Abstract
Alcohol consumption induces liver steatosis; therefore, this study investigated the possible role of adipose tissue dysfunction in the pathogenesis of alcoholic steatosis. Mice were pair-fed an alcohol or control liquid diet for 8 weeks to evaluate the alcohol effects on lipid metabolism at the adipose tissue–liver axis. Chronic alcohol exposure reduced adipose tissue mass and adipocyte size. Fatty acid release from adipose tissue explants was significantly increased in alcohol-fed mice in association with the activation of adipose triglyceride lipase and hormone-sensitive lipase. Alcohol exposure induced insulin intolerance and inactivated adipose protein phosphatase 1 in association with the up-regulation of phosphatase and tensin homolog (PTEN) and suppressor of cytokine signaling 3 (SOCS3). Alcohol exposure up-regulated fatty acid transport proteins and caused lipid accumulation in the liver. To define the mechanistic link between adipose triglyceride loss and hepatic triglyceride gain, mice were first administered heavy water for 5 weeks to label adipose triglycerides with deuterium, and then pair-fed alcohol or control diet for 2 weeks. Deposition of deuterium-labeled adipose triglycerides in the liver was analyzed using Fourier transform ion cyclotron mass spectrometry. Alcohol exposure increased more than a dozen deuterium-labeled triglyceride molecules in the liver by up to 6.3-fold. These data demonstrate for the first time that adipose triglycerides due to alcohol-induced hyperlipolysis are reverse transported and deposited in the liver.
Alcoholic liver disease may evolve through three progressive stages: steatosis (fatty liver), hepatitis, and cirrhosis. Alcoholic steatosis, characterized by lipid droplet accumulation in the cytoplasm of hepatocytes, is the critical initial metabolic disorder in the progression of alcoholic liver disease.1 Lipid droplets occupy cytoplasmic space of the hepatocytes, which may diminish cellular functions and make the hepatocytes susceptible to toxic or stress factors. Multiple mechanisms have been suggested to be involved in the pathogenesis of alcoholic steatosis, including induction of fatty acid synthesis, inhibition of fatty acid oxidation, and reduction of very low-density lipoprotein (VLDL) secretion to the blood.2–4 Increasing evidence also suggests that adipose tissue dysfunction may impact hepatic lipid metabolism.5–7
Lipid homeostasis at the liver–adipose tissue axis plays a key role in regulating whole-body energy homeostasis. White adipose tissue (WAT) is the major organ to store excess energy under positive energy balance conditions, whereas it provides energy for other organs by releasing fatty acids under negative energy balance conditions. Disordered fat storage function in WAT may cause excess fatty acid influx into the liver, leading to steatosis.5–7 A recent study demonstrated that diminishing lipid storage function in WAT by overexpressing leptin-receptor b (lpr-b) on the aP2-lpr-b promoter (aP2lepr-b transgene) in db/db mice attenuated obesity after high-fat feeding.8 However, the aP2lepr-b transgene significantly increased liver weight and triglyceride concentrations, and accelerated the development of diabetes. On the other hand, adiponectin-overexpressing ob/ob mice showed a higher level of fat mass, but a lower hepatic triglyceride level and improvement of insulin sensitivity.9 Therefore, healthy adipose tissue is required for maintaining lipid homeostasis at the liver–WAT axis.
Clinical studies have demonstrated that lower fat mass is associated with higher liver fat in alcoholics.10,11 Alcohol exposure reduced adipose mass in rodents12,13 while increased fatty acid uptake by hepatocytes.14,15 Our recent study demonstrated that suppression of alcohol-induced WAT loss by dietary zinc supplementation was associated with attenuation of steatosis in mice.13 These data suggest that alcohol may induce excess adipose lipolysis, thereby increasing fatty acid influx to the liver. The present study was designed to investigate the effects of alcohol on lipid metabolism at the adipose tissue–liver axis and to define the mechanistic link between WAT dysfunction and the development of alcoholic steatosis.
Materials and Methods
Effects of Alcohol on Lipid Metabolism at the Liver–WAT Axis in Alcoholic Steatosis (Experiment I)
Animals and Alcohol Feeding Experiments
Male C57BL/6 mice were obtained from Harlan Laboratories (Indianapolis, IN). All of the mice were treated according to the experimental procedures approved by the institutional animal care and use committee. Mice at 4 months of age were pair-fed a modified Lieber-DeCarli alcohol or control liquid diet for 8 weeks with a stepwise feeding procedure. The ethanol content (%, w/v) in the diet was 4.8 (34% of total calories) at initiation, and increased by 0.2% every 2 weeks, reaching 5.4 (38% of total calories) for the last 2 weeks. After 8 weeks of feeding, mice were anesthetized at 1:00pm, and plasma, liver and WAT samples were collected.
Plasma Alcohol, Alanine Aminotransferase, and Hormone Concentrations
Plasma alcohol concentrations were measured using an Ethanol Assay Kit (BioVision, Mountain View, CA). Plasma alanine aminotransferase (ALT) activity was colorimetrically measured using an Infinity ALT Reagent (Thermo Scientific, Waltham, MA). Plasma adrenaline and noradrenaline were measured with a 2-CAT Research ELISA kit (Labor Diagnostika, Nord, Germany), and insulin was measured with an Insulin (Mouse) Ultrasensitive ELISA kit (ALPCO Diagnostics, Salem, NH), respectively.
Insulin Tolerance Assay
Mice were fasted for 4 hours, and insulin solution (0.25 U/kg) was administrated via intraperitoneal injection. Glucose levels in tail vein blood samples were measured using an OneTouch Ultra2 Blood Glucose Meter (LifeScan, Milpitas, CA).
Assessment of WAT Lipolysis
Adipose tissue explants were obtained from the epididymal WAT after 8 weeks of feeding. Adipose tissue explants were washed in culture plates with pre-warmed Dulbecco's PBS containing 100 U/mL penicillin and 100 mg/mL streptomycin. After removing connective tissue and blood vessels by dissection, 20 mg of adipose tissue explants were placed into 24-well plates, cut into small pieces, and cultured in 200 μL of Dulbecco's modified Eagle's medium with 2 mmol/L l-glutamine, 50 U/mL penicillin, 50 mg/mL streptomycin, and 2% fatty acid-free bovine serum albumin for 2 hours. Free fatty acids released into the culture medium were determined by a Free Fatty Acid Quantification Kit (BioVision).
Histopathological Examination of WAT
Adipose tissues were fixed in 10% formalin, and processed for paraffin embedding. Paraffin tissue sections were cut at 7 μm, and processed for hematoxylin and eosin (H&E) stain. For estimating adipocyte size, diameters of adipocytes were measured under a microscope with a ×20 objective. Five tissue sections from each group were selected, and the diameters of 40 adipocytes from each tissue section were measured.
