Abstract
In this review, OVA1® (Vermillion, Inc., Austin, TX), the first in vitro diagnostic multivariate index assay (IVDMIA) of protein biomarkers cleared by the US Food and Drug Administration (FDA), is used to explain the concept behind IVDMIA, the use of multiple markers to improve clinical performance of a diagnostic tool, and the key considerations in the development of IVDMIA.
Key words: In vitro diagnostic multivariate index assay, Protein biomarkers, Ovarian cancer, CA-125
OVA1® (Vermillion, Inc., Austin, TX) is the first in vitro diagnostic multivariate index assay (IVDMIA) of protein biomarkers cleared by the US Food and Drug Administration (FDA) for clinical use. Since OVA1 clearance in 2009, a number of IVDMIA tests have been used in clinical applications. Some of these tests have sought regulatory approval/clearance, whereas others have been offered as a laboratory-developed test (LDT). In this review, OVA1 is used to explain the concept behind IVDMIA, the use of multiple markers to improve clinical performance of a diagnostic tool, and the key considerations in the development of IVDMIA.
What Is an IVDMIA?
In a 2007 draft guideline,1 the FDA defines an IVDMIA as
...a device that 1) combines the values of multiple variables using an interpretation function to yield a single, patient-specific result (e.g., a “classification,” “score,” “index,” etc.), that is intended for use in the diagnosis of disease or other conditions, or in the cure, mitigation, treatment or prevention of disease, and 2) provides a result whose derivation is non-transparent and cannot be independently derived or verified by the end user.
The use of multiple tests (biomarkers) itself is not new. In fact, it has been practiced by physicians routinely when they order multiple tests or panels to assist in their differential diagnostic decision process. A simple example is the use of leukocyte levels (white blood cell count [WBC]) to rule out bacterial infection in patients who present with high fever. A slightly more complex example is the use of the ratio of two tests: free (unbound) and total prostate-specific antigen (%free PSA). A lower %free PSA is indicative of elevated risk of prostate cancer.2 However, when the number of tests increases and/or the disease-associated patterns among the tests are complex, simple visual inspection of test results (eg, WBC) or calculation of ratios becomes inadequate. Instead, advanced mathematical and computational tools become necessary to derive multivariate models that capture the signatures or patterns of disease among an often large number of biomarkers. The tools used to derive such models as well as the resultant models that produce the indices are often not obvious or readily understandable to those who are responsible for the interpretation of the results. This can occasionally cause unease in the adoption of IVDMIA in clinical practice.
Why IVDMIA?
The Pros and Cons
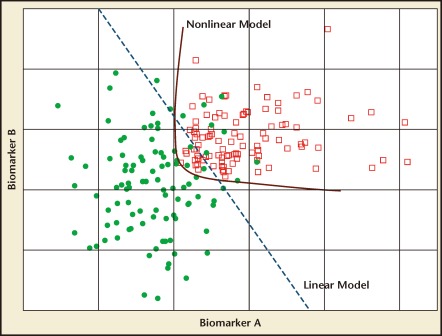
The advantages of IVDMIA in comparison with a single biomarker assay are based on the premise that the single-valued index, with its aggregated information from complementary biomarkers, will outperform each of its component biomarkers used individually. Figure 1 illustrates this concept through a simulated example. Biomarkers A and B both have a decent ability to separate the cases (red squares) from the controls (filled green circles). However, if the intended clinical use demands an extremely high sensitivity, neither of the biomarkers will be able to achieve that end without a significantly sacrificed specificity. Figure 1 illustrates that a simple linear model combining the two biomarkers using methods such as linear regression is able to achieve a higher level of sensitivity while retaining much of the specificity. The classification performance is improved further through the use of a nonlinear model.
Figure 1.
Simulated data for two biomarkers. Red squares, cases; filled green circles, controls. The blue dashed line illustrates a linear model that combines the two biomarkers to achieve better separation of the two classes of samples than either of the biomarkers used alone. The red line is an example of a nonlinear model that can further improve model performance.
Within the short span of a decade, technological advances in genomic and proteomic analysis and development of multiplex assay platforms have made it possible to analyze a large number of biomarkers and to use their information collectively to assist in clinical decision making. In order to integrate information from a large number of biomarkers and capture the patterns of expressions for a disease indication with multiple underlying molecular characteristics, advanced computational and statistical tools must be used to derive the IVDMIA models. The corresponding high dimensionality of data and complex decision boundary will be difficult to visualize. However, the basic concept behind the use of the multivariate model to improve clinical performance of tests remains the same as in the simple two-variable example illustrated in Figure 1.
