Abstract
This report provides practical recommendations for the design and execution of Multi-Center functional Magnetic Resonance Imaging (MC-fMRI) studies based on the collective experience of the Function Biomedical Informatics Research Network (FBIRN). The paper was inspired by many requests from the fMRI community to FBIRN group members for advice on how to conduct MC-fMRI studies. The introduction briefly discusses the advantages and complexities of MC-fMRI studies. Prerequisites for MC-fMRI studies are addressed before delving into the practical aspects of carefully and efficiently setting up a MC-fMRI study. Practical multi-site aspects include: (1) establishing and verifying scan parameters including scanner types and magnetic fields, (2) establishing and monitoring of a scanner quality program, (3) developing task paradigms and scan session documentation, (4) establishing clinical and scanner training to ensure consistency over time, (5) developing means for uploading, storing, and monitoring of imaging and other data, (6) the use of a traveling fMRI expert and (7) collectively analyzing imaging data and disseminating results. We conclude that when MC-fMRI studies are organized well with careful attention to unification of hardware, software and procedural aspects, the process can be a highly effective means for accessing a desired participant demographics while accelerating scientific discovery.
Keywords: Functional magnetic resonance imaging, fMRI, multi-center, multi-site, FIRST Biomedica Informatics Research Network, FBIRN
I. Introduction
Functional neuroimaging is now an indispensible tool in the study of human neuroscience as well as for the study of various neurological and psychiatric diseases. Multi-center fMRI (MC-fMRI) studies offer several advantages over single-center studies: the opportunity to increase the rate of accrual and the total number of subjects enrolled in the study, to increase the demographic diversity of the subject population(s), to include significant numbers of subjects from rare (homogeneous) subgroups within clinical populations and to pool expertise and opinions across multiple disciplines. Increasing the geographic diversity of subject enrollment may improve the generalizability of the results while increasing the number of subjects enrolled may greatly increase the statistical power of a study. Both may result in a deeper understanding of subtle cause-effect relationships, provided that unwanted variability is not introduced. In addition, lessons learned about stability and calibration in MC-fMRI studies may provide valuable insights for conducting longitudinal studies.
The current literature contains more than 40 peer-reviewed papers that focus on the topic of MC-fMRI. Early examples of MC-fMRI studies date to 1998, when Casey et al. (1) and Ojemann et al. (2) reported on studies where about 8 subjects were scanned at four and two sites, respectively. While good reproducibility was reported in both publications, and these findings suggested the promise of MC-fMRI, neither study addressed cross-site reliability using more formal methods such as intraclass correlation coefficients (3,4), and further, neither study addressed the complexities associated with including patient groups. Vlieger et al. (5) reported the first MC-fMRI reproducibility study. The same 12 healthy subjects performed a visual fMRI task for a total of three sessions on two 1.5T scanners from different vendors. This study found that the within-scanner reproducibility for one of the scanners was lower than the between-scanner reproducibility, suggesting the need for frequent monitoring of scanner quality assurance (QA) as well as for continued monitoring of scan session implementation in imaging studies.
Recently, many more MC-fMRI studies have been reported, including reproducibility studies (4,6–13) and of clinical populations such as stroke (14), multiple sclerosis (15–17,18) and first-episode (19) and chronic schizophrenia (20–26). In the latter studies, clinical populations and matched controls were recruited and imaged at multiple sites, and the data were then pooled into a single analysis. MC-fMRI studies are clearly becoming more common, and may become the norm in the next decade. With burgeoning deployment of MC-fMRI studies, questions of how to best carry out such enterprises have become more frequent.
The development and performance of any substantial fMRI study requires a diverse set of expertise in: sensory psychophysics, cognitive psychology, MRI physics, statistical methods and structural and functional neuroanatomy. In turn, each of these factors is profoundly affected by many other experimental and methodological factors that can be exacerbated in a multi-center study. Without careful planning and coordination across the participating centers there is a high likelihood of introducing undesirable inter-site variability or even error into the study data and analysis. Undesirable inter-site variability may be introduced through differences in the specific sequence parameters used to acquire the imaging data, but can also result from site differences in the ancillary components, e.g., differences in the stimulus presentation and response devices used during imaging, as well as differences in clinical and cognitive measures used to evaluate the subjects (which may be used as factors in the analysis). Such undesirable site differences may confound or obscure the desirable site differences such as variable demographics between sites. Inter-site variability can reduce the efficiency of a multi-center study compared to a single-center study with the same enrollment. However, for typical values of between-site variability, within-subject repeatability, and between-site differences in subject variation, the reduced efficiency of a multi-site study compared with a large single site study can be as little as 12% (Zhou, unpublished results). Such a small drop in efficiency is far outweighed by the increased statistical power associated with the large sample sizes that multi-center projects make possible.
There are many MR and non-MR related factors which, if properly controlled, can improve overall quality and/or reduce inter-site variability in a MC-fMRI study. Such factors include: rationalization and maintenance of scan parameters across scanners/vendors, monitoring of longitudinal stability in scanner performance, especially after hardware and software upgrades(27–29), initial and longitudinal reliability of the imaging measures within and between sites (4,6,7,30), and use of a secure Health Insurance Portability and Accountability Act (HIPAA) compliant database infrastructure that can store, retrieve, and monitor the variety of data collected (imaging, behavioral, clinical, genetic, demographic, etc.) (31). The purpose of this paper is to provide the reader with a cookbook containing the main ingredients required to set up and implement a successful MC-fMRI study, based on the collective experience of FBIRN. Although there are a number of papers that provide practical recommendations for the maximizing efficiency of functional imaging task design and the optimization of functional imaging parameters within a single center design, we believe this paper is the first to provide practical information for efficiently implementing a MC-fMRI study. While the recommendations embodied here may not apply to every large study, the goal is to provide generic, practical information to be used as a basic recipe that cuts across most multi-center fMRI studies and allows modifications based on study specifics.
In the following sections we introduce the important considerations required for planning and implementing a MC-fMRI study, describe the benefits of a traveling subjects study, and discuss the unique considerations that should go into MC-fMRI data analysis. Note that “multi-center” and “multi-site” terms are used interchangeably, since both have been used in the literature to refer to studies that collect data on multiple scanners at one or several institutions. Information referred to in this paper as Supplemental Material is found in the JMRI website XXX as well as the fBIRN site, http://www.birncommunity.org/resources/supplements.
II. Planning phase
The first step in any fMRI study is to develop an overall research design that addresses the study’s hypotheses. Apart from specifying the actual data collection and analysis methods, additional characteristics that must be considered include the number and diversity of participants and the desired rate of data accrual. If these latter constraints can not be satisfied in a single-site design, then an MC-fMRI study is required, and the number of sites that are needed will be dictated by these constraints. The MC-fMRI study will have fixed costs associated with setting up the infrastructure needed, as well as variable costs that depend on the number of sites. As the number of sites increases the cost/site decreases.
With the overall design in place, the next step in a MC-fMRI study is to develop an organizational structure that supports the overall study goals. FBIRN had both technical and scientific purposes: to develop and evaluate MC-fMRI calibration techniques and data management tools, and to test numerous hypotheses in schizophrenia. Many investigators across the FBIRN centers had hypotheses or models to test using the data collected as part of the FBIRN project, while others focused on the development of methods that optimize the reliability of inter-site data pooling. These two aspects shaped the structure necessary to achieve both types of goals.
To maximize the available expertise in each domain, FBIRN was structured around Working Groups (WGs), each of which was tasked with particular facets of the study preparation and implementation. Initially each working group had at least one representative from every center; in later phases of the study, as the focus turned from development and implementation to data acquisition, analysis and evaluation, researchers with the particular expertise or vested interest were most active in the appropriate WGs. In contrast, a study that is more centrally controlled may choose to have all study aspects decided at a single center, with researchers at the other sites implementing the set protocol and passing the data back to the central authority. In this more centralized case WGs may not be necessary. However, any MC-fMRI study will need to assemble sufficient expertise to thoroughly consider and evaluate the issues addressed by the FBIRN WGs, whether through a WG mechanism or through other methods of evaluation, review and discussion. In addition to the WGs, a steering committee should be appointed to handle all strategic issues including publication and resource allocation. A more thorough discussion of the FBIRN working groups can be found in Supplemental Material I.
Once the overall administrative structure of the study is established, several key components should be put in place.
A clear plan describing the study goals is needed to guide the project. The plan is usually based on either the research grant or clinical trial contract, but further details will ordinarily need to be discussed by the working groups and/or administrative structure.
A communication schedule involving regular conference calls with representation from every site should be established to begin the work of implementing the study plan.
Based on the study plan, Institutional Review Board applications must be drafted, redacted by participating centers as needed and submitted by each center as quickly as possible to avoid delays in launching the study. Keep in mind that Institutional Review Boards have different requirements (e.g., some sites require pregnancy tests prior to MRI scans, while others do not, etc.) and take different amounts of time to review applications. The IRB protocol must also include a HIPAA compliant data sharing agreement as well as an agreement for sharing of data with new collaborators or the wider research community as the study matures (see example of such a plan here: http://www.birncommunity.org/wp-content/uploads/2009/09/FBIRN_Data_Use_Agreement.pdf).
A list of scanner, stimulus delivery, and response collection hardware and software by site should be created to guide the study design and plan for possible hardware and software (license) purchases. Scanner information should include, but is not limited to: manufacturer, platform, software version, available coils, gradient system, whether a research agreement exists with the vendor, pulse sequence compiler, available software licenses, manufacturer representative, name and contacts of local coordinators, and available phantoms. With regard to the stimulus delivery and response systems this list should include, but is not limited to: computer hardware (PC, Mac), stimulus delivery software (visual, auditory, etc.), stimulus delivery equipment (goggles, projection, headphones, visual angle, intensity), response box (brand, number of response keys etc.), availability of and type of scanner trigger to be used to start the tasks (e.g., one trigger per 3D volume, one trigger per slice, during or after dummy scans, etc.), ability to measure physiology (e.g., respiration, heart rate, galvanic skin response, etc., and if so what equipment), and head motion restriction options.
