Abstract
In tomato, Ve is implicated in race-specific resistance to infection by Verticillium species causing crop disease. Characterization of the Ve locus involved positional cloning and isolation of two closely linked inverted genes. Expression of individual Ve genes in susceptible potato plants conferred resistance to an aggressive race 1 isolate of Verticillium albo-atrum. The deduced primary structure of Ve1 and Ve2 included a hydrophobic N-terminal signal peptide, leucine-rich repeats containing 28 or 35 potential glycosylation sites, a hydrophobic membrane-spanning domain, and a C-terminal domain with the mammalian E/DXXXLφ or YXXφ endocytosis signals (φ is an amino acid with a hydrophobic side chain). A leucine zipper-like sequence occurs in the hydrophobic N-terminal signal peptide of Ve1 and a Pro-Glu-Ser-Thr (PEST)-like sequence resides in the C-terminal domain of Ve2. These structures suggest that the Ve genes encode a class of cell-surface glycoproteins with receptor-mediated endocytosis-like signals and leucine zipper or PEST sequences.
Keywords: verticillium wilt, Ve1, Ve2, Lycopersicon esculentum, Solanum tuberosum
Verticillium wilt is a common fungal disease that causes severe yield and quality losses in many crops, including alfalfa, cotton, cucurbits, eggplant, mint, potato, tomato, strawberry, and sunflower (1). Several species of Verticillium have been reported to cause wilt and control often has relied on the use of expensive chemical fumigants that may impact health and environment adversely. In a few cases, effective control of verticillium wilt has been reported in specific crops that exhibit race-specific resistance (2, 3).
Plant resistance to viruses, bacteria, and fungi frequently involves specific host–pathogen interactions between the products of a plant resistance gene (R) and corresponding avirulence gene (Avr) in the pathogen (4, 5). Absence of either of these entities results in a susceptible response whereby plant defenses are not elicited and infection proceeds. Several plant disease resistance genes have been cloned and assigned to one of five classes based on structural features. One class includes R genes that encode a cytoplasmic serine/threonine protein kinase such as Pto in Pseudomonas syringae (6). Another class includes the P. syringae RPS2 and RPM1 resistance genes of Arabidopsis and the tomato Fusarium oxysporum resistance gene I2, which encode cytoplasmic proteins with a leucine zipper, a nucleotide-binding site (NBS), and a C-terminal leucine-rich repeat (LRR) (7–10). A third class of cytoplasmic proteins possess LRR and NBS motifs and an N-terminal domain with homology to the mammalian Toll/interleukin-1 receptor domain. This class includes the tobacco N gene for resistance against tobacco mosaic virus, the flax L6 gene for resistance to Melamspora lini, and the Arabidopsis RPP5 resistance gene for Peronospora parasitica (11–13). A fourth class consists of the tomato Cladosporium fulvum resistance genes that have an extracellular LRR, a membrane-spanning domain, and a short cytoplasmic C terminus (14). The rice Xa21 resistance gene for Xanthomonas represents a fifth class having an extracellular LRR and an intracellular serine/threonine kinase domain (15).
In tomato (Lycopersicon esculentum), resistance to race 1 of Verticillium dahliae and other species is conferred by a single dominant Ve gene that was mapped to linkage group IX (16). We identified in near-isogenic tomato germplasm a codominant random-amplified polymorphic DNA (RAPD) marker within 3.2 ± 0.3 centimorgans (cM) of Ve (17). Sequences of the RAPD were used subsequently to develop allele-specific sequence-characterized amplified regions (SCARs) determined by high-resolution mapping to be within 0.67 ± 0.49 cM or 290 kb of Ve (18).
We describe in this study the positional cloning of two inverted resistance genes from the tomato Ve locus and demonstrate that both Ve1 and Ve2 independently confer resistance to an aggressive race 1 isolate of V. albo-atrum in potato. Structures within the Ve genes suggest they encode a class of cell-surface glycoproteins with signals for receptor-mediated endocytosis and leucine zipper or Pro-Glu-Ser-Thr (PEST) sequences.
Materials and Methods
Screening of Tomato Genomic and cDNA Libraries.