Detection of Liver Steatosis
Neutral lipids in the liver were detected by Oil Red O stain. Liver cryostat sections were cut at 7 μm, fixed with 10% formalin for 5 minutes, and stained with Oil Red O in 2-propynal solution for 10 minutes. Triglyceride and cholesterol concentrations in the liver were measured using Infinity Triglyceride Reagent and Infinity Cholesterol Reagent (Thermo Scientific), respectively, after extracting hepatic lipid with chloroform/methanol (2:1).
Quantitative PCR
The total RNA was isolated from WAT and liver, and reverse transcription was conducted with the TaqMan Reverse Transcription Reagents Kit (Applied Biosystems, Foster City, CA). The forward and reverse primers (Table 1) were designed using Primer Express Software (Applied Biosystems), and quantitative PCR (qPCR) analysis with SYBR green PCR Master Mix was performed on an Applied Biosystems PRISM 7500 Sequence Detection System (Applied Biosystems). The data were normalized to β-actin for liver samples and to acidic ribosomal phosphoprotein P0 (36B4) for adipose tissue samples, and presented as fold changes, setting the values of pair-fed mice as one.
Table 1.
Sequences of Primers for Real-Time RT-PCR
| Gene | GenBank accession no. | Primer sequences (forward/reverse) |
|---|---|---|
| CD36 | NM_007643 | 5′-ATGGGCTGTGATCGGAACTG-3′ |
| 5′-GTCTTCCCAATAAGCATGTCTCC-3′ | ||
| Fatp1 | NM_011977 | 5′-CGCTTTCTGCGTATCGTCTG-3′ |
| 5′-GATGCACGGGATCGTGTCT-3′ | ||
| Atgl | AK031609 | 5′-GAGCCCCGGGGTGGAACAAGAT-3′ |
| 5′-AAAAGGTGGTGGGCAGGAGTAAGG-3′ | ||
| Hsl | NM_010719 | 5′-GCCGGTGACGCTGAAAGTGGT-3′ |
| 5′-CGCGCAGATGGGAGCAAGAGGT-3′ | ||
| Gpam | NM_008149 | 5′-ACAGTTGGCACAATAGACGTTT-3′ |
| 5′-CCTTCCATTTCAGTGTTGCAGA-3′ | ||
| Dgat1 | NM_010046 | 5′-TCCGTCCAGGGTGGTAGTG-3′ |
| 5′-TGAACAAAGAATCTTGCAGACGA-3′ | ||
| Pparg | NM_011146 | 5′-TCGCTGATGCACTGCCTATG-3′ |
| 5′-GAGAGGTCCACAGAGCTGATT-3′ | ||
| Cebpα | NM_007678 | 5′-CAAGAACAGCAACGAGTACCG-3′ |
| 5′-GTCACTGGTCAACTCCAGCAC-3′ | ||
| Insr | NM_010568 | 5′-ATGGGCTTCGGGAGAGGAT-3′ |
| 5′-GGATGTCCATACCAGGGCAC-3′ | ||
| Irs-1 | NM_010570 | 5′-CGATGGCTTCTCAGACGTG-3′ |
| 5′-CAGCCCGCTTGTTGATGTTG-3′ | ||
| Raptor | NM_028898 | 5′-CAGTCGCCTCTTATGGGACTC-3′ |
| 5′-GGAGCCTTCGATTTTCTCACA-3′ | ||
| Rictor | NM_030168 | 5′-GCTGCGCTATCTCATCCAAGA-3′ |
| 5′-CTTTCTGACTAAGCGAAGGGC-3′ | ||
| Pp1r3α | NM_080464 | 5′-TCTGTCTGATTCTGTGTGTGAGG-3′ |
| 5′-GAGGTTGGAGTGTCCAGATACA-3′ | ||
| Pten | NM_008960 | 5′-TGGATTCGACTTAGACTTGACCT-3′ |
| 5′-GCGGTGTCATAATGTCTCTCAG-3′ | ||
| Socs3 | NM_007707 | 5′-ATGGTCACCCACAGCAAGTTT-3′ |
| 5′-TCCAGTAGAATCCGCTCTCCT-3′ | ||
| Fatp2 | NM_011978 | 5′-TCCTCCAAGATGTGCGGTACT-3′ |
| 5′-TAGGTGAGCGTCTCGTCTCG-3′ | ||
| Fatp5 | NM_009512 | 5′-CTACGCTGGCTGCATATAGATG-3′ |
| 5′-CCACAAAGGTCTCTGGAGGAT-3′ | ||
| Mttp | NM_008642 | 5′-CTCTTGGCAGTGCTTTTTCTCT-3′ |
| 5′-GAGCTTGTATAGCCGCTCATT-3′ | ||
| Apob | NM_009693 | 5′-TTGGCAAACTGCATAGCATCC-3′ |
| 5′-TCAAATTGGGACTCTCCTTTAGC-3′ | ||
| 36B4/Rplp0 | NM_007475 | 5′-AAGCGCGTCCTGGCATTGTCT-3′ |
| 5′-CCGCAGGGGCAGCAGTGGT-3′ | ||
| β-Actin | NM_007393 | 5′-GGCTGTATTCCCCTCCATCG-3′ |
| 5′-CCAGTTGGTAACAATGCCATGT-3′ |
Immunoblot Analysis
Whole-protein lysates of liver or adipose tissue were extracted using 10% Nonidet P-40 lysis buffer supplemented with 1% protease inhibitor cocktail and 1% phenylmethylsulfonyl fluoride. Protein concentrations were measured with a protein assay reagent based on the Bradford method (Bio-Rad, Hercules, CA). Aliquots containing 60 μg of proteins were loaded onto a 8% to 12% sodium dodecyl sulfate–polyacrylamide gel, transblotted onto polyvinylidene difluoride membrane, blocked with 5% nonfat dry milk in Tris-buffered saline with 0.1% Tween-20, and then incubated with one of the primary antibodies listed in Table 2. The membrane was then incubated with horseradish peroxidase–conjugated donkey anti-rabbit or goat anti-mouse IgG (Cell Signaling, Danvers, MA). The bound complexes were detected with enhanced chemiluminescence (GE Healthcare, Piscataway, NJ) and quantified by densitometry analysis. The immunoblot bands were quantified by densitometry analysis, and the ratio to β-actin was calculated and presented as fold changes, setting the values of pair-fed mice as one.
Table 2.