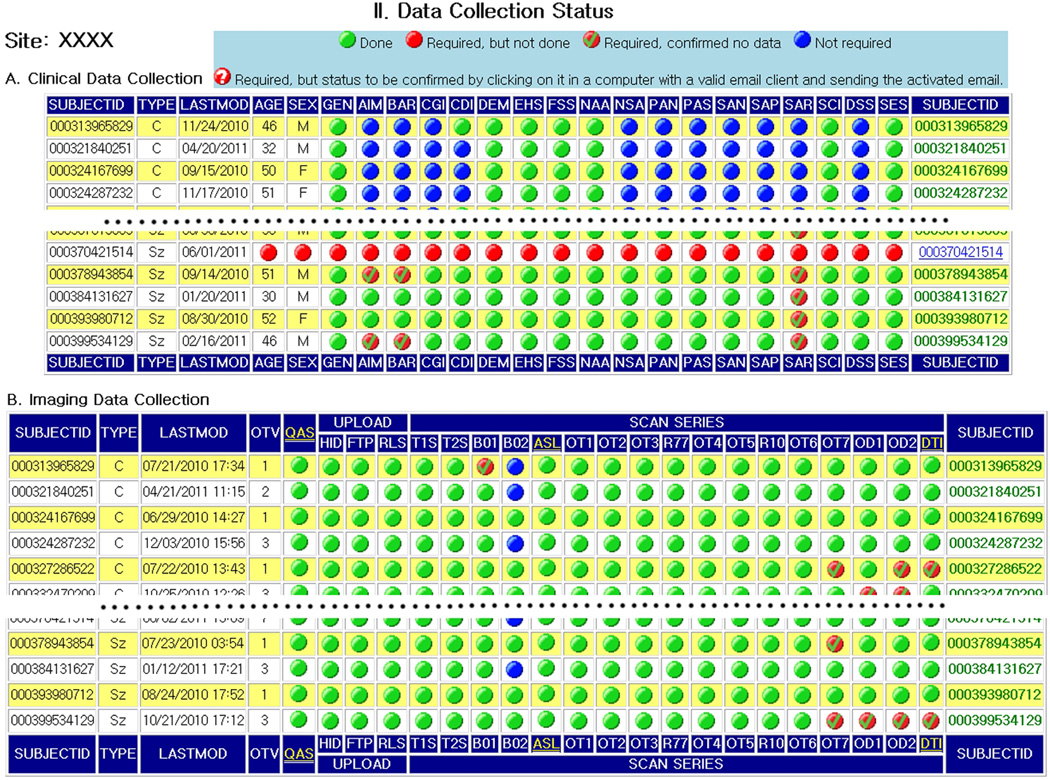
A central website should be established for disseminating study-related information, including scan parameters, inclusion and exclusion criteria, clinical scales, scan checklist, and progress reports for all data types being collected, such that participants from each site always have access to the most up-to-date study information. For example, Fig. 1 shows a screen capture of the wiki page for the most recent FBIRN study while Fig. 2 shows the FBIRN dashboard, a dynamic, web-accessible, project specific snapshot of what data have been collected by each participating site. Further details for the wiki and dashboard are found in Supplemental Material V.
A database infrastructure should be created for uploading imaging and behavioral data as well as other metadata. This technology should include the ability to monitor data quality as it is deposited so that data characteristics for each site are tracked from the beginning of the study.
Figure 1.
Example showing the FBIRN wiki page. This demonstrates an effective means for rapid sharing of critical protocol information between sites, as implemented in the most recent FBIRN study.
Figure 2.
Screen capture of FBIRN Data Tracking Dashboard, which shows status of data collection for each site.
III. Implementation of Study
The most crucial aspect of the MC-fMRI study is standardization of every component of the data acquisition across sites, particularly if multiple scanner vendors are utilized. Domains of standardization include the fMRI imaging protocol, on-going scanner image quality, session protocol implementation, stimulus presentation and behavioral hardware/software, data informatics and final site verification.
III.A Implementing the multi-Center fMRI imaging protocol
The overall goal in developing the multi-center imaging protocol is to minimize site-specific differences in the data acquisition across sites. This typically requires an iterative design process in which the project’s scientific goals and scanner capability differences must be jointly considered.
1. Field strength
One of the key initial choices is whether the study will require that all scanners be at the same magnetic field strength or whether a diversity of field strengths (e.g. 1.5T, 3T, and 7T) will be allowed. Since both the biophysics of the fMRI signal and the physics of the MR image acquisition depend on the magnitude of the static magnetic field(32,33), limiting scanners to the same field strength has the advantage of greatly minimizing systematic site differences that are difficult to calibrate out. However, field strength concerns may be outweighed if, for example, a site has a significant recruitment advantage but does not have access to a scanner with the chosen field strength. In this case calibration methods must be employed to minimize differences in BOLD contrast for the brain regions being interrogated (27,34). Invariably, however, the use of multiple field strengths may result in an overall site dependence in BOLD sensitivity even after the calibration corrections are applied to the imaging data. In addition, such methods may result in an overall performance that is set by the lowest sensitivity scanners.
2. Scan parameters
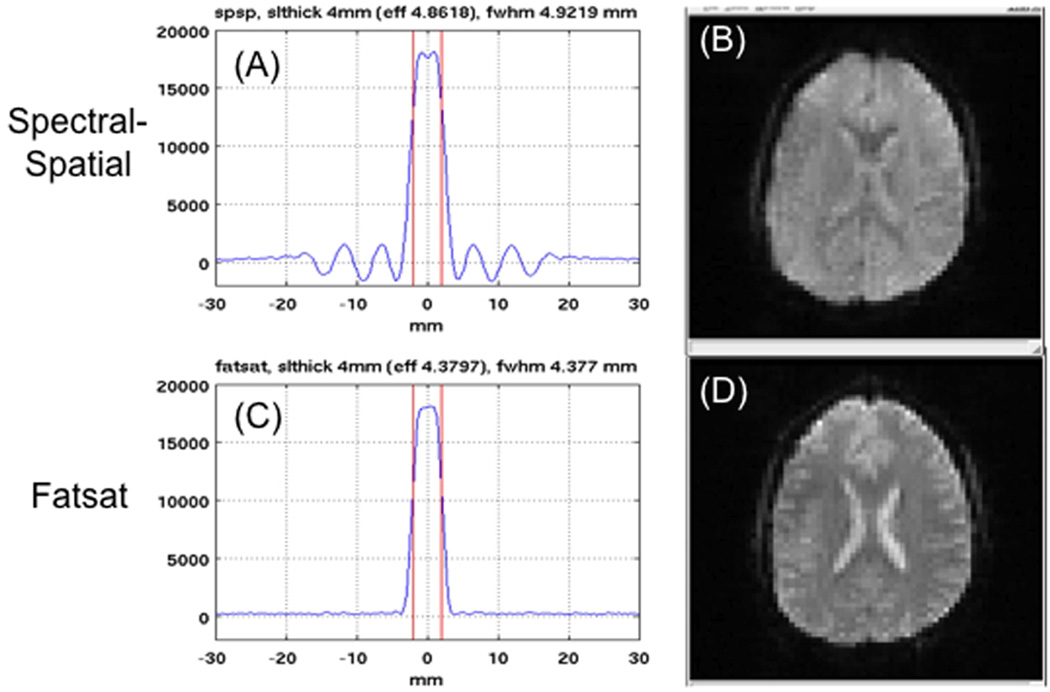
A critical step is specification of all MRI parameters, including the field of view, spatial resolution, temporal resolution and slice orientation. Most MC-fMRI studies will require whole brain coverage; however, trade-offs in coverage versus TR will lead to choices that need standardization, such as whether to cover the cerebellum. Almost all fMRI studies currently utilize some form of 2D slice acquisition, although more sophisticated acquisition schemes, such as those being developed for the Human Connectome Project (http://www.humanconnectomeproject.org/), are likely to find increased use in the near future. When using a 2D slice acquisition, the choice between interleaved and sequential slice acquisition order must be made. Interleaved acquisition can reduce inter-slice crosstalk, i.e. the interference between adjacent slices due to imperfect slice profiles, but leads to a larger time difference between the acquisitions of contiguous slices, which may impact motion correction. Sequential slice acquisition minimizes the adjacent-slice time difference at the potential cost of increased inter-slice crosstalk. Since slice profile will vary across slice excitation type and vendors (see Fig. 3, discussed below), the use of a small gap (e.g. 25% of slice thickness between slices) may be advised to reduce slice cross talk. The slice orientation (axial, coronal, sagittal, or oblique) and the direction of acquisition (e.g. inferior to superior) must also be standardized across sites. In specifying the slice acquisition, it is important to note that the method and nomenclature for specifying the orientation and slice acquisition order differs between vendors.
Figure 3.
Influence of RF excitation method on image quality, using (A,C) spectral-spatial (spsp) or (B,D) fatsat options. The measured slice profile with spsp demonstrates increased width and substantial sidelobes (A) that cause partial saturation of adjacent slices, and result in contrast changes in EPI images (B). Fatsat methods can provide better slice profiles (C) and diminished saturation effects (D). Since spsp is the default on GE scanners while fatsat is the default on Siemens, care must be used to standardize. (Single-slice profile measurements by T. Liu and K. Lu, UCSD; images acquired by B. Mueller, U. Minn. TR = 2000 ms, FA = 77°, 4 mm slice/1 mm gap prescribed).
