Abstract
For men in the United States, prostate cancer (PCa) is the most frequent malignancy and the second leading cause of cancer mortality. The metastatic spread of PCa is responsible for most deaths related to PCa. Although KISS1 functions as a metastasis suppressor in various cancers, its expression levels and functions in PCa development and progression remain undetermined. The goals of this study were to correlate the expression levels of KISS1 in PCas with clinicopathologic characteristics and to assess the biological relevance of KISS1 to the viability and motility of PCa cells. Strong KISS1 staining was detected in benign prostate tissues, but the staining was weaker in primary and metastatic PCas (both P < 0.001, t-test). Furthermore, the low expression levels of KISS1 in PCas correlated with clinical stage (P < 0.01) and with KISS1R expression (P < 0.001). Overexpression of full-length KISS1 in low KISS1-expressing PC-3M cells, but not KFMΔSS, which lacks the secretion signal sequence, induced re-sensitization of cells to anoikis, although it had no effect on either cell proliferation or apoptosis. Overexpression of KISS1 also suppressed steps in the metastatic cascade, including motility and invasiveness. Moreover, cells overexpressing KISS1 were found to enhance chemosensitivity to paclitaxel. Collectively, our data suggest that KISS1 functions as a metastasis suppressor in PCas and may serve as a useful biomarker as well as a therapeutic target for aggressive PCas.
Prostate cancer (PCa) is the most frequent malignancy and the second leading cause of cancer mortality in men 40 years of age and older. Although early detection and hormone-based therapies generally result in responses and reduce mortality, current therapies for advanced or recurrent PCa are not curative.1 Even patients who have undergone apparently successful surgical resection may experience recurrence locally or at distant sites months or years later.2,3 Metastatic spread is responsible for most PCa-related deaths. The size and histology of the primary tumor do not provide a reliable prognosis, because tumor cells may have already disseminated by the time PCa is diagnosed.2 Thus, identification and characterization of molecular signatures that provide information about clinical risk assessment are needed.
Over the past two decades, >30 molecules involved in suppression of metastasis have been recognized.4,5 These inhibit one or more steps of the metastatic cascade, including cell proliferation, apoptosis, epithelial to mesenchymal transition, adhesion, invasion, angiogenesis, and colonization of ectopic tissues. The KISS1 gene was first identified as a metastasis suppressor for human melanoma.6 It is differentially up-regulated in C8161 melanoma cells, which had lost the potential to metastasize after microcell-mediated chromosomal transfer of an intact copy of chromosome 6. Radiation hybrid mapping and fluorescence in situ hybridization showed that the KISS1 gene mapped to 1q32 as a single locus.7 The KISS1 protein can be cleaved and secreted as kisspeptin (KP54, KP14, KP13, or KP10).8,9 KISS1 binds to an G protein–coupled receptor, GPR54, now referred to as the KISS1 receptor (KISS1R).10–12 Despite the lack of KISS1R expression in many tumor cell lines, metastasis is suppressed by KISS1 re-expression, suggesting that KISS1 signals either through KISS1R or through unidentified, alternative signaling pathways.9,13
Levels of KISS1 expression have prognostic relevance and correlate with invasiveness in several human cancers, including renal cell carcinoma and melanoma and esophageal, bladder, brain, breast, ovarian, and pancreatic cancers.14–19 Low or lost KISS1 expression is generally associated with higher tumor grade, increased metastatic potential, and a poor prognosis. A function for KISS1 has been reported in xenograft models, where its re-expression in melanomas, breast cancers, and pancreatic cancers resulted in reduced growth of orthotopic tumors in breast and pancreatic cancers and metastases to other organs in all of the tested models.6,13,20 Studies with human melanomas and breast cancer xenografts show that KISS1 maintains disseminated tumor cells in a dormant state, blocks development of macroscopic metastases, and prevents cancer cells from colonizing and proliferating at secondary sites.9,13,14,20,21 Because dissemination of cancer cells may occur before clinical detection of micrometastasis, interventions that target colonization will be valuable for cancer therapy. Although KISS1 has been implicated in controlling formation of macroscopic metastases of some cancers, its function as a suppressor of metastasis for other cancers is not universal. For example, KISS1 expression was not associated with tumor size or invasion in esophageal squamous cell carcinoma, but it was a significant predictor of lymph node metastasis.17 In hepatocellular carcinoma and breast cancers, increased KISS1 levels are associated with disease progression or a worse clinical outcome.18,22,23
In this report, we show that KISS1 expression is down-regulated in PCas and correlates with clinical stage and with expression of KISS1R. Furthermore, loss of KISS1 expression correlates with enhanced metastatic capacity of commonly used PCa cell lines; stable overexpression of KISS1 induces anoikis, increased chemosensitivity, and decreased cell migration and invasion. These results indicate that loss of KISS1 expression in PCas is associated with increased cancer progression and that KISS1 regulates steps that are involved in cell metastasis.
Materials and Methods
Cells
DU-145, MDA-PC-2b, PC-3, and PC-3M cells were cultured in T-medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (HyClone Laboratories, Inc., Logan, UT) in a 37°C, 5% CO2 incubator.
Wound-Healing Cell Migration Assay
Cell migration assays were performed as previously described. Briefly, cells (2 × 105) were seeded in 6-well plates, and cells were allowed to form an 80% confluent monolayer in a six-well plate and then cultured in serum-free medium overnight before wounding. The wound was made by the tip of a 200-μL micropipette passed across the monolayer. Cell migration was induced by adding serum-free medium supplemented with 10 ng/mL epidermal growth factor. After 24 or 48 hours, the distance that cells moved was determined and quantified in Metamorph imaging software. Shown are the mean values ± SEMs of six measurements for each time point and condition. All measurements were normalized to 0-hour controls.
Invasion Assays
Cancer cell invasiveness was determined with a 24-well Transwell plate (8-μm pore polycarbonate membrane inserts; Corning Inc., Corning, NY) according to the manufacturer's protocol. For these assays, 5 × 104 cells were plated in the Matrigel-coated chamber inserts. Cells were suspended in medium without serum or growth factors, and medium with 10 ng/mL epidermal growth factor was used as a chemoattractant in the lower chamber. After incubation at 37°C for 18 hours, the noninvasive cells on the top chambers were gently removed with cotton swabs. The invading cells on the underside of the membrane were fixed in 100% methanol for 10 minutes, air-dried, stained in 0.1% crystal violet, and counted under a microscope. The numbers of invaded cells in five random, 10× microscopic fields were counted and expressed as percentages of the control. The results shown represent mean ± SD of invading cells from three independent experiments, each accomplished in triplicates.