Genomic clones (Fig. 1) were isolated by initially screening a λ EMBL3 library (CLONTECH) of the V. dahliae race 1-resistant L. esculentum germplasm VFN8 with allele-specific SCARs (18). Approximately three copies of the genome, represented by 2 × 105 recombinant plaques, were transferred to duplicate Hybond N+ membranes and were probed. Rescued clones were subcloned into pBluescript SK(−) (Stratagene) and were sequenced.
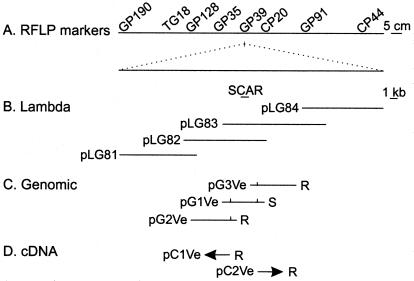
Figure 1.
Schematic genetic and physical representation of L. esculentum linkage group (A) bordering the Ve gene. Analysis of populations segregating for Ve identified closely linked codominant random-amplified polymorphic DNAs and allele-specific SCARs that map to the region of RFLP GP39. Identification of contiguous λ-genomic clones (B) facilitated the subcloning of genomic DNA (C) containing the Ve locus. Vertical lines indicate the location of the AUG initiation codon in the subgenomic clones. Expressed sequences were cloned into λ, and arrowheads depict the direction of transcription for the cDNAs (D) identified by using the genomic clone pG1Ve. Potato plants transformed with the genomic subclones pG2Ve and pG3Ve or cDNAs pC1Ve and pC2Ve exhibited in vivo complementation and resistance (R) when challenged with V. albo-atrum race 1. Potato plants transformed with the genomic pG1Ve and the binary vectors pBIN19 and pBI121 were susceptible (S) to infection.
A Stratagene cDNA cloning kit was used to prepare and unidirectionally clone cDNA as described by the manufacturer. Total RNA was isolated from detached leaves of greenhouse-propagated verticillium wilt-resistant L. esculentum cultivar Craigella, stressed in 1 mM l-serine for 48 h. Polyadenylated [poly(A)+] RNA was isolated by oligo(dT)-cellulose chromatography. First-strand cDNA synthesis was primed with an oligo(dT) linker-primer that contained an XhoI site and was reverse transcribed by using an RNase H− reverse transcriptase in the presence of 5-methyl-dCTP to hemimethylate the cDNA. Second-strand cDNA was prepared by using RNase H and DNA polymerase I, and the double-stranded DNA was treated with the Klenow fragment of DNA polymerase before ligation to EcoRI adapters. The cDNA was ligated to EcoRI- and XhoI-restricted arms of the λ Uni-ZAP XR vector. Phage were packaged and used to infect the recA− Escherichia coli XL1-Blue MRF′. Approximately 3 × 105 recombinant plaques were transferred to Hybond N+ membranes and were screened with the genomic subclone pG1Ve. Eight cDNA clones were recovered and pBluescript SK(−) phagemid with the cloned inserts were excised and recircularized.
Southern and Transcription Analyses.
Genomic and cDNA clones linked to Ve were subcloned into the pBIN19 or pBI121 binary vectors for Agrobacterium tumefaciens-mediated transformation of potato (19). Stable integration of the sequences between the T-DNA borders was confirmed by Southern analysis (20). Isolated genomic DNA was restricted, separated by gel electrophoresis, transferred to Hybond N+ membranes (Amersham Pharmacia), and hybridized with pC1Ve as recommended by the membrane manufacturer. Expression of Ve in potato was determined by reverse transcription and amplification with the PCR. Total RNA was isolated from fresh plant material by using TRIZOL (Life Technologies, Rockville, MD) and 2.5 μg of RNA treated with RNase-free DNase I (Life Technologies) before cDNA synthesis with the ThermoScript RT-PCR system (Life Technologies). Reverse transcription was performed at 68°C (Ve1) or 58°C (Ve2) with 500 nM of gene-specific primer 5′-CTGGTTTCAACTCTGAAGTATC-3′ (Ve1) or 5′-ATTTGCTGCCCCTACTATGTATCC-3′ (Ve2), complementary to 3′ untranslated sequences. Subsequent PCR was performed with an annealing temperature of 68°C (Ve1) or 66°C (Ve2) in a 50-μl volume containing 10% (vol/vol) of cDNA reaction, 0.2 μM cDNA primer, and 5′-TAACAGTCTTGTTGATCGTTTCCC-3′ (Ve1) or 5′-TGAATTGTAAGTTGTTGGAGGTCC-3′ (Ve2) primers specific to 5′ exon sequences.
