Abstract
We report a freely available software program, Pyromaker, which generates simulated traces for pyrosequencing results based on user inputs. Simulated pyrograms can aid in the analysis of complex pyrosequencing results in which various hypothesized mutations can be tested, and the resultant pyrograms can be matched with the actual pyrogram. We validated the software using the actual pyrograms for common KRAS gene mutations as well as several mutations in the BRAF, GNAS, and p53 genes. We demonstrate that all 18 possible single-base mutations within codons 12 and 13 of KRAS generate unique pyrosequencing traces and highlight the distinctions between them. We further show that all reported codon 12 and 13 complex mutations produce unique pyrograms. However, some complex mutations are indistinguishable from single-base mutations. For complicated pyrograms, Pyromaker was used in two modes, one in which hypothesis-based simulated pyrograms were pattern-matched with the actual pyrograms. In a second strategy with only the pyrogram, Pyromaker was used to identify the underlying mutation by iteratively reconstructing the mutant pyrogram. Either strategy was able to successfully identify the complex mutations, which were confirmed by cloning and sequencing. Using two examples of KRAS codon 12 mutations (specifically GGT→TTT, G12F and GGT→GAG, G12E), we report which combinations of five approaches permit unambiguous mutation identification. The most efficient approach was found to be pyrosequencing with Pyromaker.
Strategies to detect mutations in the molecular diagnostic laboratory include pyrosequencing and Sanger sequencing. Dye-terminator Sanger sequencing can provide read lengths of up to 800 bases, but has a limit of detection of only ∼20% mutant alleles, depending on sequence context. Sanger sequencing results also can be ambiguous for some mutations, for example, whether mutation of two adjacent nucleotides in a sequencing trace is due to a single allele bearing both mutations or two alleles with one mutation each. Pyrosequencing is a newer “sequencing-by-synthesis” technology, in which a DNA polymerase adds a deoxynucleotide-triphosphate (dNTP) into an elongating strand, thereby liberating a molecule of pyrophosphate.1–3 Through a series of enzymatic reactions, the pyrophosphate is converted to ATP, a co-factor in the light-producing luciferase reaction. Injection of the next dNTP then occurs, and the presence or absence of light indicates whether the polymerase was able to extend the nascent strand with the injected dNTP. Compared to Sanger sequencing, pyrosequencing is inherently more quantitative and has a superior limit of detection (∼5% minor allele frequency).4,5 However, pyrosequencing does not do as well at sizing long homopolymeric repeats (eg, differentiating eight from nine uninterrupted adenines), and its read length is shorter than with Sanger sequencing. Pyrograms of complex mutations can yield confusing patterns that are often difficult to resolve without further investigation.
The KRAS proto-oncogene encodes the Kirsten rat sarcoma viral oncogene homolog (K-ras), a small G-protein identified in the early 1980s.6,7 The K-ras protein normally switches between active and inactive conformations, but activating mutations produce a protein that is constitutively activated, and these play an important role in the pathogenesis of a variety of tumors including colorectal, lung, and pancreatic adenocarcinomas, among others.8–11 Clinical tests to detect KRAS mutations were quickly adopted into routine clinical testing because they predict the lack of response to epidermal growth factor receptor (EGFR) inhibitor therapy.12,13 Anti-EGFR monoclonal antibodies such as cetuximab or panitumumab are currently used in patients with metastatic colorectal cancer, whereas anti-EGFR tyrosine kinase inhibitors such as erlotinib and gefitinib are used to treat non–small-cell lung cancer patients. However, since the K-ras protein is distal to EGFR in the signaling cascade, activating KRAS mutations cause resistance to anti-EFGR therapy.12–15 Recently, it was reported that colorectal cancers with a specific codon 13 KRAS mutation (G13D) retain sensitivity to cetuximab, suggesting that the simple paradigm that any activating KRAS mutation causes resistance may not be correct, and emphasizing the need to identify the specific mutated base in a given patient's tumor.16
We present a software program, named Pyromaker, which produces simulated pyrograms. It was primarily developed to assist in interpreting complex pyrograms. In this report, we validate Pyromaker by comparing virtual pyrograms to actual pyrograms for several genes. For example, we show that all possible single-base substitution mutations within codons 12 and 13 of the KRAS gene generate qualitatively or quantitatively unique pyrograms. All currently reported complex mutations of these codons also generate unique pyrograms from each other; however, some complex mutations are indistinguishable from some single-base mutations. We present two complex pyrograms that were impossible to interpret initially, and used five approaches to resolving them: Sanger sequencing, Pyromaker hypothesis testing, Pyromaker iterative mutation re-creation, melting curving analysis, and TA cloning with Sanger sequencing.
Materials and Methods
Pyromaker
The general features of the software are reported in the results section. The proper peak morphology was emulated as a linear increase to the maximum followed by decay phase predicted by taking the Gaussian curve from the maximum point to four standard deviations. This piecewise function and the SD was chosen based on visual comparisons of the measured pyrograms against the virtual peak widths.
Pyromaker is a Web-based application that uses an HTML/JavaScript client interface to collect and to validate user input. If the Web browser being used does not support JavaScript or simply has JavaScript disabled, instructions in English are provided. Once the form is submitted, the data are revalidated and pre-processed using PHP. If all supplied input passes validation, PHP calls a script written in R to generate the appropriate pyrogram, which is returned to the user in PDF format. The Web version of Pyromaker is executed on a server running the latest version of Ubuntu (10.4 long-term support, 5/1/10).
