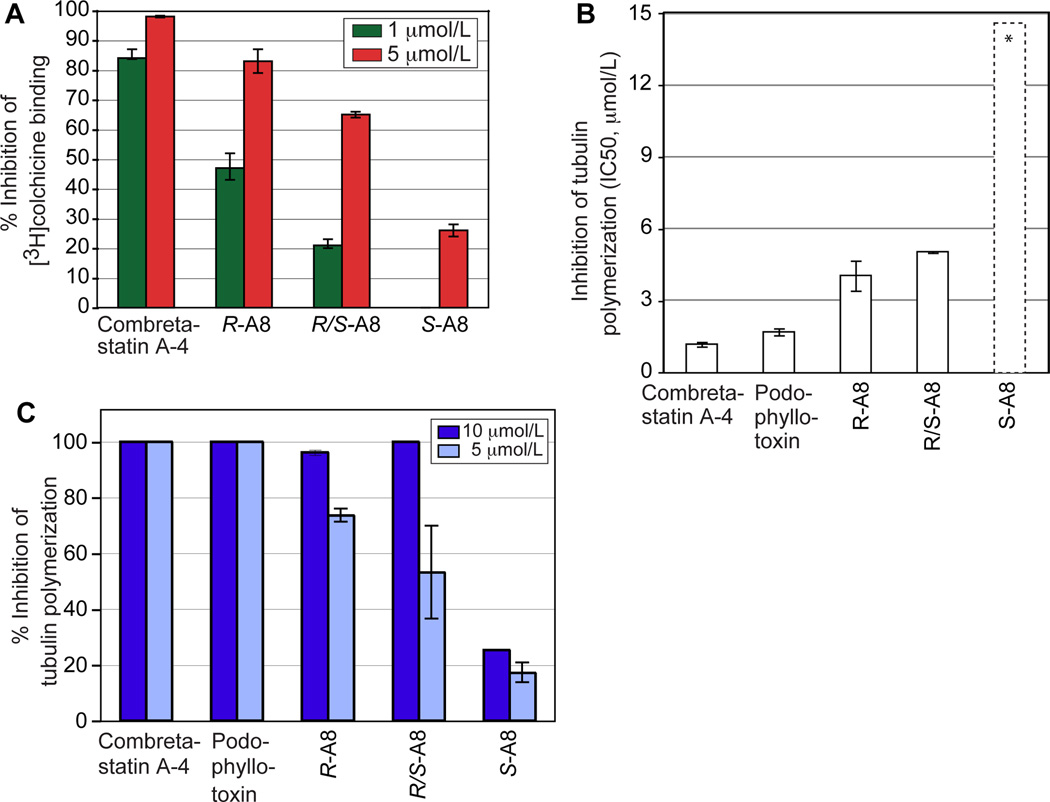
Following their separation, we measured in vitro the antitubulin activity of R- and S-A8. We first analyzed their ability to compete with [3H]colchicine (5 µmol/L) for binding to 1 µmol/L tubulin at a concentration of 1 and 5 µmol/L. Combretastatin A-4 and racemic A8 and were used as controls (Fig. 3A). Both enantiomers significantly inhibited [3H]colchicine binding to tubulin but R-A8 was 3.2-fold more potent than S-A8 (see activity values at 5 µmol/L). At 5 µmol/L, the activities of R-A8 and S-A8 were potent, and 83 ± 4% and 26 ±2 % of that of combretastatin A-4, respectively. Next, we photospectrometrically measured the ability of R-A8 and S-A8 (0.6 to 20 µmol/L) to inhibit the polymerization of 10 µmol/L tubulin. Combretastatin A-4, podophyllotoxin, and racemic A8 acted as controls. R-A8 inhibited tubulin polymerization with an IC50 value of 4 µmol/L, which is only 3-fold higher than that of our control drugs (Fig. 3B). The IC50 value of S-A8 could not be precisely determined as the compound tended to precipitate at the higher concentrations. However, when measuring the ability of our isomers to depolymerize MTs we found that at 5 µmol/L, the potencies of R-A8 and S-A8 were 72 ± 2.4% and 20 ± 3.5% of that of our control drugs, respectively (Fig. 3C). In sum, both enantiomers actively targeted tubulin, but the R-enantiomer was at least 4-fold more potent than the S-enantiomer (Figs. 3A-C).
Figure 3.