Along with the advantages of IVDMIA, there are also certain dangers in its development and application. The ability of multivariate models to capture complex patterns in high-dimensional data also means that non-disease-related artifacts that happen to confound the disease status of the samples used to train the models will also be captured. Examples of such artifacts could be the result of biases in the selection of cases and controls when the training samples are from a retrospective, case-control study or subtle differences in conditions during the collection, storage, or processing of specimens.3,4 Such biases have resulted in models that were reported to have extremely high-level clinical performance yet ultimately failed or have not had their results replicated in further independent studies.5–7 Great care needs to be taken in the design of clinical studies from which samples are drawn and the actual usage of samples during the development of IVDMIA. An example of good design is based on the concept of “prospective collection of samples and outcome ascertainment in the clinical context of interest with biomarker assays of random subsets of cases and controls,” as in PRoBE (prospective specimen collection, retrospective blind evaluation) study design. It avoids many of the common sources of biases and confounding factors.8
Considerations During the Development of an IVDMIA
With the explosive advances in genomic and proteomic research, discoveries of novel biomarkers or new applications of existing biomarkers have become frequent events reported in the literature. However, for biomarkers to make their way into an IVDMIA and eventually become part of a commercial product in clinical use, the path is often long and difficult, involving a phased process of gathering and building evidence of clinical efficacy and the development of an analytically stable assay platform.
The development of an IVDMIA must be driven by a clearly defined intended use. The intended use defines the time point along the disease path at which the IVDMIA is to be used. This in turn defines the target population of the test, the utility of the test in terms of changes in clinical intervention it may cause, and the consequences of false-positive or -negative results. For IVDMIA development, the intended use determines the inclusion and exclusion criteria of subjects of studies from which the training data are generated and the patient populations in which the final IVDMIA are to be validated, the criteria for the selection of biomarkers to be included, and the minimum requirement to establish efficacy. From the point of view of product commercialization, the definition of intended use requires compromise between the desire for a wider applicability of the IVDMIA and the ability, cost, and time required to prove its safety and efficacy for regulatory approval.
The inclusion of biomarkers in an IVDMIA requires that they are complementary and collectively outperform a single marker with respect to the test’s intended use. It is not always true that biomarkers with the highest discriminatory power individually will make the best panel of markers in an IVDMIA. For ovarian cancer, cancer antigen 125 (CA-125) remains the best tumor marker. The selection of additional biomarkers, therefore, will be based mainly on their ability to detect malignancy in cancer patients with a low serum CA-125 level or to reduce false-positive results among noncancer patients with an elevated serum CA-125 level.
The decision to develop an IVDMIA as a commercial product is a significant commitment of resources and effort, and should be based on solid scientific evidence. As discussed, biomarker data generated from clinical specimens of a retrospective, case-control study are susceptible to effect of biases and confounding factors. Due to the practical constraints on how specimens from cases and controls are handled differently in a clinical setting, some of the biases might be unavoidable. Results from a single-site study alone, no matter how strong the results might be, are often not sufficient to extrapolate an IVDMIA’s future performance at different sites. In practice, the portability of disease-associated patterns of expressions of the selected biomarkers across multiple independent clinical sites is often a more important piece of evidence in making go or no-go decisions in IVDMIA development.
The defined intended use can also be used to influence the derivation of the multivariate model in an IVDMIA. In the optimization procedures used in model derivation, it is often possible to incorporate the desired clinical performance characters, such as the need for high sensitivity, into the objective function used in optimization.