Validation of the mutations was performed by the following: i) visually comparing the virtual pyrograms against clinical pyrosequencing data for a spectrum of KRAS mutations; ii) computing the virtual traces for all theoretically possible single point mutations in codons 12 and 13; iii) generating the predicted traces for all reported complex mutations in the Catalogue Of Somatic Mutations In Cancer (COSMIC) at the Sanger Institute (http://www.sanger.ac.uk/genetics/CGP/cosmic, last accessed May 26, 2011) for KRAS codons 12 and 13; and iv) comparing an assortment of pyrograms against those generated by pyrosequencing of other genes.
All virtual pyrograms discussed here were plotted assuming heterozygosity of the mutant allele and a tumor cellularity of 50% (mutant allele frequency 25%), similar to the peak heights in the actual data from the two examples highlighted. In hypothetical examples containing two mutations, the mutations were assigned equal proportions of the mutant DNA (mutant allele frequency 12.5% each).
Sample Preparation and DNA Extraction
After H&E-stained slide review and tumor tissue selection, the corresponding tissue from five unstained, 5-μm-thick tissue sections was manually microdissected using Pinpoint reagents according to the manufacturer's protocol (ZymoResearch, Orange, CA). DNA was purified from the sample using QIAmp DNA kit (Qiagen, Valencia, CA) and quantified by OD 260 nm.
Pyrosequencing
Samples were PCR amplified using the KRAS v2.0 kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. The amplicons were sequenced using the PyroMark Q24 (Biotage, SE) with PyroMark Gold reagents (Qiagen) containing 0.3 μmol/L sequencing primer and annealing buffer. The nucleotide dispensation order for codons 12 to 14 was 5′-TACGACTCAGATCGTAG-3′, ie, where the nucleotide triphosphates (dTTP, then dATP, then dCTP, etc) are simply listed as single characters in a sequence (ie, TAC…). Dispensation order is the order in which dNTPs are sequentially injected into the reaction chamber and is optimized based on the gene sequence (eg, for KRAS to determine whether a mutation is in the first (12a) G or the second (12b) G in the codon 12 sequence GGT). A position refers to one dNTP injection/dispensation on the x axis.
Sanger Sequencing
PCR amplification reactions included 5′-AAGGCCTGCTGAAAATGACTG-3′ (forward) and 5′-GGTCCTGCACCAGTAATATGCA-3′ (reverse) and were amplified in a Veriti Thermal Cycler (Applied Biosystems/AB, Foster City, CA) as follows: 95°C for 15 minutes, 42 cycles of 95°C for 20 seconds, 53°C for 30 seconds, and 72°C for 20 seconds, and 72°C for 5 minutes. Then the products were purified using ExoSAP-IT (GE Healthcare), sequenced using the forward or reverse PCR primers and Big Dye v3.1 reagents (AB), products purified with Big Dye XTerminator reagents (AB), and automated sequencing performed by capillary electrophoresis on an AB3130xl (AB). Sequences were aligned and examined by visual inspection of the electropherogram, using Sequencher software version 4.10.1 (Gene Codes Corporation, Ann Arbor, MI).
Melting Curve Analysis
Melting curve analysis was adapted from the method reported by Wallen et al.17 Briefly, reaction mixture consisted of LightCycler FastStart DNA Master Hybridization Probes Mix (Roche, Indianapolis, IN), 500 nmol/L Forward Primer (5′-AAGGCCTGCTGAAAATGACTG-3′), 100 nmol/L Reverse Primer (5′-GGTCCTGCACCAGTAATATGC-3′), 400 nmol/L Sensor Probe (5′-Rox-TGCCTACGCCACCAGCTCCAA-Phos-3′), 200 nmol/L Anchor Probe (5′-CCACAAAATGATTCTGAATTAGCTGTATCGTCAAGGCACT-FAM-3′), and 100 nmol/L peptide nucleic acid (PNA, 5′-NH2-CCTACGCCACCAGCTCC-COOH-3′). Reactions were amplified in a Veriti Thermal Cycler as follows: 95°C for 10 minutes, and 45 cycles of 95°C for 10 seconds, 61°C for 10 seconds, and 72°C for 15 seconds. Melting curve analysis was performed on the StepOne Plus instrument (AB) through one cycle of 95°C for 20 seconds and 45°C for 60 seconds, and ramp to 78°C at 0.4°C per second. Melting curves were generated using the StepOne Plus software (AB). Controls included a wild-type sample (GGT and GGC for codons 12 and 13 respectively) and one with a G13b G→A single-base substitution (GAC, G13D).
TA Cloning and Sequencing
Samples were PCR amplified as described for Sanger sequencing. Following amplification, fresh PCR products were cloned using the pcDNA 3.3-TOPO TA cloning Kit (Invitrogen, Carlsbad, CA), transformed into One Shot TOP10 chemically competent Escherichia coli, and plated on LB-ampicillin plates according to the manufacturer's protocols. Sets of colonies from each transformation were picked and cultured overnight in LB medium containing 100 μg/mL ampicillin. Plasmid DNA was isolated by using PureLink Quick Plasmid Miniprep kit (Invitrogen), Sanger sequenced using the CMV forward primer and BigDye V3.1 reagents, and resolved on an AB3700 instrument (AB). Sequences were aligned and examined by visual inspection of the electropherogram, using Sequencher. Up to 30 colonies had to be analyzed in some cases because of the presence of primer dimers and wild-type alleles. Two or more clones bearing the KRAS mutations were identified in controls and for each case.
Results
Pyromaker was devised to accept the following parameters: the percent tumor and normal cells, the wild-type sequence, the dispensation order, and any number of mutant sequences with their relative abundances and zygosity. Using the percent tumor cells and whether the mutation is homozygous or heterozygous, it calculates the relative mutant and wild-type allele percentages. These percentages are then used to generate the expected signal at each point during the dispensation sequence and Pyromaker then produces a virtual trace of the expected pyrogram. The signal generated by addition of the ATP analog, dATPαS (hereafter simply dATP, or A) was set to 110% of the other dNTPs, as the addition of this base in actual pyrograms gives consistently higher peak heights than those measured after the addition of other bases. Graphs were produced automatically for each set of parameters by the graphical tools available in R. Additional details are provided in Materials and Methods. Pyromaker has been made available as a free, publicly available program (http://pyromaker.pathology.jhmi.edu).
