Abstract
Targeting the complement component 3a receptor (C3aR) with selective agonists or antagonists is believed to be a viable therapeutic option for several diseases such as stroke, heart attack, reper-fusion injuries, and rheumatoid arthritis. We designed a number of agonists, partial agonists, and antagonists of C3aR using our two-stage de novo protein design framework. Of the peptides tested using a degranulation assay in C3aR-transfected rat basophilic leukemia cells, two were prominent agonists (EC50 values of 25.3 and 66.2 nM) and two others were partial agonists (IC50 values of 15.4 and 26.1 nM). Further testing of these lead compounds in a calcium flux assay in U937 cells yielded similar results, although with reduced potencies compared to transfected cells. The partial agonists also displayed full antagonist activity when tested in a C3aR inhibition assay. In addition, the electrostatic potential profile was shown to potentially discriminate between full agonists and partial agonists.
Introduction
The complement system is an important part of the immune system and is involved in aiding clearance of immune complexes, debris removal, opsonization, inflammation, and cell lysis1. Improper activation of the complement system, however, can cause tissue injury in various pathological conditions and contributes to several immune diseases, including stroke, heart attack, adult respiratory distress syndrome, septic shock, inflammatory bowl disease, reperfusion injuries, asthma, rheumatoid arthritis, psoriasis, and rejection of xenotransplantation1–3. One possibility to prevent this improper activation is to target the complement component 3a receptor, or C3aR.
C3aR is a G protein-coupled receptor protein that is targeted by C3a, an anaphylatoxin that mediates the proinflammatory activities of the complement system. C3a is a 77-residue cationic peptide that is derived from the cleavage of the amino-terminus of the α-chains of C3. Besides mediating proinflammatory activities, C3a possibly has opposing immunological roles in some cellular systems4. C3a is known to be a chemoattractant and secretagogue for eosinophils and mast cells4–8 and a mediator of smooth muscle contraction4,9. In addition, in vitro studies have shown that C3a can suppress or enhance cytokine production which may indicate that C3a is involved in secondary cytokine release and thus inflammatory and immune effects at local sites of inflammation4,10–12.
C3a is a potent mediator, considering that only micromolar or sub-micromolar concentrations of C3a are sufficient to elicit its biological effects13. Earlier work by Ember et al.13 identified a potent C3aR agonist: WWGKKYRASKLGLAR. More recently Scully et al.14 have reported potent hexapeptide agonists and antagonists that are selective for C3aR: FLPLAR, FIPLAR, FWTLAR, FLTLAR (agonists) and FLTChaAR (antagonist). Given the potential immunomodulatory role of C3a in several diseases, targeting the C3aR with selective agonists or antagonists is a viable therapeutic option15.
Our de novo protein design framework was applied to the design of novel agonists and antagonists of C3aR and our best predictions were experimentally validated using a rat basophilic leukemia cell degranulation assay and a monocytic cell calcium flux assay. The de novo protein design framework is a first-principles, computational predictive framework that determines amino acid sequences that are energetically favorable in a given 3-dimensional (3D) template structure16–19. The sequences can be further validated to be candidate binders of a target protein20,21. For the design of C3aR agonists and antagonists, we utilized the 3D structure of the endogenous ligand for C3aR, C3a. The two-stage framework first generates low-energy amino acid sequences then ranks these sequences based on a fold specificity or an approximate binding affinity. Fold specificity measures how likely the amino acid sequence will adopt the template structure while approximate binding affinity determines how likely the designed peptide or protein will bind to a target protein. For this design, we used only the fold specificity validation metric because structural information of the C3a:C3aR complex, which is necessary to compute the approximate binding affinity, was unavailable in the open literature.
Key features of the de novo protein design framework include the use of deterministic global optimization for sequence selection (determining amino acid sequences that will fold into given template structure)18, a computationally efficient fold specificity metric that can sample hundreds of conformational structures in minutes19, and an approximate binding affinity metric that is a measure of the functionality of the designed sequences20,21. Both stages incorporate protein flexibility by introducing the flexibility of the template structure in the sequence selection stage and by generating thousands of flexible conformations of the designed sequences in the validation stage. Our framework has achieved numerous successes including the full sequence design of human-beta-defensin-219 and numerous experimentally validated peptides including compstatin inhibitors of human C320,22, HIV-1 gp41 inhibitors21, and Bak inhibitors of Bcl-xL and Bcl-2 23. These successes have wide applicability to many diseases, including immune diseases, AIDS, and cancer.
Results
Computational Design
The de novo protein design framework was used to design agonists and antagonists of C3aR by designing amino acid sequences that would adopt the C3a fold. In this case, since structural information of the C3a:C3aR complex was unknown, we were only able to use our fold specificity ranking metric to determine likely C3aR agonists and antagonists. A summary of this method is provided in the Materials and Methods section and details can be found in the Supporting Information.
Five separate design runs were performed, each one differing in what mutations were allowed. Table 1 presents the designed sequences from the latter two runs, along with the sequences designed by Ember et al.13 (E1 and E2) and two sequences from our preliminary work on this system (S3 and S4)24. Details on all five runs can be found in the Materials and Methods section and designed sequences from all five runs can be found in the Supporting Information. The de novo protein design method is freely available to the scientific community at www.proteinwisdom.org.
Table 1.
Sequence selection and fold specificity results for C3a agonists/antagonists. Rankings are given for sequence selection (lowest energy = 1, E) and fold specificity (highest specificity = 1, fspec). Mutations away from native C3a are indicated in bold.
| Name | Rank | Sequence | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E | fspec | 63 | 77 | ||||||||||||||
| native C3a | L | R | R | Q | H | A | R | A | S | H | L | G | L | A | |||
| S4 | W | W | T | R | R | W | R | A | S | K | L | G | L | A | R | ||
| S3 | W | W | T | R | R | Y | R | A | S | K | L | G | L | A | R | ||
| E1 | W | W | G | K | K | Y | R | A | S | K | L | G | L | A | R | ||
| E2 | W | W | R | N | R | W | R | E | N | R | L | G | L | A | R | ||
| Run 4 (using MS4) | |||||||||||||||||
| SQ074-4* | 74 | 1 | W | W | T | R | R | W | R | T | D | K | L | G | L | A | R |
| SQ062-4* | 62 | 2 | W | W | T | R | R | W | R | T | E | K | L | G | L | A | R |
| SQ110-4* | 110 | 3 | W | W | T | R | R | W | R | G | D | K | L | G | L | A | R |
| SQ018-4 | 18 | 4 | W | W | T | R | R | W | R | L | D | K | L | G | L | A | R |
| SQ101-4 | 101 | 5 | W | W | T | R | R | W | R | G | E | K | L | G | L | A | R |
| SQ022-4 | 22 | 6 | W | W | T | R | R | W | R | Y | D | K | L | G | L | A | R |
| SQ032-4 | 32 | 7 | W | W | T | R | R | W | R | C | D | K | L | G | L | A | R |
| SQ060-4* | 60 | 8 | W | W | T | R | R | W | R | A | D | K | L | G | L | A | R |
| SQ105-4 | 105 | 9 | W | W | T | R | R | W | R | V | D | K | L | G | L | A | R |
| SQ033-4 | 33 | 10 | W | W | T | R | R | W | R | A | E | K | L | G | L | A | R |
| Run 5 (using MS5) | |||||||||||||||||
| SQ007-5* | 7 | 1 | W | W | D | R | R | W | R | A | S | K | L | G | L | A | R |
| SQ002-5* | 2 | 2 | W | W | E | R | R | W | R | A | S | K | L | G | L | A | R |
| SQ006-5 | 6 | 3 | W | W | N | R | R | W | R | A | S | K | L | G | L | A | R |
| SQ003-5 | 3 | 4 | W | W | Q | R | R | W | R | A | S | K | L | G | L | A | R |
| SQ011-5 | 11 | 5 | W | W | T | R | R | W | R | A | S | K | L | G | L | A | R |
| SQ010-5* | 10 | 6 | W | W | S | R | R | W | R | A | S | K | L | G | L | A | R |
| SQ012-5 | 12 | 7 | W | W | G | R | R | W | R | A | S | K | L | G | L | A | R |
| SQ009-5 | 9 | 8 | W | W | A | R | R | W | R | A | S | K | L | G | L | A | R |
| SQ008-5 | 8 | 9 | W | W | P | R | R | W | R | A | S | K | L | G | L | A | R |
| SQ005-5 | 5 | 10 | W | W | R | R | R | W | R | A | S | K | L | G | L | A | R |
Indicates experimentally tested compound
In run 4, position 71 consists solely of negatively charged amino acids while position 70 is very varied and mutates to Thr, Tyr, Leu, Ala, Gly, Cys, and Val. Run 5 is interesting in that it ranks negatively charged amino acid mutations in position 65 first and second. This disrupts the +3 charge seen in the native sequence. In fact, the native amino acid of Arg is ranked 10 out of 12 possible sequences.