Tissue Specimens
PCa tissue microarrays were from commercially available suppliers (US Biomax, Inc., Rockville, MD; PR954 and PR2085a) and those developed by the University of Alabama at Birmingham Cancer Program Tissue Core. Included were 35 normal/benign prostatic hyperplasia (BPH) tissues, 210 prostate primary cancers, and 8 metastatic tumors. The use of all tissues was approved by the Institutional Review Boards of Tuskegee University and the University of Alabama at Birmingham. All specimens were subjected to immunohistochemical (IHC) analysis in parallel and processed simultaneously with an identical protocol. Clinicohistopathologic characteristics of the subjects in the tissue microarray study included age at surgery, grade, clinical stage, and Gleason score according to information provided by the suppliers. The expression levels were classified as negative (≤0.3), weak positive (0.3 to 1.5), or strong positive (≥1.5).
IHC Staining
Tissues were de-paraffinized in xylene and rehydrated in graded alcohols. For antigen retrieval of KISS1 and KISS1R, the slides were pressure-cooked for 10 minutes. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide for 5 minutes. Slides were blocked by 3% goat serum at room temperature for 1 hour in humidity chambers with KISS1 or KISS1R antibody. Mass spectrometry-validated human KISS1 monoclonal antibody was a gift from Dr. Danny Welch (University of Kansas). Rabbit anti-human KISS1R (375 to −398) polyclonal antibody was from Phoenix Pharmaceuticals Inc. (Burlingame, CA). The biotinylated goat anti-mouse secondary antibody and the avidin/horseradish peroxidase label were each applied for 10 minutes (BioGenex Laboratories, Inc., Fremont, CA). The antigen-antibody reaction was visualized after diaminobenzidine was applied for 7 minutes. The slides were counterstained with hematoxylin for 1 minute. Positive controls were included in each staining run; negative controls were obtained by omitting the primary antibody. Slides were then dehydrated in alcohols and cleared in three xylene baths before being mounted with Permount media. As reported earlier,24 these antigens are stable in paraffin blocks.
Generation of PC-3M Cell Lines Stably Expressing KISS1 (KFM) or KISS1ΔSS (KFMΔSS)
PC-3M cells were transfected with pcDNA3.1 constructs (pcDNA3.1 vector control; the KFM construct contains full-length KISS1 and FLAG tag; KFMΔSS lacks the secretion signal sequence) with the use of Lipofectamine 2000 (Invitrogen). DNA-containing medium was removed and replaced with T-medium. KISS1-overexpressing cells were selected by incubation in medium containing 400 μg/mL G418 (selective for eukaryotic cells) and 2.5 μg/mL puromycin for 14 days.
RNA Extraction and Quantitative Real-Time PCR
Total RNA was isolated with Trizol (Invitrogen) according to the manufacturer's protocol. For quantification, cDNA was synthesized with 1 μg of RNA in a reverse transcription reaction with random hexamers and oligo-dT primer in an unbiased manner (SABiosciences, Frederick, MD). cDNA was amplified with the SYBR Green qPCR Master Mix (SABiosciences) with a real-time PCR Applied Biosystems 7500 fast thermocycler (Applied Biosystems Inc., Foster City, CA). The expression of mRNAs (KISS1 and KISS1R) was determined with manufacturer-supplied RT2 qPCR Primer Assays (SABiosciences). The mRNA expression levels were normalized to GAPDH or β-actin housekeeping genes. Experiments were performed in triplicate.
Western Blot Analyses
Approximately 5 × 106 cells were lysed in RIPA buffer with a proteinase inhibitor cocktail (Sigma-Aldrich, St. Louis, MO), and the total protein concentrations were determined with the bicinchoninic acid assay (Pierce, Rockford, IL). Protein (30 to 50 μg) was loaded onto 10% SDS-PAGE gels. Antibodies used for Western blot or IHC analyses included polyclonal anti-actin antibody (Sigma-Aldrich; 1:5000), anti-KISS1 antibody (provided by Dr. Welch, University of Alabama at Birmingham; 1:400), and horseradish peroxidase-conjugated anti-rabbit or anti-mouse secondary antibodies (enhanced chemiluminescence-horseradish peroxidase linked secondary antibodies, 1:5000; Amersham Biosciences, Buckinghamshire, UK). Each experiment was performed in triplicate.
Cell Proliferation Assay and Chemosensitivity Assay
Cell growth was determined by the cell proliferation reagent WST-1 (water soluble tetrazolium salt-1; Roche, Mannheim, Germany). Briefly, 2.5 × 103 cells were seeded into 96-well plates in quadruplicate for each condition. At 24, 48, and 72 hours, WST reagent was added to each well at a 1:10 dilution, and the preparations were incubated for another 2 hours. Sample absorbance was measured at 560 nm/495 nm. Each experiment was performed in triplicate.
For chemosensitivity assay, the cells were cultured for the indicated number of days and exposed to paclitaxel at the concentrations indicated. The proliferation rates of cells were measured by the WST-1 assay.
Anchorage-Independent Growth (Soft Agar) Assay
Noble agar (1.5 mL of 0.6%) in growth medium was solidified in six-well plates. Cells (1 × 104) per well were plated in triplicate in 0.3% agar onto the bottom agar. The cells were incubated at 37°C in a 5% CO2 incubator for 15 days. Fresh growth medium (0.5 mL/well) was added after 1 week. At the end of incubation, colonies were stained with 0.005% crystal violet for 1 hour and photographed.
Anoikis Assay
For assay of anoikis, cells were cultured in complete medium on an ultra-low attachment six-well plate (1 × 106 per well; Corning Inc., Corning, NY). The growth rate and viability of cells cultured in suspension were measured by the trypan blue exclusion assay.
Statistical Analyses
All data presented as means ± SDs were analyzed with Prism software version 5.0 (GraphPad, La Jolla, CA) or Matlab version 7.9 software (Mathworks, Natick, MA). The significance of the observed differences was determined with the Student's t-test, the χ2 test, or Fisher's exact test. Fisher's exact test is performed when one or more cells with expected values is <5. P < 0.05 was considered significant.