Validation of Pyromaker
We first validated the Pyromaker software against actual pyrograms containing common KRAS mutations. Supplemental Table S1 (available at http://jmd.amjpathol.org) lists the 10 KRAS mutations for which the virtual and actual pyrograms have been compared. After running Pyromaker using the diagnosed mutation, the actual pyrogram and the virtual pyrogram were compared, and all were qualitatively identical for all mutations tested.
We also validated Pyromaker using the V600E mutation in BRAF, for which the actual pyrogram (see Supplemental Figure S1A at http://jmd.amjpathol.org) is qualitatively similar to the Pyromaker generated virtual pyrogram (see Supplemental Figure S1B at http://jmd.amjpathol.org). Similarly we validated the software for the R201H GNAS oncogene mutation (see Supplemental Table S1 at http://jmd.amjpathol.org, reference 38) and for two mutations in the p53 tumor suppressor gene (R248W and R282W; see Supplemental Table S1 at http://jmd.amjpathol.org, reference 39). The magnitude of the mutations detected was well above the coefficient of variation, estimated as less than 5% (see Supplemental Table S2 at http://jmd.amjpathol.org).
All Codon 12 and Codon 13 KRAS Single-Base Mutations Produce Distinct Pyrograms
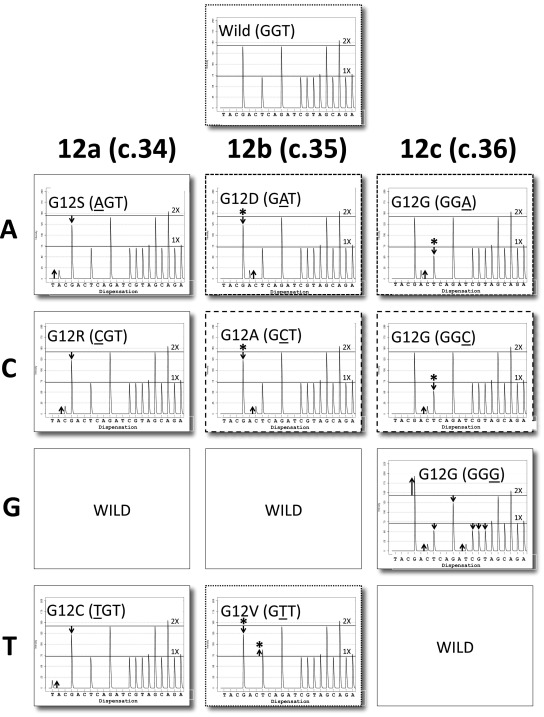
After having validated Pyromaker for the above mutations, we produced pyrograms for all possible base substitution mutations in codon 12 (Figure 1), and all mutations gave distinct patterns. We considered two pyrograms to be qualitatively similar if they contained the peaks at the same positions, albeit at different heights. Pyrograms for many of the mutations are qualitatively distinct; however GAT and GGA mutations (mutated bases underlined, Figure 1, panels with short dashed borders) are qualitatively similar, in that both have the novel A peak compared to the wild-type trace. However, they are quantitatively distinct in that, with the GAT trace, the novel A activity is associated with loss of preceding G activity (see down arrow), whereas for the GGA mutation, the A activity is associated with a reduction of the subsequent T peak (see down arrow). Asterisks highlight peaks that distinguish two qualitatively similar pyrograms. Similarly GCT and GGC (panels with long dashed borders) are qualitatively similar, but for the GCT mutation, the novel C activity derives from the preceding G, whereas for GGC mutation, the C activity is associated with loss of subsequent T activity. Finally, the GTT mutation is qualitatively similar to the wild-type pyrogram (panels with dotted borders), as no new peaks are generated, but it is recognized by the deviation between the G and T activities from the 2:1 ratio characteristic of the wild-type trace.
Figure 1.
KRAS codon 12 mutations. Virtual pyrograms for wild type (GGT) and nine mutations at the three base positions 12a, 12b, and 12c are shown. Some mutations are qualitatively distinct from wild type and all of the other mutations (AGT, CGT, GGG, and TGT) whereas others are only quantitatively distinct (GAT versus GGA, short dashed line borders); GCT versus GGC, long dashed line borders; and GTT versus wild type, dotted borders). Up arrows indicate either novel peaks that should not be present or peaks that are too high; down arrows indicate peaks that are lower than expected. Asterisks indicate peaks that distinguish qualitatively similar virtual pyrograms and are placed above the relevant arrows. (The relative height of the up arrows and down arrows should be constant, but the absolute height of the arrows can vary depending on the percent tumor cellularity in a sample). Note that all three 12c mutations are silent.