Experimental Validation
Seven of the designed sequences were selected for synthesis (SQ074-4, SQ062-4, SQ110-4, SQ060-4, SQ007-5, SQ002-5, SQ010-5,) in addition to S3, S4, and the two peptides by Ember et al.13 (E1 and E2) for comparison. Peptides were selected from runs 4 and 5. Runs 1 – 3 provided information on what positions to fix to S4 or mutate, which then motivated runs 4 and 5 (described in the Materials and Methods section). The top three sequences from run 4 based on fold specificity were selected along with the 8th ranked sequences because it retained the native Ala in position 70. The top two sequences from run 5 based on fold specificity were selected in addition to the 6th ranked sequence based upon inspection.
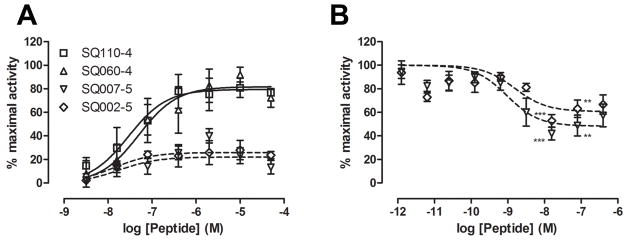
Both the agonist and antagonist activity of the designed sequences were measured. Experiments were carried out in the rat basophilic leukemia cell line (RBL-2H3), which resembles a rat serosal mast cell. This cell line is transfected with human receptors for C5a (C5aR/CD88) or C3a (C3aR). Cells that have been exposed to a stimulus (such as the C3a peptides) secrete β-hexosaminidase from intracellular granules, providing a read-out of receptor activity. The level of secretion induced by each peptide is shown relative to the maximal secretion stimulated by a high dose of recombinant human C3a (Table 2) or an analog of the C-terminus of C3a, FLPLAR (Figure 1) and is reported as EC50, the half-maximal effective concentration. Antagonist activity was measured as the inhibition of the response to the same high dose of either C3a or FLPLAR, following a pre-incubation with peptide and is reported as IC50, the half-minimal inhibitory concentration. Dose response curves for two selected agonists and two antagonists are shown in Figure 1.
Table 2.
Agonist and antagonist activities of peptide sequences. Upper panel: Agonist activity of peptides using RBL-C3aR cells is shown as Log EC50 (± SEM), EC50 in nM and as a calculated maximum (M ± SEM) relative to levels of secretion stimulated by an optimal concentration of C3a or FLPLAR. Lower panel: Antagonist activity of peptides using RBL-C3aR cells is shown as Log IC50 (± SEM), IC50 in nM and as a calculated minimum (M ± SEM) relative to levels of secretion stimulated by an optimal concentration of C3a or FLPLAR. S4-noAC was the same sequence as S4, but with no N-terminal acetylation. Significance calculated using 1 way ANOVA with Dunnetts multiple comparison post-test.
| # | Name | Log EC50 | SEM | Significance | EC50 (nM) | M (%) | SEM | Significance | n |
|---|---|---|---|---|---|---|---|---|---|
| 1 | S4 | −6.71 | 0.28 | ns | 194 | 35 | 4.1 | ns | 6 |
| 2 | S3 | −6.00 | 0.36 | ns | 1001 | 29 | 5.6 | *** | 6 |
| 3 | E1 | −6.77 | 0.36 | - | 170 | 38 | 4.9 | - | 6 |
| 4 | E2 | −6.17 | 0.20 | ns | 675 | 62 | 5.9 | *** | 6 |
| 5 | SQ074-4 | −6.47 | 0.27 | ns | 337 | 43 | 5.0 | ns | 6 |
| 6 | SQ062-4 | −6.15 | 0.25 | ns | 715 | 45 | 5.4 | ns | 6 |
| 7 | SQ110-4 | −7.60 | 0.20 | * | 25.3 | 72 | 4.7 | *** | 7 |
| 8 | SQ060-4 | −7.18 | 0.16 | ns | 66.2 | 79 | 4.7 | *** | 6 |
| 9 | SQ007-5 | −6.65 | 0.81 | ns | 225 | 14 | 4.8 | *** | 8 |
| 10 | SQ002-5 | −7.45 | 0.50 | ns | 35.5 | 22 | 3.7 | *** | 8 |
| 11 | SQ010-5 | −6.83 | 1.09 | ns | 146 | 13 | 5.8 | *** | 6 |
| 12 | S4-noAc | −7.09 | 0.30 | ns | 81.6 | 37 | 4.4 | ns | 6 |
| # | Name | Log IC50 | SEM | Significance | IC50 (nM) | M (%) | SEM | Significance | n |
|---|---|---|---|---|---|---|---|---|---|
| 1 | S4 | −7.30 | 0.24 | ns | 50.2 | 47 | 4.6 | *** | 5 |
| 2 | S3 | −7.63 | 0.27 | ns | 23.4 | 42 | 4.4 | * | 5 |
| 3 | E1 | −7.96 | 0.40 | - | 10.9 | 52 | 4.2 | - | 5 |
| 4 | E2 | −8.73 | 1.40 | ns | 1.85 | 62.5 | 4.5 | * | 5 |
| 5 | SQ074-4 | −8.30 | 0.55 | ns | 5.06 | 60 | 3.6 | ns | 5 |
| 6 | SQ062-4 | −8.11 | 0.54 | ns | 7.80 | 60 | 4.3 | ns | 5 |
| 7 | SQ110-4 | −8.04 | 0.83 | ns | 9.20 | 70 | 4.6 | *** | 5 |
| 8 | SQ060-4 | −8.34 | 1.37 | ns | 4.58 | 68 | 5.9 | *** | 5 |
| 9 | SQ007-5 | −7.81 | 0.30 | ns | 15.4 | 24 | 5.9 | *** | 4 |
| 10 | SQ002-5 | −7.58 | 0.30 | ns | 26.1 | 35 | 6.1 | *** | 5 |
| 11 | SQ010-5 | −7.20 | 0.28 | ns | 63.0 | 40 | 6.7 | ** | 5 |
| 12 | S4-noAc | −7.91 | 0.54 | ns | 12.4 | 63 | 5.2 | * | 5 |
ns > 0.05,
< 0.05,
< 0.005,
< 0.001, compared to #3, peptide E1.