Primary Antibodies for Immunoblot Analysis and Immunohistochemistry
| Primary antibody | Full name | Molecular wt. | Source | Maker |
|---|---|---|---|---|
| ATGL | Adipose triglyceride lipase | 54 kDa | Rabbit | Cell Signaling |
| p-HSL | Phospho-hormone-sensitive lipase | 81 kDa | Rabbit | Cell Signaling |
| HSL | Hormone-sensitive lipase | 81 kDa | Rabbit | Cell Signaling |
| PPAR-γ | Peroxisome proliferator-activated receptor-gamma | 53,57 kDa | Rabbit | Cell Signaling |
| CEBP-α | CCAAT enhancer-binding protein alpha | 42 kDa | Rabbit | Santa Cruz Biotechnology |
| PTEN | Phosphatase and tensin homologue | 54 kDa | Rabbit | Cell Signaling |
| SOCS3 | Suppressor of cytokine signaling 3 | 26 kDa | Rabbit | Cell Signaling |
| p-PP1 | Phospho-type 1 protein phosphatase | 38 kDa | Rabbit | Cell Signaling |
| PP1 | Type 1 protein phosphatase | 38 kDa | Rabbit | Cell Signaling |
| CD36 | Fatty acid translocase | 53 kDa | Rabbit | Novus Biologicals |
| FATP5 | Fatty acid transport protein 5 | 75 kDa | Goat | Santa Cruz Biotechnology |
| MTTP | Microsomal triglyceride transfer protein | 97 kDa | Goat | Santa Cruz Biotechnology |
| ApoB | Apolipoprotein B100 | 512 kDa | Rabbit | Santa Cruz Biotechnology |
| β-Actin | Beta-actin | 42 kDa | Mouse | Santa Cruz Biotechnology |
Immunohistochemistry
Liver cryostat sections were cut at 5 μm, and fixed with cold acetone for 5 minutes. The sections were incubated with polyclonal rabbit anti-CD36 at 4°C overnight, followed by incubation with a horseradish peroxidase–conjugated anti-rabbit IgG for 30 minutes. Diaminobenzidine was used as a horseradish peroxidase substrate for visualization. The negative controls were conducted by omitting the primary antibody.
Statistics
All data are presented as mean ± SD. The data were analyzed by Student's t-test, and differences between pair-fed and alcohol-fed groups were considered significant at P < 0.05.
Defining the Mechanistic Link between Alcoholic Lipodystrophy and Steatosis (Experiment II)
Animals and Alcohol Feeding Experiments
To determine whether alcohol exposure causes a shift of triglyceride storage from adipose tissue to the liver, lipid homeostasis at the liver–adipose tissue axis was estimated by a deuterium-labeling approach. Adipose triglycerides were first labeled with deuterium before alcohol exposure, and then deposition of deuterium-labeled adipose triglycerides in the liver were detected after alcohol exposure. To label triglyceride in adipose tissues, a common approach using heavy water (2H2O) as a stable isotope tracer was followed.16 Male C57BL/6 mice at 2 months of age were given an initial priming dose of 99.8% 2H2O via intraperitoneal injection to achieve 2.5% of body water enrichment, followed by administration of 5% 2H2O in the drinking water for 5 weeks. Mice were then fed an ethanol-containing liquid diet or pair-fed a control liquid diet for 2 weeks. At the end of 2 weeks feeding, mice were anesthetized, and liver tissues were collected for measuring lipid components labeled by deuterium.
Mass Spectrometry of Deuterium-Labeled Triglycerides
Hepatic lipid was extracted by methanol-chloroform (v/v = 2:1). The direct infusion experiments were performed on a hybrid mass spectrometer, with linear quadrupole ion trap/Fourier transform ion cyclotron mass spectrometry (MS) (Thermo Electron Corporation, Bremen, Germany) equipped with a chip-based nano-electrospray ionization ion source (Triversa NanoMate; Advion Biosciences, Ithaca, NY). The mass spectrometer was operated in positive mode. Each metabolite extract was measured for 5 minutes, covering the mass-to-charge ratio (m/z) = 100 to 1600. All mass spectra were recorded using Fourier transform ion cyclotron resonance in the profile mode, and the resolving power was set at 400,000 at m/z = 400. The maximum ion accumulation time was set at 1000 ms. The ion optics was tuned for the sodium adduct of tricaprylin [(C27H50O6+Na+)] at m/z = 493.25 using the linear ion trap. The two most important nano-electrospray ionization parameters were as follows: the spray voltage = +1.8 kV, and the nitrogen gas pressure = 0.5 psi. The MS/MS spectrum of each metabolite ion was acquired on linear trap quadrupole MS. The parameters were set as follows: parent ion m/z isolation window = ±0.5, spectrum accumulation time = 1 minute. The collision-induced dissociation voltage is a molecule-dependent parameter and ranged from 16 to 40 mV.
Data Analysis
The instrument data were first reduced into a peak list using second-order polynomial fitting and Gaussian mixture model. After peak alignment, a contrast-based method was used for normalization.17 Both the Fisher's exact test and pairwise two-tailed t-test were used to study the regulation change of each metabolite between two physiological conditions. The parameters used during analysis are as follows: parent ion m/z accuracy <5 ppm and the q-value for false discovery rate <0.3. Metabolite identification was achieved in two sequential steps, tentative assignment and confirmative identification. The metabolite tentative assignment was accomplished using the Fourier transform MS data, whereas the confirmative identification was accomplished using the linear quadrupole ion trap MS/MS data. The tentative metabolite assignment was achieved by matching the experimentally measured metabolite ion m/z value and isotopic peak profile to the corresponding theoretical information of metabolites recorded in public databases, including the Kyoto Encyclopedia of Genes and Genomes, LIPID MAPS, and the Human Metabolome Database (HMDB), resulting in 43,245 records. Possible positive-mode adduct ions include H+, Na+, K+, and NH4+. For the confirmative metabolite identification, Mass Frontier 6.0 (Thermo Scientific) was used to generate the in silico MS/MS spectrum for each of the tentatively assigned metabolite candidates, and the in silico MS/MS spectrum was then matched to the experimental MS/MS spectrum of the metabolite ion. The metabolite candidate with the best MS/MS spectrum match was considered as the metabolite present in the sample.