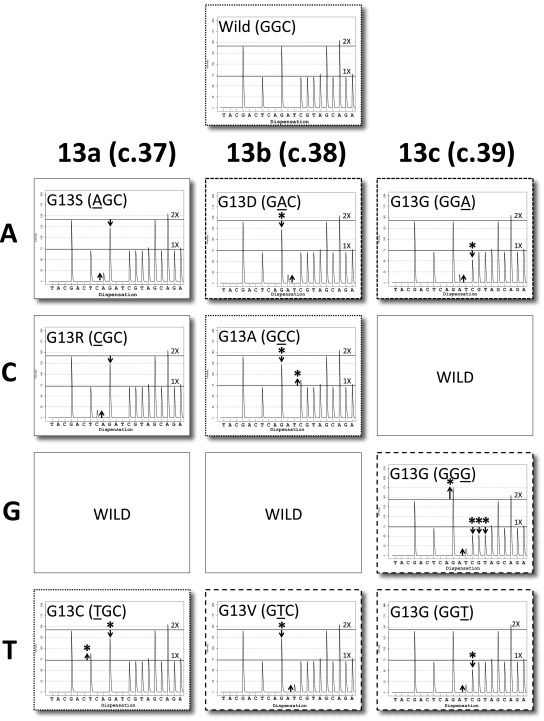
Similar to codon 12, all single-base codon 13 base substitution mutations are also either qualitatively or quantitatively distinct (Figure 2). GCC and TGC are most similar to the wild-type trace, as they do not generate any novel peaks (three panels with dotted borders). They are easily distinguished however, because with the GCC mutation there is a decrease in the G:C ratio from 2:1, due to a direct transfer of activity from the G peak to the subsequent C peak. The TGC mutation also changes that ratio; however, this is due to a shift in G activity to the preceding T position of codon 12. Both GAC (G13D) and GGA (panels with short dashed borders) have a novel A peak (compared to wild type); however, the former is associated with a decrease in the preceding G peak, whereas the latter derives from the subsequent C. As discussed, distinguishing the G13D mutation may be clinically important (as discussed in introductory section of text). The GGG, GTC, and GGT traces (panels with long dashed borders) all contain the same novel T peak, but the GGG mutation has an increase in G activity preceeding, and loss of C, G, and T activities following the novel T peak. In contrast, for the GTC mutation, the extra T activity derives from the preceding G, whereas with the GGT mutation, the extra T activity is associated with loss of subsequent C peak activity. A similar analysis was performed for the less frequently reported codon 61 (see Supplemental Figures S2 and S3 at http://jmd.amjpathol.org) and codon 146 mutations (see Supplemental Figure S4 at http://jmd.amjpathol.org).
Figure 2.
KRAS Codon 13 mutations. Virtual pyrograms for wild type (GGC) and nine mutations are shown. Some mutations are qualitatively distinct from wild-type and the other mutations (AGC and CGC), whereas others are only quantitatively distinct (GAC versus GGA, short dashed line borders; wild type versus GCC versus TGC, dotted borders; and GGG versus GTC versus GGT, long dashed line borders). Note that all three 13c mutations are silent.
All Reported Codon 12 and Codon 13 KRAS Complex Mutations Produce Pyrograms Distinct from One Another
Interrogation of the Cosmic database at the Sanger Institute for all reported complex (two-base) mutations of KRAS codons 12 and 13 revealed 11 complex codon 12 mutations and 9 complex codon 13 mutations. Virtual pyrograms for all of the codon 12 mutations (see Supplemental Figure S5 at http://jmd.amjpathol.org) demonstrate that, similar to the single-base changes, many traces are qualitatively unique, in that novel peaks are unique to that one mutation. The most qualitatively similar are GAA and GAG (panels with dotted borders), TTT and TGG (short dashed borders), in addition to ATT and AAT (long dashed borders); however, each of these pairs can be quantitatively distinguished (asterisks). These results are provided for other investigators to use for pattern matching complex pyrograms. Similarly, all of the codon 13 mutations (see Supplemental Figure S6 at http://jmd.amjpathol.org) could be distinguished from one another either qualitatively or quantitatively. Only GAA and GAG (panels with dotted borders), in addition to GTG and GTT (dashed borders) are qualitatively similar and need to be quantitatively distinguished.
Some Mutations Produce Identical Pyrograms Using the Default KRAS Dispensation Order
Comparing pyrosequencing traces for single-base changes (Figures 1 and 2) to complex mutations (see Supplemental Figures S5 and S6 at http://jmd.amjpathol.org) raises some concerns. For example, AGT (Figure 1) and AAT (see Supplemental Figure S5 at http://jmd.amjpathol.org) show the same pattern of peaks, where the increased A activity is accompanied by the same amount of decrease in G peak activity. Although these traces are slightly different using our default parameters, they can be made to look identical by varying the percent tumor cellularity (not shown). We demonstrated that these two sequences could be distinguished if an additional dTTP were dispensed after the A and before the G (not shown). A similar problem would likely occur, distinguishing CGT from CCT; however CCT for codon 12 has not yet been reported in Cosmic, and similarly, these could be distinguished by dispensing a T after the C and before the G.
Some codon 13 mutations also are indistinguishable using the current default dispensation. AGC (Figure 2) and AAC (see Supplemental Figure S6 at http://jmd.amjpathol.org) are similar and can be made to be identical by varying percent tumor cellularity or zygosity (not shown). A similar situation would be expected for TGC and TTC, although the TTC mutation has not been reported. Similar to codon 12, these mutations could be distinguished using a different dispensation order, designed to exaggerate sequence differences.
A Single Sequencing Approach Can Yield Ambiguous Results for Complex Cases
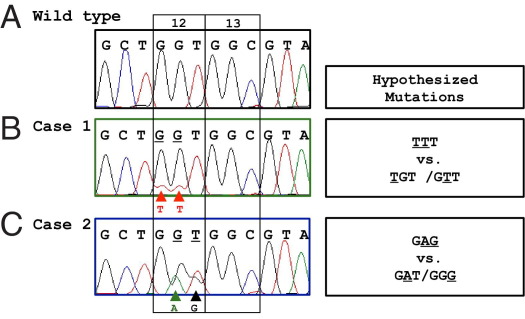
Paraffin-embedded tissue section samples from two patients with colon adenocarcinoma were tested for KRAS mutation analysis to guide chemotherapy selection. Pyrosequencing of KRAS codons 12 and 13 was initially performed, but the pyrograms could not be interpreted. Compared to the wild type (Figure 3A), there are three signature changes in the pyrogram for case 1, namely, extra T, C, and T peaks (Figure 3B, numbered). Reductions of peak heights at other positions are also noted (arrowheads). For case 2, the four signature changes are a reduction of a G peak, an extra A peak, a reduction of a T peak, and an increased G peak (Figure 3C, arrows, numbered).