Figure 1.
Agonist and antagonist activities of selected peptides. A) Agonist activity. RBL-C3aR cells were treated with the indicated concentrations of peptides and receptor activation measured by β-hexosaminidase secretion. Data are the means of a representative experiment performed in duplicate and are shown as a percentage of the maximal response. B) Antagonist activity. RBL-C3aR cells were treated with the indicated concentrations of peptides before the addition of an optimal concentration of C3aR agonist peptide FLPLAR. Data are the means ± SEM of 5 experiments performed in duplicate and are shown as a percentage of the response to FLPLAR alone. A one-sample t test was used to determine if means were significantly different to 100 %; ***: p < 0.0001; **: p < 0.001.
All the peptides had a degree of agonist activity but two peptides showed strong agonistic properties (SQ110-4, SQ060-4), with EC50 values of 25.3 nM and 66.2 nM, respectively, stimulating secretion to 72 % and 79 % of the maximal response (Table 2). The full dose-response curves for these peptides are shown in Figure 1A. In contrast, the previously reported “superagonist” peptide E113 stimulated only 38 % of the maximal response, with an EC50 of 170 nM (Table 2). We then tested the peptides for their ability to antagonize C3a activity and again, all of the peptides had some antagonist activity at low nanomolar concentrations. Two peptides (SQ007-5, SQ002-5) that suppressed C3a activity to 24 % and 35 % of the maximal response, with IC50 values of 15.4 nM and 26.1 nM, respectively, were selected for further study. Their weak agonist effects are shown in Figure 1A and their suppression of the response to FLPLAR is shown in Figure 1B. The suppression of the peptide response was less than for C3a, at 48 % and 61 % of the maximal, but with IC50 values of 0.95 nM and 1.6 nM, respectively.
When tested on RBL cells expressing human C5aR, none of the peptides had any agonist or antagonist activity at concentrations of up to 1 μM (data not shown), indicating that the peptides are selective for C3aR.
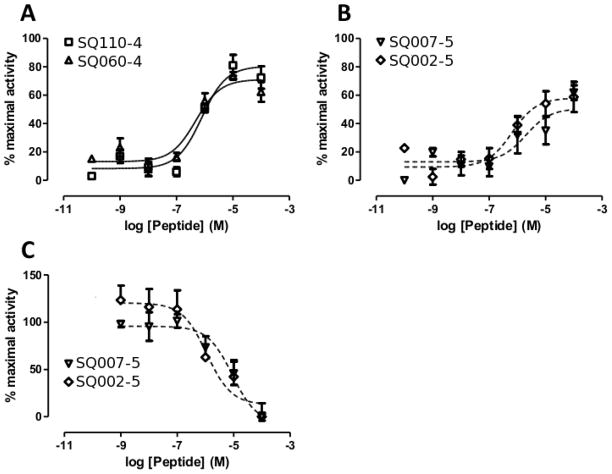
In order to further test the activities of the peptides SQ110-4, SQ060-4, SQ007-5, and SQ002-5, experiments were performed in dbcAMP differentiated U937 cells. As with the β-hexosaminidase assays in RBL-transfected cells, peptides SQ110-4 and SQ060-4 again displayed strong agonistic activity, with calculated EC50 values of 0.80 and 0.49 μM respectively and inducing 81 % and 74 % of the maximal response (Table 3, Fig. 2A). Again, similar to the transfected system, peptides SQ007-5 and SQ002-5 displayed more partial agonist effects in U937 cells, inducing 62 % and 59 % of the maximal response, with calculated EC50 values of 2.0 and 0.66 μM respectively (Table 3, Fig. 2B). Though displaying mixed partial agonist responses, both SQ007-5 and SQ002-5 had full antagonistic activity, with 100 μM of the peptides completely blocking a FLPLAR induced response. Antagonist IC50 values of 8.95 and 1.12 μM were calculated for SQ110-4 and SQ060-4 respectively (Table 3, Fig. 2C).
Table 3.
Agonist and antagonist activities of peptide sequences of interest. Upper panel: Agonist activity of peptides in U937 cells is displayed as Log EC50 (± SEM), EC50 in nM and as a calculated maximum (M) relative to the response of the cells to 10 μM of the C3aR agonist, FLPLAR. Lower panel: Antagonists activity of peptides in U937 cells is displayed as Log IC50 (± SEM), IC50 in nM and as a calculated minimum (M) relative to the response of the cells to 10 μM of the C3aR agonist, FLPLAR, following treatment with the peptides of interest. n = 3–6
| # | Name | Log EC50 | EC50 (nM) | M (%) |
|---|---|---|---|---|
| 7 | SQ110-4 | −6.10 (0.18) | 797 | 81 |
| 8 | SQ060-4 | −6.31 (0.22) | 487.4 | 74 |
| 9 | SQ007-5 | −4.58 (0.39) | 2002 | 62 |
| 10 | SQ002-5 | −6.18 (0.31) | 654.9 | 59 |
| # | Name | Log IC50 | IC50 (nM) | M (%) |
|---|---|---|---|---|
| 9 | SQ007-5 | −5.05 (0.21) | 8946 | 0 |
| 10 | SQ002-5 | −5.95 (0.34) | 1121 | 6 |
Figure 2.
Agonist and antagonist activities of selected peptides. Agonist activity was measured for peptides SQ110-4 and SQ060-4 (A) and SQ007-5 and SQ002-5 (B). Dibutyryl cAMP differentiated U937 cells were treated with the peptides of interest and receptor activation measured by intracellular calcium mobilization. Following treatment with peptides SQ007-5 and SQ002-5, 10 μM of the C3aR agonist, FLPLAR, was added and antagonist activity of the peptides measured (C). Data is displayed as the mean ± SEM. n = 3–6.
Electrostatic Contributions to the Mechanism of Binding
Charge plays significant role in the function of C3a. First, C3a is a cationic protein, and second, C3a function is mediated by its C-terminal amino acid Arg77, which is positively charged. It is known that the C-terminal arginine of C3a (and homologous C5a) is indispensable for binding and biological activity (e.g. Morikis et al.3, Ember et al.13, Scully et al.14, and references therein). In addition, Ember et al.13 points out the importance of charge because of the presence of conserved arginines at the C-terminus of C3a from various species. These observations indicate the significance of considering charge in the design of our peptides. In addition, in a study of short cyclic peptides derived from the C-terminus of C5a, the C-terminal arginine was replaced by the neutral arginine-like amino acid citruline in two peptides, PMX200 and PMX201. This replacement in PMX200 resulted in 87-fold loss of binding affinity and 335-fold loss of antagonist activity compared to its parent peptide PMX205. The same replacement in PMX201 resulted in 16-fold loss of binding affinity and 293-fold loss of antagonist activity compared to its parent peptide PMX5325.