Resistance Complementation Assays.
Plants propagated in the greenhouse were inoculated with aggressive isolates of V. albo-atrum race 1 (18) or Phytophthora infestans US8 mating type A2 (21). Disease reactions were obtained by challenging a minimum of 10 plants from at least three independent lines of transgenic potato plants for each construct. Plants ≈15 cm in height were inoculated with V. albo-atrum by removing the lower roots before submerging the remaining roots for 10 min in a 5 × 107 conidia per milliliter suspension. Alternatively, leaves and stems were immersed in a 5 × 104 sporangia and zoospore suspension of P. infestans. Plants were rated 3 weeks postinoculation as either susceptible to the pathogen, as indicated by chlorosis and necrosis of the leaves and stunting of the plant, or as resistant if there were no disease symptoms and appearance was similar to the uninoculated plants.
Sequence Analysis.
Genomic DNA and cDNA sequences were determined with a Sequitherm Long-Read Cycle Sequencing kit (Epicentre Technologies, Madison, WI) and an ABI 377 automated sequencer (PE Biosystems, Foster City, CA) by using primers derived from the genomic sequences and the polylinker cloning site of the vectors. Various versions of the blast algorithm (22) were used to search DNA and protein databases for similarity. Motifs were identified with the pcgene (IntelliGenetics) program version 6.8.
Results
Positional Cloning of Ve and Genomic Complementation in Transgenic Potato Plants.
To proceed with map-based cloning of Ve, we used the SCAR sequences as hybridization probes to identify λ clones that possessed contiguous, overlapping inserts of genomic DNA (Fig. 1) from resistant L. esculentum VFN8 germplasm. Identification of Ve involved in vivo functional complementation within the potato (Solanum tuberosum ssp. tuberosum) cultivar Désirée, which is highly susceptible to verticillium wilt. In vivo complementation and specificity were observed initially in potato plants transformed with the 4-kb genomic sequences of λ-subclone pG2Ve and subsequently pG3Ve (Fig. 1). These plants exhibited delayed and reduced disease symptoms after inoculation with V. albo-atrum race 1 (Fig. 2) but no resistance to P. infestans (Table 1). All untransformed plants and control plants transformed with the binary vector pBIN19 or pG1Ve displayed wilt, chlorosis, and necrosis within a few weeks of V. albo-atrum race 1 inoculation.
Figure 2.
Genetic complementation in potato plants transformed with Ve. Disease symptoms were recorded 3 weeks after plants containing pG2Ve∷pBIN19 (left) (resistant) or pBIN19 (right) (susceptible) were inoculated with V. albo-atrum race 1. Similar disease resistance was observed in plants transformed with pG3Ve∷pBIN19, pC1Ve∷pBI121, and pC2Ve∷pBI121.
Table 1.
Disease incidence in potato and tomato genomes challenged with aggressive isolates of V. albo-atrum race 1 and P. infestans A2 US8
| Line* |
V. albo-atrum race
1
|
P. infestans A2 US8
|
||
|---|---|---|---|---|
| R | S | R | S | |
| Potato | ||||
| pG1Ve | 0 | 31 | 0 | 8 |
| pG2Ve | 48 | 0 | 0 | 8 |
| pG3Ve | 83 | 0 | 0 | 8 |
| pC1Ve | 30 | 0 | 0 | 8 |
| pC2Ve | 78 | 0 | 0 | 8 |
| pBIN | 0 | 48 | 0 | 8 |
| pBI122 | 0 | 48 | 0 | 8 |
| Désirée | 0 | 56 | 0 | 8 |
| Tomato | ||||
| Ailsa Craig | 0 | 32 | 0 | 32 |
| Craigella | 32 | 0 | 0 | 32 |
Plants were rated 3 weeks postinoculation as resistant (R, no disease symptoms) or susceptible (S, advanced necrosis). Complementation was observed only in plants challenged with V. albo-atrum race 1 expressing full-length genomic DNA or cDNA of Ve1 (pG2Ve and pC1Ve) or Ve2 (pG3Ve and pC2Ve).
