Abstract
An important precondition for the successful development of diagnostic assays of cerebrospinal fluid (CSF) biomarkers of age-related neurodegenerative diseases is an understanding of the dynamic nature of the CSF proteome during the normal aging process. In this study, a novel proteomic technology was used to quantify hundreds of proteins simultaneously in the CSF from 90 cognitively normal adults 21 to 85 years of age. SomaLogic's highly multiplexed proteomic platform can measure more than 800 proteins simultaneously from small volumes of biological fluids using novel slow off-rate modified aptamer (SOMAmer) protein affinity reagents with sensitivity, specificity, and dynamic ranges that meet or exceed those of enzyme-linked immunosorbent assays. In the first application of this technology to CSF, we detected 248 proteins that possessed signals greater than twofold over background. Several novel correlations between detected protein concentrations and age were discovered that indicate that both inflammation and response to injury in the central nervous system may increase with age. Applying this powerful proteomic approach to CSF provides potential new insight into the aging of the human central nervous system that may have utility in discovering new disease-related changes in the CSF proteome.
Research focused on understanding and treating neurodegenerative disorders such as Alzheimer's disease (AD) or Parkinson's disease (PD) will benefit greatly from validated laboratory diagnostic methods to aid in diagnosing, quantifying progression, and assessing response to therapeutics. Laboratory diagnostic approaches to central nervous system (CNS) disorders are hampered by sampling issues. It is dangerous, impractical, and costly to directly sample CNS tissue by surgical biopsy; and, even when performed, the results of brain biopsy are often uninformative for directing a change in therapy.1 On the other hand, sampling blood or urine is relatively simple, but informative biomarkers of CNS disease are not always transmitted from the CNS to the peripheral circulation or may not be observed because of dilution. Efforts to find blood or urine tests for degenerative CNS disease have not yet yielded any clinically useful assays.2 Thus, efforts to diagnose neurological disorders such as AD have concentrated on cerebrospinal fluid (CSF),3,4 a fluid that is in direct contact with CNS tissue, yet is relatively easy to sample in a safe procedure (lumbar puncture) that can be performed in the outpatient setting.5
CSF is by no means a perfect diagnostic sample. Aside from considerable public concern over the so-called “spinal tap,” there are several reasons why the fluid itself is not an optimal substrate from which to recover diagnostically informative biomarkers. First, although CSF is partly derived from brain interstitial fluid, it is largely a transudate of plasma produced by choroid plexus.6,7 Second, CSF is recycled up to six times per day, possibly limiting the persistence of potential biomarkers that can be removed from CSF. Finally, although sampling CSF is safe and relatively straightforward in skilled hands, blood contamination is possible and can complicate collection and confound analysis.
Despite these potential limitations, the evidence supporting use of CSF biomarker-based diagnosis of CNS disease is increasing, especially for AD. CSF concentrations of Aβ42 and tau have been found to significantly decrease and increase, respectively, in patients who have AD or are at increased risk for obtaining a future diagnosis of AD.8–10 Research assays for these analytes are available, as is a commercial clinical assay (Athena Diagnostics ADmark assay, which also measures phosphorylated tau).
There is now an intense research effort to discover additional biomarkers to diagnose other age-related neurodegenerative diseases, to quantify progression, and to assess therapeutic response. Because neurodegenerative diseases like AD occur predominantly in older individuals, a key component of disease-focused biomarker discovery efforts will be a full understanding of the age-related changes in CSF that occur independently of disease. In addition, understanding the age-related changes in the CSF proteome will likely provide insight into the biology of aging in the CNS.
Although there have been numerous reports investigating the CSF proteome,11–15 as well as age-related changes in specific CSF biomarkers during the course of other studies,16–18 we are only aware of only one previous study that has taken an unbiased approach toward studying the entire CSF proteome in aging.19 Using an isotope labeling strategy paired with mass spectrometry (ICAT, for Isotope-Coded Affinity Tags), Zhang et al identified peptides corresponding to CSF proteins. Relative quantitation was implied by the mass spectrometry technique used, and confirmed by Western blot in a selected small subset of proteins, as a mass spectrometric proteomic approach could not be easily translated into a high-throughput assay. This limitation exemplifies a problem with current mass spectrometry–based approaches to proteomics; namely, proteomic biomarker discoveries made with mass spectrometry are often difficult to translate into clinically feasible assays. Furthermore, although mass spectrometry–based approaches have great promise in clinical proteomics, many challenges remain including issues of sensitivity (typically nmol/L in current approaches), quantification, specificity, reproducibility, throughput, and cost.20–25
Although immunoassays are currently preferred for targeted clinical assays, they often are not used in primary biomarker discovery efforts. Instead, the traditional pipeline for biomarker discovery has focused on mass spectrometric discovery of biomarkers followed by validation on targeted immunoassay platforms. This is because it is currently not possible to run immunoassays in high enough multiplex, ie, hundreds or thousands of analytes per assay, to effectively query a sufficient number of potential biomarkers in each clinical sample.26
Slow off-rate modified aptamers (SOMAmers) are a new class of aptamer binding reagents that contain novel chemically modified nucleotides and are selected with a novel systematic evolution of ligands by exponential enrichment (SELEX) process to have slow dissociation rates.27 SOMAmers enable multiplex quantitative measurements of thousands of proteins with sensitivity, specificity, and dynamic range that meet or exceed those of enzyme-linked immunosorbent assays.27 In these assays, SOMAmers are used in high multiplex (∼1000 simultaneous measurements presently) as binding reagents to quantify proteins in biological samples. In contrast to mass spectrometric assays, SOMAmer arrays use target-binding reagents that are not interfered with by noninformative, high-concentration proteins such as albumin. Because SOMAmer target affinities are on par with monoclonal antibodies (sub-nM Kd), SOMAmers can detect proteins at far lower concentrations than is possible in traditional discovery-mode shotgun mass spectrometric techniques while still detecting many proteins at once. Another distinct advantage of SOMAmer-based discovery is that SOMAmer-derived biomarker discoveries can, in principle, easily be converted to targeted assays because the reagents used for discovery and targeted assays are identical,28 obviating the need for prolonged efforts to translate findings to a separate platform. Novel SOMAmer discovery is also automated, taking a matter of days to weeks to generate candidate reagents for validation, as opposed to antibody generation in animals or cell culture, which can take weeks to months to generate reagent candidates.
Materials and Methods
Patient Samples
The Human Subjects Division at the University of Washington approved this study to collect specimens and analyze results from human subjects. Subjects were compensated community volunteers in good health, with no signs or symptoms suggesting cognitive decline or neurological disease. After providing written informed consent, all individuals underwent evaluation that consisted of medical history; physical and neurological examinations; laboratory tests (complete blood count, serum electrolytes, blood urea nitrogen, creatinine, glucose, vitamin B12, and thyroid-stimulating hormone), as well as neuropsychological assessment that consisted of the Mini-Mental State Examination (MMSE),29 memory tests including immediate and delayed Paragraph Recall as well as Category Fluency and Letter Fluency30,31 and parts A and B of the Trail Making Test as a measure of attention.32 All results were within normal limits for samples included in this study. Exclusion criteria were heavy cigarette smoking (>10 packs/yr), alcohol use other than social, or any psychotherapeutic use.