Results
Clinical Significance of KISS1 and KISS1R Expression in Noncancerous Prostate and PCa Tissues
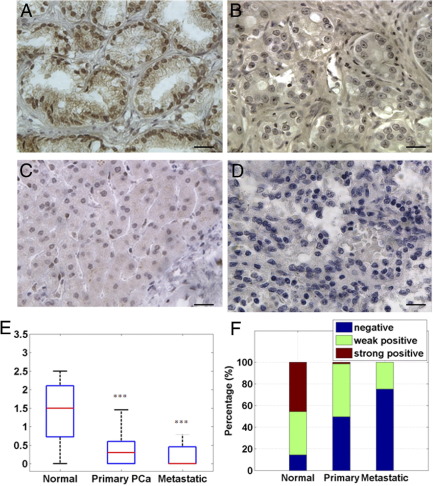
To determine the clinical significance of KISS1 and KISS1R in human PCas, analysis of their expression was conducted by IHC staining. Tissue microarrays, derived from 253 samples of prostate tissues from patients with BPH, uninvolved normal prostate, and PCa (as described in Tissue Specimens) were evaluated for expression of KISS1 and KISS1R. The clinicopathologic characteristics of the cohort are described in Table 1. Benign (BPH or uninvolved normal prostate tissues) human prostate cells showed intense KISS1 expression in both the cytoplasmic and nuclear compartments (Figure 1A). Primary tumors showed decreased KISS1 staining and a pattern of progressively reduced KISS1 levels with increasing tumor grades (Figure 1, B and C). A lack of KISS1 staining in metastatic PCa tumors was observed (Figure 1D). The mean intensity score of KISS1 was 1.34 ± 0.87 in benign tumors (n = 35); primary and metastatic tumors exhibited a lower expression [the mean intensity scores were 0.35 ± 0.36 in primary PCa tissues (n = 210) and 0.21 ± 0.36 in metastasis tissues (n = 8); Figure 1E; both P values ≤ 0.0001]. The expression levels were also categorized as negative (≤0.3), weak positive (0.3 to 1.5), or strong positive (≥1.5). KISS1 expression in primary PCas (49% negative, 48% weak positive, and 3% strong positive) and metastatic PCa tissues (77% negative, 23% weak positive, and 0% strong positive) were significantly lower relative to BPH tissues (18% negative, 39% weak positive, and 43% strong positive) (Figure 1F). Although no significant difference in KISS1 expression was observed between primary and metastatic tissues, (P = 0.32; Figure 1, E and F), loss of KISS1 expression positively correlated with clinical stages II/III and IV and metastatic tumors (P < 0.01; Table 2), and the KISS1 expression is significantly lower (P < 0.01) in metastatic tissues than in earlier stage primary PCa (clinical stages II/III), indicating loss of KISS1 expression correlated with PCa progression. KISS1R expression was weak and only mildly positive in normal prostate tissues, and decreased expression was observed in primary and metastatic tissues (see Supplemental Figure S1 available at http://ajp.amjpathol.org). This decrease in KISS1 expression positively correlated with the decrease of KISS1R expression in PCas, as determined by the χ2 test (Table 2).
Table 1.
Clinicopathologic Characteristics of the Cohort (n = 253)
| Parameter | n (%) |
|---|---|
| Age | |
| 28 to 50 years | 11 (4.4) |
| 51 to 60 years | 29 (11.5) |
| 61 to 70 years | 120 (47.4) |
| 71 to 88 years | 93 (36.7) |
| Clinical stage | |
| 1 | 0 (0.0) |
| 2 | 7 (15.9) |
| 3 | 24 (54.5) |
| 4 | 13 (29.6) |
| Gleason score | |
| ≤6 | 35 (25.5) |
| 7 | 26 (19) |
| ≥8 | 76 (55.5) |
Gleason score or clinical stage may not be available in some cases. Therefore, the total number of patients varies.
Figure 1.
Expression of KISS1 in human noncancerous prostate tissues and PCa tissues. Representative immunostaining photomicrographs: Strong immunoreactivity of KISS1 was detected in control prostate benign tissues (A) and reduced staining in low-grade (Grade II) (B) and high-grade (Grade IV) (C) PCas. The KISS1 protein was not evident in metastatic PCas (D). Original magnification, ×400. Immunostaining was accomplished with anti-human KISS1 antibody. Scale bar = 20 μm. E: Compared with noncancerous prostate tissues, the overall expression of KISS1 was lower in PCa tissues and metastatic tissues (both P ≤ 0.001). F: Frequency distribution of KISS1 staining intensity in prostate tissues. The expression levels were classified as negative (≤0.3), weak positive (0.3 to 1.5), or strong positive (≥1.5).
Table 2.
Relation between KISS1 and Clinical Features in Patients with Prostate Cancer
| KISS1 score |
||||
|---|---|---|---|---|
| Variable | Analyzable cases, n⁎ | Negative, n (%) | Positive, n (%) | P† |
| Total | 218 | 109 (50.0) | 109 (50.0) | |
| Age | ||||
| 60 years | 34 | 16 (47.0) | 18 (53) | 0.7845 |
| >60 and ≤70 years | 91 | 44 (48.4) | 47 (51.6) | |
| >70 years | 93 | 49 (52.7) | 44 (47.3) | |
| Gleason score | ||||
| ≤6 | 35 | 15 (42.9) | 20 (58.1) | 0.3466 |
| 7 | 26 | 12 (46.2) | 14 (53.8) | |
| ≥8 | 76 | 43 (56.6) | 33 (43.4) | |
| Clinical stage | ||||
| II/III | 31 | 7 (22.6) | 24 (77.4) | 0.0035 |
| IV | 13 | 8 (61.5) | 5 (38.5) | |
| Metastatic PCa | 8 | 6 (75) | 2 (25) | |
| KISS-1R | ||||
| Negative | 130 | 80 (61.5) | 50 (38.5) | <0.0001 |
| Positive | 65 | 21 (32.3) | 44 (67.7) | |
Gleason score or clinical stage may not be available in some cases. Therefore, the total number of patients varies.
P values were obtained with the χ2 test or Fisher's exact test when one or more cells had expected values < 5.
Re-Expression of KISS1 in Prostate PC-3M Cell Line
To assess whether KISS1/KISS1R expression is involved in cancer metastasis, their expression was determined, by real-time PCR analysis, in a panel of human PCa cell lines with varying degrees of metastatic capacity (low to high: DU-145, MDA-PC-2b, PC-3, PC-3M). Decreased expression of KISS1/KISS1R correlated with the metastatic propensity of the cell lines, with PC-3M, which is highly metastatic, exhibiting the lowest expression (see Supplemental Figure S2 available at http://ajp.amjpathol.org). Thus, PC-3M cells were selected to determine the biological significance of re-expressing KISS1.