Results
Chronic Alcohol Exposure Reduces WAT Mass and Adipocyte Size
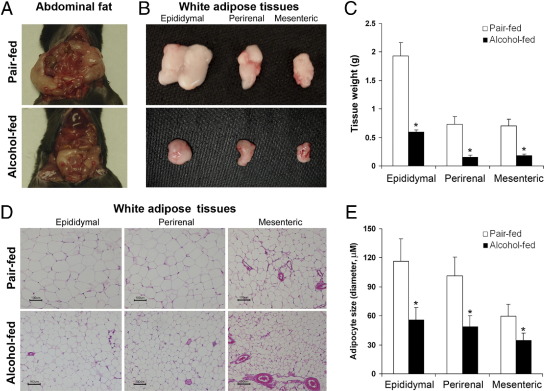
The plasma alcohol concentration after 8 weeks alcohol feeding was 148 ± 31 mg/dL, and the plasma ALT level was significantly higher in alcohol-fed mice (68.8 ± 17.0 U/L) than pair-fed mice (28.4 ± 6.7 U/L). The average body weight of alcohol-fed mice (27.2 ± 1.17 g) was significantly lower than the pair-fed mice (32.7 ± 1.52 g). Accordingly, the alcohol-fed mice exhibited less abdominal fat (Figure 1A). Alcohol exposure reduced the masses of all of the three major abdominal fat depots, including epididymal, perirenal, and mesenteric WAT (Figure 1B), and the weight loss was 69%, 79%, and 74%, respectively (Figure 1C). Histopathological observation found that alcohol exposure reduced the adipocyte size in all of the three abdominal fat depots (Figure 1D). Adipocyte diameters measured under microscope were reduced in alcohol-fed mice by 52%, 51%, and 42% in epididymal, perirenal, and mesenteric WAT (Figure 1E), respectively.
Figure 1.
Reduction of WAT mass and adipocyte size in mice chronically fed alcohol for 8 weeks. A: Abdominal fat. B: Abdominal WAT depots. C: Weight of abdominal WAT depots. D: Histopathology of WAT depots. E: Adipocyte size. Diameter of adipocytes on H&E tissue sections was measured under a microscope with a ×20 objective. Five tissue sections from each group were selected, and diameters of 40 adipocytes from each tissue section were measured. Results are means ± SD (n = 5 to 8 in C; n = 100 in D). Significant differences between alcohol-fed and pair-fed mice were determined by t-test. *P < 0.05.
Chronic Alcohol Exposure Enhances Adipose Tissue Lipolysis
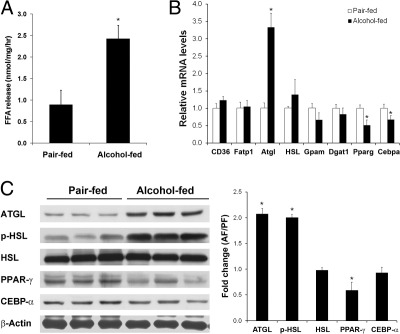
To understand how alcohol exposure reduces adipose tissue mass, lipolysis capacity was determined by incubating freshly isolated epididymal WAT. Adipose explants from alcohol-fed mice showed a twofold increase in fatty acid release during 2 hours incubation, in comparison with that of pair-fed mice (Figure 2A). Genes and proteins/enzymes involved in lipid metabolism were measured by qPCR and immunoblot analysis, respectively. As shown in Figure 2B, alcohol exposure did not affect fatty acid transport genes, but up-regulated adipose triglyceride lipase (ATGL) and down-regulated peroxisome proliferator activated receptor γ (PPAR-γ) and CCAAT-enhancing binding protein-α (C/EBP-α). Chronic alcohol exposure significantly increased the protein levels of ATGL and phosphorylated hormone sensitive lipase (p-HSL), whereas the total HSL protein level was not affected (Figure 2C).
Figure 2.
WAT hyperlipolysis in mice chronically fed alcohol for 8 weeks. A: Fatty acid release from epididymal WAT. Epididymal adipose explants from alcohol-fed mice were isolated, cut into small pieces, and cultured in Dulbecco's modified Eagle's medium for 2 hours. Free fatty acids released into the culture medium were determined. B: qPCR analysis of gene expression of epididymal adipose tissue. C: Immunoblot analysis of protein levels of epididymal WAT. The immunoblot bands were quantified by densitometry analysis, and the ratio to β-actin was calculated by setting the value of controls as one. Results are means ± SD (n = 5 in A; n = 4 in B; n = 3 in C). Significant differences between alcohol-fed and pair-fed mice were determined by t-test. *P < 0.05. AF, alcohol-fed; FFA, free fatty acid; PF, pair-fed.
Insulin Resistance Is Associated with Alcohol-Induced Adipose Dysfunction
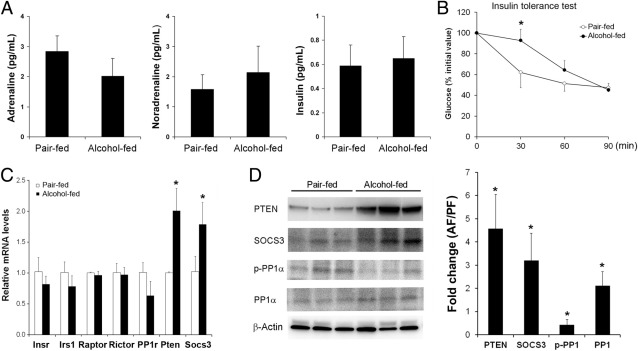
To explore the mechanisms by which alcohol stimulates lipolysis, plasma hormones involved in lipolysis regulation were measured. As shown in Figure 3A, neither positive regulators, adrenaline and nonadrenaline, nor negative regulator, insulin, were affected by chronic alcohol exposure. To determine whether alcohol exposure causes insulin resistance, an insulin tolerance test was performed. As shown in Figure 3B, alcohol-fed mice had a significantly higher blood glucose level at 30 minutes after insulin administration. Alterations of insulin signaling molecules in epididymal WAT were measured by qPCR and immunoblot, respectively. Chronic alcohol exposure did not affect genes of insulin receptor (Insr), insulin receptor substrate 1 (Irs1), raptor, rector, and protein phosphatase 1 (PP1), but significantly up-regulated phosphatase and tensin homolog (PTEN) and suppressor of cytokine signaling 3 (SOCS3) (Figure 3C). Immunoblot analysis demonstrated that chronic alcohol exposure increased the protein levels of PTEN and SOCS3. The protein level of p-PP1 was reduced by alcohol, whereas the total PP1 protein level was increased (Figure 3D).
Figure 3.
Insulin resistance in mice chronically fed alcohol for 8 weeks. A: Plasma hormone levels. Quantitative assay of adrenaline, noradrenaline, and insulin was performed using commercial enzyme-linked immunosorbent assay kits. B: Insulin tolerance assay. C: qPCR analysis of genes related to insulin signaling. D: Immunoblot analysis of proteins related to insulin signaling. The immunoblot bands were quantified by densitometry analysis, and the ratio to β-actin was calculated by setting the value of controls as one. Results are means ± SD (n = 5 to 8 in A and B; n = 4 in C; n = 3 in D). Significant differences between alcohol-fed and pair-fed mice are determined by t-test. *P < 0.05. AF, alcohol-fed; PF, pair-fed.