Transformants pG1Ve, pG2Ve, and pG3Ve contain genomic DNA with the intergenic region and N terminus of Ve1 and Ve2, a full-length clone of Ve1, or a full-length clone of Ve2, respectively. Plants transformed with pC1Ve (Ve1) and pC2Ve (Ve2) express full-length cDNA clones under the transcriptional control of the cauliflower mosaic virus 35S promoter.
Identification of Expressed Sequences and cDNA Complementation in Transgenic Potato Plants.
To identify expressed sequences and the Ve locus, the λ subclones were sequenced and pG1Ve was used to probe an L. esculentum cDNA library of the verticillium wilt-resistant tomato variety Craigella. Genomic sequences confirmed that pG1Ve possessed the SCAR sequence linked to the resistant Ve allele and revealed inverted terminal ORFs in pG1Ve homologous to the N-terminal domain of plant and animal receptors that possess LRRs. The cDNA clones, pC1Ve and pC2Ve, corresponding to the ORF sequences detected in the genomic subclone pG1Ve, were isolated and designated Ve1 and Ve2 (Fig. 1). To confirm complementation observed with the genomic clones, the cDNA of pC1Ve and pC2Ve was cloned into the binary vector pBI121 in a sense orientation under transcriptional control of the cauliflower mosaic virus 35S promoter for plant transformation. All plants expressing pC1Ve and pC2Ve cDNA were resistant to V. albo-atrum race 1 but not P. infestans, whereas untransformed germplasm and plants transformed with the vector alone were susceptible to both pathogens (Table 1).
Deduced Primary Structure of Ve1 and Ve2.
Sequence analysis of the isolated cDNA and corresponding genomic clones did not detect any introns within the Ve ORFs. However, an amino acid identity of 84% and several structural domains were observed within the deduced Ve proteins (Fig. 3). A hydrophobic N terminus in the Ve proteins (domain A), indicative of a signal peptide that may target the protein to the cytoplasmic membrane (23), contains a leucine zipper-like motif with four contiguous amphipathic heptad repeats in Ve1. Domain A precedes an LRR with 38 imperfect copies of a 24-aa consensus [XXIXNLXXLXXLXLSXNXLSGXIP (domain B)] that is often associated with protein–protein interactions and ligand binding. The presence of a glycine within the consensus sequence is consistent with that of extracytoplasmic proteins, a location that would facilitate the recognition of an extracellular pathogen ligand (14, 15). Within the predicted LRR region, 28 or 35 sequences matching the N-glycosylation consensus sequence NX(S/T) were observed in Ve1 and Ve2, respectively (Fig. 3).
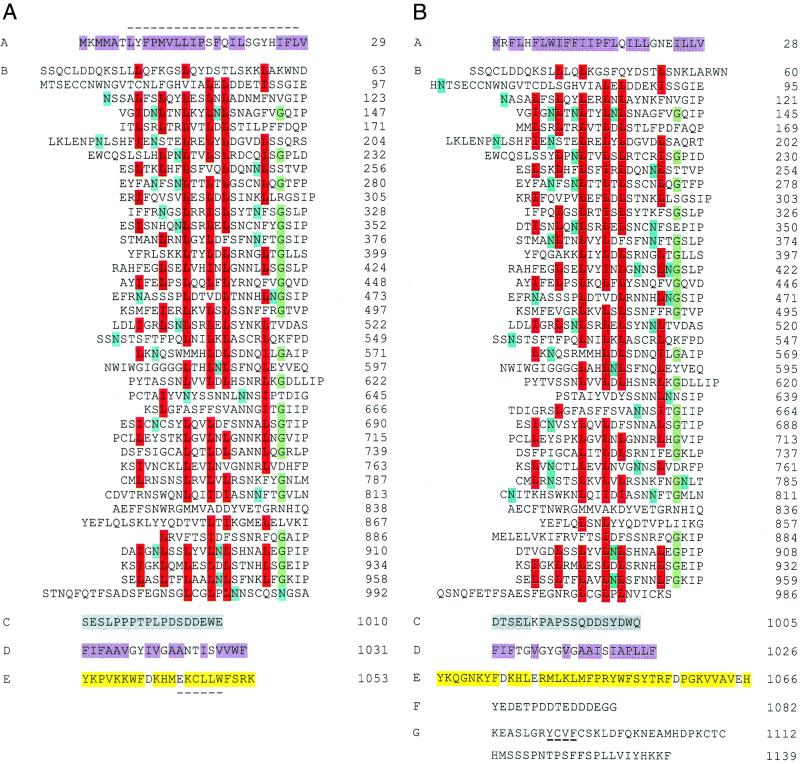
Figure 3.