Figure 3.
Complex pyrosequencing results for two KRAS mutation cases. A: Pyrogram for wild-type KRAS. B: Pyrogram for case 1. Note the three novel “signature peaks,” T, C, and T (arrows, numbered) that are not present in the wild-type pyrogram. Reduction of peak height at other positions is noted (arrowheads). 1X and 2X are determined by the peak height of bases where the mutant and normal alleles are in register (distal to the region shown in the figure). C: Pyrogram for case 2. Note the four signature peaks (arrows, numbered) unique to this mutation. Horizontal lines designate the expected 1X and 2X activities. Wild-type activity is present for both cases (due to stromal cells and the remaining wild-type allele in the cancer cells).
To clarify the complex pyrosequencing data, the samples were reamplified and sequenced by the Sanger dye-terminator method. Analysis of Sanger sequencing revealed mutations of two adjacent nucleotides of codon 12 for both cases (Figure 4, B and C, arrowheads), compared to the wild type (Figure 4A). Wild-type bases are also present at these positions, which is not surprising, as tumors contain non-neoplastic stromal cells and because activating KRAS mutations are usually heterozygous in malignant cells. However, Sanger sequencing alone cannot elucidate whether the mutations are on the same allele or on two different alleles. For case 1, there could be a two-base TTT mutation on a single allele (from wild-type codon 12 GGT) or a mixture of two single nucleotide mutations (TGT and GTT) on two separate alleles (Figure 4B, hypotheses shown on the right). Similarly, the Sanger sequencing results for case 2 may be due to a two-base mutation producing a single GAG allele or due to a mixture of two alleles (GAT and GGG), as illustrated in Figure 4C. Repeat analysis of the pyrogram, with the knowledge of the Sanger sequencing, indicates the two base mutation on one allele is the only solution, as described for case 1 in Supplemental Figures S7 and S8 (available at http://jmd.amjpathol.org). However, an unambiguous solution from either sequencing strategy alone, is either impossible (Sanger sequencing) or extremely difficult (pyrosequencing).
Figure 4.
Sanger sequencing results and hypotheses generated. A: Sanger sequence for wild-type KRAS. B: Sanger sequence for case 1 and alternate hypotheses to explain the sequencing results are listed. Mutant bases are underlined. C: Sanger sequence for case 2 and hypotheses. Arrowheads indicate the mutant peaks compared to the wild-type sequence. Codons 12 and 13 are bracketed.
Software Analysis
Hypothesis-Based Pattern Matching
To resolve the ambiguity that exists for the pyrosequencing results, we used Pyromaker, in hypothesis testing mode. If one is entertaining two alternative hypotheses, one can enter each of them and then pattern-match the computer-generated pyrograms to the actual data. The wild-type pyrosequencing pattern generated by the computer program (Figure 5A) closely matched the actual wild-type pyrogram (Figure 3A). We then tested the two hypotheses for the first case, and the pyrosequencing pattern for TTT generated by the software qualitatively matched the actual clinical data where all three novel peaks in the actual pyrogram are present (compare Figure 5B, left panel, versus Figure 3B). The alternative hypothesis of two single-base mutations, TGT/GTT, on two separate alleles (Figure 5B, right panel) generated a pattern clearly distinct from Figure 3B. Similarly, for the second case, the GAG mutation (Figure 5C, left) qualitatively matched the four signature peaks (generated by the mutation) in the pyrogram for case 2 (Figure 3C), whereas a mixture of GAT/GGG alleles clearly did not (Figure 5C, right).
Figure 5.
Pyromaker-generated pyrosequencing traces, hypothesis-testing mode. Simulated pyrosequencing traces for wild-type KRAS (A), the two hypotheses for case 1 (B), and the two hypotheses for case 2 (C). Only those peaks that correspond to signature peaks identified in Figure 3 are numbered. Arrowheads indicate peaks that are not consistent with the experimental data (peaks that are either novel or not the appropriate height).
Iterative Approach
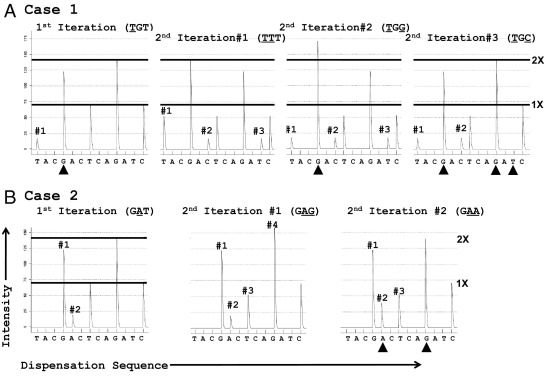
Alternatively, Pyromaker can be used to identify the mutated bases where the user iteratively adds putative mutated bases to the wild-type pyrogram to ultimately reproduce a given pyrogram. This can be done from just the pyrosequencing result by itself, without using Sanger sequencing data to first generate hypotheses. As discussed, the pyrogram for the first case contains three signature peaks in the pyrogram (Figure 3B). To obtain the first T peak (#1) ahead of the wild-type G, the minimum mutation needed is TGT (Figure 6A, far left); however, the resultant pyrogram does not include the other two signature peaks. To advance the polymerase and to obtain the C peak (#2 in Figure 3B) ahead of the wild-type T, and without inducing any other new peaks or frameshifting the gene, one must replace the G in the 12b position (TGT) with a second T to move the G activity (which otherwise blocks the polymerase from advancing) into the T position (ie, TTT, Figure 6A, middle-left). Alternatively, the T in the 12c position (TGT) can be replaced with a G to produce a run of 4 Gs (TGG/GGC, codons 12/13, Figure 6A, middle-right), thereby moving the T activity into the G position. Both of these produce the three signature peaks in the clinical pyrogram (Figure 3A), but only the TTT pyrogram contains the novel peaks at the correct peak height ratios (3:1:1, respectively). A third potential solution is TGC (Figure 6A, far right), which produces peaks #1 and #2, but not peak #3.