The matrix size (number of readout points by number of phase encode lines) in conjunction with the field of view (FOV) determines the in-plane spatial resolution, which of course must be kept constant across sites. However it is also crucial to control the phase encode direction between sites to avoid differences in the axis of distortion, which is more pronounced in the phase encode direction. With the phase encode direction chosen in the anterior-posterior (A/P) dimension, off-resonance distortion is manifested as stretching or compression of the image in the A/P direction, which is more desirable than asymmetric distortion in the R/L direction. In addition, the direction of the phase encode traversal (e.g., from the bottom to top of k-space) must be the same across sites to avoid further differences in stretching vs. compression distortion.
The degree of geometric distortion arising from off-resonance depends on the time required to traverse k-space, which is the product of the number of phase encode lines and the echo spacing. Thus, for a specified number of phase-encode lines, it is important to ensure that the echo spacing is the same across sites.
The echo spacing is inversely proportional to the receiver bandwidth, but the specification of bandwidth is not always sufficient to uniquely specify the echo spacing. For example, on the GE systems, an additional parameter (CV taratio) must be modified to achieve the desired echo spacing. Some systems also have “forbidden zones”, which indicate ranges of echo spacing that are prohibited in order to avoid the excitation of mechanical resonances in the gradient coils. In addition, gradient strength and peripheral nerve stimulation considerations can set lower limits on the achievable echo spacing. Thus, it is critical to perform test scans to rationalize the effective bandwidth setting for each type of vendor’s scanner.
Additional methods for reducing distortion and speeding up the acquisition time are (a) the use of partial Fourier acquisitions and (b) the use of parallel imaging. Unfortunately, it can be difficult to standardize either method across sites because of differences in implementation across vendors. For example, the specification of TE can result in the use of partial Fourier under circumstances that differ across vendors. Sites should standardize on the use of either full or partial Fourier, and if partial Fourier is used, the number of phase encode lines acquired should be the same across sites. If parallel imaging is chosen, the acceleration factor and general approach (i.e. SENSE vs. GRAPPA) should be the same across sites.
In fMRI EPI acquisitions, the reduction of signal from fat in the scalp is important because this off-resonance signal will shift into the brain region and interfere with the accurate measurement of the BOLD response. There are two main approaches: (1) the use of fat suppression pulses prior to the RF slice excitation pulses that can excite both water and fat, and (2) the use of RF slice excitation pulses designed to excite only the water signal (typically denoted as spatial-spectral pulses or water excitation pulses). The use of fat suppression pulses has the advantage of providing relatively sharp slice profiles, but has the disadvantage of slightly increasing the time to acquire each slice and also of leaving some residual fat signal (depending on the specifics of the implementation). On the other hand, spatial-spectral pulses typically result in less fat signal at the cost of a broader slice profile with pronounced out-of-slice ripples, which cause partial saturation of adjacent slices, as illustrated in Fig. 3. Due to the difference in slice profiles, it is important to ensure that all sites use the same approach to dealing with the fat signal. As scanner vendors have a different default choices (e.g. Siemens uses fat suppression while GE uses a spatial-spectral pulse) and the method for choosing the other option is not always obvious, special attention must be paid to this standardization step.
Other parameter choices that must be kept constant include reconstruction image smoothing, the slice prescription method and prospective motion correction. The application of imaging smoothing during reconstruction, which increases SNR at a loss of in-plane resolution (34), is not applied by default on Siemens systems but the default on GE systems is to apply a Fermi k-space filter during reconstruction. The GE Fermi filter can be turned off at scan time, which allows for a better matching between the GE and Siemens acquisitions. Some vendors have an automatic slice prescription feature that may result in better reproducibility of scan plane alignment within and across sites than would result from manual alignment by each site’s technician. However, if automatic slice prescription is not available at all sites, this may introduce additional site/vendor effects and should therefore be avoided. Similarly, on-line prospective motion correction algorithms should not be employed unless uniformly available across all sites in a single-vendor study.
Supplemental Material III contains a summary of the FBIRN scan parameters and other protocol information.
III.B Implementing the MC-fMRI scanner quality assurance program
Shortly after determining the fMRI scan protocol the MC-fMRI study should design and implement a standard QA program that will be performed routinely on each scanner. During any substantial fMRI study, hardware and software changes are inevitable, particularly in studies that include multiple sites. Such changes can dramatically affect the ability to measure longitudinal change in subjects or differences among subjects. A QA program with scans that are performed and monitored regularly will help identify hardware and software changes that are likely to affect the data analysis and interpretation.
A poorly performing scanner will produce suboptimal data and will very likely introduce site variance. Minimizing image noise is important in fMRI because BOLD contrast is a small fraction of total signal and many imaging volumes must be acquired over time to track cognitive processes. MRI scanners must have excellent time series stability to measure accurately the small BOLD signal within the long time series. fMRI methods, however, are particularly demanding on the scanner hardware, using fast imaging methods such as EPI or spiral acquisitions that typically push the scanner hardware to the limits to achieve high temporal resolution. High temporal stability sustained over the course of a long study (up to two hours) is required to obtain quality data within a single subject, sustained stability is required to obtain consistent data throughout the course of an experiment, and consistent stability across centers is needed to allow data from the sites to be combined within a MC-fMRI study.
There is a diverse set of QA methods used on MRI scanners. Some sites may rely exclusively on vendor supplied QA programs; however in our experience vendor QA programs do not adequately test a scanner’s temporal stability. Other centers may rely on QA programs that focus on assessing issues such as ghosting, image uniformity, signal contrast and geometric distortion. While such QA programs are useful, they are not designed to test scanners under the demands of high temporal resolution imaging present during an fMRI study, and may not uncover problems that may only appear when scanners are pushed in the way fMRI experiments stress a system.
Several examples of scanner QA programs can be found in the literature (10,28–30,35). It is recommended that MC-fMRI studies design and implement a uniform QA program on every scanner within the study consortium. A complete description of the QA program used during the FBIRN MC-fMRI studies is beyond the scope of this paper but is found in Friedman et al.(30) and Greve, et al., (29) and important elements are introduced in Supplemental Material IV. Briefly, the FBIRN QA program has three parts: a custom phantom duplicated at each site, a specific fMRI protocol, and software to analyze, detect deviations and share the QA data and processed results with the consortium.
Scan parameters used in the QA acquisition should match those that will be used in the human data. Ideally, QA scans are run and analyzed on a daily basis, but if that is impractical they should at least be run contemporaneously with the subject scan acquisitions to provide a timely alert to changes in scanner performance. Baseline data from each scanner in the study should be acquired at least one month prior to human data collection to allow for a normative database of QA metrics to be established. The values of specific QA metrics will depend on the specific scan parameters used in the experiment (voxel size, slice select method, hardware, image reconstruction, etc.) in ways that are hard to predict without actual measurement. The normative database allows the study to establish minimum acceptable values for the QA metrics based on the needs, hardware, and imaging parameters of the specific study. Scanners that fail to meet these standards should be referred to the manufacturer for repair before human data collection occurs at that site. Once a scanner is considered to be at standard condition, QA acquisition and monitoring should continue throughout the lifetime of the experiment to ensure the instrument maintains acceptable performance. Ensuring each scanner is working at optimal performance is a simple yet important step towards avoiding unnecessary inter-site variability in the study.
III.C Protocol implementation issues
1. Distributing scan protocols
Efficient and well-controlled methods for distributing the protocols to each site must be used to avoid implementation errors. Each of the major vendors (Siemens, GE, and Philips) now has tools to export and import imaging protocols that contain most scan parameters in documentation files. Once the scan protocol is established it should be translated for each scanner type in the study, and the scanner-specific protocol files should be made available to all participating sites during the course of the study. At each site, the implementation should then be verified by generating a documentation file from the scanner that is sent to a central study curator, who can validate the local protocol’s accuracy. Note, however, that the documentation files may not contain all of the relevant scan protocol information, particularly if non-product features are being used (e.g. setting a scan parameter through a research CV). In addition, we recommend that the documentation file be checked against the scan parameters shown on the console, as occasional errors in file creation can occur. New scanner documentation files should be created and sent to the curator for protocol adherence evaluation after any hardware or software upgrades. In general, protocol files are needed for as many different hardware (scanner vendor, head coils) and software platforms as present in the study.
2. Consistent naming of protocols
The use of imported scan protocol files also ensures consistent scan naming (including capitalization) and parameters across sites, which helps minimize errors in multi-center data retrieval and analysis. While FBIRN’s image upload tools allow the mapping of locally-used scan naming schemes to study-consistent naming schemes before upload to the shared data repository, not all imaging databases have this feature. The ability to recognize scan type based on consistent information in the DICOM ProtocolName(0018,1030) and SeriesDescription(0008,103e) fields can therefore be very useful.
3. Scan protocol notes
Important reminders to the technologist can be added as comments in the protocol by adding notes to the scan sequence. Information may include: (1) slice prescription details (e.g., a note stating “AC-PC orientation, first two slices above the brain”); (2) special instructions (e.g. “No Angulation”); and (3) specification of Research and User CVs for GE scanners (e.g. setting the taratio CV to obtain the correct echo spacing). Such information should also be noted in the MR scan checklist. There are many details to remember while performing scans and providing reminders in multiple locations reduces error. Examples of FBIRN scanner protocol documentation files (pdf and edx files from Siemens, pdf and text files from GE) and an MR scan checklist can be downloaded from the supplemental material site (http://www.birncommunity.org/resources/supplements).
4. Left-Right orientation fiducial
Despite all good intentions, differences between image conversion and display tools can cause uncertainty in the left-right image orientation. We therefore recommend the use of a fiducial marker, e.g., vitamin E capsule or commercial equivalent, positioned in the same hemisphere for each subject at each site during each scan. Taping the marker to the specified temple appears to work well and is tolerated by most study participants, although it can also be attached inside the coil. This provides for a good solution to left-right issues, but all centers must make sure the fiducials show up in the scans (i.e., are placed within the FOV) and that the centers consistently place the fiducial on the correct side of all subjects scanned.