Up-Regulation of Fatty Acid Uptake Genes/Proteins Is Associated with Alcohol-Induced Hepatic Lipid Dyshomeostasis
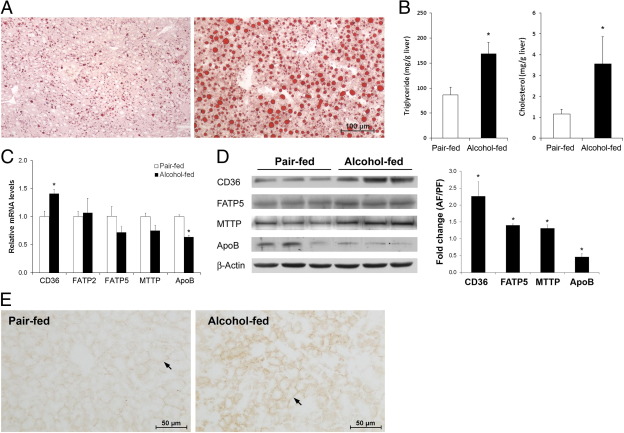
Alcohol exposure caused lipid accumulation in the liver as indicated by Oil Red O stain (Figure 4A) and increased triglyceride and cholesterol levels (Figure 4B). The mRNA and protein levels of major proteins that involve in hepatic fatty acid uptake and VLDL export were measured. Among the three proteins/enzymes related to fatty acid uptake, alcohol exposure increased the mRNA level (Figure 4C) of CD36/fatty acid translocase (FAT) and the protein levels (Figure 4D) of CD36/FAT and fatty acid transport protein 5 (FATP5). Alcohol exposure increased the protein level of microsomal triglyceride transfer protein (MTTP), but decreased the mRNA and protein levels of apolipoprotein B100 (ApoB100). Immunohistochemistry showed that CD36 mainly locates on the plasma membrane of hepatocytes, and chronic alcohol exposure increased CD36 distribution (Figure 4E).
Figure 4.
Hepatic lipid dyshomeostasis in mice chronically fed alcohol for 8 weeks. A: Lipid accumulation in the liver. Micro- and macrolipid droplets in hepatocytes are stained red with Oil Red O stain. B: Biochemical assay of hepatic triglyceride and cholesterol concentrations. C: qPCR analysis of genes related to lipid uptake and export. D: Immunoblot analysis of proteins related to lipid uptake and export. The immunoblot bands were quantified by densitometry analysis, and the ratio to β-actin was calculated by setting the value of controls as one. E: Immunohistochemical localization of CD36 in the liver. Arrows indicate plasma localization of CD36 on hepatocytes. Results are means ± SD (n = 5 to 8 in B; n = 4 in C; n = 3 in D). Significant differences between alcohol-fed and pair-fed mice were determined by t-test. *P < 0.05.
Identification of Hepatic Triglycerides Reverse Transported from the Adipose Tissue after Alcohol Exposure
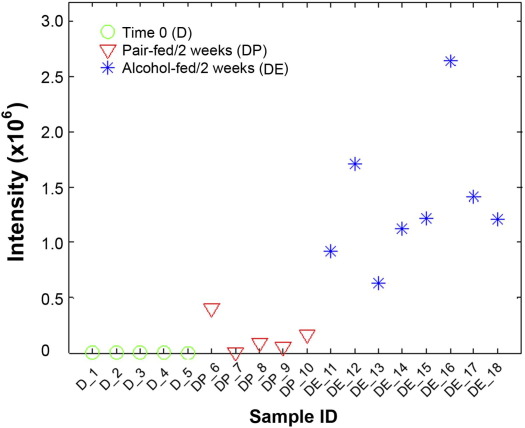
To determine whether fatty acids released from adipose tissue due to excess lipolysis may contribute to lipid accumulation in the liver, adipose triglycerides were first labeled by deuterium for 5 weeks (time 0) and then hepatic accumulation of deuterium-labeled metabolites after 2 weeks of control or alcohol liquid diet feeding was identified by Fourier transform MS and linear trap quadrupole MS/MS. Figure 5 shows the level of a deuterium-labeled triglyceride TG(16:0/18:1(9Z)/20:4(5Z,8Z,11Z,14Z))[iso6] (LIPID MAPS id: LMGL03010355 and HMDB id: HMDB05385) in each liver sample. In comparison with the baseline at time 0, the level of this deuterium-labeled triglyceride was increased in the liver of pair-fed mice. Alcohol feeding stimulated hepatic accumulation of this deuterium-labeled triglyceride in the liver as indicated by up to 6.3-fold increase, compared to the pair-fed mice. Table 3 lists all of the triglyceride molecules identified with significant regulation changes between the control cohort and the test cohort at 2 weeks, and the number of deuterium incorporated in each triglyceride molecule varied from one to four. Fourteen triglyceride molecules, identified via confirmative identification using MS/MS spectra, were recognized with a significant increase in the alcohol-fed mice, of which 2 molecules were detected by Fisher's exact test and 12 were detected by pairwise two-tailed t-test.
Figure 5.
Abundance of a deuterium-labeled triglyceride molecule in the liver of mice fed alcohol for 2 weeks. Mice were exposed to deuterium for 5 weeks (Group D), and then fed an ethanol liquid diet (Group DE) or pair-fed a control liquid diet (Group DP) for 2 weeks. A triglyceride molecule, TG(16:0/18:1(9Z)/20:4(5Z,8Z,11Z,14Z)), was identified by MS/MS analysis. The mean values of the peak area of this metabolite for Group D, DP, and DE are 3991.0, 198,341.1, and 1,260,999.2, respectively. The abundance test (pairwise two-tailed t-test) shows that the level of this triglyceride molecule after 2 weeks of alcohol feeding was up-regulated by up to 6.3-fold with a P value of 4.0 × 10−6, compared to pair-fed mice.
Table 3.