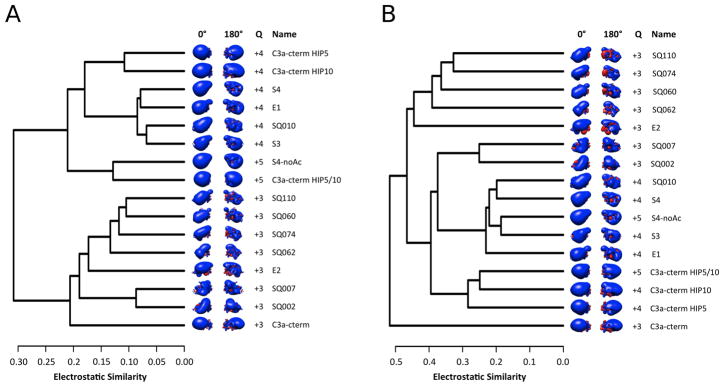
The 15-amino acid C-terminal segment of C3a, used for our peptide design, is highly cationic with side chain net charge +4 across residues 63 – 77 and +3 across residues 63 – 72 (assuming His67 and His72 are neutral). The +3 charge across residues 63 – 72 was maintained in runs 1 –4 of our design by imposing a physicochemical constraint, however this constraint was dropped for run 5. Because of the role of charge in the function of C3a and in the agonist/antagonist activities of C3a-derived peptides, we present here an analysis of electrostatic potentials. Table 1 –3 show that the location of charged amino acids in the peptide discriminates agonist from partial agonist activities. For example, the two full agonists, SQ110-4 and SQ060-4, have a negatively charged amino acid at position 71, whereas the two partial agonists, SQ007-5 and SQ002-5, have a negatively charged amino acid at position 65. In both cases, full and partial agonists, the side chain net charge is +4 across residues 63 –77, which makes charge non-discriminatory for agonism/antagonism. Electrostatic potentials are more informative than net charge or the location of individual charges within the peptide sequence. This is because electrostatic potentials account for the variable dielectric environment of the protein-solvent system and for the ionic environment of the solvent, with both dielectric and ionic properties influencing (screening) electrostatic interactions within the peptides. In addition, proximity of charges within the structure can produce additive or subtractive effects in the electrostatic potentials, which affect ligand-receptor recognition. This is information not available when examining charges alone. Figure 3 shows hierarchical clustering of the electrostatic potentials of the designed peptides at ionic strengths corresponding to salt concentrations of 0 mM and 150 mM. By performing electrostatic clustering analysis with no counterions, electrostatic potentials are more exaggerated presenting global differences in electrostatics, which can affect long-range recognition. Where electrostatic clustering performed with 150 mM of counterions, on the other hand, captures local differences that may affect specific interactions involved in binding. Figure 3 depicts the two full agonists, SQ110-4 and SQ060-4, cluster together and the two partial agonists, SQ007-5 and SQ002-5, cluster together, but separately from the full agonists, at both 0 mM and 150 mM. Since the agonists and partial agonists cluster separately in both the 0 and 150 mM clustering, this indicates that both local and global effects contribute to agonist and partial agonist activities, and suggests a key role for electrostatics. Therefore, the electrostatic potential profile has potentially predictive value, together with the locations of specific amino acid types within the peptide sequences, for the determination of agonist or partial agonist activity. For completion, we have included in Figure 3, all 12 peptides of Table 2 for which we collected experimental data, and 3 additional sequences of the native peptide in which each or both histidines, His67 and His72, are protonated (charged). The alternative charged states of the native C3a sequence were also analyzed to identify any similarities between the designed analogs and the four potential charged states of the parent peptide. Histidine protonation can result in peptide net charge increase to +5 (single protonated histidine) or +6 (two protonated histidines). Based on the electrostatic clustering results, the electrostatic potentials of all of the newly designed sequences are distinct from that of the native sequence, even when considering alternative charge states of histidine.
Figure 3.
Hierarchical clustering dendrogram of spatial distributions of electrostatic potentials for the 12 experimentally tested peptides of Table 2 and four native peptides with different combinations of histidine tautomers. Electrostatic potentials were calculated at 0 mM ionic strength (A) and 150 mM ionic strength (B). Isopotential surfaces are shown at two different orientations rotated about the vertical axis and are plotted at ± 2.5 kBT/e (0 mM) and ± 1 kBT/e (150 mM), with blue and red denoting positive and negative electrostatic potential, respectively. The net charge (Q) of the sequences is given and is calculated taking into account the charged side chains and the negatively charged backbone at the unblocked C-termini (and the positively charged backbone at the N-terminus of S4-noAc). Native peptide His67 and His72 protonation is as follows: (i) C3a-cterm, His67/His72 neutral; (ii) C3a-cterm HIP5, His67 charged, His72 neutral; (iii) C3a-cterm HIP10, His67 neutral, His72 charged; and (iv) C3a-cterm HIP5/10, His67/His72 charged.
Discussion
The de novo protein design framework was applied to the design of C3aR agonists and antagonists. Since structural information on the C3a:C3aR complex was unknown, the design employed the structure of C3a and identified short sequences (15-residues) that were favorable in the C3a folded structure.
The computational results provided a number of strong patterns in the mutations of C3a. In particular, the introduction of negatively charged amino acids in positions 65 and 71 elucidated a number of potent agonists and partial agonists. For the majority of the computational runs, a charge of +3 across residues 63 – 69 was imposed to mimic the charge of the native peptide. However, the best antagonists have either Asp or Glu in position 65, bringing the side chain net charge across residues 63 – 69 down to +1.
Of the seven designed peptides initially tested in our transfected cell system, two were prominent agonists while two others were partial agonists with prominent antagonist activity. These peptides were selected for further testing using a more direct measure of receptor activation with a cell line natively expressing C3aR26. Both systems were able to distinguish between the prominent agonists and partial agonists, although potency of these compounds in the native C3aR expressing cell line was lower than that in the transfected cell system.
The two partial agonist peptides were also able to inhibit the activity of both intact C3a and an analog of the C-terminus of C3a. Ligand binding to C3aR and C5aR involves the cooperation of at least two sites on the receptor. For C3aR, one site comprises charged residues in the very large second extracellular loop but binding here does not lead to receptor activation. Instead, a second binding site located in the pore formed by the helical transmembrane domains must be engaged27. For C5aR, the first site is located at the receptor N-terminus but the second site has similarities with that of C3aR and some charged residues have been identified that are common to both receptors27. For C5aR, it is clear that the C-terminus of C5a binds in the transmembrane pores, and so C-terminal peptides of C5a will activate the receptor even when the N-terminus of the receptor has been removed. By analogy, we would expect the C-terminus of C3a to interact with C3aR in a similar way and there is some experimental data to support this view27. It is thus likely that the family of peptides described here interact with residues in the transmembrane domains of C3aR, because they are similar to the C-terminus of C3a and also can antagonize the activity of another peptide (FLPLAR) derived from the C3a C-terminus.
Examination of the sequences of the 4 most potent peptides (#’s 7, 8, 9 and 10) allowed us to gain insight into specific physicochemical properties of their function. In addition to the LGLAR C-terminal sequence, the potent agonists (#’s 7 and 8) contained the N-terminal sequence WWTR-RWR and only differed in position 70 with # 7 mutating to the small amino acid Gly and # 8 retaining the native Ala. The presence of two N-terminal Trp and an additional aromatic amino acid (Trp) at position 68 suggests the possibility for aromatic ring π-stacking that stabilizes the structure of the peptides. This type of interaction may be necessary to stabilize the C-terminal sequence LGLAR, while allowing some flexibility, in a conformation that facilitates interaction with C3aR. It is likely that these aromatic amino acids act as a scaffold, in the absence of the remaining C3a sequence, to orient Arg77 in a proper conformation for interaction with C3aR amino acids. The aromatic amino acids may form hydrophobic and/or π-cation contacts with C5aR. We expect the remaining amino acids of LGLAR to form favorable contacts, since they are part of the C3a binding site. In the case of the two better partial agonists (#’s 9 and 10), the only difference was Asp65 for #9 and Glu65 for #10. Despite this difference, both sequences had a negatively charged amino acid in position 65.