CSF was obtained by lumbar puncture as previously described.33 Briefly, after providing written informed consent, individuals were placed in the lateral decubitus position and the L4 to L5 interspace was infiltrated with 1% lidocaine. Lumbar puncture was performed with a 20-gauge or 24-gauge spinal needle, after which individuals remained at bed rest for 1 hour. All lumbar punctures were performed in the morning to limit potential circadian fluctuation in CSF protein concentrations. All CSF for analysis was obtained from the 15th to 25th milliliter collected, stored in 0.5-mL aliquots at −80°C, and never thawed before this study. Red blood cell (RBC) counts from samples averaged 14 ± 42 RBC/μL, and 83% of samples had RBC counts <10 RBC/μL (see Supplemental Figure S1 at http://ajp.amjpathol.org).
Assay Methodology
Details of the SOMAmer array technology and assay methodology have been reported elsewhere in great detail,27 but specific modifications used to adopt the technique to CSF follow. Briefly, oligonucleotides bearing modified nucleotide bases (SOMAmers) were produced in vitro to bind to 813 human proteins27 comprising a wide variety of different functional classes: 20% cytokines, 13% growth factors, 21% receptors, 17% proteases, 5% protease inhibitors, 20% kinases, 1% structural proteins, and 3% hormones. The protein targets were chosen based on literature reports of involvement in a wide variety of biological and pathophysiological processes (for description of protein targets, see Supplemental Table S1 and Supplemental Figure S2, available at http://ajp.amjpathol.org). Of note, the targets were not chosen to represent the entire CSF proteome, nor were they chosen for their specific relevance to CNS tissue, neurodegeneration, or aging. Specifically, although a SOMAmer to tau is included in the target list, no SOMAmer to Aβ42 is currently available.
Sample Thawing and Plating
Aliquots of 100% CSF, stored at −80°C, were thawed by incubating in a 25°C water bath for 10 minutes. After thawing, the samples were stored on ice before assay. An 80% sample solution was prepared by transferring 64 μL of thawed sample into the well of a 96-well plate (Hybaid Omnitube 0.3 mL, Fisher Scientific) containing 16 μL per well of the appropriate sample diluent at 4°C. Sample diluent was 0.2x SB17 (8 mmol/L HEPES, pH 7.5, 25 mmol/L NaCl, 1 mmol/L KCl, 0.2 mmol/L MgCl2, 0.2 mmol/L CaCl2). with 2.56 mmol/L MgCl2, 2 μmol/L Z-Block_2 (the modified nucleotide sequence (AC-(BnBn)7-AC),27 and 0.05% Tween.
Preparation of 40%, 4%, and 0.4% SOMAmer Solutions
Because target analyte concentrations vary widely in CSF, analyses were performed on three different CSF dilutions. To determine which SOMAmers should be used at each dilution, SOMAmer targets were assayed in a serum dilution series to define pools of SOMAmers giving the largest linear range of signal at each dilution. Pools of SOMAmers defined in this manner were then adapted to CSF by ensuring large linear signal ranges on diluted CSF. In this manner, segregating SOMAmers and mixing them with different sample dilutions (40%, 4%, and 0.4%) allowed the assay to span an expected range of protein concentrations in CSF.
Signal Quantitation
The signals for each protein were quantitated as relative fluorescence units (RFUs) on the oligonucleotide array used to make the final measurement, as indicated.27 Although calibration curves relating protein concentrations to RFU values can be derived, the purpose of this study was to identify relative, rather than absolute, changes in protein concentrations with age, and thus all analysis was performed on raw RFU values. In general, SOMAmer RFU signals are nearly linear with respect to target concentration for the targets discussed here (data not shown).
Spike and Recovery
Spike and recovery experiments were performed by the same assay methodology on samples prepared from spiked proteins added to two separate pooled lumbar CSF samples (Bioreclamation, Hicksville, NY). Spiked proteins included a selection of biomarkers from Table 1: BCAM, BMP-14, C7, cystatin C, Ephrin-A4, ERBB3, Factor D, HGF, IGFBP-4, PTN, TIMP-1, TNF sR-II, and Troponin I. Additional information for source proteins (commercial vendor, city, state, part number, recombinant/purified, biological source) is listed here: TNF sR-II (R & D Systems, Minneapolis, MN, 726-R2-050, recombinant, murine myeloma cell line, NS0 derived), BCAM (R & D Systems, 148-BC-100/CF, recombinant, murine myeloma cell line, NS0 derived), BMP-14 (Pepro Tech, Rocky Hill, NJ, 120-01X, recombinant, Escherichia coli derived), Troponin I (Research Diagnostics, Acton, MA, RDICARDTROPIB, purified, human cardiac tissue), ERBB3 (R & D Systems, 348-RB-050, recombinant, murine myeloma cell line, NS0 derived), Ephrin-A4 (R & D Systems, 369-EA-200/CF, recombinant, murine myeloma cell line, NS0 derived), HGF (R & D Systems, 294-HGN-025/CF, recombinant, murine myeloma cell line, NS0 derived), Factor D (VWR, Radnor, PA, 80051–580, purified, human), C7 (Quidel, San Diego, CA, A405, purified, human), PTN (Pepro Tech, 450–15, recombinant, E. coli derived), TIMP-1 (R & D Systems, 970-TM-010, recombinant, murine myeloma cell line, NS0 derived), IGFBP-4 (R & D Systems, 804-GB-025, recombinant, murine myeloma cell line, NS0 derived), and Cystatin C (R & D Systems, 1196-PI-010, recombinant, murine myeloma cell line, NS0 derived).
Table 1.