List of Deuterium-Labeled Triglycerides with Significant Regulation Changes between Control Cohort and Test Cohort in the Liver
| m/z | P value | Fold change (T/C) | Metabolite common name | Adduction | No. 2H |
|---|---|---|---|---|---|
| 878.7338 | 7.8 × 10−3 | 2.1 | TG(16:1(9Z)/18:2(9Z,12Z)/20:4(5Z,8Z,11Z,14Z))[iso6] | H+ | 1 |
| 904.7407 | 1.4 × 10−5 | 2.8 | TG(16:0/18:2(9Z,12Z)/20:4(5Z,8Z,11Z,14Z))[iso6] | Na+ | 3 |
| 904.7504 | 4.0 × 10−6 | 6.3 | TG(16:0/18:1(9Z)/20:4(5Z,8Z,11Z,14Z))[iso6] | Na+ | 1 |
| 907.7698 | 2.8 × 10−3 | 2.8 | TG(17:2(9Z,12Z)/17:2(9Z,12Z)/20:0)[iso3] | Na+ | 2 |
| TG(16:0/18:0/20:4(5Z,8Z,11Z,14Z))[iso6] | Na+ | 4 | |||
| TG(16:0/20:4(5Z,8Z,11Z,14Z)/20:4(5Z,8Z,11Z,14Z))[iso3] | H+ | 4 | |||
| 926.736 | 6.9 × 10−3 | 4.2 | TG(16:0/20:4(5Z,8Z,11Z,14Z)/20:4(5Z,8Z,11Z,14Z))[iso3] | Na+ | 1 |
| TG(18:3(9Z,12Z,15Z)/18:2(9Z,12Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z))[iso6] | H+ | 1 | |||
| 927.7376 | 2.3 × 10−3 | 1.9 | TG(16:0/20:4(5Z,8Z,11Z,14Z)/20:4(5Z,8Z,11Z,14Z))[iso3] | Na+ | 2 |
| 928.7407 | 5.7 × 10−3 | 1.9 | TG(16:0/20:4(5Z,8Z,11Z,14Z)/20:4(5Z,8Z,11Z,14Z))[iso3] | Na+ | 3 |
| 928.7513 | 6.8 × 10−2 | 2.9 | TG(18:3(9Z,12Z,15Z)/18:2(9Z,12Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z))[iso6] | H+ | 3 |
| 929.7534 | 1.6 × 10−2 | 1.7 | TG(16:0/20:4(5Z,8Z,11Z,14Z)/20:4(5Z,8Z,11Z,14Z))[iso3] | Na+ | 4 |
| TG(18:3(9Z,12Z,15Z)/18:2(9Z,12Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z))[iso6] | H+ | 4 | |||
| 930.7666 | 3.1 × 10−2 | 3.1 | TG(18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)/20:0)[iso3] | Na+ | 1 |
| 952.7521 | 8.6 × 10−3 | 6.3 | TG(18:1(9Z)/20:4(5Z,8Z,11Z,14Z)/20:4(5Z,8Z,11Z,14Z))[iso3] | Na+ | 1 |
| TG(18:3(9Z,12Z,15Z)/20:4(5Z,8Z,11Z,14Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z))[iso6] | H+ | 3 | |||
| 954.7666 | 9.1 × 10−3 | 3.0 | TG(20:4(5Z,8Z,11Z,14Z)/18:1(9Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z))[iso6] | H+ | 1 |
| 903.7797 | 7.2 × 10−3 | NA⁎ | TG(16:0/18:0/18:0)[iso3] | K+ | 2 |
| 905.7533 | 4.8 × 10−3 | NA⁎ | TG(16:0/18:2(9Z,12Z)/20:4(5Z,8Z,11Z,14Z))[iso6] | Na+ | 4 |
| TG(16:0/20:4(5Z,8Z,11Z,14Z)/20:4(5Z,8Z,11Z,14Z))[iso3] | H+ | 2 |
C, control cohort; T, test cohort.
NA refers to a metabolite that was detected only in the test cohort (deuterium/alcohol-fed), not in the control (deuterium/pair-fed) cohort. Therefore, the values of fold change for these metabolites are not available.
Discussion
The present study demonstrated a significant loss of WAT in a mouse model of alcoholic steatosis. Although reduction of adipocyte size is likely a major cause of WAT loss, alcohol-induced hyperlipolysis due to insulin resistance accounts for the reduction of adipocyte size. On the other hand, alcohol exposure up-regulated hepatic fatty acid uptake proteins in association with development of steatosis. A deuterium tracer approach revealed that adipose triglycerides labeled with deuterium were deposited in the liver after alcohol exposure. These data demonstrated a mechanistic link between WAT loss and hepatic lipid gain in alcoholic steatosis. Although disturbance of adipokine secretion has been implicated in the dysregulation of hepatic lipid metabolism in alcoholic steatosis,18–20 the present study suggests that WAT dysfunction can directly impact hepatic lipid homeostasis by reverse triglyceride transport.
Adipocytes take up triglycerides from VLDL or chylomicrons, and release fatty acids via ATGL/HSL-mediated lipolysis.21 Reduction of adipose mass could result from either reduced triglyceride uptake or increased adipose lipolysis. A previous study demonstrated that chronic alcohol exposure increased triglyceride synthesis and degradation by 1.4-fold and 2.3-fold, respectively, suggesting an increased triglyceride turnover in WAT.12 The present study found that chronic alcohol exposure enhanced WAT lipolysis in association with activation of the major adipose triglyceride hydrolases, ATGL and HSL. Alcohol exposure did not affect WAT genes involved in triglyceride uptake and triglyceride synthesis, although PPAR-γ and C/EBP-α, the key adipose transcription factors, were reduced. These data suggest that hyperlipolysis is like the major functional defect of WAT after chronic alcohol exposure.
Adipose lipolysis is regulated positively by catecholamine and negatively by insulin.21 Previous reports have shown that chronic alcohol–stimulated triglyceride turnover in adipose tissue was not due to catecholamine-mediated lipolysis.12,22 Instead, chronic alcohol exposure impaired insulin-mediated negative regulation of lipolysis.12 Insulin signaling up-regulates PDE3B, which reduces cellular cAMP level, thereby suppressing the lipolysis pathway.23 However, previous reports showed that neither adipose PDE3B nor insulin-induced tyrosine phosphorylation of phosphoinositide 3-kinase (PI3K, p85 subunit) and phosphorylation of Akt was affected by chronic alcohol feeding.12,24 The antilipolysis action of insulin may also be mediated by activation of PP1. Insulin activates PP1 through phosphorylation of its regulating subunit, and activated PP1, in turn, dephosphorylates and deactivates HSL.23 Although the plasma catecholamine and insulin levels were not affected by alcohol exposure, the present study demonstrated that chronic alcohol exposure inactivated adipose PP1, suggesting dysregulation of intracellular insulin signal transduction. Further analysis demonstrated that chronic alcohol exposure up-regulated the insulin signaling negative regulators PTEN and SOC3. Our data suggest that inhibition of insulin signal transduction is likely a key mechanism underlying alcohol-induced adipose insulin resistance and hyperlipolysis.