Of particular interest is the net charge of the potent peptides. All four peptides had a side chain net charge of +4 across the entire 15-residue peptide. However the agonists had a charge of +3 across residues 63 – 69 while the partial agonists had a charge of +2 across these positions. This difference in charged areas was seen in hierarchical clustering of electrostatic potentials, where the two agonists clustered together and the two partial agonists clustered together, but separately from the agonists.
Based on the above analysis, the main characteristics for agonist activity were: (i) C-terminal sequence LGLAR; (ii) N-terminal sequence WWTRRWR; (iii) Gly or Ala in position 70; and (iv) side chain net charge of +4 with a charge of +3 across positions 63 – 69. Similarly, the main characteristics for partial agonist activity were: (i) C-terminal sequence LGLAR, (ii) N-terminal sequence WW, (iii) negatively charged amino acid (D/E) in position 65, and (iv) side chain net charge of +4 with a charge of +2 across positions 63 – 69. These findings are important for further sequence optimization using our computational protocol, in which sequence segments W63W(D/E)65, and L73GLAR77 may be considered indispensable for activity if the side chain net charge is +4, whereas the remaining seven amino acids may be amenable to further optimization. Incorporation of non-natural amino acids may be necessary to improve antagonist activity.
This work highlights the success of the protein design framework. In this case, structural information on the binding site was unknown, so the design was driven by fold specificities. Since structure often implies function, this is a good measure to use when structural binding data is unavailable.
Materials and Methods
Computational De Novo Design
A two-stage de novo protein design framework16–21 was applied to the design of C3a analogs. The first stage selects amino acid sequences using either a single template or multiple templates defined only by either the Cα positions or the side-chain centroids of the template(s). The second stage ranks the sequences from stage one based upon their fold specificity, a measure of how well the sequences assume the template fold. In addition to the design template, mutation sets, biological constraints, and a forcefield are also specified for the calculations. Components specific to the design of C3aR agonists and antagonists are described below while details about the framework can be found in the Supporting Information.
Five separate design runs were performed. For each run, the design template and forcefield were kept constant but the allowed mutations were varied. For runs 1 – 3, 1000 sequences were generated in stage one. For run 4 and run 5, all sequences were generated since the computational complexity was so small (132 and 12 sequences total, respectively). Once the sequences were generated in stage one, fold specificity calculations from stage two were performed in order to rank the sequences based upon the likelihood of assuming the template fold28–34.
Design Template
The three-dimensional structure of C3a was first elucidated by Huber et al.35 using X-ray crystallography at a relatively low resolution of 3.2 Å. However, the structure did not encompass residues 1–12, owing to the presence of either a disordered or or mobile ordered structure with respect to the rest of C3a3.
The overall structure of C3a resembles a drumstick that has very little core. Disulfide bridges exist between Cys22-Cys49, Cys23-Cys56, and Cys36-Cys57. These S-S bonds keep the shape of the head of the drumstick structure and the long carboxy-terminal helix (residues 47 – 69) forms the stick of the drumstick. Besides the one present in the carboxy-terminus, helices are also found in residue segments 17–23 and 37–41 in the crystal structure. Salt bridges that contribute to packing interactions in C3a are: Glu24-Arg20, Asp25-Arg28, Glu62-Arg65, and weaker ones are observed in Asp25-Arg39, Asp55-Lys51, Glu24-Lys17, and Glu47-Lys50.
A flexible design template was generated using molecular dynamics (MD) simulations with explicit water molecules as solvent. The initial structure for the MD simulations was constructed as a composite of the C3a domain of the crystal structure of C3 for amino acids Val1 - Ala69 (PDB code: 2A73)36 and the crystal structure of C3a for amino acids Ser70 - Arg76 (coordinates kindly provided by Prof. Huber; in electronic supplementary material of Morikis et al.3). Construction of the composite structure was necessary because the C3a structure is missing the first alpha-helix, whereas the C3 structure is missing the 7 C-terminal amino acids. The MD simulation was performed for 10 ns, using the program CHARMM37. MD snapshots were saved every 1 ns.
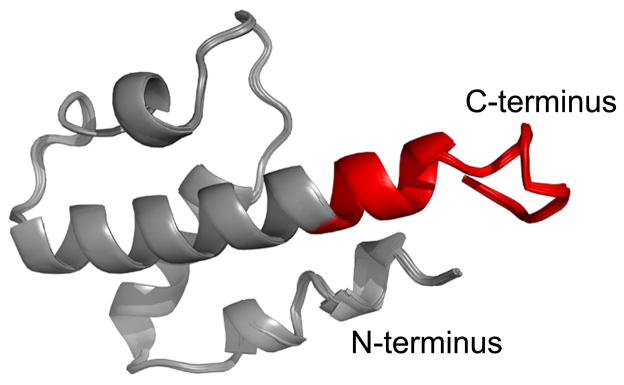
Figure 4 shows the flexible design template. Since previous potent analogues of C3a consisted of only the last 21 or 15 residues of C3a13 and the current binding model of C3a to C3aR suggests interaction between residues 69 – 77 of C3a to helices 4 and 5 – 7 of C3aR24, only these last residues were considered for the design. These last 15 residues are shown in red in Figure 4.
Figure 4.
Flexible design template of C3a (superposition of 10 MD snapshots). The last 15 residues are colored in red.
Mutation Sets
The mutation set specifies which positions can mutate and to what set of amino acids. In the most general sense, every position could be allowed to mutate to all 20 amino acids. In order to decrease the computational complexity, solvent accessible surface areas of each position or experimental knowledge are used to constrain the number of mutations allowed at each position. As previously stated, only positions 63 – 77 were designed. The last five residues (LGLAR) of C3a are highly conserved among animal species13 and so were fixed to the native sequence. These residues are considered to be the primary binding site of C3a to C3aR. Previous computational work by Fung24 elucidated potential candidates for C3a agonists/antagonists with sequences of WWTR-RYRASKLGLAR (S3) and WWTRRWRASKLGLAR (S4). Based upon these two sequences, the two mutations to Trp in positions 63 and 64 were imposed and the remaining positions (65 – 72) were allowed to mutate in five separate computational runs.
The first mutation set (MS1) allowed positions 65 – 68 to mutate. The second mutation set (MS2) allowed positions 70 – 72 to mutate. The third mutation set (MS3) allowed positions 65 and 70 – 71 to mutate. The fourth and fifth mutation sets (MS4 and MS5, respectively), split MS3 into two separate runs: MS4 allowed only positions 70 and 71 to mutate while MS5 allowed only position 65 to mutate. The allowable amino acids at each mutable position were determined by solvent accessible surface area. If a position was more than 50% exposed to solvent, hydrophilic amino acids were allowed, if a position was less than 20% exposed to solvent, hydrophobic amino acids were allowed. Finally, if the exposure was between 20% and 50%, all amino acids were allowed. Positions that were not allowed to mutate were fixed to the sequence of S4. Table 4 provides details about each mutation set along with the native sequence of C3a, the sequence of S4 and the sequence by Ember et al.13 (E1). Positions 1 – 62 were fixed as the native sequence.
Table 4.