Candidate Biomarkers of Aging Identified with the Slow Off-Rate Modified Aptamer (SOMAmer) Platform
| SOMAmer target | Pearson correlation coefficient | UniProt accession ID |
|---|---|---|
| TNFsR.II | 0.81 | P20333 |
| Activin A | 0.76 | P08476 |
| Factor D | 0.74 | P00746 |
| SLPI | 0.72 | P03973 |
| BCAM | 0.70 | P50895 |
| C7 | 0.69 | P10643 |
| vWF | 0.69 | P04275 |
| TNFsR.I | 0.67 | P19438 |
| ERBB3 | 0.67 | P21860 |
| CD30 | 0.66 | P28908 |
| Ephrin.A4 | 0.64 | P52798 |
| HGF | 0.63 | P14210 |
| IGFBP.4 | 0.63 | P22692 |
| TIMP-1 | 0.61 | P01033 |
| Trypsin | 0.61 | P07477 |
| Laminin | 0.60 | P25391 |
| HCC.1 | 0.60 | Q16627 |
| suPAR | 0.60 | Q03405 |
| α1-Antichymotrypsin | 0.59 | P01011 |
| JAM.C | 0.59 | Q9BX67 |
| sTie.1 | 0.59 | P35590 |
| Endostatin | 0.58 | P39060 |
| CXCL16.soluble | 0.57 | Q9H2A7 |
| IL.1sRI | 0.56 | P14778 |
| ART | 0.56 | O00253 |
| Fibrinogen | 0.55 | P02671 |
| Siglec.7 | 0.54 | Q9Y286 |
| C1q | 0.53 | P02747 |
| Myoglobin | 0.53 | P02144 |
| Angiopoietin.1 | 0.52 | Q15389 |
| GFAP | 0.52 | P14136 |
| DC.SIGN | 0.51 | Q9NNX6 |
| C4b | 0.51 | P0C0L4 |
| Lysozyme | 0.51 | P61626 |
| C3 | 0.51 | P01024 |
| ROR1 | 0.51 | Q01973 |
| Siglec.3 | 0.50 | P20138 |
| iC3b | 0.50 | P01024 |
| Ceruloplasmin | 0.50 | P00450 |
| APRIL | 0.50 | O75888 |
| C9 | 0.50 | P02748 |
| Ephrin.A5 | 0.49 | P52803 |
| C3adesAr | 0.49 | P01024 |
| Cadherin.5 | 0.49 | P33151 |
| LBP | 0.49 | P18428 |
| PARC | 0.49 | P55774 |
| Sonic Hedgehog | 0.48 | Q15465 |
| PIGF | 0.48 | P49763 |
| C3a | 0.48 | P01024 |
| ESAM | 0.47 | Q96AP7 |
| Angiopoietin.2 | 0.47 | O15123 |
| Protein C | 0.46 | P04070 |
| TWEAK | 0.46 | O43508 |
| MMP.8 | 0.46 | P22894 |
| Dtk | 0.46 | Q06418 |
| TIG2 | 0.45 | Q99969 |
| TGF.bRIII | 0.44 | Q03167 |
| IGFBP.5 | 0.44 | P24593 |
| HCC.4 | 0.44 | O15467 |
| SCGF.alpha | 0.43 | Q9Y240 |
| IL.1RAcP | 0.43 | Q9NPH3 |
| ERBB1 | 0.43 | P00533 |
| Factor I | 0.43 | P05156 |
| Protein S | 0.42 | P07225 |
| DAN | 0.42 | P41271 |
| SCGF.beta | 0.41 | Q9Y240 |
| Lipocalin2 | 0.40 | P80188 |
| bFGF.R | 0.40 | P11362 |
| Angiogenin | 0.40 | P03950 |
| HMW kininogen | 0.39 | P01042 |
| TRAILR4 | 0.39 | Q9UBN6 |
| FSH | 0.38 | P01215 |
| NovH | 0.38 | P48745 |
| Gr. V Phospholipase A2 | −0.38 | P39877 |
| Cystatin C | −0.39 | P01034 |
| DRR1 | −0.44 | O95990 |
| Troponin I | −0.45 | P19429 |
| IGF.I | −0.45 | P01343 |
| BMP.14 | −0.45 | P43026 |
| Pleiotrophin | −0.47 | P21246 |
| MIP.3b | −0.52 | Q99731 |
Briefly, a master mix of all proteins at 2 nmol/L was added to pooled CSF, yielding the following added concentrations: 2000 pmol/L, 400 pmol/L, 80 pmol/L, 16 pmol/L, 3.2 pmol/L, 0.64 pmol/L, 0.128 pmol/L, and 0 pmol/L. Because the individual proteins have widely varying endogenous concentrations, the CSF pool was diluted at 40%, 3%, or 0.2% in SB buffer before spiking so that the spiked concentration would be comparable to the endogenous concentration somewhere over the assay's linear range. Of the spiked proteins, seven proteins were spiked into 40% diluted CSF (BCAM, BMP-14, Ephrin-A4, ERBB3, HGF, TNF sR-II, Troponin I); five proteins were spiked into 3% diluted CSF (C7, Factor D, IGFBP-4, TIMP-1, PTN); and one protein was spiked into 0.2% diluted CSF (Cystatin C). Corresponding dilutions into buffer only were performed as well and used as a standard curve. Standard curve data points were collected in triplicate, and spike data points were collected in singlicate on each pool.
The target concentration measured in the 0-pmol/L spiked sample (no added proteins) was subtracted from the target concentrations measured in the other samples to yield the recovered amount (for the ratio of the recovered amount to the added amount, given as the percent recovery, see Supplemental Table S2 available at http://ajp.amjpathol.org). Recovery was only calculated for data points that were within the linear range of the assay for each analyte.
Statistical Analysis
RFU signals for each protein target were tabulated and analyzed using the R statistical package. Pearson correlation coefficients of target RFUs to patient ages were derived from the data, including the P values of the correlation coefficients, and a Bonferroni correction (α = 0.05, 248 simultaneous analyses) was used to select significant correlations among the potential biomarkers. In addition, principal component analysis and principal component regression were used to assess for patterns of variation in the log-transformed data that correlated with biological age. Principal component regression was validated with the “leave one out” method. To highlight patterns of change due to aging, a mathematical manipulation known as disease-specific genomic analysis (DSGA34) was used to remove patterns of variation seen in young patients from the middle and older aged data sets.
Bioinformatics Analysis of Biomarkers
The DAVID bioinformatics database of the National Institute of Allergy and Infectious Diseases (http://david.abcc.ncifcrf.gov/home.jsp) was used to group biomarkers that were found to vary significantly with age into classifications based on gene ontology. Proteins were entered into the DAVID tool using the Uniprot Accession ID.
Results
Detecting Proteins in CSF with a SOMAmer Array
Of the 813 biomarker candidates assayed in this study, 248 were observed with signals greater than twofold over background levels in the 90 subjects. Spike and recovery experiments performed on pooled lumbar CSF yielded a median recovery of 104% (range 75.3% to 177.3%) across various concentrations of selected targets (see Supplemental Figure S3 and Supplemental Table S2 at http://ajp.amjpathol.org). Although quantitation, in terms of mass or moles of analyte per volume of CSF, is feasible with this technique, many hundreds of standards would have been needed to calibrate RFU values for individual proteins. Because we were interested in defining relative trends in biomarker concentration with age rather than with absolute concentration, we chose not to include absolute quantitation in the current study. We chose to use a twofold-over-background cutoff for signals by examining the variance from negative control SOMAmers (either SOMAmers with scrambled sequences or targeted to non-human proteins), and chose a twofold cutoff as a compromise between increasing the number of SOMAmer signals detected and decreasing the false-discovery rate.