The liver plays a central role in whole-body lipid homeostasis. Although the effects of alcohol exposure on liver lipogenesis and fatty acid oxidation have been implicated in the pathogenesis of alcoholic steatosis,24–26 increasing evidence indicates that alcohol increases the hepatic capacity in uptake of exogenous fatty acids. A HepG2 cell culture study showed that alcohol treatment increased 3H-oleic acid uptake in a dose-dependent manner.14 Primary hepatocyte culture also showed that 3H-oleic acid uptake was increased by 2.6-fold in rats chronically fed alcohol.15 One-dose alcohol exposure to rats increased the incorporation of 14H-palmitate to the liver triglyceride by 50%.27 Chronic alcohol feeding to rats induced a twofold increase in the incorporation of 3H-palmitate to triglyceride or total lipid in the liver.28 In agreement with these reports, the present study demonstrated up-regulation of hepatic proteins related to fatty acid uptake, in particular, CD36, which is not expressed predominantly in the normal liver.29 Most importantly, the present study demonstrated for the first time that chronic alcohol exposure stimulated reverse transport and deposition of deuterium-labeled adipose triglycerides in the liver. These data suggest that adipose triglyceride reverse transport is a causal factor in the pathogenesis of alcoholic steatosis. Unexpectedly, an increase in deuterium-labeled triglycerides was also found in the liver of pair-fed mice. Because the control liquid diet used in the present study contains high fat (35% fat calories), these data suggest that reverse triglyceride transport mechanism may also apply to high fat–induced fatty liver.
Liver exports lipid in the form of VLDL for utilization by various tissues or storage in the WAT. Alcohol exposure has been shown to reduce hepatic VLDL secretion rate, and the levels of cholesterol and triglyceride in VLDL was reduced in alcohol-fed rats.30,31 Normalization of VLDL secretion was associated with attenuation of alcoholic steatosis by zinc, PPARα agonist, or betaine.13,32,33 VLDL is assembled from triglycerides, cholesterol, and apolipoproteins by MTTP with ApoB100 as an essential protein.34 The present study found that alcohol exposure increased hepatic MTTP protein, but decreased ApoB100 protein, suggesting that impaired AopB100 production is likely a limiting factor in alcohol-reduced VLDL secretion. Indeed, up-regulation of ApoB100 protein was associated with reduction of hepatic lipid by hepatocyte growth factor or PPAR-γ agonist.35,36 These results suggest that AopB100 is a potential molecular target in correcting alcohol-induced hepatic lipid dyshomeostasis.
In conclusion, the present study demonstrated that reduction of WAT mass and adipocyte size was associated with alcoholic steatosis. Activation of ATGL and HSL due to adipose insulin resistance is likely the major cause in alcohol-induced WAT reduction. Chronic alcohol exposure also caused an imbalance in gene/protein expression between lipid import and export toward lipid accumulation in the liver. The approach of deuterium incorporation into adipose triglycerides demonstrated, for the first time, that the triglycerides, which are stored in the adipose tissue before alcohol exposure, were reverse transported and deposited in the liver after alcohol exposure. These data provide solid evidence to support an organ–organ interaction mechanism underlying alcohol-induced lipid accumulation in the liver.
Acknowledgments
We thank Marion McClain for review of the manuscript.
Footnotes
Supported in part by the NIH (R01AA018844, R01GM087735, 1RC2AA019385, P01AA017103, P30AA019360, R01AA015970, R37AA010762, R01AA018016, R01AA018869, R01DK7071765) and the Veterans Administration. C.J.M. is a Distinguished University Scholar of the University of Louisville.
CME Disclosure: None of the authors disclosed any relevant financial relationships.
Contributor Information
Xiang Zhang, Email: xiang.zhang@louisville.edu.
Zhanxiang Zhou, Email: z_zhou@uncg.edu.
References
- 1.Hall P. Pathological spectrum of alcoholic liver disease. In: Hall P., editor. Alcoholic Liver Disease. 2nd ed. Edward Arnold; London: 1995. pp. 41–88. [Google Scholar]
- 2.Lakshman M.R. Some novel insights into the pathogenesis of alcoholic steatosis. Alcohol. 2004;34:45–48. doi: 10.1016/j.alcohol.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Purohit V., Russo D., Coates P.M. Role of fatty liver, dietary fatty acid supplements, and obesity in the progression of alcoholic liver disease: introduction and summary of the symposium. Alcohol. 2004;34:3–8. doi: 10.1016/j.alcohol.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 4.You M., Crabb D.W. Recent advances in alcoholic liver disease II: Minireview: molecular mechanisms of alcoholic fatty liver. Am J Physiol Gastrointest Liver Physiol. 2004;287:G1–G6. doi: 10.1152/ajpgi.00056.2004. [DOI] [PubMed] [Google Scholar]
- 5.Lafontan M., Girard J. Impact of visceral adipose tissue on liver metabolism. Part I: heterogeneity of adipose tissue and functional properties of visceral adipose tissue Diabetes Metab. 2008;34:317–327. doi: 10.1016/j.diabet.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Cusi K. The role of adipose tissue and lipotoxicity in the pathogenesis of type 2 diabetes. Curr Diab Rep. 2010;10:306–315. doi: 10.1007/s11892-010-0122-6. [DOI] [PubMed] [Google Scholar]
- 7.Wree A., Kahraman A., Gerken G., Canbay A. Obesity affects the liver: the link between adipocytes and hepatocytes. Digestion. 2011;83:124–133. doi: 10.1159/000318741. [DOI] [PubMed] [Google Scholar]
- 8.Wang M.Y., Grayburn P., Chen S., Ravazzola M., Orci L., Unger R.H. Adipogenic capacity and the susceptibility to type 2 diabetes and metabolic syndrome. Proc Natl Acad Sci U S A. 2008;105:6139–6144. doi: 10.1073/pnas.0801981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J.Y., van de Wall E., Laplante M., Azzara A., Trujillo M.E., Hofmann S.M., Schraw T., Durand J.L., Li H., Li G., Jelicks L.A., Mehler M.F., Hui D.Y., Deshaies Y., Shulman G.I., Schwartz G.J., Scherer P.E. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. J Clin Invest. 2007;117:2621–2637. doi: 10.1172/JCI31021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Addolorato G., Capristo E., Greco A.V., Stefanini G.F., Gasbarrini G. Energy expenditure, substrate oxidation, and body composition in subjects with chronic alcoholism: new findings from metabolic assessment. Alcohol Clin Exp Res. 1997;21:962–967. [PubMed] [Google Scholar]