KISS1-containing constructs with (KFM) or without (KFMΔSS) a secretion signal sequence were used (the pcDNA3.1 vector control; KFM, contains full-length KISS1; KFMΔSS lacks the secretion signal construct, as described previously9). These constructs were transfected into PC-3M-RFP cells. After transfection, neomycin- and puromycin-resistant PC-3M clones were selected, and KISS1 expression was confirmed by real-time PCR and Western blot analyses (see Supplemental Figure S3 available at http://ajp.amjpathol.org). Transfected clones were selected and identified according to the highest levels of KISS1 expression at the RNA and protein levels (see Supplemental Figure S3 available at http://ajp.amjpathol.org). From each transfection, the clone with the highest KISS1 expression was selected for experiments focused on factors affecting metastases.
KISS1 Expression Inhibits Anchorage-Independent Growth but Not Anchorage-Dependent Growth of PCa Cells
To determine whether KISS1 expression affects growth of PCa cells, we used cell proliferation reagent WST-1 to assay cell proliferation and cell viability of PC-3M KFM and PC-3M KFMΔSS stable clones. Overexpression of KISS1 in PC-3M cells (PC-3M KFM and PC-3M KFMΔSS) had no effect on cell growth in monolayer cultures (Figure 2A), even under serum-free conditions (data not shown). An essential characteristic of metastatic cancer cells is resistance to anoikis.25,26 This attribute allows the survival of metastatic cancer cells during systemic circulation and facilitates their metastasis to distant organs. Therefore, the influence of KISS1 on anoikis in PC-3M KFM and PC-3M KFMΔSS cells was determined by measuring cell viability in the absence of matrix attachment. Under nonadherent conditions (cells cultured in ultra-low attachment plates), PC-3M KFM cells exhibited a reduction in cell viability greater than that of PC-3M vector control cells, as determined by trypan blue viability experiments and crystal violet staining compared with cells recovered from normal culture plates (Figure 2B). PC-3M KFMΔSS cells exhibited almost no difference from vector control cells. These results indicate that KISS1 expression and secretion have no effect on cell proliferation but that both promote sensitization to anoikis under anchorage-independent conditions.
Figure 2.
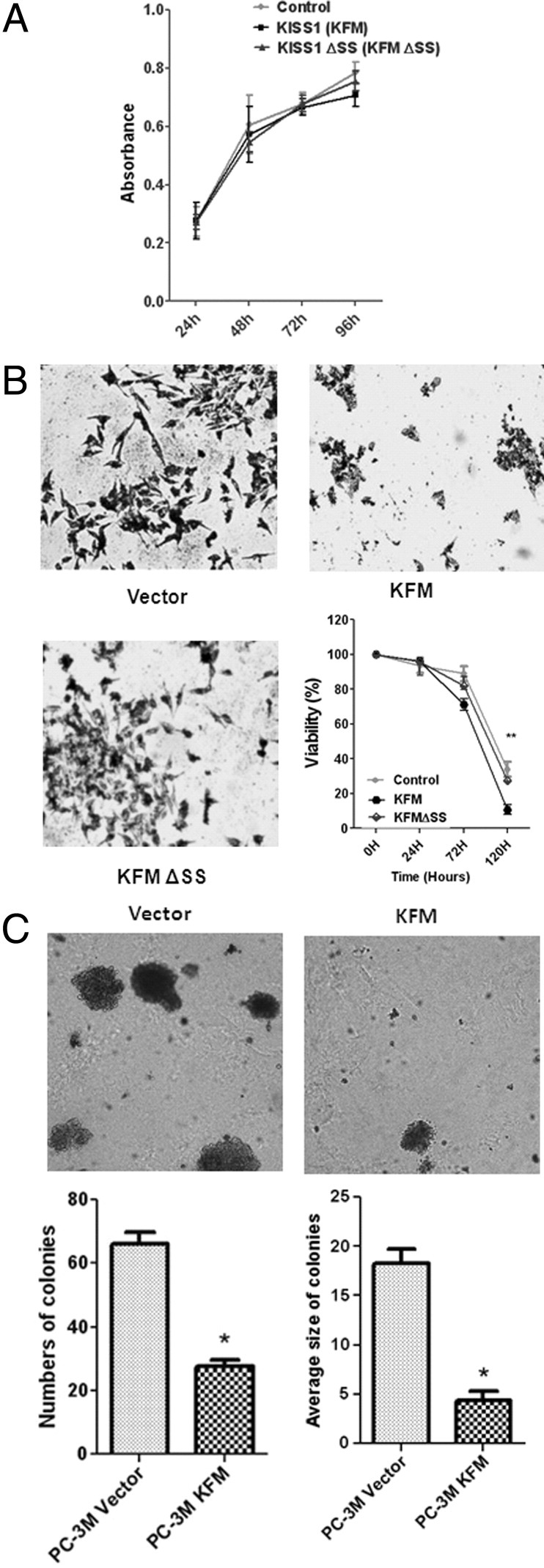
Effect of KISS1 expression on cell proliferation and viability. A: KISS1 does not affect anchorage-dependent growth of PCa cells. The cells were cultured for the indicated number of days, and proliferation of KISS1-expressing cells was measured by the WST-1 assay. B: KISS1 reduces cell viability under anchorage-independent conditions compared with vector controls and KFM and KFMΔSS expression. At each time point, cell viability was determined by trypan blue staining. Points indicate mean; bars, SD. **P < 0.01, ***P < 0.001. Photomicrographs show representative viable cells from KISS1 and KFMΔSS-expressing and vector control, PC-3M cells regrown in normal plates after 96 hours of independent culture, stained with crystal violet. C: KISS1 overexpression inhibits anchorage-independent growth of PCa cells. Photographs show representative colonies formed from PC-3M vector and PC-3M KFM-transformed cells grown in soft agar. KISS1 reduced the numbers and sizes of colonies compared with the vector control (points indicate mean; bars, SD). ***P < 0.001, lower panels.