Mutation sets for C3a. Two sequences (native and S4) are given for reference. A dash (−) indicates that position is fixed with respect to the amino acid in S4. Sequence numbering is with respect to the full C3a sequence. Mutations away from the native sequence of C3a are indicated in bold. For MS1 - MS5, B indicates hydrophobic amino acids (A, C, G, I, L, M, F, T, W, Y, V) and Z indicates hydrophilic amino acids (A, R, N, D, Q, E, G, H, K, P, S, T). The native residue was also always allowed at each mutable position. Computational complexity is defined as bnzm, where b and z are the number of amino acids in each hydrophobic and hydrophilic set, respectively, and n and m the number of their respective mutable sequence positions.
| Name | Positions | Computational Complexity | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 63 | 77 | |||||||||||||||
| native C3a | L | R | R | Q | H | A | R | A | S | H | L | G | L | A | R | |
| E1 | W | W | G | K | K | Y | R | A | S | K | L | G | L | A | R | |
| S4 | W | W | T | R | R | W | R | A | S | K | L | G | L | A | R | |
| MS1 | - | - | Z | Z | Z | B | - | - | - | - | - | - | - | - | - | 1.9 × 104 |
| MS2 | - | - | - | - | - | - | - | B | Z | Z | - | - | - | - | - | 1.6 × 103 |
| MS3 | - | - | Z | - | - | - | - | B | Z | - | - | - | - | - | - | 1.6 × 103 |
| MS4 | - | - | - | - | - | - | - | B | Z | - | - | - | - | - | - | 1.3 × 102 |
| MS5 | - | - | Z | - | - | - | - | - | - | - | - | - | - | - | - | 1.2 × 101 |
MS4 and MS5 were motivated by the fact that some of the results using MS1 exhibited the mutations seen in the S4 and S3 sequences from Fung24. Using MS1, positions 66 and 67 showed a predominance of Arg, which appears in positions 66 and 67 in both S4 and S3. Thus we decided to fix these two positions to Arg. Position 68 was fixed to Trp since a large hydrophobic amino acid was present in this position in S3, S4, and the “superagonist” elucidated by Ember et al.13. MS2 showed mutations to Asp and Glu in position 71 and some retention of the native amino acid in position 70. We decided to allow these two positions to mutate but fixed position 72 to Lys, which is the mutation identified by Ember et al.13, S3, and S4. The remaining mutable positions (65, 70, and 71) were allowed to vary in MS3, which was then split into two separate runs, allowing only position 65 to mutate in MS5 and only positions 70 and 71 to mutate in MS4. Table 1 provides the top ten sequences from runs 4 and 5 (using MS4 and MS5, respectively) ranked according to fold specificity. Additionally, the sequences of native C3a, S3, S4, and two potent sequences from Ember et al.13 (E1 and E2) are provided for comparison.
Biological Constraints
One biological constraint was imposed on the design. For runs 1 – 4 (corresponding to MS1 –MS4) a charge of +3 between residues 63 and 69 was imposed. This is the charge between those residues in the native sequence.
Peptide Synthesis for Experimental Studies
The peptides were synthesized by GenScript (Piscataway, NJ) with acetylated N-termini, unless otherwise noted, and in quantities of 1 – 4 mg at > 95% purity. Purity was confirmed with HPLC. The C-terminus was unblocked in all peptides.
Rat Basophilic Leukemia Cell Degranulation assay
RBL-2H3 cells transfected with either human C5aR or human C3aR38 were routinely cultured in Dulbeccos modified Eagles medium (DMEM), 10 % (v/v) fetal calf serum and 400 mg/liter G-418 at 37C in 5 % CO2. Degranulation was measured by assaying for β-hexosaminidase activity in the cell supernatant, as described previously39. In agonist assays, degranulation was calculated as a percentage of maximal activity in response to 200 nM recombinant human C3a, human C5a38 or 10 nM agonist hexapeptide, FLPLAR14. For antagonist assays, cells were pre-treated with peptides for 10 min prior to the addition of C3a, C5a or FLPLAR. EC50, IC50 and standard error values were obtained by iterative curve fitting in GraphPad Prism v5.0.
Calcium Flux assay
The human monocytic lymphoma cell line, U937, was maintained in RPMI-1640 medium supplemented with 10 % v/v fetal calf serum at 37°C and 5 % CO2. Prior to experimentation, U937 cells were incubated for 48 h in 1 mM dibutyryl cAMP (Sigma-Aldrich, USA) in order to differentiate the cells into a C3a-responsive macrophage-like cell14. Receptor activation was assessed using a Flexstation 3 microplate reader (Molecular Devices, USA) and Fluo-4 dye (Invitrogen, USA) to detect increases in intracellular calcium indicative of GPCR activation. For the agonist assays, responses were calculated as a percentage of maximal activity compared to 10 μM of the C3aR agonist, FLPLAR. For antagonist assays, cells were pre-treated with peptides for 10 min prior to the addition of 10 μM FLPLAR. Dose response curves were plotted and EC50 and IC50 values calculated using GraphPad Prism v5.0.
Poisson-Boltzmann Electrostatics
For electrostatic clustering, the structures of the experimentally tested C3a peptide analogs were generated using homology modeling with the Automodel module of Modeller 9v940. The template used for the homology modeling consisted of the 15 C-terminal residues from the last snapshot of the 10-ns MD simulation of whole C3a, described above. Sequence alignments between the native peptide (named C3a-cterm) and the 12 analogs were generated using the R41 package Bio3D42, and supplied as input to Modeller. The generated models were then prepped using PDB2PQR43, in order to add atomic radii and partial charges to the coordinate files according to the PARSE44 forcefield. When adding charges in PDBPQR, the N-termini of all peptides were neutralized because of the addition of the acetyl group, with the exception of S4-noAc which was positively charged, whereas all C-termini were negatively charged. PDB2PQR was also used to generate four alternative charged states of the native peptide by different combinations of charges for His67 and His72. The number of peptide structures used for electrostatic calculations was 16, including 8 newly designed sequences, 4 known sequences, and 4 native peptides with alternative protonation state combinations for His67 and His72. Electrostatic potentials were calculated using the linearized Poisson-Boltzmann equation implemented in APBS45. The dielectric surface was defined as the contact surface using a probe sphere with 1.4 Å radius, resembling a water molecule, and the dielectric coefficients were 20 for the protein interior and 78.54 for the solvent (justification for dielectric of 20 for the protein interior is given in Gorham et al.46). Electrostatic potentials were calculated by embedding the peptides in a grid with 65 × 65 × 65 grid points and dimensions of 52 Å × 38 Å × 40 A. The peptide structures were superimposed and the same centering was used ° in the grid prior to performing the calculations. Two sets of electrostatic potentials were generated, corresponding to counterions at 0 and 150 mM ionic strength. The ion accessibility surface was defined using a probe sphere with a 2 Å radius, resembling a monovalent ion. Electrostatic similarity calculations and clustering were performed for each set of electrostatic potentials using the AESOP (Analysis of Electrostatic Similarities of Proteins) framework47–49. A 16 × 16 electrostatic distance matrix was determined for each set of potentials using the pairwise localized difference (LD) measure (Eq. 1)49.
| (1) |
where LDAB is the similarity between proteins A and B, N is the number of grid points, and φA(i, j, k) and φB(i, j, k) refer to the electrostatic potentials of proteins A and B, respectively, at identical grid points (i, j, k). The summation is over all grid points. This LD corresponds to the average of the normalized error between all corresponding grid points. According to this scheme, two peptides are identical if they have LD 0 and totally different if they have LD 2. Hierarchical clustering was performed based on the generated electrostatic distance matrices using average linkage and the clustering results are presented as dendrograms, using R. The spatial distributions of electrostatic potential were visualized and plotted as isopotential contours using the molecular graphics program Chimera50.