Figure 6.
Use of Pyromaker-generated pyrosequencing traces, iterative mode. A: Iterative approach to interpret case 1. The first iteration (TGT) is shown on the far left, followed by the three alternative second iterations. Only the TTT mutation produces the three signature peaks at the correct height ratios. B: Iterative approach to interpret case 2. Only those peaks corresponding to signature peaks identified in Figure 1 are numbered. Arrowheads indicate peaks that are not consistent with the experimental data (either missing peaks or ones that are not the appropriate height).
Similarly for case 2, the simplest way to reduce the G peak (Figure 3C, #1) and create the A peak (#2) before the T peak (#3) is a 12b G→A to produce a GAT (Figure 6B, left). (A 12a G→A, AGT, would manifest as an A peak ahead of the first G peak.) However, this single change to GAT does not produce changes #3 and #4 in Figure 3C. To reduce the T (#3) and increase the G activity (#4), a 12c T→G might occur to produce a GAG (Figure 6B, middle). Alternatively, a 12c T→A producing a GAA (Figure 6B, right) also reduces the T peak, but does not increase the subsequent G peak. In summary, one can identify a specific mutation in a gene by sequentially adding mutations to the wild-type sequence to iteratively reproduce a complex pyrogram.
Melting Curve Analysis and TA Cloning with Sequencing
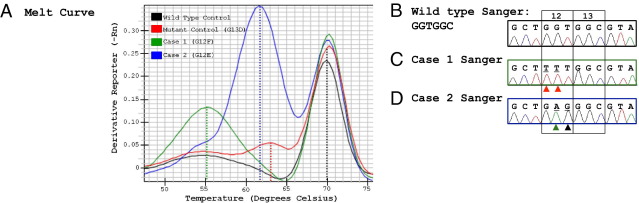
We also performed melt curve analysis. For case 1, the mutant allele demonstrated a melting temperature substantially lower than the wild-type (∼15°C) and single mutation (∼8°C) control (Figure 7A), consistent with a single allele with two adjacent mutated bases (TTT) that are presumably highly destabilizing to the DNA duplex. However, for case 2 there was a smaller difference (∼1.5°C) between the melting temperature for the double-base mutant allele (GAG) and the single-base mutant control (G13D, GAC). Two other single-base mutation controls (TGT, G12C; and TGC, G13C) melted in the same region as the single-base mutant control (data not shown).
Figure 7.
Melting curve analysis and TA cloning/sequencing. A: Melting curves for wild-type KRAS, a single mutant control, case 1 (TTT, G12F) and case 2 (GAG, G12E). Note that for case 1, the melt curve for the mutant allele is ∼9°C lower than the single-base GAC, G13D mutant control, whereas for case 2, there is only about a 1.5°C difference. B–D: TA cloning/sequencing results of single representative plasmids for wild-type KRAS (GGT) (B), case 1 (TTT, G12F) (C), and case 2 (GAG, G12E) (D). Red arrowheads indicate mutant T bases. Green arrowhead indicates mutant A base. Black arrowhead indicates mutant G base.
To unequivocally determine the KRAS mutation sequences, we performed TA cloning of PCR products, and Sanger sequencing of the cloned mutant KRAS plasmids. Double mutant TTT and GAG mutations were confirmed for these two cases as expected (Figure 7, B–D).
Some clinical molecular diagnostics laboratories rely on Qiagen's KRAS pyrosequencing interpretation software (“KRAS plug-in report”) for analysis of KRAS mutations. This software is able to identify mutant peaks and is proposed to assist and/or to automate the interpretation of KRAS mutants. When these two complex cases were tested, the software was unable to correctly identify the mutations (not shown).
Discussion
Sequencing technologies such as pyrosequencing or Sanger sequencing occasionally encounter complex results that are either not interpretable or difficult to interpret. In this report, we provide three strategies to resolve these issues using KRAS mutation detection as an example. We present Pyromaker as a validated tool that generates virtual pyrograms, and demonstrate that all codon 12 and 13 single and complex mutations generate unique pyrograms. Notably, some single-base mutations are indistinguishable from some complex ones (using the default dispensation sequence), suggesting that such complex mutations may be underreported.
Pyromaker allows for user-directed hypothesis testing by generating virtual traces that can be compared with the actual data. This distinction is important when difficult or confusing data are encountered for which common mutants do not offer a sufficient explanation of all of the features in the pyrogram. In addition, there are no limitations to the kinds of sequences that can be input into Pyromaker, so the same uses can be applied to pyrograms from other assays as the need arises. Given the fact that the common KRAS mutations are recognized easily by experienced molecular pathologists, the automation of simple mutations is less critical than the tools offered by Pyromaker that are necessary to understand more complex mutations. Pyromaker may be a useful tool when designing dispensation sequences for other genes, in that it allows one to confirm that a proposed dispensation detects all of the mutations.