In addition, PC-3M KFM cells exhibited either no change or a slight decrease in total cell number when cultured in ultra-low attachment plates, whereas the number of PC-3M vector control cells cultured in suspension continued to increase (data not shown). To determine whether KISS1 overexpression affects the anchorage-independent growth of PC-3M cells, the capacity of KISS1-expressing cells to form colonies in soft agar was assessed (Figure 2C). After 15 days of culture, image analysis showed that vector control cells had formed a greater number (1.5-fold) of colonies of larger size (1.9-fold) relative to PC-3M KFM cells (Figure 2C). This indicates that KISS1 expression reduces anchorage-independent growth of PCa cells, an essential step for metastasis.
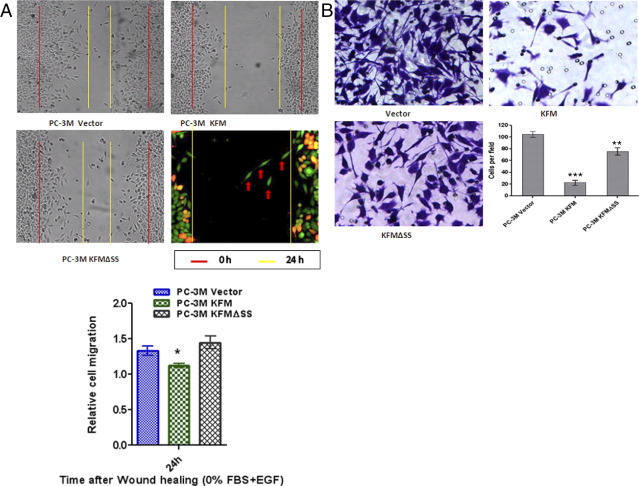
KISS1 Re-Expression Decreases PCa Cell Motility and Invasiveness
Because KISS1 functions as a metastasis suppressor, the effect of its overexpression on cell migration, a feature necessary for the motility of cancer cells, was tested. The migration of PC-3M cells was delayed by expression of KFM (full-length KISS1) but not with KFMΔSS (Figure 3A), indicating that KISS1 secretion is required for inhibition of cancer cell migration. To determine the effect of KISS1 expression on the invasive potential of PC-3M cells, the invasiveness of KISS1 and vector control clones was evaluated with Matrigel chambers. Figure 3B shows a representative image of PC-3M cells stained with crystal purple on the bottom of a Matrigel-coated membrane. A greater number of cells transfected with the empty vector invaded through the Matrigel, relative to those with PC-3M KFM and KFMΔSS expression (Figure 3B), indicating that KISS1 expression inhibits invasion and that KISS1 secretion is required for inhibiting of motility.
Figure 3.
Effects of KISS1 overexpression on migration and invasion of PC-3M cells. A: Measurement of cell migration of stably overexpressing KISS1 cells that used wound healing assay. PC-3M vector only, PC-3M KFM, and PC-3M KFMΔSS cells were wounded and treated with 10 ng/mL epidermal growth factor (EGF) 24 hours. Red vertical bars indicate starting point on day 0, and yellow vertical bars indicate the leading edge of migrating cells after 24 hours. PC-3M-RFP KFM cells and PC-3M GFP cells, (middle right panel) were mixed, and migration of denuded area was measured. PC-3M cells stably expressing KISS1 (RFP; red) migrated slower than PC-3M cells stably expressing green fluorescent protein (GFP; green; arrow). Bar graph (bottom) indicates quantification of area migrated after 24 hours in six individual experiments ± SD. *P < 0.01, analyzed by t-test. B: Effects of KISS1 overexpression on invasion by PC-3M cells. Cells that penetrated the Matrigel-coated membrane were fixed in formaldehyde and stained with crystal violet. Photos are representative fields of invasive cells on the membrane. Original magnification, ×100. Bar graphs represent the average number of cells on the underside of the membrane ± SD. **P < 0.01, ***P < 0.001, analyzed by t-test.
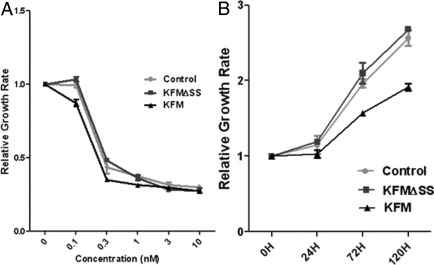
KISS1 Overexpression Enhances Chemosensitivity of Cancer Cells
Increased susceptibility of cancer cells to chemotherapeutic agents would represent an important advancement in the treatment of patients with advanced or metastatic PCa.27 Therefore, the effect of KISS1 levels on the chemosensitivity of PCa cells was determined. PCa cells (PC-3M control, PC-3M KFM, and PC-3M KFMΔSS) were exposed for 72 hours to various concentrations (0, 0.1, 0.3, 1, 3, 10, 30 nmol/L) of paclitaxel, a chemotherapeutic agent commonly used for treatment of PCas. As determined with the WST-1 assay at 72 hours after exposure, paclitaxel inhibited cell growth in a dose-dependent manner (Figure 4A). For the PC-3M control and PC-3M KFMΔSS cells, 0.1 nmol/L paclitaxel had no effect on growth, whereas for cells with KISS1 overexpression both 0.1 and 0.3 nmol/L paclitaxel decreased the number of viable cells. WST-1 assays for cell proliferation were also conducted on cells (days 1, 3, and 5) receiving 0.1 nmol/L paclitaxel treatment over a 5-day period (Figure 4B). KISS1 overexpression decreased the total number of viable PCa cells relative to PC-3M controls at 72 and 120 hours (Figure 4B). These data indicate that KISS1 overexpression sensitizes PC-3M cells to the effects of paclitaxel.
Figure 4.
The effects of KISS1 overexpression on chemosensitivity of PCa cells. The proliferation of PC-3M cells in the presence of paclitaxel or control was measured by the WST-1. A: Growth curves of PC-3M vector control, KFM, or KFMΔSS cells exposed to increasing concentrations of paclitaxel (0 nmol/L, 0.1 nmol/L, 0.3 nmol/L, 1 nmol/L, 3 nmol/L, or 10 nmol/L) for 72 hours. B: Growth curves of PC-3M vector control, KFM, or KFMΔSS cells exposed to 0.1 nmol/L paclitaxel over a 5-day periods. WST-1 assays were conducted at 0, 24, 72, and 120 hours. Data are reported as the percentage of growing cells compared with vector controls and represent mean values ± SDs of at least three independent experiments.