Primary structure of the Ve1 (A) and Ve2 (B) proteins deduced from cDNA sequence. The polypeptides have been divided into domains A–G as described in the text. A dashed line occurs above the putative N-terminal leucine zipper in domain A of Ve1 and below the endocytosis signals in domain E of Ve1 and domain G of Ve2. Highlighted are the hydrophobic amino acids (purple) of the putative signal peptide domain A and membrane-associated domain D; conserved L/I (red), G (green), and potential N-glycosylation sites (blue) within the LRR domain B; neutral and acidic amino acids (gray) of domain C; and neutral and basic amino acids (yellow) of domain E. The PEST sequence of Ve2 is shown in domain F.
As frequently observed with membrane-spanning proteins, a hydrophobic sequence with a predicted α-helix secondary structure (domain D) is flanked by a negatively charged extracytoplasmic domain C and a positively charged cytoplasmic domain E. Each cytoplasmic domain possesses the dileucine E/DXXXLφ or tyrosine YXXφ signal sequences, where φ is an amino acid with a hydrophobic side chain, that stimulate receptor-mediated endocytosis and degradation of mammalian cell-surface receptors (24). In Ve2, the C terminus also contains a PEST-like sequence found in proteins with cytoplasmic half-lives of only a few hours (25) and concludes with the residues KKF, similar to the KKX motif that signals endoplasmic reticulum retention in mammalian and plant cells (26, 27).
Discussion
Verticillium wilt resistance has been incorporated into most commercial tomato varieties and has proven to be very durable. We report in this study the positional cloning of the verticillium wilt Ve resistance genes from tomato. Identification of two closely linked inverted genes independently conferring resistance to the same pathogen was unexpected. The only other example of two closely linked functional disease resistance genes is the Cf2 locus, which consists of direct repeats differing by only three nucleotides (28). Like Cf2, the Ve genes likely resulted from relatively recent gene duplication and homologous recombination events that are believed to contribute to R gene evolution. It is possible that the Ve receptors recognize different ligands that would require a pathogen to possess at least two virulence products before inciting disease.
Homology to the Ve genes was observed in genes encoding several plant proteins with LRRs, including disease resistance genes that probably produce cytoplasmic proteins (6–13) and the Xa and Cf genes (14, 15) reported to encode proteins with an extracytoplasmic domain that interacts with an extracellular ligand. Unlike Xa21, the Ve genes do not include a protein kinase and are unique, because they are the only disease-resistant receptors containing endocytosis-like signals and leucine zipper or PEST sequences. Leucine zippers have been reported in the cytoplasmic class of Arabidopsis resistance genes RPS2 and RPM1 for P. syringae and can facilitate dimerization of proteins through the formation of coiled-coil structures (29). PEST sequences are often involved in ubiquitinization, internalization, and degradation of proteins (30). Discovery of a leucine zipper, PEST, and endocytosis signals in the Ve receptors expands the available motifs and complexity of plant cell receptors.