Antitubulin activities and tubulin-interaction profiles of compound A8. A, racemic R/S-A8 and its enantiomers localize to the β-tubulin colchicine site as shown by inhibition of [3H]colchicine binding. Combretastatin A-4 acted as the positive control. B, inhibition of tubulin polymerization by R/S-A8 and its enantiomers. Combretastatin A-4 and podophyllotoxin acted as positive controls. Compound concentrations ranged from 0.6 to 20 µmol/L. The asterisks and dashed bar indicate that the IC50 value for S-A8 is higher than 20 µmol/L but could not be measured as S-A8 precipitated at concentrations above 20 µmol/L. C, percentage of inhibition of tubulin polymerization at 5 and 10 µmol/L by compound A8, its enantiomers, and control agents.
The A8 enantiomers inhibit all NCI60 cancer cell lines and are not antagonized by Pgp
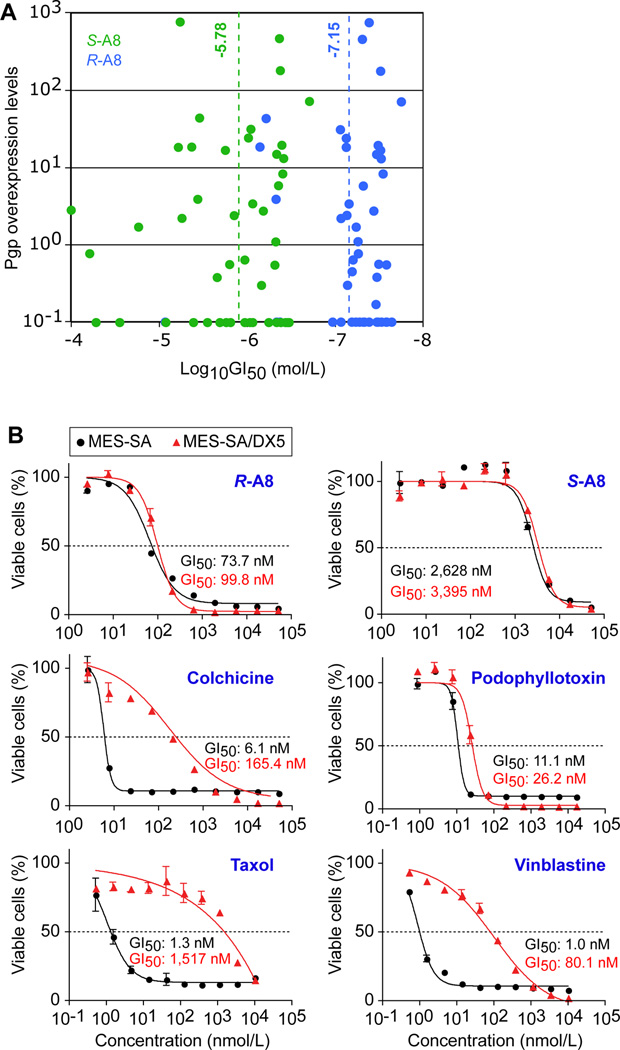
To determine whether the in vitro observed discrepancies in activities of R- and S-A8 also translated to human cells, we examined the effect of both enantiomers on the growth of 60 primary tumor cell lines (NCI Development Therapeutics Program), representing leukemias, non-small cell lung cancers, colon cancers, central nervous system tumors, lymphomas, melanomas, ovarian cancers, renal cancers, prostate cancers and breast cancers (19). Both enantiomers inhibited the growth of all cell lines (Fig. 4A; Supplementary Figs S2 and S3). The average GI50 value of R-A8 was 71 ± 9.5 nmol/L, that of S-A8 was 1.66 ± 0.26 µmol/L (dashed lines in Fig. 4A), indicating a ~23-fold greater activity of R-A8 over the S isomer in human cells. As suggested by the small standard deviation of the population average, both isomers were active in most tumor types. As most of the above cell lines are not isogenic and express different levels of the Pgp drug-efflux pump (20), we determined whether the GI50 activity values and Pgp expression levels in the cell lines were correlated. For both isomers, no correlation was found (Fig. 4A), suggesting that our enantiomers were not antagonized by Pgp. To further corroborate this conclusion, we evaluated the growth inhibition of the isogenic MES-SA and MES-SA/DX5 uterine carcinoma cell lines by R- and S-A8. The latter cell line is resistant to a large number of chemotherapeutic agents due to a 106-fold overexpression of Pgp (21). As activity controls, we analyzed colchicine, podophyllotoxin, taxol and vinblastine (Fig. 4B). R-A8 was 35-fold more potent than S-A8 in killing both cell lines. The ability of each isomer to inhibit the growth of both cell lines was statistically the same, confirming that Pgp did not antagonize our compounds. In contrast, the efficacy of colchicine and the clinically used vinblastine and taxol significantly dropped, with a factor of 27, 80, and 1,116; respectively, in the Pgp overexpressing cells (Fig. 4B).