Age-Dependent Changes in CSF Biomarkers
Correlation coefficients of those biomarkers that changed significantly with age, as well as average change between ages 21 and 85 years, are listed in Table 1. Interestingly, many more biomarkers were found to increase than to decrease with increasing age.
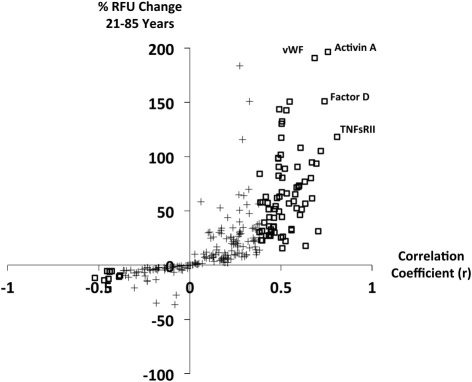
In Figure 1, the estimated percent change in biomarker RFU signal from age 21 to age 85 years (derived from linear regression) is plotted against the Pearson correlation coefficient for biomarker versus age. Note that RFU does not scale perfectly linearly with target concentration, so this percentage change is only a rough estimate of the magnitude of change. Because of the true log-linear or sigmoidal relationship between RFU and concentration, the relative changes in concentration of some biomarkers with age may be larger than the RFU changes suggest, such that a 100% increase in RFU could correspond to more or less than a 100% increase in concentration. Although many biomarkers with significant age-dependent changes showed RFU signal elevations of 30% to 75% between younger and older subjects, several of the most highly age-correlated biomarkers increased in RFU by more than 100% between younger and older individuals.
Figure 1.
Percent change in relative fluorescence unit signal between subjects 21 and 85 years of age, estimated from linear regression, plotted against Pearson correlation coefficients for biomarkers. Analytes with significant age correlation after Bonferroni correction are shown with open boxes, and remaining analytes are shown with crosses. Points corresponding to four biomarkers with large and significant age-dependent changes are labeled.
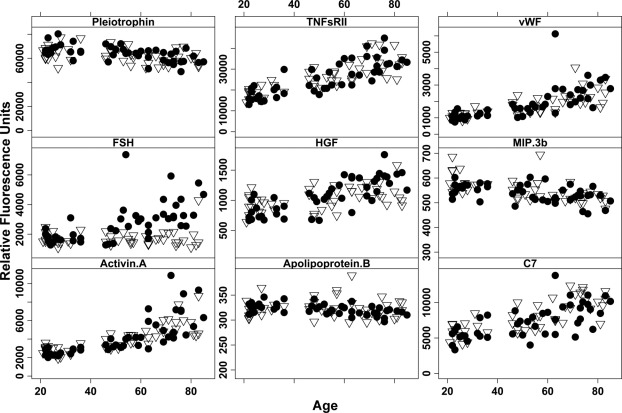
Plots of target RFU from the SOMAmer array versus age are shown in Figure 2 for some representative biomarkers that changed in concentration with age. No subjects 37 to 45 years of age were represented in this cohort, explaining the bimodal appearance of the data. Male and female subjects are shown with different symbols to highlight that most biomarker concentration changes were independent of sex, with the notable exception of follicle-stimulating hormone (FSH). Production of FSH in the pituitary increases around the time of menopause (40–60 years of age), indicating that this approach is able to detect expected and biologically significant age-dependent changes in CSF.
Figure 2.
Plots of SOMAmer signal (y axis, relative fluorescence units) versus age (x axis, years) for selected biomarkers. Female subjects (closed circles) and male subjects (open triangles) are identified in each plot.
Principal Component Analysis
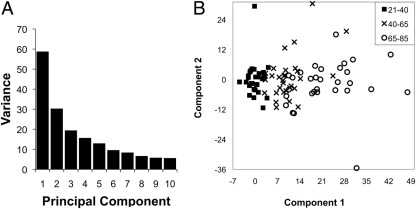
Principal component analysis of the log-transformed biomarker data set was performed, with results shown in Figure 3. Figure 3A is a scree plot demonstrating that much, but not all, of the variation in the data were explained by a single principal component. After using DSGA to remove the variation observed in subjects 21 to 40 years of age, principal components 1 and 2 were plotted (Figure 3B). Subjects classified as younger (21 to 40 years), middle-aged (40 to 65 years), and older (65 to 85 years) were clearly separated by principal component 1, further indicating that the variation present in the first principal component was correlated with age.
Figure 3.
A: Screeplot of principal component analysis from entire biomarker dataset. The first 10 principal components, which cumulatively account for 70% of the total variance in the data set, are shown. B: The first two principal components are plotted after DSGA to remove variation observed in subjects 20 to 40 years of age.
Principal Component Regression
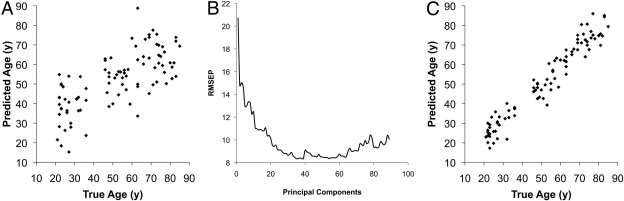
Regression analysis of the of the first principal component of biomarker data against subject age again showed that the first principal component explained much, but not all, of the age-dependent variance (Figure 4A) in the data. The error of prediction rapidly decreased when incorporating more principal components into the regression analysis (Figure 4B), reaching a global minimum of prediction error when including 38 principal components in the model (Figure 4C). These results indicated that there are multiple effects other than age that explain the variance in biomarker signatures.
Figure 4.
A: Principal component regression prediction of age by biomarker signature using only the first principal component. B: Results of leave-one-out (LOO) cross-validation of principal component regression. Root-mean-squared error of prediction (y axis) is plotted versus the number of components (x axis) included in the model. C: Principal component regression prediction of age by biomarker signature using the first 38 principal components, showing improved model prediction.
Bioinformatic Analysis of Age-Dependent CSF Biomarkers
Gene Ontology classification categories derived from this data set are shown in Table 2. By this preliminary analysis, the most significant ontological relationships among the age-dependent biomarkers involved inflammation and response to injury. In addition, significant alterations in several complement components indicated that complement activation may affect the biomarker profile in aging CSF. Additional biomarkers are found in gene ontology categories related to proteolysis, response to oxidative stress, and autophagy.
Table 2.