All eukaryotic cells exhibit receptor-mediated endocytosis as a mechanism to communicate or respond to external stimuli. In mammalian cells, ligand-dependent (e.g., insulin and cytokine CD4 receptors) or constitutive (e.g., transferrin and low density lipoprotein receptors) endocytosis is stimulated by tyrosine or dileucine motifs located within the cytoplasmic C terminus, concentrating cell-surface receptors into clathrin-coated pits that are internalized and degraded in the lysosome (31). In plant cells, clathrin-coated pits and other indirect evidence of receptor-mediated endocytosis has been observed (32). Identification of the EKCLLW and YCVF sequences in the short cytoplasmic domains of two homologous but independent protein sequences indicates that plant and mammalian cell-surface receptors may share similar endocytosis signals. In Ve, receptor-mediated endocytosis could provide a mechanism through which cells selectively capture ligands and remove signaling receptors from their surfaces, thereby actively responding to changing disease pressures.
Cytoplasmic signaling by Ve may be analogous to that of the erythropoietin cytokine receptor. Preformed dimers on the cell surface facilitate transmission of a ligand-induced conformational change from the extracytoplasmic to the cytoplasmic domain and subsequent signal transduction (33). The cytoplasmic domain interacts with kinases that link ligand binding to tyrosine phosphorylation of various signaling proteins and transcription-activation factors. A similar model has been proposed for the kinase encoded by the Pto resistance gene that lacks a receptor domain (6) and Cf extracytoplasmic receptors that lack a kinase (14, 28). Alternatively, receptor-mediated endocytosis may allow the extracellular domains and ligands of the Ve proteins to directly stimulate signal transduction.
Resistance to different pathogen species is contrary to the traditional view of a highly specific interaction with race-defining R genes. Our results demonstrate that, although the tomato Ve genes have the specificity to distinguish races 1 and 2 of V. dahliae, the genes also possess the capacity to recognize another Verticillium species in a different host. This pleiotropic resistance resembles that observed with the Mi gene, which confers resistance to nematodes and aphids (34, 35) and shares the ability of R genes to retain biological activity in other plant genera (8, 36). Several Verticillium species infect many agricultural plants and this pleiotropic, host-independent complementation should be of considerable value.
Acknowledgments
We thank K. Toohey, M. Kokko, S. Smienk, E. Lyon, C. Mueller, and C. Verhaeghe for technical assistance. Research was supported in part by grants from the Alberta Research Council and Canada-Saskatchewan Agri-Food Innovation Fund.
Abbreviations
- LRR
leucine-rich repeat
- SCAR
sequence-characterized amplified regions
- PEST
Pro-Glu-Ser-Thr
Footnotes
References
- 1.Domsch K H, Gams W, Traute-Heidi A. Compendium of Soil Fungi. Vol. 1. New York: Academic; 1980. pp. 829–845. [Google Scholar]
- 2.Schaible L, Cannon O S, Waddoups V. Phytopathology. 1951;41:986–990. [Google Scholar]
- 3.Lynch D R, Kawchuk L M, Hachey J, Bains P S, Howard R J. Plant Dis. 1997;81:1011–1014. doi: 10.1094/PDIS.1997.81.9.1011. [DOI] [PubMed] [Google Scholar]
- 4.Leister D, Ballvora A, Salamini F, Gebhardt C. Nat Genet. 1996;14:421–429. doi: 10.1038/ng1296-421. [DOI] [PubMed] [Google Scholar]
- 5.Flor H H. J Agric Res. 1946;73:241–262. [Google Scholar]
- 6.Martin G B, Brommonschenkel S H, Chunwongse J, Frary A, Ganal M W, Spivey R, Wu T, Earle E D, Tanksley S D. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- 7.Bent A F, Kunkel B N, Dahlbeck D, Brown K L, Schmidt R, Giraudat J, Leung J, Staskawicz B J. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- 8.Mindrinos M, Katagiri F, Yu G L, Ausubel F M. Cell. 1994;78:1089–1099. doi: 10.1016/0092-8674(94)90282-8. [DOI] [PubMed] [Google Scholar]