Supplementary Material
Acknowledgments
This work was supported by grants from NSF (CAF and DM), NIH (R01 GM52032, CAF; R24 GM069 736, CAF and DM), University of California Tobacco-Related Disease Research Program (DM and CAK), the Beckman Initiative for Macular Research (DM), and the US Environmental Protection Agency, EPA (R 832721-010, CAF). Although the research described in the article has been funded in part by the US Environmental Agency’s STAR program, it has not been subjected to any EPA review and therefore does not necessarily reflect the views of the Agency, and no official endorsement should be inferred. A portion of this research was made possible with Government support by DoD, Air Force Office of Scientific Research, National Defense Science and Engineering Graduate (NDSEG) Fellowship, 32 CFR 168a (MLBP).
Footnotes
Abbreviations List: C3a, complement component 3a receptor; C3aR, complement component 3a receptor; 3-D, 3-dimensional; MS, mutation set; RBL-2H3, rat basophilic leukemia cell line; C5a, complement component 5a; C5aR, complement component 5a receptor; SEM, standard error of the mean; MD, molecular dynamics; HPLC, high-performance liquid chromatography.
Supporting Information Availability
Details about the mathematical model and formulations for the de novo design framework and sequence results from all five computational design runs are available as Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
Contributor Information
Meghan L. Bellows-Peterson, Department of Chemical and Biological Engineering, Princeton University, Princeton, NJ
Ho Ki Fung, Department of Chemical and Biological Engineering, Princeton University, Princeton, NJ.
Christodoulos A. Floudas, Department of Chemical and Biological Engineering, Princeton University, Princeton, NJ.
Chris A. Kieslich, Department of Bioengineering, University of California at Riverside, Riverside, CA
Li Zhang, Department of Bioengineering, University of California at Riverside, Riverside, CA.
Dimitrios Morikis, Department of Bioengineering, University of California at Riverside, Riverside, CA.
Kathryn J. Wareham, Department of Infection and Immunity, The University of Sheffield Medical School, Sheffield, UK
Peter N. Monk, Department of Infection and Immunity, The University of Sheffield Medical School, Sheffield, UK
Owen A. Hawksworth, School of Biomedical Sciences, The University of Queensland, Brisbane, Australia
Trent Woodruff, School of Biomedical Sciences, The University of Queensland, Brisbane, Australia.
References
- 1.Tamamis P, Morikis D, Floudas CA, Archontis G. Species specificity of the complement inhibitor compstatin investigated by all-atom molecular dynamics simulations. Proteins: Struct, Funct Bioinf. 2010;78(12):2655–2667. doi: 10.1002/prot.22780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sahu A, Lambris J. Structure and biology of complement protein C3, a connecting link between innate and acquired immunity. Immunol Rev. 2001;180:35–48. doi: 10.1034/j.1600-065x.2001.1800103.x. [DOI] [PubMed] [Google Scholar]
- 3.Morikis D, Holland MCH, Lambris JD. Structure of the anaphylatoxins C3a and C5a. In: Morikis D, Lambris JD, editors. Structural Biology of the Complement System. Chp 7. CRC Press; 2005. pp. 161–177. [Google Scholar]
- 4.Proctor LM, Moore TA, Monk PN, Sanderson SD, Taylor SM, Woodruff TM. Complement factors C3a and C5a have distinct hemodynamic effects in the rat. Int Immunopharmacol. 2009;9:800–806. doi: 10.1016/j.intimp.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Daffern PJ, Pfeifer PH, Ember JA, Hugli TE. C3a is a chemotaxin for human eosinophils but not for neutrophils. I. C3a stimulation of neutrophils is secondary to eosinophil activation. J Exp Med. 1995;181(6):2119–2127. doi: 10.1084/jem.181.6.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elsner J, Oppermann M, Czech W, Dobos G, Schöpf E, Norgauer J, Kapp A. C3a activates reactive oxygen radical species production and intracellular calcium transients in human eosinophils. Eur J Immunol. 1994;24(3):518–522. doi: 10.1002/eji.1830240304. [DOI] [PubMed] [Google Scholar]
- 7.Hartmann K, Henz BM, Krüger-Krasagakes S, Köhl J, Burger R, Guhl S, Haase I, Lippert U, Zuberbier T. C3a and C5a stimulate c hemotaxis of human mast cells. Blood. 1997;89(8):2863–2870. [PubMed] [Google Scholar]
- 8.Takafuji S, Tadokoro K, lto K, Dahinden CA. Degranulation from human eosinophils stimulated with C3a and C5a. Int Arch Allergy Immunol. 1994;104(Suppl 1):27–29. doi: 10.1159/000236743. [DOI] [PubMed] [Google Scholar]
- 9.Marceau F, Lundberg C, Hugli TE. Effects of the anaphylatoxins in circulation. Immunopharmacology. 1987;14(2):67–84. doi: 10.1016/0162-3109(87)90031-2. [DOI] [PubMed] [Google Scholar]
- 10.Fischer WH, Jagels MA, Hugli TE. Regulation of IL-6 synthesis in human peripheral blood mononuclear cells by C3a and C3adesArg. J Immunol. 1999;162(1):453–459. [PubMed] [Google Scholar]
- 11.Takabayashi T, Vannier E, Burke JF, Tompkins RG, Gelfand JA, Clark BD. Both C3a and C3a desArg regulate Interleukin-6 synthesis in human peripheral blood mononuclear cells. J Infect Dis. 1998;177(6):1622–1628. doi: 10.1086/515316. [DOI] [PubMed] [Google Scholar]
- 12.Takabayashi T, Vannier E, Clark B, Margolis N, Dinarello C, Burke J, Gelfand J. A new biologic role for C3a and C3a desArg: regulation of TNF-alpha and IL-1 beta synthesis. J Immunol. 1996;156(9):3455–3460. [PubMed] [Google Scholar]
- 13.Ember J, Johansen N, Hugli T. Designing synthetic superagonists of C3a anaphylatoxin. Biochemistry. 1991;30:3603–3612. doi: 10.1021/bi00229a003. [DOI] [PubMed] [Google Scholar]
- 14.Scully CCG, Blakeney JS, Singh R, Hoang HN, Abenante G, Reid RC, Fairlie DP. Selective hexapeptide agonists and antagonists for human complement C3a receptor. J Med Chem. 2010;53:4938–4948. doi: 10.1021/jm1003705. [DOI] [PubMed] [Google Scholar]
- 15.Holland MCH, Morikis D, Lambris J. Synthetic small-molecule complement inhibitors. Curr Opin Invest Drugs. 2004;5(11):1164–1173. [PubMed] [Google Scholar]
- 16.Klepeis J, Floudas C, Morikis D, Tsokos C, Argyropoulos E, Spruce L, Lambris J. Integrated computational and experimental approach for lead optimization and design of compstatin variants with improved activity. J Am Chem Soc. 2003;125:8422–8423. doi: 10.1021/ja034846p. [DOI] [PubMed] [Google Scholar]
- 17.Klepeis JL, Floudas CA, Morikis D, Tsokos CG, Lambris JD. Design of peptide analogs with improved activity using a novel de novo protein design approach. Ind Eng Chem Res. 2004;43:3817–3826. [Google Scholar]
- 18.Fung HK, Taylor MS, Floudas CA. Novel formulations for the sequence selection problem in de novo protein design with flexible templates. Optimization Methods and Software. 2007;22:51–71. [Google Scholar]