Before performing the analysis, it was not immediately obvious that all of the pyrograms for each of the different codon 12 or 13 mutations must be unique. In fact, some are qualitatively highly similar (eg, Figure 1, GAT versus GGA). Because we have highlighted all of the peaks that are increased and decreased for each of the KRAS codon 12 and 13 mutations (Figures 1 and 2), quantitative differences that distinguish two qualitatively similar mutations can be easily identified. We also show that the pyrograms for all of the currently reported complex mutations (see Supplemental Figures S5 and S6 at http://jmd.amjpathol.org) are distinct from one another and distinct from the single-base changes. These may serve as reference materials for those signing out clinical results. In fact, we hypothesize that there are only three types of mutations that cannot be resolved by pyrosequencing: i) the exact location of lost base within a homopolymeric run (eg, if A9→A8 is due to loss of the second adenine versus the sixth adenine); ii) whether a particular pyrogram is due to a heterozygous mutation in a tumor with 60% cancer cells or a homozygous mutation in a tumor with 30% cancer cells; and iii) certain mutations with a dispensation order not designed to distinguish them (eg, AGT versus AAT as discussed above with the default dispensation).
Because of the sequencing by synthesis nature of pyrosequencing, cases with more than a single point mutation can produce complex pyrograms, and additional methods are needed to definitively identify the mutation in these cases. Sanger sequencing is clearly one of the options. For example, with the two cases presented in this report, Sanger sequencing data allowed us to list the alternative hypotheses of nucleotide alterations. With the possibilities suggested by Sanger sequencing in mind, we were able to interpret the pyrosequencing pattern thoroughly and accurately (see Supplemental Figure S7 at http://jmd.amjpathol.org). Supplemental Figure S8 (available at http://jmd.amjpathol.org) demonstrates how the advancing polymerase sequentially adds bases and generates signals. Thus pyrosequencing and Sanger sequencing are one combination that permit unambiguous interpretation (Table 1), although this adds to the cost and turn-around time of the testing.
Table 1.
Approaches to Resolving Complex Cases
| Technology | Unambiguous interpretation |
|---|---|
| Pyro alone | No |
| Sanger alone | No |
| Pyro + Sanger | Yes |
| Pyro + software⁎ | Yes |
| Pyro or Sanger + MCA | +/−† |
| Pyro or Sanger + C/S | Yes |
C/S, cloning and sequencing; MCA, melting curve analysis; Pyro, pyrosequencing; Sanger, Sanger sequencing.
Software used in iterative mode.
MCA was clearly distinct for the TTT mutation, but was less obvious for the GAG mutation.
Alternatively, Pyromaker is extremely helpful for quickly and efficiently testing the possibilities that can explain a complicated pyrosequencing result. As discussed, the hypothesis testing mode requires previous Sanger sequencing, and thus the iterative mode is preferred. With only the pyrogram in hand, one can add mutations to the wild-type sequence sequentially to reproduce the actual pyrogram (Figure 6, Table 1). This is the most efficient approach because it can be done with only the experimental pyrogram and the Pyromaker traces and does not require Sanger sequencing or any other data. It is readily and freely available online and can be accessed from any computer with Internet access.
In addition, melting curve analysis may supply helpful information. The effect of two-base mutations on melting temperature has not been studied in detail, but it does not seem to be easily extrapolated from data with single-base mispairs,18 and it is interesting that the two complex mutations have very different effects. In this regard, SantaLucia et al have studied in detail the effects of nearest neighbors on melting temperature for several specific mispairs.19 We note that additional optimization on a more advanced platform might permit this distinction, especially by using the delta Tm method.20 In this regard, TA cloning and sequencing provided an unequivocal interpretation; however, although it is substantially easier than in the past, it remains somewhat labor intensive, risks plasmid contamination of the laboratory, may delay reporting, and is not routinely used in most clinical molecular diagnostics laboratories. Without access to pyrosequencing, TA cloning and sequencing would probably be the best method for resolving ambiguous Sanger sequencing results, although limiting dilution PCR should also work. Other methods can also be used, but interestingly, some methods such as allele-specific PCR may not detect these complex mutations, which may be reported as wild type.
In summary, although pyrosequencing and Sanger sequencing are both powerful tools to resolve most mutations, for certain complex cases, neither of them alone is enough to provide a definitive interpretation. Additional methods, such as Pyromaker analysis or TA cloning/sequencing, allow one to definitively diagnose the mutant alleles. Iterative Pyromaker analysis is the least expensive and the fastest method for resolving these cases.
Acknowledgments
We thank Dr. Hanno Matthaei for the GNAS pyrogram and Drs. Michael Goggins and Mitsuro Kanda for the p53 pyrograms. We acknowledge Drs. Athanasios Tsiatis, Kathleen Murphy, and Susan Eshleman, in addition to Jacki Huckins (Qiagen) and Matthew Cousins and Kellie Feck (Applied Biosystems), for helpful discussions. We also acknowledge Lisa Haley and Stacy Mosier for expert technical assistance.
Footnotes
This work was funded in part by the Sol Goldman Pancreatic Cancer Research Center, the Michael Rolfe Foundation, the Dick Knox Foundation, and the NIH (CA130938).
G.C. and M.T.O. contributed equally to this work.
Supplemental material for this article can be found at http://jmd.amjapthol.org or at doi: 10.1016/j.jmoldx.2011.12.001.