- 9.Grant M R, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes R W, Dangl J L. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- 10.Ori N, Eshed Y, Paran I, Presting G, Aviv D, Tanksley S, Zamir D, Fluhr R. Plant Cell. 1997;9:521–532. doi: 10.1105/tpc.9.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitham S, McCormick S, Baker B. Proc Natl Acad Sci USA. 1996;93:8776–8781. doi: 10.1073/pnas.93.16.8776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawrence G J, Finnegan E J, Ayliffe M A, Ellis J G. Plant Cell. 1995;7:1195–1206. doi: 10.1105/tpc.7.8.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parker J E, Coleman M J, Szabo V, Frost L N, Schmidt R, van der Biezen E A, Moores T, Dean C, Daniels M J, Jones J D. Plant Cell. 1997;9:879–894. doi: 10.1105/tpc.9.6.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones D A, Thomas C M, Hammond-Kosack K E, Balint-Kurti P J, Jones J D. Science. 1994;266:789–793. doi: 10.1126/science.7973631. [DOI] [PubMed] [Google Scholar]
- 15.Song W Y, Wang G L, Chen L L, Kim H S, Pi L Y, Holsten T, Gardner J, Wang B, Zhai W X, Zhu L H, et al. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- 16.Diwan N, Fluhr R, Eshed Y, Zamir D, Tanksley S D. Theor Appl Genet. 1999;98:315–319. [Google Scholar]
- 17.Kawchuk L M, Lynch D R, Hachey J, Bains P S. Theor Appl Genet. 1994;89:661–664. doi: 10.1007/BF00223701. [DOI] [PubMed] [Google Scholar]
- 18.Kawchuk L M, Hachey J, Lynch D R. Genome. 1998;41:91–95. [PubMed] [Google Scholar]
- 19.De Block M. Theor Appl Genet. 1988;76:767–774. doi: 10.1007/BF00303524. [DOI] [PubMed] [Google Scholar]
- 20.Southern E M. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 21.Helgeson J P, Pohlman J D, Austin S, Haberlach G T, Wielgus S M, Ronis D, Zambolim L, Tooley P, McGrath J M, James R V, et al. Theor Appl Genet. 1998;96:738–742. [Google Scholar]
- 22.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Heijne G. EMBO J. 1984;3:2315–2318. doi: 10.1002/j.1460-2075.1984.tb02132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Letourneur F, Klausner R D. Cell. 1992;69:1143–1157. doi: 10.1016/0092-8674(92)90636-q. [DOI] [PubMed] [Google Scholar]
- 25.Rogers S, Wells R, Rechsteiner M. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 26.Jackson M R, Nilsson T, Peterson P A. EMBO J. 1990;9:3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benghezal M, Wasteneys G O, Jones D A. Plant Cell. 2000;12:1179–1202. doi: 10.1105/tpc.12.7.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dixon M S, Jones D A, Keddie J S, Thomas C M, Harrison K, Jones J D G. Cell. 1996;84:451–459. doi: 10.1016/s0092-8674(00)81290-8. [DOI] [PubMed] [Google Scholar]
- 29.Lupas A, Van Dyke M, Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 30.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 31.Goldstein J L, Brown M S, Anderson R G W, Russell D E, Schneider W J. Annu Rev Cell Biol. 1985;1:1–39. doi: 10.1146/annurev.cb.01.110185.000245. [DOI] [PubMed] [Google Scholar]
- 32.Battey N H, James N C, Greenland A J, Brownlee C. Plant Cell. 1999;11:643–660. doi: 10.1105/tpc.11.4.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Remy I, Wilson I A, Michnick S W. Science. 1999;283:990–993. doi: 10.1126/science.283.5404.990. [DOI] [PubMed] [Google Scholar]
- 34.Rossi M, Goggin F L, Milligan S B, Kaloshian I, Ullman D E, Williamson V M. Proc Natl Acad Sci USA. 1998;95:9750–9754. doi: 10.1073/pnas.95.17.9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vos P, Simons G, Jesse T, Wijbrandi J, Heinen L, Hogers R, Frijters A, Groenendijk J, Diergaarde P, Reijans M, et al. Nat Biotechnol. 1998;16:1365–1369. doi: 10.1038/4350. [DOI] [PubMed] [Google Scholar]
- 36.Tai T H, Dahlbeck D, Clark E T, Gajiwala P, Pasion R, Whalen M C, Stall R E, Staskawicz B J. Proc Natl Acad Sci USA. 1999;96:14153–14158. doi: 10.1073/pnas.96.24.14153. [DOI] [PMC free article] [PubMed] [Google Scholar]