- 11.Addolorato G., Capristo E., Greco A.V., Stefanini G.F., Gasbarrini G. Influence of chronic alcohol abuse on body weight and energy metabolism: is excess ethanol consumption a risk factor for obesity or malnutrition. J Intern Med. 1998;244:387–395. doi: 10.1046/j.1365-2796.1998.00381.x. [DOI] [PubMed] [Google Scholar]
- 12.Kang L., Chen X., Sebastian B.M., Pratt B.T., Bederman I.R., Alexander J.C., Previs S.F., Nagy L.E. Chronic ethanol and triglyceride turnover in white adipose tissue in rats: inhibition of the anti-lipolytic action of insulin after chronic ethanol contributes to increased triglyceride degradation. J Biol Chem. 2007;282:28465–28473. doi: 10.1074/jbc.M705503200. [DOI] [PubMed] [Google Scholar]
- 13.Kang X., Zhong W., Liu J., Song Z., McClain C.J., Kang Y.J., Zhou Z. Zinc supplementation reverses alcohol-induced steatosis in mice through reactivating hepatocyte nuclear factor-4alpha and peroxisome proliferator-activated receptor-alpha. Hepatology. 2009;50:1241–1250. doi: 10.1002/hep.23090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou S.L., Gordon R.E., Bradbury M., Stump D., Kiang C.L., Berk P.D. Ethanol up-regulates fatty acid uptake and plasma membrane expression and export of mitochondrial aspartate aminotransferase in HepG2 cells. Hepatology. 1998;27:1064–1074. doi: 10.1002/hep.510270423. [DOI] [PubMed] [Google Scholar]
- 15.Berk P.D., Zhou S., Bradbury M.W. Increased hepatocellular uptake of long chain fatty acids occurs by different mechanisms in fatty livers due to obesity or excess ethanol use, contributing to development of steatohepatitis in both settings. Trans Am Clin Climatol Assoc. 2005;116:335–344. [PMC free article] [PubMed] [Google Scholar]
- 16.Pouteau E., Beysen C., Saad N., Turner S. Dynamics of adipose tissue development by 2H2O labeling. Methods Mol Biol. 2009;579:337–358. doi: 10.1007/978-1-60761-322-0_17. [DOI] [PubMed] [Google Scholar]
- 17.Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 18.Xu A., Wang Y., Keshaw H., Xu L.Y., Lam K.S., Cooper G.J. The fat-derived hormone adiponectin alleviates alcoholic and nonalcoholic fatty liver diseases in mice. J Clin Invest. 2003;112:91–100. doi: 10.1172/JCI17797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.You M., Considine R.V., Leone T.C., Kelly D.P., Crabb D.W. Role of adiponectin in the protective action of dietary saturated fat against alcoholic fatty liver in mice. Hepatology. 2005;42:568–577. doi: 10.1002/hep.20821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.You M., Rogers C.Q. Adiponectin: a key adipokine in alcoholic fatty liver. Exp Biol Med (Maywood) 2009;234:850–859. doi: 10.3181/0902-MR-61. [DOI] [PubMed] [Google Scholar]
- 21.Large V., Peroni O., Letexier D., Ray H., Beylot M. Metabolism of lipids in human white adipocyte. Diabetes Metab. 2004;30:294–309. doi: 10.1016/s1262-3636(07)70121-0. [DOI] [PubMed] [Google Scholar]
- 22.Kang L., Nagy L.E. Chronic ethanol feeding suppresses beta-adrenergic receptor-stimulated lipolysis in adipocytes isolated from epididymal fat. Endocrinology. 2006;147:4330–4338. doi: 10.1210/en.2006-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaworski K., Sarkadi-Nagy E., Duncan R.E., Ahmadian M., Sul H.S. Regulation of triglyceride metabolism: IV. Hormonal regulation of lipolysis in adipose tissue. Am J Physiol Gastrointest Liver Physiol. 2007;293:G1–G4. doi: 10.1152/ajpgi.00554.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crabb D.W., Galli A., Fischer M., You M. Molecular mechanisms of alcoholic fatty liver: role of peroxisome proliferator-activated receptor alpha. Alcohol. 2004;34:35–38. doi: 10.1016/j.alcohol.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 25.You M., Crabb D.W. Molecular mechanisms of alcoholic fatty liver: role of sterol regulatory element-binding proteins. Alcohol. 2004;34:39–43. doi: 10.1016/j.alcohol.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 26.Sozio M.S., Liangpunsakul S., Crabb D. The role of lipid metabolism in the pathogenesis of alcoholic and nonalcoholic hepatic steatosis. Semin Liver Dis. 2010;30:378–390. doi: 10.1055/s-0030-1267538. [DOI] [PubMed] [Google Scholar]
- 27.Abrams M.A., Cooper C. Quantitative analysis of metabolism of hepatic triglyceride in ethanol-treated rats. Biochem J. 1976;156:33–46. doi: 10.1042/bj1560033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baraona E., Leo M.A., Borowsky S.A., Lieber C.S. Alcoholic hepatomegaly: accumulation of protein in the liver. Science. 1975;190:794–795. doi: 10.1126/science.1198096. [DOI] [PubMed] [Google Scholar]
- 29.Febbraio M., Hajjar D.P., Silverstein R.L. CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J Clin Invest. 2001;108:785–791. doi: 10.1172/JCI14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lakshmanan M.R., Felver M.E., Veech R.L. Alcohol and very low density lipoprotein synthesis and secretion by isolated hepatocytes. Alcohol Clin Exp Res. 1980;4:361–365. doi: 10.1111/j.1530-0277.1980.tb04832.x. [DOI] [PubMed] [Google Scholar]
- 31.Venkatesan S., Ward R.J., Peters T.J. Effect of chronic alcohol feeding on the hepatic secretion of very-low-density lipoproteins. Biochim Biophys Acta. 1988;960:61–66. doi: 10.1016/0005-2760(88)90009-4. [DOI] [PubMed] [Google Scholar]
- 32.Fischer M., You M., Matsumoto M., Crabb D.W. Peroxisome proliferator-activated receptor alpha (PPARalpha) agonist treatment reverses PPARalpha dysfunction and abnormalities in hepatic lipid metabolism in ethanol-fed mice. J Biol Chem. 2003;278:27997–28004. doi: 10.1074/jbc.M302140200. [DOI] [PubMed] [Google Scholar]
- 33.Kharbanda K.K., Todero S.L., Ward B.W., Cannella J.J., 3rd, Tuma D.J. Betaine administration corrects ethanol-induced defective VLDL secretion. Mol Cell Biochem. 2009;327:75–78. doi: 10.1007/s11010-009-0044-2. [DOI] [PubMed] [Google Scholar]
- 34.Hussain M.M., Shi J., Dreizen P. Microsomal triglyceride transfer protein and its role in apoB-lipoprotein assembly. J Lipid Res. 2003;44:22–32. doi: 10.1194/jlr.r200014-jlr200. [DOI] [PubMed] [Google Scholar]
- 35.Tahara M., Matsumoto K., Nukiwa T., Nakamura T. Hepatocyte growth factor leads to recovery from alcohol-induced fatty liver in rats. J Clin Invest. 1999;103:313–320. doi: 10.1172/JCI4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomita K., Azuma T., Kitamura N., Nishida J., Tamiya G., Oka A., Inokuchi S., Nishimura T., Suematsu M., Ishii H. Pioglitazone prevents alcohol-induced fatty liver in rats through up-regulation of c-Met. Gastroenterology. 2004;26:873–885. doi: 10.1053/j.gastro.2003.12.008. [DOI] [PubMed] [Google Scholar]