5. Protocol length and what to do when a study runs out of time
Careful consideration should be given when establishing the total length of the scanning session. Session duration cannot be based solely on the total scan time for the protocol or even a single site’s experience in running the study. Protocol duration should take into account vendor software differences, and particularly for fMRI studies of clinical populations, the time needed to get the participants in and out of the scanner (which may differ according to local participant differences), time to set up stimulus delivery equipment, time to provide instructions and practice the tasks prior to the start of scans, time needed for pre-scans (to determine the dynamic data range and to perform auto-shimming), and differential expertise at each center.
Even with the best preparation and the best-trained team of MR technicians, issues inevitably arise during scans that result in delays. Because sites may have different customs in such situations, it is important to establish and follow a uniform protocol for cases when the study runs out of time before the protocol is completed. Possibilities include: calling subjects back to finish the protocol, skipping scans, repeating the entire scan session using alternate task versions, etc. fMRI studies often suffer from the challenges associated with cognitive tasks; simply repeating an uncompleted scan may lead to questions of differential habituation, exposure, or over learning the task or stimuli, which may or may not be an issue in the context of the clinical sample in the broader study. If the scan order is not counterbalanced across subjects, ordering the scans from most to least important can help ensure that the most important data are collected on the majority of subjects.
6. Data collection priorities
In large-scale MC-fMRI studies, it is strongly recommended that subject recruitment be coordinated across sites, so that an equal number of patients and controls are recruited at each site and that data is acquired on all groups in parallel. For example, if all patients are scanned first and controls later, possibly undetected changes in acquisition characteristics over time can introduce biases in the resulting data.
III.D Multi-Center task paradigm stimulus presentation and behavioral data collection
The tasks and the stimulus presentation and behavioral data collection software/hardware that are employed must be capable of addressing the research goals across the entire study population and must be duplicated accurately across all sites. When designing the tasks by committee, one of the lessons learned by the FBIRN Cognitive WG is that extra care must be taken to discuss and make explicit all non-assumed and assumed aspects of task design prior to actual programming of the stimuli. This includes counterbalancing across sites and within subject scans, number of stimuli per condition, number of targets and foils per condition, etc.
Behavioral task paradigms are a critical component of any fMRI study and present special challenges for multi-center studies. Whereas there are relatively few types of MRI scanners, every site in a multi-center fMRI study is likely to have a different combination of stimulus computers, video and audio stimulus transducers, and response devices. Each site will also tend to have their preferred stimulus software packages and personnel who are accustomed to training and preparing subjects in particular ways. Every subject also introduces behavioral variability in the way they adapt to the scanner environment and how they perform the tasks. Standardizing the behavioral environment and the task paradigm experience across subjects at every site therefore requires special experimental design consideration. Moreover, because there are so many factors that can potentially affect subject behavior, it is especially important in multi-center studies to ensure that as much as possible of what happens during each scan is accurately recorded.
Intrinsic and random subject variability should be carefully considered. Subjects can present differently on different days or even at different times during the same day. Natural variation of the condition under study or medication-induced variations can lead to inconsistent results in different scans. Issues that need to be routinely considered and should be included in a pre-scan clinical QA are: time of the day of clinical/behavioral assessments and time of the scan, time interval between clinical/behavioral assessments and scan, food intake, sleep and medications, including over the counter drugs, herbals, supplements and vitamins. There should be clear guidelines about the use (dose and timing) of any medications, tobacco, alcohol and caffeine prior to scanning. For at-risk populations, a rapid drug test (with results available prior to scanning) can eliminate problem subjects to reduce variability.
In the early phases of the FBIRN experience, many of the most common and persistent problems we encountered were associated with unanticipated procedural, behavioral, or configurational variations at different sites or for different sessions, which were not fully documented in the acquired data. In the later phases of the study we implemented a number of design changes in our behavioral software and procedures, which significantly improved both consistency and data quality across all sites. We also developed scripts that study personnel read to the subjects as well as a video/slide presentation, and we had collaborative training sessions for all of the experimenters (and provided the same scanning checklist and reference documents at all sites) to further minimize potential differences in participant treatment/interactions across sites. Please see Supplemental Material II for expanded descriptions of FBIRN experiences in dealing with issues described below.
The specific issues that need to be addressed in collaborative multi-center studies are:
The behavioral paradigm software needs to be flexible enough to cope with different scanner trigger interfaces, stimulus environments, and recording devices. These include either TTL pulses or serial input devices for scanner synchronization, video projectors or goggles with screens of different sizes, and a variety of different button box devices. Sites are typically reluctant to change hardware for a new study, so the study needs to be able to accommodate the equipment that is available.
Accurate synchronization of scanner and task timing is a recurrent problem because scanners differ in when they send out synchronization pulses relative to image acquisition. For example, Siemens scanners send a trigger signal after an initial set of discarded TR cycles, whereas GE scanners trigger for every TR, whether discarded or not. Any study that involves different types of scanners, therefore, needs to provide options to properly identify the timing of the first real MR image. In the later FBIRN studies all trigger pulses were recorded throughout each scan to verify the correct start and end of every acquisition series.
Accurate recording of button press responses is important, particularly if response time is used as a behavioral measure. Ensuring that buttons work and every press is recorded is fairly straightforward. Small differences in device responsiveness are likely to be more difficult to assess, so balancing experimental conditions within site is probably the safest way to avoid a site bias in response time results.
If recording cardiac, respiration, galvanic skin response or similar psychometric data, it is important to standardize across sites. The FBIRN study found that the quality of vendor-supplied cardiac and respiration data was inconsistent and implemented independent recording instruments (see Supplemental Material II).
Variable data quality or missing behavioral data was another recurrent problem in early stages of our multi-center study. By the time data were reviewed at central collation sites it was difficult or impossible to resolve problems associated with poor task performance, mislabeled data files, or missing or questionable data values. Behavioral paradigm software and procedures can reduce such problems by enabling run-time quality assessment, providing highly automated standardized procedures, checking important parameters for invalid or missing values, and recording detailed documentation and log files of everything that happens during a session. Implementing run- time quality assessments in the FBIRN study was very valuable as it allowed for corrections in subject performance (e.g. performing at chance levels, use of wrong buttons) or hardware (e.g., loose connections to be addressed and remedied during the scanning).
Finally, the logistics of distributing, installing, and updating paradigm software at multiple sites can lead to another source of problems. Ideally, a single package of task software with stimulus files could be distributed once and used for an entire study, but in practice most software requires occasional updates to fix problems or add features. Recording which version of each software component was used for each subject at each site is critical for properly interpreting the data. The FBIRN used a simple version synchronization program to make software updates easy to install, in which every component is labeled with a version number and data files are tagged with the software version that created them.
III.E Informatics: data uploading, storing, and monitoring
Given the volume of data collected and importance of standardized processing, the informatics system requires careful planning. While multi-site imaging projects significantly increase the speed of data collection, they add complexity to the data management environment that is responsible for ensuring accurate representations of the data and timely access for participating sites. Methods for data upload and download, conversion, scoring, and quality assurance must be in place prior to commencing the study to avoid massive data loss and/or data collection errors. In federated data management systems where each site maintains their own storage and database, there are more opportunities to make mistakes. The FBIRN Neuroinformatics working group has developed FIRE (Federated Informatics Research Environment), an integrated suite of tools and methods to address data collection, storage, tracking, and access in a federated research environment (36). FIRE consists of tools for data management and QA; namely, the Human Imaging Database (HID) (37), the XML-based Clinical and Experimental Data Exchange schema (XCEDE) (38), data publication and retrieval tools, data validation tools, and data integration and curation tools. The suite was designed in the context of neuroimaging studies but is generally applicable to multi-site data collection and federated data management. While the FIRE for FBIRN is set up as a federation, the tools can also function in the context of a data warehouse in which data are stored at a single location and access is controlled by a single site. All of FBIRN’s tools are based on freeware and available to the research community (http://www.nitrc.org/).
1. Environment
A supporting infrastructure is important for multi-site federated projects because it relieves scientists and informaticians from the burden of maintaining generic data access and movement services and instead allows them to focus on domain specific tool building. The BIRN coordinating center provides a number of capabilities in support of multisite studies(39). The FBIRN FIRE environment builds domain specific informatics tools on top of the core data access and movement functionality supplied by the coordinating center. Please see Supplemental Material V for further details.
2. Data publication workflow
Each site in the collaboratory is responsible for data publication to their local database, ideally within 24 hours of data acquisition. Data publication, in this context, refers to making data collected at a particular site available to other sites in the project consortium by publishing it to the local node in the federation. The within-site workflow consists of inputting the clinical assessment data into the database either via web-accessible data entry forms or via Tablet PCs that connect to the database via the internet, transferring the imaging data from the scanner to a local staging area, preparing images for publication, and uploading the images into the distributed file system. The data collected for fMRI experiments consist of not only the imaging data but also behavioral and/or physiological data collected during the scans. Furthermore, some parameters required for fMRI data analysis, such as slice acquisition order, are often not contained within the standard DICOM image header. A tool is needed to augment the image headers with the additional metadata and to organize the imaging data with the behavioral and physiological data in a consistent manner across sites, without the tight control over data that is often in place when using a centralized data warehouse. Additionally, to support automated computation and quality control measures over the distributed data, a consistent and structured “global” data storage hierarchy is essential; but, mapping the local data storage hierarchy to the global one is non-trivial due to site heterogeneity.
To address each of these requirements, tools must be developed that each site uses to publish their data to the global storage facility, ideally automatically and with sufficient provenance to guarantee data integrity. For example, the FBIRN consortium has defined a global data storage hierarchy, defined an XML format for augmenting the dataset with additional metadata and mapping between local storage elements and global storage elements, and implemented a data publication (upload) tool. The global data storage hierarchy organizes the acquired data on the file system such that tools for computation and quality assurance can find datasets and parse relevant subject, project, visit, and series identifiers from the directory hierarchy itself while also making the hierarchy readable by humans. In practice we have found that both data access through the web-accessible database (including autodownloads) and direct access to the images on the file system are important to researchers, and therefore having a human readable component to the directory names is prudent. Once the data have been uploaded, they are immediately available to participating sites and to automated QA and data validation tools.