Discussion
KISS1 expression has been measured in clinical samples, including melanomas and breast, urinary bladder, and pancreatic cancers, showing its diagnostic and prognostic capacity.14–18 This report provides an extensive evaluation of KISS1 expression in human PCa tissues in a large cohort and evaluates the effect of KISS1 expression on PCa cell lines. As hypothesized, KISS1 was strongly expressed in normal and BPH tissues. In contrast, decreased expression was observed in early stage primary tumors, with a progressive loss of expression in late stage and metastatic tumors. The loss of KISS1 expression correlated with both advanced clinical cancer stages and the loss of KISS1R. Although a statistical correlation was not observed, there was an increasing percentage of patients with cancers that were KISS1 negative and a decreasing percentage of patients with cancers that were KISS1 positive in relation to increasing Gleason score and tumor grade. Given the significant inverse correlation between KISS1 expression and clinical stage, measurement of KISS1 expression levels may be useful for improving the overall risk assessment of patients with PCa in combination with clinical staging, Gleason score, and tumor grade.
We have re-expressed full-length KISS1 and have prepared a deletion mutant, KFMΔSS, which lacks capacity to be secreted.9 Re-expression of full-length KISS1 resulted in changes in cellular structure, with associated decreases in cell migration, invasion, and anchorage-dependent survival, without affecting cellular proliferation. Cells containing the PC-3M KFMΔSS deletion mutation, however, failed to exhibit these features, and their capacity for cell invasion was similar to that of control cells. Our findings are similar to metastatic melanoma cell line C8161.9 KFM and C8161.9 KFMΔSS cells, whereby KFM-transfected but not KFMΔSS-transfected cells were suppressed for metastasis to lung, eye, kidney, and bone.9 These facts, coupled with the correlation of decreased KISS1 expression with increasing metastasis capacity in commonly used PCa cell lines, suggest a clinical and biological role for KISS1 as a metastasis suppressor in PCa.
KISS1 re-expression in ovarian, breast, pancreatic, and melanoma cancer cells reduced metastases (50% for ovarian cancers, 90% for melanomas, ≥50% for breast cancer, 50% for pulmonary metastasis, and 95% for hepatic metastasis in pancreatic cancer).14–19 This is hypothesized to be through reestablishment of autocrine or paracrine KISS1R signaling (see Beck and Welch14 for review). Through KISS1R, KISS1 activates a variety of signals, including phospholipase C, protein kinase C, intracellular Ca2+ mobilization, and the mitogen-activated protein kinase and phosphatidylinositol 3-kinase/Akt pathways.14 The KISS1/KISS1R pair also affects signaling events by interacting with other receptors, such as the chemokine receptor, CXCR4, and the gonadotrophin-releasing hormone receptor.28,29 These signals are selectively triggered by kisspeptin by KISS1R in a manner dependent on the cell type. Our data indicated that effects of KISS1 expression were not likely to be regulated through KISS1R, because PC-3M before and after transfection with KFM express low levels of receptors (Wang et al, 2012, unpublished observation). Similarly, this has been observed in most cell lines in which KISS1 suppresses metastasis,9 suggesting an alternative mechanism, such as an intracrine or a paracrine signaling mechanism, or the existence of other unidentified KISS1 receptors. A recent report of plasma kisspeptin levels in PCa suggest that circulating levels of kisspeptin are not elevated in patients with prostatic carcinoma,30 although KISS1R was not measured. It deserves mention that plasma levels in this study were compared with patients with other malignancies. Because loss of KISS1 has been found in numerous cancer types, a comparison from normal healthy persons should be conducted before conclusions are drawn. More investigation in this area is warranted to determine the mechanisms by which KISS1 functions in PCa.
Kisspeptins are thought to work through the hypothalamus-pituitary-gonadal axis, which is the current target of multiple hormone suppressive therapies.31 Although it is not clear whether KISS1 regulates gonadotropin-releasing hormone receptors or vise-versa, a recent report in breast cancer reported that KISS1 and GPR54 are estrogen regulated, and their expression levels depend on the status of estrogen receptor α.23,32 A similar study that correlates KISS1 expression in relation to androgen receptor status should be conducted to further define a clinical role for KISS1 expression in PCa.
Mimetics of kisspeptin have been proposed for clinical administration.14,33 The observation that re-expression of KISS1 increases sensitivity to proliferation-targeting agents such as paclitaxel provides a rationale for the clinical use of kisspeptins or small molecules having the properties of KISS1. These results are consistent with the re-expression of KISS1 in head and neck squamous cell carcinoma chemo-resistant cells, restoring chemosensitivity through nuclear factor-κB-associated poly (ADP-ribose) polymerase-1 cleavage.34 Nuclear factor κB signaling in PCa is implicated in regulation of various genes responsible for cell proliferation, epithelial to mesenchymal transition-associated metastasis, and resistance to chemotherapy.35 Because decreased chemosensitivity is associated with cancer progression to metastasis, reversion of these cellular processes should enhance chemosensitivity.
In conclusion, this study suggests that loss of KISS1 and KISS1R expression is associated with clinical progression of PCas. Re-expressed KISS1 exhibits properties of metastasis suppressors, that is, suppression of cell migration and invasion and increasing cell sensitization to anoikis and chemotherapeutic drugs. Thus, these findings suggest KISS1 functions as a metastasis suppressor in PCas and will be useful for the diagnosis, risk assessment of PCa progression, and a therapeutic target for aggressive PCas.
Acknowledgments
We thank members of the Yates Laboratory for their comments and discussions.
Footnotes
Supported by the Department of Defense Prostate Cancer Research Program (PC073977 to C.Y.), NIH (RCMI grant G12 RR03059-21A1 to C.Y.), a pilot project on U54 CA118623 (NIH/National Cancer Institute to C.Y. and D.R.W.; RO1-CA134981 to D.R.W.); Susan G. Komen for the Cure (SAC110037 to D.R.W.), and the National Foundation for Cancer Research-Center for Metastasis Research (D.R.W.).
Supplemental material for this article can be found at http://ajp.amjpathol.org or at doi: 10.1016/j.ajpath.2011.11.020.
Contributor Information
Honghe Wang, Email: wangh@mytu.tuskegee.edu.
Clayton Yates, Email: cyates@mytu.tuskegee.edu.
Supplementary data
Expression of KISS1R in human noncancerous prostate tissues and PCa tissues. Representative immunostaining pictures. Immunoreactivity of KISS1R was detected in control prostate benign tissues (A) and reduced staining in PCas (B). The KISS1R protein was not evident in metastatic PCas (C). Original magnification: ×400. Immunostaining was accomplished with anti-human KISS1R antibody.