Figure 4.
Inhibition of primary tumor cell lines by R- and S-A8. A, Correlation between P-glycoprotein (Pgp) expression levels in the NCI60 cancer cell lines (20) and the degree of growth inhibition (GI50) of the cell lines by S-A8 (green) and R-A8 (blue) (original data in Supplementary Figs S2 and S3). B, growth inhibition by R- and S-A8 of the isogenic uterus cancer cell lines MES-SA and MES-SA/DX5 (overexpresses Pgp 106-fold (21)). Colchicine, podophyllotoxin, taxol and vinblastine acted as controls.
Structural basis for in vitro activities of A8 enantiomers
To delineate a structural basis for the antitubulin activities of the A8 enantiomers, we modeled their binding modes and showed that they occupy the β-tubulin colchicine site in distinct manner. In the models, the R-isomer takes on the molecular conformation of colchicine, and the S-isomer that of podophyllotoxin (Fig. 5A). The R-isomer localized into the colchicine-binding site with its furan moiety mapped over the trimethoxy motif of colchicine and its ester side chain in the conformational space of the tropolone functionality (Figs. 5B,C). In contrast, the S-isomer localized into the colchicine-binding site with its ester side chain functioning as a bioisostere of the trimethoxy group of podophyllotoxin and its furan overlapping with the dioxolane ring of podophyllotoxin (Fig. 5C). In essence, the models indicate that the opposite handedness of the enantiomers flips the relative orientation of the ester side chain of the two isomers in the colchicine site (Fig. 5C). This allows the A8 enantiomers to occupy relatively the same conformational space in the colchicine site as colchicine and podophyllotoxin. The calculated binding affinities of the R- and S-isomers were −35.6 and −22.5 kcal/mol, respectively. Those of the control compounds combretastatin A-4, colchicine, and podophyllotoxin were −39.4, −37.1, and −34.9 kcal/mol, respectively. When the calculated binding energies were plotted against the biochemically measured in vitro activities of the compounds, we obtained excellent correlations (Fig. 5D). These suggested that the lower level of antitubulin activity of S-A8 relative to R-A8 is due to an energetically less favorable occupancy of β-tubulin.
Figure 5.
A, Compound A8 and superimposition of the conformations of its R- and S-enantiomers derived from docking studies with the binding modes of colchicine and podophyllotoxin, obtained from their crystal structures in complex with tubulin (13). A8 is rendered in ball-and-stick with its C atoms colored green, while colchicine and podophyllotoxin are similarly rendered with their C atoms colored cyan. The N, O, and S atoms are colored blue, red, and yellow, respectively. B, Binding models of R- and S-A8, colchicine and podophyllotoxin. β-tubulin is shown as a brown ribbon, with the key residues shown in stick mode with C atoms colored grey for residues in β-tubulin and yellow for residues in α-tubulin. A8, colchicine and podophyllotoxin are rendered as in panel B. C, representation of how R-A8 and S-A8 bind to the colchicine site of β-tubulin (brown curve). The regions in R- and S-A8 (encircled) that mimic those of colchicine and podophyllotoxin are indicated with letters. D, Correlation between the calculated free binding energies of the S-A8, podophyllotoxin, colchicine, R-A8 and combretastatin A4 and their in vitro measured antitubulin activity ([3H]colchicine competition, inhibition of tubulin polymerization). The activity values were determined at compound concentrations of 5µmol/L (Figs. 3A,C). E, Modeled binding trajectories of R- and S-A8. A plot of the hydrophobic-polar interaction constants versus the distance (Å) of the ligand to its colchicine binding site (from 10 Å away from the binding site to a final docked pose at 0 Å) as calculated by the HINT program. More favorable interaction scores are indicated by more positive values on the y-axis.
Discussion
Anticancer compounds that target MTs act at four major binding sites: the taxane and laulimalide/peloruside A sites for MT-stabilizing agents, and the vinca and colchicine sites for MT-destabilizing agents (2). Taxanes and vinca alkaloids have had great success in cancer chemotherapy. So far, no colchicine-site agents have been approved for use against cancer. However, analogs of combretastatin A-4, which act at the colchicine site, are currently in clinical trials as they also exhibit antivascular activity leading to the rapid collapse of tumor vasculature (2). This observation has refueled the search for novel colchicine-site chemotypes. In addition, emerging tumor resistance to taxanes and vinca alkaloids was recently linked to their binding sites and expression levels of the βIII tubulin isotype. It was proposed that these resistance mechanisms may be circumvented by colchicine-site agents (22). In addition, the efficacy of the clinically used antitubulin drugs suffers from overexpression of the Pgp drug-efflux pump in tumors. Hence, identifying novel colchicine-site chemotypes that are not detected by Pgp provide exciting opportunities for new scaffold design and drug development.