3. Data validation
As described previously, distributing scanner protocols electronically between sites can reduce implementation errors. However, it is as important to verify after the fact that a particular scanning session was acquired correctly. The FBIRN consortium has implemented several levels of protocol validation in the automated upload infrastructure. These include the use of an automated upload template file, project-specific Schematron-based (http://www.schematron.com/) validation of acquisition and other parameters and other adaptive code that can de-identify images, handle behavioral data, etc. Please see Supplemental Material V for further details.
4. Data monitoring
Because the data are physically distributed across all participating sites, automated tools are vitally important for each aspect of monitoring and QA of all data after upload to the local data storage system. FBIRN developed a QA tool that automatically checks the image and other data and generates a report that is available for data curators. In addition, a Dashboard (Fig. 2) has been implemented that provides an up-to-date study status overview showing the percent of the target sample collected by site and the state of the data in the federation. The dashboard also links to QA pages and has been an invaluable tool for integration and coordination among sites. (See Supplemental Material V.)
III.F Final site verification: The role of the visiting fMRI expert in site standardization
The use of fMRI entails the ability to present stimuli and collect responses from the subject in the MRI scanner while maintaining careful control over the timing of the scanner and cognitive task machinery, as noted in III.D. While the acquisition or analysis software can be modified to do automatic error-checking as in the structural imaging ADNI study (40), some characteristics of a center’s scanning methods are only identified when a knowledgeable researcher participates in the scan. Examples:
Visual presentation. FMRI visual presentation can be via goggles or via projector and mirror present from either the head or the foot of the subject. Are the brightness, sharpness, contrast and solid angle of the visual stimuli sufficiently similar across sites?
Is the response device and positioning equally comfortable across sites or are subjects more likely to become uncomfortable at specific sites?
If the study uses auditory stimuli, are they equally easy to hear over the scanner noise across sites?
Do all sites understand the task instructions and are they conveying the instructions to the subjects in an identical manner?
Are sites training the subjects with a rigorous adherence to the task instructions?
Are sites using similar head restraints and physiological measurement techniques?
Are sites executing the protocol in the correct manner (e.g., slice prescription, scan order, pausing in specific spots if necessary, etc.)?
To address these questions, FBIRN recommends sending a traveling fMRI expert to each center once human test data have been collected that passes all acceptance criteria but prior to collection of participant data. That is, the traveling expert visits the site only after these steps have been completed and confirmed:
The phantom QA program is functional, and the scanner is shown to have acceptably stable performance.
The final protocol parameters are set up correctly at the site.
Local test subject(s) have been successfully scanned at the site without any major problems.
Imaging data are artifact-free and show the expected activation patterns.
The ancillary equipment (stimulation computer, button box, visual display system, sound system, physiological monitoring equipment, etc) is functional and producing good data.
The clinical and cognitive assessment batteries are defined and the local staff is trained to administer them.
The data-handling infrastructure is in place and has been successfully tested.
The role of the visiting fMRI expert is to provide the final piece in the site standardization and certification process. Many potential sources of intersite variability such as subject task training, subject comfort in the scanner, head restraint, and stimulus presentation differences are difficult or impossible to standardize remotely. In our experience the best way to reduce these sources of inter-site variability is to send the same highly experienced MRI expert to every site to provide training as needed, answer remaining protocol questions, observe the set up of the imaging acquisition, and to undergo the imaging protocol at each site to experience the tasks as the study participants will experience them. The resulting imaging data should then be uploaded by the site to the central data repository, where the images are processed and evaluated for site differences. Please see Supplemental Material VI for further details.
IV. Traveling Subjects Study
As a final test of site standardization effectiveness, an MC-fMRI consortium may decide to perform a traveling subjects study, wherein a fixed cohort of subjects visits each site and performs the complete MRI protocol on every scanner. The first of three aims of performing such a study is to analyze imaging data from each site to identify sites inconsistently implementing the standardization and to determine the level of residual site variance after successful standardization. Assessing inconsistent implementation of study methods, identifying sites producing inadequate imaging data, and determining the level of residual variance after standardization involve differences in the types of information to be acquired, in the sample sizes needed and in the cost. Some multi center fMRI studies will not choose to perform a traveling subject study that determines precisely between-site reliability because of travel costs and because of the tremendous logistical challenges associated with subject transportation and lodging, the variability in scanner availability, etc. However, even multi-center consortia not performing a formal reliability study should consider the traveling subject design to assess whether sites are consistently implementing standardized study procedures recommended by the visiting fMRI expert or to identify sites producing inadequate imaging data prior to the initiation of the main study.
Participant self-report is a simple yet informative approach to assess inconsistent implementation of standardized study procedures. For this purpose, research assistants or other individuals experienced with fMRI methods can be utilized. Sample sizes required to assess variation in the standardized study procedures may be small (2 to 5 individuals), and not all subjects need to go to all sites though some overlap of site experiences would be useful for the different traveling cohorts. The small sample sizes required and the use of geographically determined cohorts can help contain costs.
A second aim of traveling subjects is to provide a warning sign that human imaging data at a site is not adequate. Often the warning signal is apparent in basic descriptive statistics from coarse regions of interest, such as cortical perfusion or dorsolateral prefrontal cortex percent signal change. When identifying sites producing inadequate data, traveling subject studies take advantage of within-subject designs, which control for possible cohort differences that could confound designs using site-specific subjects to compare the adequacy of imaging data across sites. Because a consistent warning sign of a problem is sought rather than a precise estimate of a sample statistic, sample sizes can be kept small when probing for an outlier. Under the simple assumption, for example, that sites produce equivalent effects and that site deviations are due to chance, the probability that all of six subjects produced their most deviant value at the same site in a four site study is about 0.001. Less stringent outlier patterns, such as four deviant values at one site, two at another and none at the remaining two sites are also uncommon under the assumptions.
The third aim of a traveling subjects study is to compute between-site reliability estimates at a reasonable level of precision. This aim requires larger sample sizes than the other two aims and is the most expensive, though an adequately powered between-site reliability study obtains all of the information described in the two aims above, as well as quantitative information about the impact of site differences on data variability when analyzing data combined from the different study centers. In some cases, between-site reliability studies provide information about the magnitude of site by subject variation – an unwanted source of variability. Sample sizes for such studies will need to be sufficiently large to produce useful estimates of reliability statistics, such as the intraclass correlation (ICC). Several factors that determine the width of an ICC confidence interval include the number of subjects studied, the number of magnet sites, the anticipated ICC value, the ratio of person variance to error variance, the ratio of site variance to error variance, and the confidence level (usually denoted as 1-α) (41). If the impact of these factors on the width of the confidence interval for the estimated ICC is not carefully considered, the confidence interval surrounding the observed ICC might include zero. Using simulations, Doros and Lew (41) show that the confidence interval for a commonly used form of the ICC appreciably narrows as the number of raters (sites in our example) increases from three to five. They also found that the number of subjects should range from 16 to 32 to obtain 0.95 confidence interval widths in the 0.3 to 0.4 range for intraclass correlations between 0.60 and 0.70 over five magnet sites. The results of the Doros and Lew analysis generally match the experience of the FBIRN group. The FBIRN traveling-subjects study of 18 individuals revealed many spatially coherent clusters of brain activation where between-site reliability equaled or exceeded 0.60 (4). In addition to the cost and logistical challenges of traveling subjects designs, these studies need to consider methods to mitigate variance related to the repeated use of a behavioral task. Training subjects until they reach a plateau of performance on the activation task and the use of alternate forms can reduce the practice effects associated with repeated testing. Appropriate spacing study sessions to prevent burnout while sustaining study interest should be considered. In the fBIRN traveling subjects study the session intervals typically ranged from one to four weeks. Any remaining adverse repetition effects can be minimized by randomly sequencing or permuting site order.
If the between-site variance is found to be small compared to the between subject variance, subsequent large-scale study data can be confidently combined. If the traveling subject study shows a substantial amount of site or site by subject variation, calibration methods to account for the inter-site variability can be developed from traveling subject data and applied to the main study data1,2,5. Although the quality of information obtained to derive calibration values or between-site reliability estimates benefits from the within-subjects design made possible by traveling subjects, the subject sampling strategy for the two types of studies differ. Calibration studies aim to precisely measure between-magnet system variation. The magnet systems, therefore, are the object of study and other potential sources of variation, including between-subject variation, should be minimized. Between-site reliability studies, on the other hand, examine consistency in the ordering and magnitude of between subject differences across sites(3). Because subjects are the object of investigation in between-site reliability studies, subject variation should not be constrained by the sampling procedure. When magnets are the object of study, as in a calibration effort, confidence intervals formed about magnet site statistics will be reduced if study subjects are as similar as possible. When subjects are the object of study, reliability coefficients will be artificially attenuated when subjects are similar. When reliability estimates are the aim of the traveling subjects study, subject variation should reflect the anticipated subject variation in the subsequent large-scale study, possibly necessitating that clinical subjects are included as part of the reliability study, greatly increasing the complexity of the study.
In practice, to study reliability the same tasks that are to be employed in the actual multi-site study must be used in the traveling subjects study, since repeatability of task performance depends on the task. For calibration, one approach is to obtain fMRI data on a very simple task that requires minimal cognitive processing, such as breath holding (42) or a sensorimotor task (7), with the hypothesis that motivated participants will provide higher levels of reproducibility with such a task than with the study’s cognitive tasks. With this assumption, the acquired data may then be used to normalize between-site differences in several ways (27,34).