KISS1/KISS1R expression in prostate cancer cell lines. Real-time PCR were used to quantify the relative expression of KISS1 and KISS1R in the prostate cell lines DU-145, MDA-PC-2b, PC-3, PC-3M.
Characterization of KISS1-overexpressing PC-3M cells. PC-3M cells and the stable transfectants were grown as previously described in Materials and Methods. KISS1 expression was characterized by real-time RT-PCR (A) or Western blotting analysis (B).
References
- 1.Buchan N.C., Goldenberg S.L. Intermittent androgen suppression for prostate cancer. Nat Rev Urol. 2010;7:552–560. doi: 10.1038/nrurol.2010.141. [DOI] [PubMed] [Google Scholar]
- 2.Plantade A., Massard C., de Crevoisier R., Fizazi K. Locally advanced prostate cancer: definition, prognosis and treatment [in French] Bull Cancer. 2007;94:F50–F61. [PubMed] [Google Scholar]
- 3.Ramiah V., George D.J., Armstrong A.J. Clinical endpoints for drug development in prostate cancer. Curr Opin Urol. 2008;18:303–308. doi: 10.1097/MOU.0b013e3282fb7807. [DOI] [PubMed] [Google Scholar]
- 4.Stafford L.J., Vaidya K.S., Welch D.R. Metastasis suppressors genes in cancer. Int J Biochem Cell Biol. 2008;40:874–891. doi: 10.1016/j.biocel.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida B.A., Sokoloff M.M., Welch D.R., Rinker-Schaeffer C.W. Metastasis-suppressor genes: a review and perspective on an emerging field. J Natl Cancer Inst. 2000;92:1717–1730. doi: 10.1093/jnci/92.21.1717. [DOI] [PubMed] [Google Scholar]
- 6.Lee J.H., Miele M.E., Hicks D.J., Phillips K.K., Trent J.M., Weissman B.E., Welch D.R. KiSS-1, a novel human malignant melanoma metastasis-suppressor gene. J Natl Cancer Inst. 1996;88:1731–1737. doi: 10.1093/jnci/88.23.1731. [DOI] [PubMed] [Google Scholar]
- 7.West A., Vojta P.J., Welch D.R., Weissman B.E. Chromosome localization and genomic structure of the KiSS-1 metastasis suppressor gene (KISS1) Genomics. 1998;54:145–148. doi: 10.1006/geno.1998.5566. [DOI] [PubMed] [Google Scholar]
- 8.Takino T., Koshikawa N., Miyamori H., Tanaka M., Sasaki T., Okada Y., Seiki M., Sato H. Cleavage of metastasis suppressor gene product KiSS-1 protein/metastin by matrix metalloproteinases. Oncogene. 2003;22:4617–4626. doi: 10.1038/sj.onc.1206542. [DOI] [PubMed] [Google Scholar]
- 9.Nash K.T., Phadke P.A., Navenot J.M., Hurst D.R., Accavitti-Loper M.A., Sztul E., Vaidya K.S., Frost A.R., Kappes J.C., Peiper S.C., Welch D.R. Requirement of KISS1 secretion for multiple organ metastasis suppression and maintenance of tumor dormancy. J Natl Cancer Inst. 2007;99:309–321. doi: 10.1093/jnci/djk053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohtaki T., Shintani Y., Honda S., Matsumoto H., Hori A., Kanehashi K., Terao Y., Kumano S., Takatsu Y., Masuda Y., Ishibashi Y., Watanabe T., Asada M., Yamada T., Suenaga M., Kitada C., Usuki S., Kurokawa T., Onda H., Nishimura O., Fujino M. Metastasis suppressor gene KiSS-1 encodes peptide ligand of a G-protein-coupled receptor. Nature. 2001;411:613–617. doi: 10.1038/35079135. [DOI] [PubMed] [Google Scholar]
- 11.Kotani M., Detheux M., Vandenbogaerde A., Communi D., Vanderwinden J.M., Le Poul E., Brezillon S., Tyldesley R., Suarez-Huerta N., Vandeput F., Blanpain C., Schiffmann S.N., Vassart G., Parmentier M. The metastasis suppressor gene KiSS-1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. J Biol Chem. 2001;276:34631–34636. doi: 10.1074/jbc.M104847200. [DOI] [PubMed] [Google Scholar]
- 12.Muir A.I., Chamberlain L., Elshourbagy N.A., Michalovich D., Moore D.J., Calamari A., Szekeres P.G., Sarau H.M., Chambers J.K., Murdock P., Steplewski K., Shabon U., Miller J.E., Middleton S.E., Darker J.G., Larminie C.G., Wilson S., Bergsma D.J., Emson P., Faull R., Philpott K.L., Harrison D.C. AXOR12, a novel human G protein-coupled receptor, activated by the peptide KiSS-1. J Biol Chem. 2001;276:28969–28975. doi: 10.1074/jbc.M102743200. [DOI] [PubMed] [Google Scholar]
- 13.Lee J.H., Welch D.R. Suppression of metastasis in human breast carcinoma MDA-MB-435 cells after transfection with the metastasis suppressor gene: KiSS-1. Cancer Res. 1997;57:2384–2387. [PubMed] [Google Scholar]
- 14.Beck B.H., Welch D.R. The KISS1 metastasis suppressor: a good night kiss for disseminated cancer cells. Eur J Cancer. 2010;46:1283–1289. doi: 10.1016/j.ejca.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagai K., Doi R., Katagiri F., Ito T., Kida A., Koizumi M., Masui T., Kawaguchi Y., Tomita K., Oishi S., Fujii N., Uemoto S. Prognostic value of metastin expression in human pancreatic cancer. J Exp Clin Cancer Res. 2009;28:9. doi: 10.1186/1756-9966-28-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shengbing Z., Feng L.J., Bin W., Lingyun G., Aimin H. Expression of KiSS-1 gene and its role in invasion and metastasis of human hepatocellular carcinoma. Anat Rec (Hoboken) 2009;292:1128–1134. doi: 10.1002/ar.20950. [DOI] [PubMed] [Google Scholar]
- 17.Ikeguchi M., Yamaguchi K., Kaibara N. Clinical significance of the loss of KiSS-1 and orphan G-protein-coupled receptor (hOT7T175) gene expression in esophageal squamous cell carcinoma. Clin Cancer Res. 2004;10:1379–1383. doi: 10.1158/1078-0432.ccr-1519-02. [DOI] [PubMed] [Google Scholar]
- 18.Martin T.A., Watkins G., Jiang W.G. KiSS-1 expression in human breast cancer. Clin Exp Metastasis. 2005;22:503–511. doi: 10.1007/s10585-005-4180-0. [DOI] [PubMed] [Google Scholar]