Gene Ontology Analysis of Age-Varying Biomarker Candidates
| Gene ontology classification⁎ | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 |
| TNFsR.II | • | • | • | ||||||||||||||
| Activin A | • | • | |||||||||||||||
| Factor D | • | • | • | • | • | • | • | ||||||||||
| SLPI | |||||||||||||||||
| BCAM | |||||||||||||||||
| C7 | • | • | • | • | • | • | • | ||||||||||
| vWF | • | • | • | • | |||||||||||||
| TNFsR.I | • | • | • | • | |||||||||||||
| ERBB3 | • | • | • | ||||||||||||||
| CD30 | • | ||||||||||||||||
| Ephrin.A4 | |||||||||||||||||
| HGF | • | ||||||||||||||||
| IGFBP.4 | • | • | • | ||||||||||||||
| TIMP-1 | • | • | |||||||||||||||
| Trypsin | • | ||||||||||||||||
| Laminin | • | • | • | ||||||||||||||
| HCC-1 | • | • | |||||||||||||||
| suPAR | • | • | • | • | • | ||||||||||||
| α1-antichymotrypsin | • | • | • | ||||||||||||||
| JAM.C | |||||||||||||||||
| sTie.1 | • | ||||||||||||||||
| Endostatin | • | • | • | • | |||||||||||||
| CXCL16.soluble | • | • | • | ||||||||||||||
| IL.1sRI | • | • | |||||||||||||||
| ART | |||||||||||||||||
| Fibrinogen | • | • | • | • | • | ||||||||||||
| Siglec.7 | |||||||||||||||||
| C1q | • | • | • | • | • | • | • | ||||||||||
| Myoglobin | • | • | |||||||||||||||
| Angiopoietin.1 | • | ||||||||||||||||
| GFAP | |||||||||||||||||
| DC.SIGN | |||||||||||||||||
| C4b | • | • | • | • | • | • | • | ||||||||||
| Lysozyme | • | • | • | ||||||||||||||
| C3 | • | • | • | • | • | • | • | ||||||||||
| ROR1 | |||||||||||||||||
| Siglec.3 | • | ||||||||||||||||
| iC3b | • | • | • | • | • | • | • | ||||||||||
| Ceruloplasmin | |||||||||||||||||
| APRIL | • | • | • | ||||||||||||||
| C9 | • | • | • | • | • | • | • | • | • | • | |||||||
| Ephrin.A5 | |||||||||||||||||
| C3adesAr | • | • | • | • | • | • | • | ||||||||||
| Cadherin.5 | • | • | |||||||||||||||
| LBP | • | • | • | • | |||||||||||||
| PARC | • | • | • | ||||||||||||||
| Sonic Hedgehog | • | • | • | • | • | • | • | • | • | • | • | ||||||
| PIGF | • | • | • | ||||||||||||||
| C3a | • | • | • | • | • | • | • | ||||||||||
| ESAM | |||||||||||||||||
| Angiopoeitin.2 | • | ||||||||||||||||
| Protein C | • | • | • | • | • | ||||||||||||
| TWEAK | • | • | • | ||||||||||||||
| MMP.8 | • | • | |||||||||||||||
| Dtk | |||||||||||||||||
| TIG2 | |||||||||||||||||
| TGF.bRIII | • | • | • | • | |||||||||||||
| IGFBP.5 | • | • | • | ||||||||||||||
| HCC.4 | • | • | • | ||||||||||||||
| SCGF.alpha | • | • | |||||||||||||||
| IL.1RAcP | • | • | • | • | |||||||||||||
| ERBB1 | • | • | • | • | |||||||||||||
| Factor I | • | • | • | • | • | • | • | ||||||||||
| Protein S | • | • | • | • | |||||||||||||
| DAN | |||||||||||||||||
| SCGF.beta | • | • | |||||||||||||||
| Lipocalin2 | |||||||||||||||||
| bFGF.R | • | • | • | ||||||||||||||
| Angiogenin | • | • | • | ||||||||||||||
| HMW kininogen | • | • | • | • | • | • | • | ||||||||||
| TRAILR4 | |||||||||||||||||
| FSH | • | ||||||||||||||||
| NovH | |||||||||||||||||
| Gr. V Phospholipase A2 | |||||||||||||||||
| Cystatin C | • | ||||||||||||||||
| DRR1 | |||||||||||||||||
| Troponin I | • | ||||||||||||||||
| IGF.I | • | • | • | • | • | • | |||||||||||
| BMP.14 | • | ||||||||||||||||
| PTN | • | • | |||||||||||||||
| MIP.3b | • | • | • | ||||||||||||||
1 = GO:0009611 response to wounding; 2 = GO:0006954 inflammatory response; 3 = GO:0006952 defense response; 4 = GO:0045087 innate immune response; 5 = GO:0006956 complement activation; 6 = GO:0042127 regulation of cell proliferation; 7 = GO:0040007 growth; 8 = GO:0042060 wound healing; 9 = GO:0007596 blood coagulation; 10 = GO:0008284 positive regulation of cell proliferation; 11 = GO:0016485 protein processing; 12 = GO:0030334 regulation of cell migration; 13 = GO:0001944 vasculature development; 14 = GO:0050878 regulation of body fluid levels; 15 = GO:0006508 proteolysis; 16 = GO:0006979 response to oxidative stress; 17 = GO:0006914 autophagy.
Discussion
CSF Proteome Detected by SOMAmer Array
This study represents the first broad survey of CSF proteins using an oligonucleotide SOMAmer array. The menu of SOMAmers used in this study was not initially intended to be used specifically for studies of the CNS, nor was the distribution of targets intended to reflect the relative distribution of classes of proteins in the entire proteome. Nonetheless, we fortuitously detected 248 proteins in CSF samples of normal, aging adults 21 to 85 years of age and identified significant trends in abundance over this age range for 82 proteins. Although the age dependencies of some of these CSF biomarkers, such as Activin A,16 have been reported previously in smaller studies, most are novel biomarkers of aging that are prominently associated with immune system activation or response to injury.
There are several advantages of this approach over mass spectrometry and immunoassays. First, all of the analyses performed on these samples required only 64 μL of CSF, whereas mass spectrometric studies achieving similarly detailed characterizations would probably require greater than 100 times as much sample. Second, no existing immunoassay platform has been demonstrated to operate at such a high multiplex in CSF as this approach. However, there are also potential pitfalls inherent in interpreting any data derived from an assay based on novel technology. It is not known, for example, how protein posttranslational modifications (PTMs) will affect aptamer binding in all cases, and heavily phosphorylated or glycosylated proteins may not be detected with aptamers used here that were created to bind to native proteins. Robustness to PTMs is a problem for any assay based on affinity reagents, however, and not just for aptamer-based assays. Therefore, work is ongoing to create PTM-specific SOMAmers, analogous to PTM-specific antibodies, to address more specifically the question of PTMs in studies such as this.
A total of 248 proteins were quantified at signals at least two-fold over background out of the 813 proteins targeted by the array. The remaining proteins were presumably not present in samples at concentrations greater than the lower limit of quantification (median limit of 1 pmol/L, ranging from 10fM-1 μmol/L for the array; data for all analytes detailed in Gold et al27). Further optimization of the assay for CSF could improve sensitivity and quantify more proteins, and this work is ongoing. The menu of SOMAmers to measure specific proteins is expanding, and more CNS-specific proteins have been added to the menu of >1000 targets in the months since these experiments were performed.