Supplementary data
References
- 1.Hyman E.D. A new method of sequencing DNA. Anal Biochem. 1988;174:423–436. doi: 10.1016/0003-2697(88)90041-3. [DOI] [PubMed] [Google Scholar]
- 2.Ahmadian A., Ehn M., Hober S. Pyrosequencing: history, biochemistry and future. Clin Chim Acta. 2006;363:83–94. doi: 10.1016/j.cccn.2005.04.038. [DOI] [PubMed] [Google Scholar]
- 3.Nyren P. Enzymatic method for continuous monitoring of DNA polymerase activity. Anal Biochem. 1987;167:235–238. doi: 10.1016/0003-2697(87)90158-8. [DOI] [PubMed] [Google Scholar]
- 4.Ogino S., Kawasaki T., Brahmandam M., Yan L., Cantor M., Namgyal C., Mino-Kenudson M., Lauwers G.Y., Loda M., Fuchs C.S. Sensitive sequencing method for KRAS mutation detection by pyrosequencing. J Mol Diagn. 2005;7:413–421. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsiatis A.C., Norris-Kirby A., Rich R.G., Hafez M.J., Gocke C.D., Eshleman J.R., Murphy K.M. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. J Mol Diagn. 2010;12:425–432. doi: 10.2353/jmoldx.2010.090188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellis R.W., Defeo D., Shih T.Y., Gonda M.A., Young H.A., Tsuchida N., Lowy D.R., Scolnick E.M. The p21 src genes of Harvey and Kirsten sarcoma viruses originate from divergent members of a family of normal vertebrate genes. Nature. 1981;292:506–511. doi: 10.1038/292506a0. [DOI] [PubMed] [Google Scholar]
- 7.Parada L.F., Tabin C.J., Shih C., Weinberg R.A. Human EJ bladder carcinoma oncogene is homologue of Harvey sarcoma virus ras gene. Nature. 1982;297:474–478. doi: 10.1038/297474a0. [DOI] [PubMed] [Google Scholar]
- 8.Bos J.L., Fearon E.R., Hamilton S.R., Verlaan-de Vries M., van Boom J.H., van der Eb A.J., Vogelstein B. Prevalence of ras gene mutations in human colorectal cancers. Nature. 1987;327:293–297. doi: 10.1038/327293a0. [DOI] [PubMed] [Google Scholar]
- 9.Der C.J., Cooper G.M. Altered gene products are associated with activation of cellular rasK genes in human lung and colon carcinomas. Cell. 1983;32:201–208. doi: 10.1016/0092-8674(83)90510-x. [DOI] [PubMed] [Google Scholar]
- 10.Jones S., Zhang X., Parsons D.W., Lin J.C., Leary R.J., Angenendt P., Mankoo P., Carter H., Kamiyama H., Jimeno A., Hong S.M., Fu B., Lin M.T., Calhoun E.S., Kamiyama M., Walter K., Nikolskaya T., Nikolsky Y., Hartigan J., Smith D.R., Hidalgo M., Leach S.D., Klein A.P., Jaffee E.M., Goggins M., Maitra A., Iacobuzio-Donahue C., Eshleman J.R., Kern S.E., Hruban R.H., Karchin R., Papadopoulos N., Parmigiani G., Vogelstein B., Velculescu V.E., Kinzler K.W. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood L.D., Parsons D.W., Jones S., Lin J., Sjoblom T., Leary R.J., Shen D., Boca S.M., Barber T., Ptak J., Silliman N., Szabo S., Dezso Z., Ustyanksky V., Nikolskaya T., Nikolsky Y., Karchin R., Wilson P.A., Kaminker J.S., Zhang Z., Croshaw R., Willis J., Dawson D., Shipitsin M., Willson J.K., Sukumar S., Polyak K., Park B.H., Pethiyagoda C.L., Pant P.V., Ballinger D.G., Sparks A.B., Hartigan J., Smith D.R., Suh E., Papadopoulos N., Buckhaults P., Markowitz S.D., Parmigiani G., Kinzler K.W., Velculescu V.E., Vogelstein B. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 12.Pao W., Wang T.Y., Riely G.J., Miller V.A., Pan Q., Ladanyi M., Zakowski M.F., Heelan R.T., Kris M.G., Varmus H.E. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lievre A., Bachet J.B., Le Corre D., Boige V., Landi B., Emile J.F., Cote J.F., Tomasic G., Penna C., Ducreux M., Rougier P., Penault-Llorca F., Laurent-Puig P. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 14.Massarelli E., Varella-Garcia M., Tang X., Xavier A.C., Ozburn N.C., Liu D.D., Bekele B.N., Herbst R.S., Wistuba I.I. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res. 2007;13:2890–2896. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]
- 15.Karapetis C.S., Khambata-Ford S., Jonker D.J., O'Callaghan C.J., Tu D., Tebbutt N.C., Simes R.J., Chalchal H., Shapiro J.D., Robitaille S., Price T.J., Shepherd L., Au H.J., Langer C., Moore M.J., Zalcberg J.R. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 16.De Roock W., Jonker D.J., Di Nicolantonio F., Sartore-Bianchi A., Tu D., Siena S., Lamba S., Arena S., Frattini M., Piessevaux H., Van Cutsem E., O'Callaghan C.J., Khambata-Ford S., Zalcberg J.R., Simes J., Karapetis C.S., Bardelli A., Tejpar S. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304:1812–1820. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 17.Wallen M., Tomas E., Visakorpi T., Holli K., Maenpaa J. Endometrial K-ras mutations in postmenopausal breast cancer patients treated with adjuvant tamoxifen or toremifene. Cancer Chemother Pharmacol. 2005;55:343–346. doi: 10.1007/s00280-004-0923-x. [DOI] [PubMed] [Google Scholar]
- 18.Werntges H., Steger G., Riesner D., Fritz H.J. Mismatches in DNA double strands: thermodynamic parameters and their correlation to repair efficiencies. Nucleic Acids Res. 1986;14:3773–3790. doi: 10.1093/nar/14.9.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peyret N., Seneviratne P.A., Allawi H.T., SantaLucia J., Jr. Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A·A, C·C, G·G, and T·T mismatches. Biochemistry. 1999;38:3468–3477. doi: 10.1021/bi9825091. [DOI] [PubMed] [Google Scholar]
- 20.Montgomery J.L., Sanford L.N., Wittwer C.T. High-resolution DNA melting analysis in clinical research and diagnostics. Expert Rev Mol Diagn. 2010;10:219–240. doi: 10.1586/erm.09.84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.