V. Multi-Center Functional Imaging Analysis
In the FBIRN Image Processing Stream (FIPS), we have used FSL FEAT (www.fmrib.ox.ac.uk/fsl) to perform all first-level and second-level (i.e., cross-run) analyses. These analyses in and of themselves do not require any special modifications for multi-site studies. Briefly, the data are motion corrected, slice-time corrected, and spatially smoothed. The time series is fit to a model that includes hemodynamic responses and nuisance variables to estimate the amplitudes and variances of the responses to the conditions under study. The functional images are registered to the MNI152 space. This registration is applied to the contrasts to bring all data into alignment across all subjects. The individual runs within a subject are combined using a fixed-effects model, using FEAT to estimate the contrast amplitudes for each subject. These estimates and their variances are passed up to the next higher level where they are correlated with demographic or other variables to draw inferences at a group level.
When combining data across multiple sites, there are several steps that need special attention. In our phase 1 study(34) we discovered that the GE scanners were spatially smoothing the data through the use of a Fermi k-space filter. This made the GE data much smoother than the data from Siemens machines. Friedman et al. (34) introduced a method whereby the data were smoothed to a target level of smoothness rather than simply applying a smoothing kernel of a given size. This allowed all the data from the different sites to have the same level of smoothness. This is important because spatial smoothing can have such a drastic effect on the outcome of fMRI analysis (43). In general, this step should not need to be applied if the individual sites are careful to assure that no filters are turned on during acquisition. If this technique is needed because data smoothness is found to vary between sites, then it is recommended that different levels of smoothing be used between sites but that the level be fixed within a site.
In general, subject movement issues and corrective approaches for multi-site studies are similar to those in single-site studies (for a general discussion see Poldrack, et al., pp 43–49) (44). However, it is also important to protect against between-site differences in subject movement. Such differences may result from the use of different types of head restraints (e.g., bite bar, vacuum pillows, or other), positioning in the scanner (use of pillows under the knees, padding of arms, etc.) and differences in subject demographics. Standardizing head restraints and positioning at each site and employing a balanced design with an equal number of patients and controls can help protect against confounding effects of between-site differences in movement. Nevertheless, it is wise to verify that there is no effect of site on movement parameters. If an interaction is found, the consortium must decide on data exclusion or other corrective measures.
When the data are analyzed at the higher level, two important effects must be taken into account. First, the amplitudes of the hemodynamic responses can be systematically different (45) between sites due to different B0 distortion and intensity inhomogeneity. This can be compensated by performing B0 and intensity correction and by including site as a fixed effect in the higher-level GLM. B0 distortion correction requires a B0 map, which is not readily available on GE platforms (at the time of this manuscript). Second, the different sites can have very different levels of noise(29), which can cause heteroscedasticity at higher-level analyses and cause p-values to be underestimated. This can be compensated for by performing a mixed effects analysis that includes estimates of the variance from the lower level (46).
No matter how much care investigators take to minimize between-site variation, the data collection center should be included as an explanatory variable in the higher statistical model, even for data with excellent between-site reliability. The site effect not only accounts for variation that otherwise would be incorporated into the error term, it allows for the study of the interaction of site with other explanatory variables(47). When including site in the higher-level design, investigators must decide whether to treat site as a fixed or random effect or something in between(47). Fixed effects models assume that information about activation magnitude at one site conveys no information about activation magnitude at other sites. Such models can be used easily with any number of sites. For a random effects model, which assumes sites are being sampled from the same population, data from all sites are used to estimate a common site population variance (47). The random effects approach is most appropriate when the number of sites is moderate to large (i.e., six or more) for reasons similar to those discussed above in the context of the traveling-subjects studies. A moderate or large number of sites allows for the reliable estimation of between site variance. Hierarchical Bayesian models permit models that use random effects parameters to estimate the site population variance while also drawing inferences about individual site effects thus providing a sort of intermediate approach(48). Because of their flexibility, the FBIRN group recommends that hierarchical Bayesian models be given primary consideration when analyzing multisite fMRI data (Zhou, unpublished results). One present limitation in applying hierarchical Bayesian models is that such models are not integrated in standard imaging software (i.e., SPM or FSL). Applications to date have applied Bayesian approaches off-line (using Matlab or R) to data output from imaging software, either voxel-level summary (49) or region of interest summary statistics (Zhou, unpublished results).
VI. Discussion
Multi-center fMRI studies are becoming the method of choice for investigating neural function in both healthy and clinical populations of large size, allowing for unprecedented sample sizes, data collection rates, and clinical, cultural and geographic diversity. Recent MC-fMRI reproducibility studies (4,6–9,12,13,50) have proven that utilizing careful experimental design considerations can accrue significant benefits in a multi-center study: e.g., between-site variability can be reduced to as little as 10% of the between subject variability. Results from several large MC-fMRI studies of clinical populations have also been reported recently, yielding group comparisons of heretofore-unequalled size and allowing for new insight into a variety of illnesses. One large MC-fMRI study used a finger tapping task to investigate the abnormal connectivity of the sensorimotor network in patients with Multiple Sclerosis (15–18). Another large MC-fMRI study examined the neural correlates of working memory dysfunction in first-episode schizophrenia (19). Chronic schizophrenia has been investigated by two large MC-fMRI collaborations, FBIRN (http://www.birncommunity.org) and MIND (http://themindinstitute.org), allowing both groups to accumulate fMRI data on substantial population sizes. Analysis of the large data samples acquired by the MIND (21,51,52) and FBIRN (22,24–26,53) consortia have lead to significant advancement in our understanding of the differences in the brains of persons with schizophrenia compared to those of healthy controls. Kim et al. (23) have extended the MC-fMRI paradigm by combining imaging data from the FBIRN and MIND studies into a single mega-analysis that included 115 participants with schizophrenia and 130 controls.
As MC-fMRI becomes more common, resources developed through such studies are increasing. Van Horn and Toga (54) recently published a comprehensive discussion on multi-center neuroimaging trial design. Multiple authors have published recommendations for scanner QA methods for MC-fMRI studies (28–30). Friedman et al. (7), Gountouna et al. (8), Yendiki, et al. (13), Brown, et al. (4) and others have published descriptions of analysis considerations specific to MC-fMRI studies. Finally, the FBIRN website (http://www.birncommunity.org) contains draft “best practice” recommendations along with freely available resources developed specifically to make multicenter fMRI studies easier to execute. The resources include tools to track, store, share, and analyze multi-site fMRI data. FBIRN also has made functional imaging, clinical and cognitive data from several MC-fMRI studies performed by the network available for download on its website.
In addition to addressing basic neuroscience questions and clinical cross-sectional comparisons, well-designed multi-center longitudinal functional imaging studies are also uniquely positioned to efficiently probe cognitive changes over time (e.g., development, progression, pre/post disease onset or treatment, recovery) given the ability to collect data on large samples in a short period of time. However, detection of longitudinal changes in imaging measures requires sufficiently reliable within-as well as between-sites measures and careful control of nuisance variables that may change with time. As with other MC-fMRI studies, standardization at all levels of the longitudinal study is essential.
In the initial phases of the FBIRN project, it was hypothesized that calibration methods could be used to mitigate site differences in magnetic field strength and other hardware choices (such as coil type) and acquisition parameter choices (such as EPI vs. Spiral k-space trajectories). While methods were developed that indeed diminish intersite differences in BOLD sensitivity (27,34), the overall performance of the consortium was thereby determined by the characteristics of the weakest site. Therefore, as an overall take-home message we strongly recommend the strategy later adopted of unifying every hardware, software and procedural variable across sites to the greatest extent possible, rather than depending on post-acquisition procedures to correct for inherent site differences. Nevertheless, the subtlety of the effects being investigated and demographic considerations regarding inclusion of sites with non-standard configuration may make calibration an important step.
In this paper we have provided practical recommendations and pointed to useful tools for the design and execution of Multi-Center functional Magnetic Resonance Imaging (MC-fMRI) studies, based on the collective experience of the function Biomedical Informatics Research Network (FBIRN). Best practice recommendations are still evolving because the technical methods are improving, scanner hardware and software are changing, and our basic understanding of cognitive neuroscience is increasing. The trajectory is clear: cognitive neuroscience is evolving toward more and larger multi-center fMRI studies, given their relatively fast returns on the investment and ability to speed up scientific discovery. Future fMRI studies will include dozens of imaging centers (as in the ADNI structural study (55)) and may enroll many thousands of subjects. Such studies will be successful only if critical attention is given to the details of organization, planning and inter-site execution of all aspects.
Multi-site functional imaging studies are a complex enterprise. Together with careful planning, a good management team, thoughtful task selection, robust quality assurance procedures, the necessary domain expertise, and cooperation from all contributing sites, the recommendations put forward in this article should provide a good basis for MC-fMRI collaborators to efficiently plan and execute their study using methods that minimize undesirable inter-site variability in results.
Supplementary Material
Acknowledgements
The authors are grateful to Liv McMillan (UC Irvine), who served initially as research coordinator, and then as senior research coordinator for the FBIRN following Dr. J.A. Turner's departure as Project Manager in 2010. Kun Lu (UCSD) provided assistance with slice profile measurements. The inclusion of the Function Biomedical Informatics Research Network as an author represents the efforts of many otherwise unlisted researchers who also contributed over the years to the conception, design, and/or implementation of the study.
Grant Support: U24-RR025736, U24-RR021992, U24-RR021760, as well as individual grants to co-investigators: P41-RR009784 (GHG), R01NS051661 and R01MH084796 (TTL), RO1 MH067080-01A2 (CGW) and The Department of Veterans Affairs Research Enhancement Award Program and VA Medical Center of Excellence (CGW).