- 19.Fung HK, Floudas CA, Taylor MS, Zhang L, Morikis D. Toward full-sequence de novo protein design with flexible templates for human beta-defensin-2. Biophys J. 2008;94:584–599. doi: 10.1529/biophysj.107.110627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bellows ML, Fung HK, Floudas CA, Lopez de Victoria A, Morikis D. New comp-statin variants through two de novo protein design frameworks. Biophys J. 2010;98(10):2337–2346. doi: 10.1016/j.bpj.2010.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bellows ML, Taylor MS, Cole PA, Shen L, Siliciano RF, Fung HK, Floudas CA. Discovery of entry inhibitors for HIV-1 via a new de novo protein design framework. Biophys J. 2010;99:3445–3453. doi: 10.1016/j.bpj.2010.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.López de Victoria A, Gorham RD, Jr, Bellows-Peterson ML, Ling J, Lo DD, Floudas CA, Morikis D. A new generation of potent complement inhibitors of the compstatin family. Chem Biol Drug Des. 2011;77:431–440. doi: 10.1111/j.1747-0285.2011.01111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun JJ, Abdeljabbar DM, Clarke NL, Bellows ML, Floudas CA, Link AJ. Reconstitution and engineering of apoptotic protein interactions on the bacterial cell surface. J Mol Biol. 2009;394:297–305. doi: 10.1016/j.jmb.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fung HK. PhD thesis. Princeton University; 2008. Computational De Novo Protein Design with Flexible Templates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Proctor L, Woodruff T, Sharma P, Shiels I, Taylor S. Transdermal pharmacology of small molecule cyclic C5a antagonists. Adv Exp Med Biol. 2006;586:329–345. doi: 10.1007/0-387-34134-X_22. [DOI] [PubMed] [Google Scholar]
- 26.Burg M, Martin U, Bock D, Rheinheimer C, Kohl J, Bautsch W, Klos A. Differential regulation of the C3a and C5a receptors (CD88) by IFN- gamma and PMA in U937 cells and related myeloblastic cell lines. J Immunol. 1996;157:5574–5581. [PubMed] [Google Scholar]
- 27.Sun J, Ember JA, Chao TH, Hugli TE, Fukuoka Y, Ye RD. Identification of ligand effector binding sites in transmembrane regions of the human G protein-coupled C3a receptor. Protein Sci. 1999;8(11):2304–2311. doi: 10.1110/ps.8.11.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klepeis JL, Floudas CA. Free energy calculations for peptides via deterministic global optimization. J Chem Phys. 1999;110:7491–7512. [Google Scholar]
- 29.Klepeis JL, Floudas CA, Morikis D, Lambris J. Predicting peptide structures using NMR data and deterministic global optimization. J Comput Chem. 1999;20:1354–1370. [Google Scholar]
- 30.Klepeis JL, Floudas CA. Ab initio prediction of helical segments of polypeptides. J Comput Chem. 2002;23:246–266. doi: 10.1002/jcc.10002. [DOI] [PubMed] [Google Scholar]
- 31.Klepeis JL, Floudas CA. Prediction of beta-sheet topology and disulfide bridges in polypeptides. J Comput Chem. 2003;24:191–208. doi: 10.1002/jcc.10167. [DOI] [PubMed] [Google Scholar]
- 32.Klepeis J, Floudas C. ASTRO-FOLD: A combinatorial and global optimization framework for ab initio prediction of three-dimensional structures of proteins from the amino acid sequence. Biophys J. 2003;85:2119–2146. doi: 10.1016/s0006-3495(03)74640-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klepeis J, Pieja M, Floudas C. A new class of hybrid global optimization algorithms for peptide structure prediction: Integrated hybrids. Comput Phys Commun. 2003a;151:121–140. [Google Scholar]
- 34.Klepeis J, Pieja M, Floudas C. Hybrid global optimization algorithms for protein structure prediction : Alternating hybrids. Biophys J. 2003b;84:869–882. doi: 10.1016/S0006-3495(03)74905-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huber R, Scholze H, Paques E, Deisenhofer J. Crystal structure analysis and molecular model of human C3a anaphylatoxin. Hoppe Seylers Zeitschrifft fur Physiologische Chemie. 1980;361:1389–1399. doi: 10.1515/bchm2.1980.361.2.1389. [DOI] [PubMed] [Google Scholar]
- 36.Janssen BJC, Huizinga EG, Raaijmakers HCA, Roos A, Daha MR, Nilsson-Ekdahl K, Nilsson B, Gros P. Structures of complement component C3 provide insights into the function and evolution of immunity. Nature. 2005;437:505–511. doi: 10.1038/nature04005. [DOI] [PubMed] [Google Scholar]
- 37.Brooks B, Bruccoleri R, Olafson B. CHARMM - a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 38.Cain S, Monk P. The orphan receptor C5L2 has high affinity binding sites for complement fragments C5a and C5a des Arg74. J Biol Chem. 2002;277:7165–7169. doi: 10.1074/jbc.C100714200. [DOI] [PubMed] [Google Scholar]
- 39.Monk P, Pease J, Barker M. C5a stimulus-secretion couplng in rat basophilic leukemia (RBL-2H3) cells transfected with the human C5a receptor is mediated by pertussis and cholera toxin-sensitive G-proteins. Biochem Mol Biol Int. 1994;32(1):13–20. [PubMed] [Google Scholar]
- 40.Eswar N, Marti-Renom M, Webb B, Madhusudhan M, Eramian D, Shen M, Pieper U, Sali A. Comparative protein structure modeling with MODELLER. Current Protocols in Protein Science. 2007;50:2.9.9–2.9.30. doi: 10.1002/0471140864.ps0209s50. [DOI] [PubMed] [Google Scholar]
- 41.R Development Core Team. R: A language and environment for statistical computing. 2009. [Google Scholar]
- 42.Grant B, Rodrigues A, ElSawy K, McCammon J, Caves L. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2009;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 43.Dolinsky T, Czodrowski P, Li H, Nielsen J, Jensen J, Klebe G, Baker N. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–W525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sitkoff D, Sharp K, Honig B. Accurate calculation of hydration free energies using macroscopic solvent models. J Phys Chem. 1994;98:1978–88. [Google Scholar]
- 45.Baker N, Sept D, Joseph S, Holst M, McCammon J. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gorham R, Jr, Kieslich C, Nichols A, Sausman N, Foronda M, Morikis D. An evaluation of poisson-boltzmann electrostatic free energy calculations through comparison with experimental mutagenesis. Biopolymers. 2011;94:746–754. doi: 10.1002/bip.21644. [DOI] [PubMed] [Google Scholar]
- 47.Kieslich C, Morikis D, Yang J, Gunopulos D. Automated computational framework for the analysis of electrostatic similarities of proteins. Biotechnol Prog. 2011;27:316–325. doi: 10.1002/btpr.541. [DOI] [PubMed] [Google Scholar]
- 48.Gorham R, Jr, Kieslich C, Morikis D. Electrostatic clustering free energy calculations provide a foundation for protein design optimization. Ann Biomed Eng. 2011;39:1252–1263. doi: 10.1007/s10439-010-0226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kieslich C, Gorham R, Jr, Morikis D. Is the rigid-body assumption reasonable? Insights into the effects of dynamics on the electrostatic analysis of barnase-barstar. J Non-Cryst Solids. 2011;357:707–716. [Google Scholar]
- 50.Pettersen E, Goddard T, Huang C, Couch G, Greenblatt D, Meng E, Ferrin T. UCSF chimera - A visualization system for exploratory research and analysis. J Comp Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.