Biology and Pathology of CSF Aging Biomarkers
Before interpreting the biological significance of the results of the SOMAmer array, preanalytical variables should be examined, to minimize bias. Clinically, the subjects who donated CSF for this study were thoroughly examined physically and cognitively, supporting the contention that the data collected here represents “normal aging” rather than pathological changes. Still, it is possible that some of the elderly patients in the study could have had undetected, preclinical neurodegenerative disease, and it is thus possible (but unlikely) that some of these age-correlated biomarkers are instead disease-correlated biomarkers. A second possible source of preanalytical variation in any study of CSF is blood contamination caused by bleeding into the thecal space during the collection procedure. A reasonable check for this confounder is assessment of CSF apolipoprotein B (apoB), as it is present in blood but not in normal CSF.19 Although absolute concentration of apoB was not determined in the samples used in this study, the relative RFU signals corresponding to apoB concentrations in all samples were measured (Figure 2). Because the relative apoB signal in all samples were near the assay's limit of detection and did not vary appreciably with age, any blood contamination in these samples would appear to be small and uniform on average across ages, and would thus not confound attempts to identify apparent age-dependent trends in other biomarkers. It is possible, however, that an age-invariant rate of blood contamination could lead to spurious age-related signals if a putative biomarker varied in concentration in the blood; but the absolute level of blood contamination in these samples, as assessed by red blood cell counts, was low enough to make this possibility unlikely (see Supplemental Figure S1 at http://ajp.amjpathol.org). In addition, although several of the biomarkers reported here to increase with increasing age in CSF have been reported to behave similarly in plasma (Activin A,16 von Willebrand factor35), others displayed changes in CSF opposite from the expected age-related changes in serum (Troponin I,36 Cystatin C37), indicating that simple contamination with blood cannot be responsible for all of the CSF results. Finally, no target analyte concentration was significantly correlated with RBC counts (data not shown), indicating that contamination with blood was not reliably associated with increased biomarker signals.
Assuming that preanalytical variation is thus minimal, the data clearly indicate that several CSF proteins correlate with chronologic age, and could reflect normal CNS aging. In specific, Gene Ontology analysis indicated that markers of inflammation and wound repair are highly represented in the age-correlated proteins. Apart from supporting previous work indicating a role for inflammatory processes in CNS aging and degeneration,38,39 this finding should support the generation of many hypotheses about the interplay of tissue injury, inflammation, and the process of CNS injury, as well as suggest biomarkers that can be measured to interrogate the system.
Several of the age-correlated proteins detected here have been reported in CSF in a variety of clinical contexts but not always correlated with age, such as HGF (aging,17 neoplasia,40 vascular disorders,41 inflammation42), von Willebrand factor (infarction43), complement component C3a (aging18), MIP-3b (inflammation44,45), and SLPI (infection46). The increase in FSH observed in the female subjects is likewise in accord with well-known pituitary increases in FSH expression resulting from menopause.
von Willebrand factor demonstrated the largest average RFU signal increase between young and old subjects, with average RFU signals increasing almost 200% between 21 and 85 years of age, based on regression analysis. von Willebrand factor is a very large molecule, forming multimers with a monomer molecular weight of 260 kDa, so it is very unlikely that it crosses the blood–brain barrier into the CSF. Instead, it may derive from platelets or subendothelial connective tissue in response to tissue injury; the finding of increased fibrinogen in CSF with increasing age (Table 1) corroborates this interpretation, because fibrinogen plays a role in the coagulation cascade after tissue injury. These findings alone, however, do not prove that a chronic pattern of low-grade tissue or vascular injury accompanies normal aging, so further targeted experiments will be needed to support this hypothesis.
It is important to consider that although a number of biomarkers were significantly correlated with chronologic age, there was still substantial variation in the data that was not explained by age alone, as evidenced by the results of principal component analysis and principal component regression. This raises the question of whether a biomarker regression model can predict a “biological” CNS age, rather than just chronological CNS age. If the factors responsible for the observed variance in this model are reproducible and correlated with real biological effects, then regression-predicted age may, in fact, indicate whether one's “biological” and chronological CNS ages are discrepant even if the hidden effects are not yet understood. Such a biomarker profile, if appropriately validated, could thus be useful in studies of normal and pathological CNS aging.
Several approaches are currently underway to explain more completely the variance in correlations between proteins identified here and chronologic age. First, a number of other pathological processes, such as oxidative stress, could be responsible; thus we are beginning to correlate the findings of this SOMAmer array with measures of oxidative stress, such as F2-isoprostanes. Second, although the subjects included in this study were judged to be cognitively normal by clinical testing, it would be useful to understand whether mild variation in normal performance on cognitive tests is correlated with discrepancies between a subject's chronological age and the “biological” CNS age extrapolated from biomarker data; these studies are currently underway. Finally, the age-associated proteins discovered in this approach are being quantified in subject cohorts who have suspected or confirmed, early- or late-stage neurodenegerative disorders such as AD and PD. If these patients demonstrate significant biological and chronological CNS age discrepancies, this approach may eventually serve as an adjunct to studying and diagnosing suspected or overt neurodegenerative disease.
Footnotes
Supported by funding from SomaLogic, Inc., the Nancy and Buster Alvord Endowment, the Department of Veterans Affairs, an anonymous foundation, and NIH grant AG05136.
Disclosures: G.S.B. received research support from SomaLogic, Inc. S.K.N., T.R.K., A.S., S.W., and S.K. are employees of SomaLogic, Inc., and the assays reported in this manuscript were performed using reagents supplied by SomaLogic, Inc. None of the other authors disclosed any conflicts of interest.
Supplemental material for this article can be found at http://ajp.amjpathol.org or at doi: 10.1016/j.ajpath.2011.10.024.
Supplementary data
Histogram of red blood cell counts (RBCs/μL) in all samples used in this analysis.
Graphical representation of proteins measured in this assay.
Spike and recovery experiments on CSF pools. Green points/line: spikes into pool 1; blue points/line: spikes into pool 2; red points/line: spikes into buffer. The left-most data point in all plots (designated by a circled data point) is the 0 pmol/L-spike sample.