References
- 1.Casey BJ, Cohen JD, O'Craven K, Davidson RJ, Irwin W, Nelson CA, Noll DC, Hu X, Lowe MJ, Rosen BR, Truwitt CL, Turski PA. Reproducibility of fMRI results across four institutions using a spatial working memory task. Neuroimage. 1998;8(3):249–261. doi: 10.1006/nimg.1998.0360. [DOI] [PubMed] [Google Scholar]
- 2.Ojemann JG, Buckner RL, Akbudak E, Snyder AZ, Ollinger JM, McKinstry RC, Rosen BR, Petersen SE, Raichle ME, Conturo TE. Functional MRI studies of word-stem completion: reliability across laboratories and comparison to blood flow imaging with PET. Hum Brain Mapp. 1998;6(4):203–215. doi: 10.1002/(SICI)1097-0193(1998)6:4<203::AID-HBM2>3.0.CO;2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barch D, Mathalon D. Using brain imaging measures in studies of procognitive pharmacologic agents in schizophrenia: psychometric and quality assurance considerations. Biol Psychiatry. 2011;70(1):13–18. doi: 10.1016/j.biopsych.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown GG, Mathalon DH, Stern H, Ford J, Mueller B, Greve DN, McCarthy G, Voyvodic J, Glover G, Diaz M, Yetter E, Ozyurt IB, Jorgensen KW, Wible CG, Turner JA, Thompson WK, Potkin SG. Multisite reliability of cognitive BOLD data. Neuroimage. 2011;54(3):2163–2175. doi: 10.1016/j.neuroimage.2010.09.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vlieger EJ, Lavini C, Majoie CB, den Heeten GJ. Reproducibility of functional MR imaging results using two different MR systems. AJNR Am J Neuroradiol. 2003;24(4):652–657. [PMC free article] [PubMed] [Google Scholar]
- 6.Costafreda SG, Brammer MJ, Vencio RZ, Mourao ML, Portela LA, de Castro CC, Giampietro VP, Amaro E., Jr Multisite fMRI reproducibility of a motor task using identical MR systems. J Magn Reson Imaging. 2007;26(4):1122–1126. doi: 10.1002/jmri.21118. [DOI] [PubMed] [Google Scholar]
- 7.Friedman L, Stern H, Brown GG, Mathalon DH, Turner J, Glover GH, Gollub RL, Lauriello J, Lim KO, Cannon T, Greve DN, Bockholt HJ, Belger A, Mueller B, Doty MJ, He J, Wells W, Smyth P, Pieper S, Kim S, Kubicki M, Vangel M, Potkin SG. Test-retest and between-site reliability in a multicenter fMRI study. Hum Brain Mapp. 2008;29(8):958–972. doi: 10.1002/hbm.20440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gountouna VE, Job DE, McIntosh AM, Moorhead TW, Lymer GK, Whalley HC, Hall J, Waiter GD, Brennan D, McGonigle DJ, Ahearn TS, Cavanagh J, Condon B, Hadley DM, Marshall I, Murray AD, Steele JD, Wardlaw JM, Lawrie SM. Functional Magnetic Resonance Imaging (fMRI) reproducibility and variance components across visits and scanning sites with a finger tapping task. Neuroimage. 2010;49(1):552–560. doi: 10.1016/j.neuroimage.2009.07.026. [DOI] [PubMed] [Google Scholar]
- 9.Suckling J, Ohlssen D, Andrew C, Johnson G, Williams SC, Graves M, Chen CH, Spiegelhalter D, Bullmore E. Components of variance in a multicentre functional MRI study and implications for calculation of statistical power. Hum Brain Mapp. 2008;29(10):1111–1122. doi: 10.1002/hbm.20451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sutton BP, Goh J, Hebrank A, Welsh RC, Chee MW, Park DC. Investigation and validation of intersite fMRI studies using the same imaging hardware. J Magn Reson Imaging. 2008;28(1):21–28. doi: 10.1002/jmri.21419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zivadinov R, Cox JL. Is functional MRI feasible for multi-center studies on multiple sclerosis? Eur J Neurol. 2008;15(2):109–110. doi: 10.1111/j.1468-1331.2007.02030.x. [DOI] [PubMed] [Google Scholar]
- 12.Zou KH, Greve DN, Wang M, Pieper SD, Warfield SK, White NS, Manandhar S, Brown GG, Vangel MG, Kikinis R, Wells WM., 3rd Reproducibility of functional MR imaging: preliminary results of prospective multi-institutional study performed by Biomedical Informatics Research Network. Radiology. 2005;237(3):781–789. doi: 10.1148/radiol.2373041630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yendiki A, Greve DN, Wallace S, Vangel M, Bockholt J, Mueller BA, Magnotta V, Andreasen N, Manoach DS, Gollub RL. Multi-site characterization of an fMRI working memory paradigm: reliability of activation indices. Neuroimage. 2010;53(1):119–131. doi: 10.1016/j.neuroimage.2010.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cramer SC, Benson RR, Himes DM, Burra VC, Janowsky JS, Weinand ME, Brown JA, Lutsep HL. Use of functional MRI to guide decisions in a clinical stroke trial. Stroke. 2005;36(5):e50–e52. doi: 10.1161/01.STR.0000163109.67851.a0. [DOI] [PubMed] [Google Scholar]
- 15.Mancini L, Ciccarelli O, Manfredonia F, Thornton JS, Agosta F, Barkhof F, Beckmann C, De Stefano N, Enzinger C, Fazekas F, Filippi M, Gass A, Hirsch JG, Johansen-Berg H, Kappos L, Korteweg T, Manson SC, Marino S, Matthews PM, Montalban X, Palace J, Polman C, Rocca M, Ropele S, Rovira A, Wegner C, Friston K, Thompson A, Yousry T. Short-term adaptation to a simple motor task: a physiological process preserved in multiple sclerosis. Neuroimage. 2009;45(2):500–511. doi: 10.1016/j.neuroimage.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Manson SC, Wegner C, Filippi M, Barkhof F, Beckmann C, Ciccarelli O, De Stefano N, Enzinger C, Fazekas F, Agosta F, Gass A, Hirsch J, Johansen-Berg H, Kappos L, Korteweg T, Polman C, Mancini L, Manfredonia F, Marino S, Miller DH, Montalban X, Palace J, Rocca M, Ropele S, Rovira A, Smith S, Thompson A, Thornton J, Yousry T, Frank JA, Matthews PM. Impairment of movement-associated brain deactivation in multiple sclerosis: further evidence for a functional pathology of interhemispheric neuronal inhibition. Experimental brain research Experimentelle Hirnforschung. 2008;187(1):25–31. doi: 10.1007/s00221-008-1276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rocca MA, Absinta M, Valsasina P, Ciccarelli O, Marino S, Rovira A, Gass A, Wegner C, Enzinger C, Korteweg T, Sormani MP, Mancini L, Thompson AJ, De Stefano N, Montalban X, Hirsch J, Kappos L, Ropele S, Palace J, Barkhof F, Matthews PM, Filippi M. Abnormal connectivity of the sensorimotor network in patients with MS: a multicenter fMRI study. Hum Brain Mapp. 2009;30(8):2412–2425. doi: 10.1002/hbm.20679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wegner C, Filippi M, Korteweg T, Beckmann C, Ciccarelli O, De Stefano N, Enzinger C, Fazekas F, Agosta F, Gass A, Hirsch J, Johansen-Berg H, Kappos L, Barkhof F, Polman C, Mancini L, Manfredonia F, Marino S, Miller DH, Montalban X, Palace J, Rocca M, Ropele S, Rovira A, Smith S, Thompson A, Thornton J, Yousry T, Matthews PM. Relating functional changes during hand movement to clinical parameters in patients with multiple sclerosis in a multi-centre fMRI study. Eur J Neurol. 2008;15(2):113–122. doi: 10.1111/j.1468-1331.2007.02027.x. [DOI] [PubMed] [Google Scholar]
- 19.Schneider F, Habel U, Reske M, Kellermann T, Stocker T, Shah NJ, Zilles K, Braus DF, Schmitt A, Schlosser R, Wagner M, Frommann I, Kircher T, Rapp A, Meisenzahl E, Ufer S, Ruhrmann S, Thienel R, Sauer H, Henn FA, Gaebel W. Neural correlates of working memory dysfunction in first-episode schizophrenia patients: an fMRI multi-center study. Schizophrenia research. 2007;89(1–3):198–210. doi: 10.1016/j.schres.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 20.Demirci O, Stevens MC, Andreasen NC, Michael A, Liu J, White T, Pearlson GD, Clark VP, Calhoun VD. Investigation of relationships between fMRI brain networks in the spectral domain using ICA and Granger causality reveals distinct differences between schizophrenia patients and healthy controls. Neuroimage. 2009;46(2):419–431. doi: 10.1016/j.neuroimage.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim DI, Sui J, Rachakonda S, White T, Manoach DS, Clark VP, Ho BC, Schulz SC, Calhoun VD. Identification of Imaging Biomarkers in Schizophrenia: A Coefficient-constrained Independent Component Analysis of the Mind Multi-site Schizophrenia Study. Neuroinformatics. 