Our present study provides the first structural, functional, and computational analysis of a novel antitubulin chemotype, named furan metotica, which satisfies the above properties for new colchicine-site drugs. Furan metoticas were identified as inhibitors of the binding of the Ndc80 kinetochore complex to taxol-stabilized MTs. However, control proteins including the MT-regulating protein EB1 and antitubulin antibodies also were prevented from binding to the MTs, suggesting that our chemotype targeted the MTs rather than the Ndc80 complex. MT-depolymerizing compounds such as nocodazole and colchicine did not prevent the Ndc80 kinetochore complex from binding to MTs (5), suggesting a unique mode of interaction between furan metoticas and β-tubulin. Our docking studies of the R- and S-isomers of the most active chemotype compound A8 showed that they bound at the colchicine site in stereospecific fashion pointing to stereoselectivity of the colchicine-binding site (this was the case for the other chemotype members as well, data not shown). We found that R-A8 assumed a colchicine-like fold in which its furan ring overlaid with the trimethoxybenzene group of colchicine. S-A8 assumed a podophyllotoxin-like pose with its ester side chain folded over the trimethoxybenzene group of podophyllotoxin. The binding models of A8 indicate that the compound could indeed function as a racemic drug. To the best of our knowledge, no other colchicine-site agent exists whose isomers mimic the binding modes of two different MT-destabilizing drugs. Consistent with the differences in calculated binding energies of their binding poses, the R-A8 was at least 4-fold more active than S-A8 in vitro, and was on average 25–35 fold more cytotoxic than S-A8 in cancer cells. The in vitro and in vivo results show that both enantiomers contribute to the overall activity of the racemic compound. However, the biological activity of a compound depends not only on its static occupation of its binding site, but also on how it behaves on its way to the target. To obtain insight into this process, we previously employed a dynamic modeling method in which the binding of close congeners was simulated over a time course. The individual frames of the trajectory were analyzed revealing structural insights into activity differences found among the closely related compounds (14, 22). Would this methodology also prove useful in differentiating between the binding dynamics of stereoisomers from their initial positions in the solvent front to their final docked poses? To investigate this, we modeled the binding trajectories of R- and S-A8 to the colchicine site and applied analytical algorithms to quantify atom-atom interactions that may be crucial to affinity. The docking trajectories were created by undocking the ligand from its docked pose using constrained molecular dynamics and reversing the resulting trajectory to achieve a simulation of the docking trajectory. At each 0.2 Å, the translation of the ligand towards its docked pose in β-tubulin was saved and the hydropathic interaction values of the receptor-ligand complex calculated. The docking trajectories showed that R-A8 had a generally more favorable binding trajectory than S-A8 (Fig. 5E), whose flexible ester side chain suffers from steric clashes with Leu248 at the entrance of the colchicine-binding site.
In summary, our study provides structural and functional insights into the unique behavior of a novel stereoisomeric colchicine-site chemotype. Our observations will support a more comprehensive drug design of potent new antitubulin agents that are not antagonized by Pgp. Indeed, our compounds proved to be 80- to 1,000-fold more active against drug-resistant cancer cells than the antitubulin drugs presently used in the clinic.
Supplementary Material
Acknowledgments
This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported in part by the Developmental Therapeutics Program in the Division of Cancer Treatment and Diagnosis of the National Cancer Institute.
Grant Support
This research was supported in part by grant 08-0465 from the Association of International Cancer Research to PDW. The research was also sponsored in part by the U.S. Army Medical Research and Material Command Research Plan #02-4-3U-057 and IAA #Y3-CM-100505 (MRMC and NCI).