References
- 1.Warren J., Schott J., Fox N., Thom M., Revesz T., Holton J., Scaravilli F., Thomas D., Plant G., Rudge P., Rossor M. Brain biopsy in dementia. Brain. 2005;128:2016–2025. doi: 10.1093/brain/awh543. [DOI] [PubMed] [Google Scholar]
- 2.Schneider P., Hampel H., Buerger K. Biological marker candidates of Alzheimer's disease in blood, plasma, and serum. CNS Neurosci Ther. 2009;15:358–374. doi: 10.1111/j.1755-5949.2009.00104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verbeek M., De Jong D., Kremer H. Brain-specific proteins in cerebrospinal fluid for the diagnosis of neurodegenerative diseases. Ann Clin Biochem. 2003;40:25–40. doi: 10.1258/000456303321016141. [DOI] [PubMed] [Google Scholar]
- 4.Fagan A., Holtzman D. Cerebrospinal fluid biomarkers of Alzheimer's disease. Biomarkers Med. 2010;4:51–63. doi: 10.2217/BMM.09.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peskind E., Nordberg A., Darreh-Shori T., Soininen H. Safety of lumbar puncture procedures in patients with Alzheimer's disease. Curr Alzheimer Res. 2009;6:290–292. doi: 10.2174/156720509788486509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wood J. Physiology, pharmacology, and dynamics of cerebrospinal fluid. In: Wood J., editor. Neurobiology of Cerebrospinal Fluid. Plenum Press; New York: 1980. pp. 17–25. [Google Scholar]
- 7.Milhorat T. Cerebrospinal fluid as a reflection of internal milieu of brain. In: Wood J., editor. Neurobiology of Cerebrospinal Fluid 2. Plenum Press; New York: 1983. pp. 1–16. [Google Scholar]
- 8.De Meyer G., Shapiro F., Vanderstichele H., Vanmechelen E., Engelborghs S., De Deyn P., Coart E., Hansson O., Minthon L., Zetterberg H., Blennow K., Shaw L., Trojanowski J. Initiative ftAsDN: Diagnosis-independent Alzheimer disease biomarker signature in cognitively normal elderly people. Arch Neurol. 2010;67:949–956. doi: 10.1001/archneurol.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J., Sokal I., Peskind E., Quinn J., Jankovic J., Kenney C., Chung K., Millard S., Nutt J., Montine T. CSF multianalyte profile distinguishes Alzheimer and Parkinson diseases. Am J Clin Pathol. 2008;129:526–529. doi: 10.1309/W01Y0B808EMEH12L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li G., Sokal I., Quinn J., Leverenz J., Brodey M., Schellenberg G., Kaye J., Raskind M., Zhang J., Peskind E., Montine T. CSF tau/Abeta42 ratio for increased risk of mild cognitive impairment: a follow-up study. Neurology. 2007;69:631–639. doi: 10.1212/01.wnl.0000267428.62582.aa. [DOI] [PubMed] [Google Scholar]
- 11.Wenner B.R., Lovell M.A., Lynn B.C. Proteomic analysis of human ventricular cerebrospinal fluid from neurologically normal, elderly subjects using two-dimensional LC-MS/MS. J Proteome Res. 2004;3:97–103. doi: 10.1021/pr034070r. [DOI] [PubMed] [Google Scholar]
- 12.Sickmann A., Dormeyer W., Wortelkamp S., Woitalla D., Kuhn W., Meyer H.E. Identification of proteins from human cerebrospinal fluid, separated by two-dimensional polyacrylamide gel electrophoresis. Electrophoresis. 2000;21:2721–2728. doi: 10.1002/1522-2683(20000701)21:13<2721::AID-ELPS2721>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 13.Maccarrone G., Milfay D., Birg I., Rosenhagen M., Holsboer F., Grimm R., Bailey J., Zolotarjova N., Turck C.W. Mining the human cerebrospinal fluid proteome by immunodepletion and shotgun mass spectrometry. Electrophoresis. 2004;25:2402–2412. doi: 10.1002/elps.200305909. [DOI] [PubMed] [Google Scholar]
- 14.Yuan X., Desiderio D.M. Proteomics analysis of prefractionated human lumbar cerebrospinal fluid. Proteomics. 2005;5:541–550. doi: 10.1002/pmic.200400934. [DOI] [PubMed] [Google Scholar]
- 15.Schutzer S.E., Liu T., Natelson B.H., Angel T.E., Schepmoes A.A., Purvine S.O., Hixson K.K., Lipton M.S., Camp D.G., Coyle P.K., Smith R.D., Bergquist J. Establishing the proteome of normal human cerebrospinal fluid. PLoS One. 2010;5:e10980. doi: 10.1371/journal.pone.0010980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ebert S., Phillips D., Jenzewski P., Nau R., O'Connor A., Michel U. Activin A concentrations in human cerebrospinal fluid are age-dependent and elevated in meningitis. J Neurol Sci. 2006;250:50–57. doi: 10.1016/j.jns.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 17.Kern M., Bamborschke S., Nekic M., Schubert D., Rydin C., Lindholm D., Schirmacher P. Concentrations of hepatocyte growth factor in cerebrospinal fluid under normal and different pathological conditions. Cytokine. 2001;14:170–176. doi: 10.1006/cyto.2001.0875. [DOI] [PubMed] [Google Scholar]
- 18.Loeffler D., Brickman C., Juneau P., Perry M., Pomara N., Lewitt P. Cerebrospinal fluid C3a increases with age, but does not increase further in Alzheimer's disease. Neurobiol Aging. 1997;18:555–557. doi: 10.1016/s0197-4580(97)00110-3. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J., Goodlett D., Peskind E., Quinn J., Zhou Y., Wang Q., Pan C., Yi E., Eng J., Aebersold R., Montine T. Quantitative proteomic analysis of age-related changes in human cerebrospinal fluid. Neurobiol Aging. 2005;26:207–227. doi: 10.1016/j.neurobiolaging.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 20.Pan S., Aebersold R., Chen R., Rush J., Goodlett D., McIntosh M., Zhang J., Brentnall T. Mass spectrometry based targeted protein quantification: methods and applications. J Proteome Res. 2009;8:787–797. doi: 10.1021/pr800538n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Service R.F. Proteomics: Proteomics ponders prime time. Science. 2008;321:1758–1761. doi: 10.1126/science.321.5897.1758. [DOI] [PubMed] [Google Scholar]
- 22.Silberring J., Ciborowski P. Biomarker discovery and clinical proteomics. Trends Anal Chem. 2010;29:128. doi: 10.1016/j.trac.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell P. Proteomics retrenches. Nature Biotechnol. 2010;28:665–670. doi: 10.1038/nbt0710-665. [DOI] [PubMed] [Google Scholar]
- 24.Bell A.W., Deutsch E.W., Au C.E., Kearney R.E., Beavis R., Sechi S., Nilsson T., Bergeron J.J., Group H.T.S.W. A HUPO test sample study reveals common problems in mass spectrometry-based proteomics. Nat Methods. 2009;6:423–430. doi: 10.1038/nmeth.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aebersold R. A stress test for mass spectrometry-based proteomics. Nat Methods. 2009;6:411–412. doi: 10.1038/nmeth.f.255. [DOI] [PubMed] [Google Scholar]