2010;8(4):213–229. doi: 10.1007/s12021-010-9077-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ford JM, Roach BJ, Jorgensen KW, Turner JA, Brown GG, Notestine R, Bischoff-Grethe A, Greve D, Wible C, Lauriello J, Belger A, Mueller BA, Calhoun V, Preda A, Keator D, O'Leary DS, Lim KO, Glover G, Potkin SG, Mathalon DH. Tuning in to the voices: a multisite FMRI study of auditory hallucinations. Schizophr Bull. 2009;35(1):58–66. doi: 10.1093/schbul/sbn140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim DI, Manoach DS, Mathalon DH, Turner JA, Mannell M, Brown GG, Ford JM, Gollub RL, White T, Wible C, Belger A, Bockholt HJ, Clark VP, Lauriello J, O'Leary D, Mueller BA, Lim KO, Andreasen N, Potkin SG, Calhoun VD. Dysregulation of working memory and default-mode networks in schizophrenia using independent component analysis, an fBIRN and MCIC study. Hum Brain Mapp. 2009;30(11):3795–3811. doi: 10.1002/hbm.20807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Potkin SG, Ford JM. Widespread cortical dysfunction in schizophrenia: the FBIRN imaging consortium. Schizophr Bull. 2009;35(1):15–18. doi: 10.1093/schbul/sbn159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Potkin SG, Turner JA, Brown GG, McCarthy G, Greve DN, Glover GH, Manoach DS, Belger A, Diaz M, Wible CG, Ford JM, Mathalon DH, Gollub R, Lauriello J, O'Leary D, van Erp TG, Toga AW, Preda A, Lim KO. Working memory and DLPFC inefficiency in schizophrenia: the FBIRN study. Schizophr Bull. 2009;35(1):19–31. doi: 10.1093/schbul/sbn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wible CG, Lee K, Molina I, Hashimoto R, Preus AP, Roach BJ, Ford JM, Mathalon DH, McCarthey G, Turner JA, Potkin SG, O'Leary D, Belger A, Diaz M, Voyvodic J, Brown GG, Notestine R, Greve D, Lauriello J. fMRI activity correlated with auditory hallucinations during performance of a working memory task: data from the FBIRN consortium study. Schizophr Bull. 2009;35(1):47–57. doi: 10.1093/schbul/sbn142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedman L, Glover GH. Reducing interscanner variability of activation in a multicenter fMRI study: controlling for signal-to-fluctuation-noise-ratio (SFNR) differences. Neuroimage. 2006;33(2):471–481. doi: 10.1016/j.neuroimage.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 28.Stocker T, Schneider F, Klein M, Habel U, Kellermann T, Zilles K, Shah NJ. Automated quality assurance routines for fMRI data applied to a multicenter study. Hum Brain Mapp. 2005;25(2):237–246. doi: 10.1002/hbm.20096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greve DN, Mueller BA, Liu T, Turner JA, Voyvodic J, Yetter E, Diaz M, McCarthy G, Wallace S, Roach BJ, Ford JM, Mathalon DH, Calhoun VD, Wible CG, Brown GG, Potkin SG, Glover G. A novel method for quantifying scanner instability in fMRI. Magn Reson Med. 2011;65(4):1053–1061. doi: 10.1002/mrm.22691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedman L, Glover GH. Report on a multicenter fMRI quality assurance protocol. J Magn Reson Imaging. 2006;23(6):827–839. doi: 10.1002/jmri.20583. [DOI] [PubMed] [Google Scholar]
- 31.Keator DB. Management of information in distributed biomedical collaboratories. Methods Mol Biol:Biomedical Informatics. 2009;569:1–23. doi: 10.1007/978-1-59745-524-4_1. [DOI] [PubMed] [Google Scholar]
- 32.Kruger G, Kastrup A, Glover GH. Neuroimaging at 1.5 T and 3.0 T: comparison of oxygenation-sensitive magnetic resonance imaging. Magn Reson Med. 2001;45(4):595–604. doi: 10.1002/mrm.1081. [DOI] [PubMed] [Google Scholar]
- 33.Shmuel A, Yacoub E, Chaimow D, Logothetis NK, Ugurbil K. Spatio-temporal point-spread function of fMRI signal in human gray matter at 7 Tesla. Neuroimage. 2007;35(2):539–552. doi: 10.1016/j.neuroimage.2006.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedman L, Glover GH, Krenz D, Magnotta V. Reducing inter-scanner variability of activation in a multicenter fMRI study: role of smoothness equalization. Neuroimage. 2006;32(4):1656–1668. doi: 10.1016/j.neuroimage.2006.03.062. [DOI] [PubMed] [Google Scholar]
- 35.Olsrud J, Nilsson A, Mannfolk P, Waites A, Stahlberg F. A two-compartment gel phantom for optimization and quality assurance in clinical BOLD fMRI. Magn Reson Imaging. 2008;26(2):279–286. doi: 10.1016/j.mri.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 36.Keator DB, Grethe JS, Marcus D, Ozyurt B, Gadde S, Murphy S, Pieper S, Greve D, Notestine R, Bockholt HJ, Papadopoulos P. A national human neuroimaging collaboratory enabled by the Biomedical Informatics Research Network (BIRN) IEEE Trans Inf Technol Biomed. 2008;12(2):162–172. doi: 10.1109/TITB.2008.917893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozyurt IB, Keator DB, Wei D, Fennema-Notestine C, Pease KR, Bockholt J, Grethe JS. Federated web-accessible clinical data management within an extensible neuroimaging database. Neuroinformatics. 2010;8(4):231–249. doi: 10.1007/s12021-010-9078-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gadde S, Aucoin N, Grethe JS, Keator DB, Marcus DS, Pieper S. XCEDE: An Extensible Schema for Biomedical Data. Neuroinformatics. 2011 Apr 9; doi: 10.1007/s12021-011-9119-9. [Epub ahead of print] PMID: 21479735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Helmer KG, Ambite JL, Ames J, Ananthakrishnan R, Burns G, Chervenak AL, Foster I, Liming L, Keator D, Macciardi F, Madduri R, Navarro JP, Potkin S, Rosen B, Ruffins S, Schuler R, Turner JA, Toga A, Williams C, Kesselman C. Enabling collaborative research using the Biomedical Informatics Research Network (BIRN) J Am Med Inform Assoc. 2011;18(4):416–422. doi: 10.1136/amiajnl-2010-000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mortamet B, Bernstein MA, Jack CR, Jr, Gunter JL, Ward C, Britson PJ, Meuli R, Thiran JP, Krueger G. Automatic quality assessment in structural brain magnetic resonance imaging. Magn Reson Med. 2009;62(2):365–372. doi: 10.1002/mrm.21992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doros G, Lew R. Design based on intra-class correlation coefficients. American Journal of Biostatistics. 2010;6(1):1–8. [Google Scholar]
- 42.Thomason ME, Foland LC, Glover GH. Calibration of BOLD fMRI using breath holding reduces group variance during a cognitive task. Hum Brain Mapp. 2007;28(1):59–68. doi: 10.1002/hbm.20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strother S, La Conte S, Kai Hansen L, Anderson J, Zhang J, Pulapura S, Rottenberg D. Optimizing the fMRI data-processing pipeline using prediction and reproducibility performance metrics: I. A preliminary group analysis. Neuroimage. 2004;23(Suppl 1):S196–S207. doi: 10.1016/j.neuroimage.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 44.Poldrack R, Mumford J, Nichols T. Handbook of Functional MRI Data Analysis. New York: Cambridge University Press; 2011. [Google Scholar]
- 45.Greve DN, Mueller BA, GG Brown G, Liu TT, Glover GH FBIRN. Processing methods to reduce intersite variability in fMRI. Barcelona, Spain: OHBM Proceedings; 2010. [Google Scholar]
- 46.Beckmann CF, Jenkinson M, Smith SM. General multilevel linear modeling for group analysis in FMRI. Neuroimage. 2003;20(2):1052–1063. doi: 10.1016/S1053-8119(03)00435-X. [DOI] [PubMed] [Google Scholar]
- 47.Winer BJ, Brown DR, Cichels KM. Statistical principles in experimental design. Boston: 1991. [Google Scholar]
- 48.Gelman A, Carlin JB, Stern HS, Rubin DB. Bayesian data analysis. New York: Chpman & Hall/CRC; 2004. [Google Scholar]
- 49.Kim S, Smyth P, Stern H. A Bayesian mixture approach to modeling spatial activation patterns in multisite fMRI data. IEEE Trans Med Imaging. 2010;29(6):1260–1274. doi: 10.1109/TMI.2010.2044045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bosnell R, Wegner C, Kincses ZT, Korteweg T, Agosta F, Ciccarelli O, De Stefano N, Gass A, Hirsch J, Johansen-Berg H, Kappos L, Barkhof F, Mancini L, Manfredonia F, Marino S, Miller DH, Montalban X, Palace J, Rocca M, Enzinger C, Ropele S, Rovira A, Smith S, Thompson A, Thornton J, Yousry T, Whitcher B, Filippi M, Matthews PM. Reproducibility of fMRI in the clinical setting: implications for trial designs. Neuroimage. 2008;42(2):603–610. doi: 10.1016/j.neuroimage.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 51.Abbott C, Juarez M, White T, Gollub RL, Pearlson GD, Bustillo J, Lauriello J, Ho B, Bockholt HJ, Clark VP, Magnotta V, Calhoun VD. Antipsychotic dose and diminished neural modulation: a multi-site fMRI study. Progress in neuro-psychopharmacology & biological psychiatry. 2011;35(2):473–482. doi: 10.1016/j.pnpbp.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Demirci O, Clark VP, Magnotta VA, Andreasen NC, Lauriello J, Kiehl KA, Pearlson GD, Calhoun VD. A Review of Challenges in the Use of fMRI for Disease Classification / Characterization and A Projection Pursuit Application from Multi-site fMRI Schizophrenia Study. Brain imaging and behavior. 2008;2(3):147–226. doi: 10.1007/s11682-008-9028-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown GG, McCarthy G, Bischoff-Grethe A, Ozyurt B, Greve D, Potkin SG, Turner JA, Notestine R, Calhoun VD, Ford JM, Mathalon D, Manoach DS, Gadde S, Glover GH, Wible CG, Belger A, Gollub RL, Lauriello J, O'Leary D, Lim KO. Brain-performance correlates of working memory retrieval in schizophrenia: a cognitive modeling approach. Schizophr Bull. 2009;35(1):32–46. doi: 10.1093/schbul/sbn149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Horn JD, Toga AW. Multisite neuroimaging trials. Curr Opin Neurol. 2009;22(4):370–378. doi: 10.1097/WCO.0b013e32832d92de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jack CR, Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, J LW, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DL, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27(4):685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.