- 19.Prentice L.M., Klausen C., Kalloger S., Kobel M., McKinney S., Santos J.L., Kenney C., Mehl E., Gilks C.B., Leung P., Swenerton K., Huntsman D.G., Aparicio S.A. Kisspeptin and GPR54 immunoreactivity in a cohort of 518 patients defines favourable prognosis and clear cell subtype in ovarian carcinoma. BMC Med. 2007;5:33. doi: 10.1186/1741-7015-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNally L.R., Welch D.R., Beck B.H., Stafford L.J., Long J.W., Sellers J.C., Huang Z.Q., Grizzle W.E., Stockard C.R., Nash K.T., Buchsbaum D.J. KISS1 over-expression suppresses metastasis of pancreatic adenocarcinoma in a xenograft mouse model. Clin Exp Metastasis. 2010;27:591–600. doi: 10.1007/s10585-010-9349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welch D.R., Hunter K.W. A new member of the growing family of metastasis suppressors identified in prostate cancer. J Natl Cancer Inst. 2003;95:839–841. doi: 10.1093/jnci/95.12.839. [DOI] [PubMed] [Google Scholar]
- 22.Ikeguchi M., Hirooka Y., Kaibara N. Quantitative reverse transcriptase polymerase chain reaction analysis for KiSS-1 and orphan G-protein-coupled receptor (hOT7T175) gene expression in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2003;129:531–535. doi: 10.1007/s00432-003-0469-z. [DOI] [PubMed] [Google Scholar]
- 23.Marot D., Bieche I., Aumas C., Esselin S., Bouquet C., Vacher S., Lazennec G., Perricaudet M., Kuttenn F., Lidereau R., de Roux N. High tumoral levels of Kiss1 and G-protein-coupled receptor 54 expression are correlated with poor prognosis of estrogen receptor-positive breast tumors. Endocr Relat Cancer. 2007;14:691–702. doi: 10.1677/ERC-07-0012. [DOI] [PubMed] [Google Scholar]
- 24.Manne U., Myers R.B., Srivastava S., Grizzle W.E. Re: loss of tumor marker-immunostaining intensity on stored paraffin slides of breast cancer. J Natl Cancer Inst. 1997;89:585–586. doi: 10.1093/jnci/89.8.585. [DOI] [PubMed] [Google Scholar]
- 25.Sakamoto S., Kyprianou N. Targeting anoikis resistance in prostate cancer metastasis. Mol Aspects Med. 2010;31:205–214. doi: 10.1016/j.mam.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geiger T.R., Peeper D.S. Metastasis mechanisms. Biochim Biophys Acta. 2009;1796:293–308. doi: 10.1016/j.bbcan.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 27.Seruga B., Ocana A., Tannock I.F. Drug resistance in metastatic castration-resistant prostate cancer. Nat Rev Clin Oncol. 2011;8:12–23. doi: 10.1038/nrclinonc.2010.136. [DOI] [PubMed] [Google Scholar]
- 28.Navenot J.M., Wang Z., Chopin M., Fujii N., Peiper S.C. Kisspeptin-10-induced signaling of GPR54 negatively regulates chemotactic responses mediated by CXCR4: a potential mechanism for the metastasis suppressor activity of kisspeptins. Cancer Res. 2005;65:10450–10456. doi: 10.1158/0008-5472.CAN-05-1757. [DOI] [PubMed] [Google Scholar]
- 29.Votsi E., Roussos D., Katsikis I., Karkanaki A., Kita M., Panidis D. Kisspeptins: a multifunctional peptide system with a role in reproduction, cancer and the cardiovascular system. Hippokratia. 2008;12:205–210. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Curtis A.E., Murphy K.G., Chaudhri O.B., Ramachandran R., Young A.M., Waxman J., Nijher G.M., Bewick G.A., Ghatei M.A., Bloom S.R., Dhillo W.S. Kisspeptin is released from human prostate cancer cell lines but plasma kisspeptin is not elevated in patients with prostate cancer. Oncol Rep. 2010;23:1729–1734. doi: 10.3892/or_00000818. [DOI] [PubMed] [Google Scholar]
- 31.Colledge W.H. Kisspeptins and GnRH neuronal signalling. Trends Endocrinol Metab. 2009;20:115–121. doi: 10.1016/j.tem.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 32.Huijbregts L., de Roux N. KISS1 is down-regulated by 17beta-estradiol in MDA-MB-231 cells through a nonclassical mechanism and loss of ribonucleic acid polymerase II binding at the proximal promoter. Endocrinology. 2010;151:3764–3772. doi: 10.1210/en.2010-0260. [DOI] [PubMed] [Google Scholar]
- 33.Nash K.T., Welch D.R. The KISS1 metastasis suppressor: mechanistic insights and clinical utility. Front Biosci. 2006;11:647–659. doi: 10.2741/1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiffar T., Yilmaz T., Lee J., Hanna E., El-Naggar A., Yu D., Myers J.N., Kupferman M.E. KiSS1 mediates platinum sensitivity and metastasis suppression in head and neck squamous cell carcinoma. Oncogene. 2011;30:3163–3173. doi: 10.1038/onc.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paule B., Terry S., Kheuang L., Soyeux P., Vacherot F., de la Taille A. The NF-kappaB/IL-6 pathway in metastatic androgen-independent prostate cancer: new therapeutic approaches? World J Urol. 2007;25:477–489. doi: 10.1007/s00345-007-0175-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of KISS1R in human noncancerous prostate tissues and PCa tissues. Representative immunostaining pictures. Immunoreactivity of KISS1R was detected in control prostate benign tissues (A) and reduced staining in PCas (B). The KISS1R protein was not evident in metastatic PCas (C). Original magnification: ×400. Immunostaining was accomplished with anti-human KISS1R antibody.
KISS1/KISS1R expression in prostate cancer cell lines. Real-time PCR were used to quantify the relative expression of KISS1 and KISS1R in the prostate cell lines DU-145, MDA-PC-2b, PC-3, PC-3M.
Characterization of KISS1-overexpressing PC-3M cells. PC-3M cells and the stable transfectants were grown as previously described in Materials and Methods. KISS1 expression was characterized by real-time RT-PCR (A) or Western blotting analysis (B).