References
- 1.Tobias JS, Hochhauser D, Souhami RL. Cancer and its management. 6th ed. Chichester, West Sussex, UK; Hoboken, NJ: Wiley-Blackwell; 2010. [Google Scholar]
- 2.Dumontet C, Jordan MA. Microtubule-binding agents: a dynamic field of cancer therapeutics. Nat Rev Drug Discov. 2010;9:790–803. doi: 10.1038/nrd3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goda K, Bacso Z, Szabo G. Multidrug Resistance Through the Spectacle of P-Glycoprotein. Curr Cancer Drug Tar. 2009;9:281–297. doi: 10.2174/156800909788166493. [DOI] [PubMed] [Google Scholar]
- 4.Jackson JR, Patrick DR, Dar MM, Huang PS. Targeted anti-mitotic therapies: can we improve on tubulin agents? Nat Rev Cancer. 2007;7:107–117. doi: 10.1038/nrc2049. [DOI] [PubMed] [Google Scholar]
- 5.Screpanti E, Santaguida S, Nguyen T, Silvestri R, Gussio R, Musacchio A, et al. A screen for kinetochore-microtubule interaction inhibitors identifies novel antitubulin compounds. Plos One. 2010;5:e11603. doi: 10.1371/journal.pone.0011603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blessing RH. An Empirical Correction for Absorption Anisotropy. Acta Crystallogr A. 1995;51:33–38. doi: 10.1107/s0108767394005726. [DOI] [PubMed] [Google Scholar]
- 7.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 8. http://www.ccdc.cam.ac.uk/data_request/cif.
- 9.Kang GJ, Getahun Z, Muzaffar A, Brossi A, Hamel E. N-Acetylcolchinol O-Methyl Ether and Thiocolchicine, Potent Analogs of Colchicine Modified in the C-Ring - Evaluation of the Mechanistic Basis for Their Enhanced Biological Properties. J Biol Chem. 1990;265:10255–10259. [PubMed] [Google Scholar]
- 10. http://dtp.nci.nih.gov/branches/btb/ivclsp.html.
- 11.Hopert A, Uphoff CC, Wirth M, Hauser H, Drexler HG. Specificity and sensitivity of polymerase reaction (PCR) in comparison with other methods for the detection of mycoplasma contamination in cell lines. J Immunol Methods. 1993;164:91–100. doi: 10.1016/0022-1759(93)90279-g. [DOI] [PubMed] [Google Scholar]
- 12.Kellogg GE, Semus SF, Abraham DJ. Hint - a New Method of Empirical Hydrophobic Field Calculation for Comfa. J Comput Aid Mol Des. 1991;5:545–552. doi: 10.1007/BF00135313. [DOI] [PubMed] [Google Scholar]
- 13.Ravelli RBG, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A, et al. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen TL, McGrath C, Hermone AR, Burnett JC, Zaharevitz DW, Day BW, et al. A common pharmacophore for a diverse set of colchicine site inhibitors using a structure-based approach. J Med Chem. 2005;48:6107–6116. doi: 10.1021/jm050502t. [DOI] [PubMed] [Google Scholar]
- 15.Ikai T, Yamamoto C, Kamigaito M, Okamoto Y. Immobilized polysaccharide-based chiral stationary phases for HPLC. Polym J. 2006;38:91–108. [Google Scholar]
- 16.Stringham RW. The use of polysaccharide phases in the separation of enantiomers. Adv Chromatogr. 2006;44:257–290. doi: 10.1201/9781420027150.ch6. [DOI] [PubMed] [Google Scholar]
- 17.Ali I, Saleem K, Hussain I, Gaitonde VD, Aboul-Enein HY. Polysaccharides Chiral Stationary Phases in Liquid Chromatography. Sep Purif Rev. 2009;38:97–147. [Google Scholar]
- 18.Flack HD. On Enantiomorph-Polarity Estimation. Acta Crystallogr A. 1983;39:876–881. [Google Scholar]
- 19.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 20.Alvarez M, Paull K, Monks A, Hose C, Lee JS, Weinstein J, et al. Generation of a Drug-Resistance Profile by Quantitation of Mdr-1/P-Glycoprotein in the Cell-Lines of the National-Cancer-Institute Anticancer Drug Screen. J Clin Invest. 1995;95:2205–2214. doi: 10.1172/JCI117910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munic V, Kelneric Z, Mikac L, Haber VE. Differences in assessment of macrolide interaction with human MDR1 (ABCB1, P-gp) using rhodamine-123 efflux, ATPase activity and cellular accumulation assays. Eur J Pharm Sci. 2010;41:86–95. doi: 10.1016/j.ejps.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 22.Bai RL, Nguyen TL, Burnett JC, Atasoylu O, Munro MHG, Pettit GR, et al. Interactions of Halichondrin B and Eribulin with Tubulin. J Chem Inf Model. 2011;51:1393–1404. doi: 10.1021/ci200077t. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.