- 26.Taussig M.J., Landegren U. Progress in antibody arrays. Targets. 2003;2:169–176. [PubMed] [Google Scholar]
- 27.Gold L., Ayers D., Bertino J., Bock C., Bock A., Brody E.N., Carter J., Dalby A.B., Eaton B.E., Fitzwater T., Flather D., Forbes A., Foreman T., Fowler C., Gawande B., Goss M., Gunn M., Gupta S., Halladay D., Heil J., Heilig J., Hicke B., Husar G., Janjic N., Jarvis T., Jennings S., Katilius E., Keeney T.R., Kim N., Koch T.H., Kraemer S., Kroiss L., Le N., Levine D., Lindsey W., Lollo B., Mayfield W., Mehan M., Mehler R., Nelson S.K., Nelson M., Nieuwlandt D., Nikrad M., Ochsner U., Ostroff R.M., Otis M., Parker T., Pietrasiewicz S., Resnicow D.I., Rohloff J., Sanders G., Sattin S., Schneider D., Singer B., Stanton M., Sterkel A., Stewart A., Stratford S., Vaught J.D., Vrkljan M., Walker J.J., Watrobka M., Waugh S., Weiss A., Wilcox S.K., Wolfson A., Wolk S.K., Zhang C., Zichi D. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraemer S., Vaught J., Bock C., Gold L., Katilius E., Keeney T., Kim N., Saccomano N., Wilcox S., Zichi D., Sanders G. From SOMAmer-based biomarker discovery to diagnostic and clinical applications: A SOMAmer-based, streamlined multiplexed proteomic assay. PLoS One. 2011;6(10):e26332. doi: 10.1371/journal.pone.0026332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Folstein M., Folstein S., McHugh P. Mini-mental state: A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 30.Wechsler W. Psychological Corporation; San Antonio, TX: 1987. Wechsler Memory Scale—Revised. [Google Scholar]
- 31.Craft S., Newcomer J., Kanne S., Dagogo-Jack S., Cryer P., Sheline Y., Luby J., Dagogo-Jack A., Alderson A. Memory improvement following induced hyperinsulinemia in Alzheimer's disease. Neurobiol Aging. 1996;17:123–130. doi: 10.1016/0197-4580(95)02002-0. [DOI] [PubMed] [Google Scholar]
- 32.Reitan R., Wolfson D. The Halstead-Reitan neuropsychological test battery. In: Wedding D., editor. The Neuropsychology Handbook: Behavioral and Clinical Perspectives. Springer; New York: 1986. pp. 134–160. [Google Scholar]
- 33.Peskind E., Riekse R., Quinn J., Kaye J., Clark C., Farlow M., Decarli C., Chabal C., Vavrek D., Raskind M., Galasko D. Safety and acceptability of the research lumbar puncture. Alzheimer Dis Assoc Disord. 2005;19:220–225. doi: 10.1097/01.wad.0000194014.43575.fd. [DOI] [PubMed] [Google Scholar]
- 34.Nicolau M., Tibshirani R., Børresen-Dale A., Jeffrey S. Disease-specific genomic analysis: identifying the signature of pathologic biology. Bioinformatics. 2007;23:957–965. doi: 10.1093/bioinformatics/btm033. [DOI] [PubMed] [Google Scholar]
- 35.Coppola R., Mari D., Lattuada A., Franceschi C. Von Willebrand factor in Italian centenarians. Haematologica. 2003;88:39–43. [PubMed] [Google Scholar]
- 36.Clerico A., Fortunato A., Ripoli A., Prontera C., Zucchelli G.C., Emdin M. Distribution of plasma cardiac troponin I values in healthy subjects: pathophysiological considerations. Clin Chem Lab Med. 2008;46:804–808. doi: 10.1515/CCLM.2008.162. [DOI] [PubMed] [Google Scholar]
- 37.Odden M.C., Tager I.B., Gansevoort R.T., Bakker S.J., Katz R., Fried L.F., Newman A.B., Canada R.B., Harris T., Sarnak M.J., Siscovick D., Shlipak M.G. Age and cystatin C in healthy adults: a collaborative study. Nephrol Dial Transplant. 2010;25:463–469. doi: 10.1093/ndt/gfp474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson C.J., Finch C.E., Cohen H.J. Cytokines and cognition—the case for a head-to-toe inflammatory paradigm. J Am Geriatr Soc. 2002;50:2041–2056. doi: 10.1046/j.1532-5415.2002.50619.x. [DOI] [PubMed] [Google Scholar]
- 39.Bodles A.M., Barger S.W. Cytokines and the aging brain—what we don't know might help us. Trends Neurosci. 2004;27:621–626. doi: 10.1016/j.tins.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Navarrete R., Garcia E., Arrieta O., Sotelo J. Hepatocyte growth factor in cerebrospinal fluid is associated with mortality and recurrence of glioblastoma, and could be of prognostic value. J Neurooncol. 2010;97:347–351. doi: 10.1007/s11060-009-0037-8. [DOI] [PubMed] [Google Scholar]
- 41.Nanba R., Kuroda S., Ishikawa T., Houkin K., Iwasaki Y. Increased expression of hepatocyte growth factor in cerebrospinal fluid and intracranial artery in moyamoya disease. Stroke. 2004;35:2837–2842. doi: 10.1161/01.STR.0000148237.13659.e6. [DOI] [PubMed] [Google Scholar]
- 42.Sy C., Tsai H., Wann S., Lee S., Liu Y., Chen Y. Cerebrospinal fluid hepatocyte growth factor level in meningitis. J Microbiol Immunol Infect. 2008;41:301–306. [PubMed] [Google Scholar]
- 43.Wen F., Yu S. A study on the content of CSF vWF in patients with cerebral infarction at different location. Thromb Haemost. 2002;88:1067–1068. [PubMed] [Google Scholar]
- 44.Pashenkov M., Söderström M., Link H. Secondary lymphoid organ chemokines are elevated in the cerebrospinal fluid during central nervous system inflammation. J Neuroimmunol. 2003;135:154–160. doi: 10.1016/s0165-5728(02)00441-1. [DOI] [PubMed] [Google Scholar]
- 45.Press R., Pashenkov M., Jin J., Link H. Aberrated levels of cerebrospinal fluid chemokines in Guillain-Barré syndrome and chronic inflammatory demyelinating polyradiculoneuropathy. J Clin Immunol. 2003;23:259–267. doi: 10.1023/a:1024532715775. [DOI] [PubMed] [Google Scholar]
- 46.Amin D., Ngoyi D., Nhkwachi G., Palomba M., Rottenberg M., Büscher P., Kristensson K., Masocha W. Identification of stage biomarkers for human African trypanosomiasis. Am J Trop Med Hyg. 2010;82:983–990. doi: 10.4269/ajtmh.2010.09-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Histogram of red blood cell counts (RBCs/μL) in all samples used in this analysis.
Graphical representation of proteins measured in this assay.
Spike and recovery experiments on CSF pools. Green points/line: spikes into pool 1; blue points/line: spikes into pool 2; red points/line: spikes into buffer. The left-most data point in all plots (designated by a circled data point) is the 0 pmol/L-spike sample.