Development of OVA1
OVA1 is the first IVDMIA of protein biomarkers cleared by the FDA to further assess the likelihood of malignancy in women presenting with an ovarian adnexal mass prior to planned surgery. OVA1 combines results from five tests—CA-125 II, prealbumin, apolipoprotein A-1, β2-microglobulin, and transferrin—into a single-valued index between 0 and 10; a higher value corresponds to a higher risk of malignancy. Two cutoffs at 5.0 and 4.4, for pre- and postmenopausal women, respectively, are used to classify a woman into higher or lower probability of malignancy. In a large-scale, multicenter, prospective clinical study, it was reported that, among the 516 patients who had both physician assessment and OVA1 values, the addition of OVA1 to physician assessment improved sensitivity from 72.2% (52/72) to 91.7% (66/72) for non-gynecologic oncologists and from 77.5% (69/89) to 98.9% (88/89) for gynecologic oncologists.9 Such noticeable improvement in sensitivity translates into a high negative predictive value (NPV), which is a clinically important measure to assure physicians and patients that the risk of malignancy will be low for patients who have a negative result by OVA1. In fact, the 92.5% (149/161) sensitivity of OVA1 itself will produce an NPV of 92.9% (156/168). Further details of this study and a companion analysis using OVA1 in place of CA-125 in the American Congress of Obstetricians and Gynecologists ovarian tumor referral guidelines have been previously reported.9,10
The target population for OVA1, women who have planned to have surgery due to suspected risk of ovarian cancer yet have not been referred to a gynecologic oncologist, represents a real clinical need. A number of clinical studies have indicated that ovarian cancer patients referred to a gynecologic oncologist for their surgeries are more likely to have a better outcome, including surgical staging, optimal debulking, and improved median and overall 5-year survival.11–16 However, currently only about one-third of ovarian cancer patients are referred to a gynecologic oncologist for primary surgery.17,18 OVA1 provides additional information to help guide the referral decision process.
The addition of OVA1 to clinical assessment brings significant improvement in sensitivity. This is, however, at the cost of a reduced specificity. Ideally, one would like to have an assay that is both highly sensitive and specific. Unfortunately, a study that systematically evaluated a large number of reported ovarian cancer biomarkers using samples from the National Cancer Institute’s PLCO (Prostate, Lung, Colorectal, and Ovarian Cancer) screening trial concluded that none of these biomarkers are likely to offer such an ideal level of performance.19 During the construction of the OVA1 multivariate model and the choice of cutoff values, a conscious decision was made to emphasize the need for a high sensitivity. This decision took into consideration the need to mitigate the safety concern of OVA1 with respect to its predefined intended use. Because OVA1 is to be used prior to the decision to refer to a specialist, a high sensitivity minimizes the risk of false-negative results for patients who actually have malignant diseases.
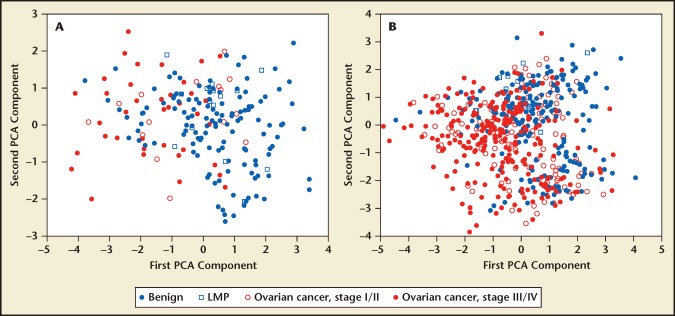
Except for CA-125, the biomarkers in the OVA1 panel were part of seven biomarkers discovered through large-scale proteomic analysis of clinical serum samples from multiple centers.20,21 Statistically sound designs and robust bioinformatics tools were used to alleviate the impact of biases and confounding factors.21 Prior to the derivation of the actual OVA1 model and commitment to start a multicenter clinical study to seek regulatory clearance of OVA1, these biomarkers were further validated for the abovementioned evidence of portability of the biomarkers’ discriminatory power across multiple independent clinical sites. In Figure 2(A), prospectively collected clinical samples from patients of benign or malignant ovarian tumors at a single clinical site were clustered using principal component analysis (PCA) and plotted in the first two PCA dimensions. Because PCA is an unsupervised method in which the samples’ clinical labels were not used, the two-dimensional PCA plot shows the natural separation of cancer and benign cases due to the discriminatory power of the seven biomarkers. In Figure 2B, retrospective samples from five additional clinical sites were plotted using the same PCA projection coefficients as for the plot in Figure 2A. It can be seen that the pattern of separation persists from site to site, representing geographically extremely distant locations. Although for assay analytical performance reasons only four of the seven biomarkers were added to CA-125 to form the OVA1 panel, and the final assay forms are not the same as those used for the plots, this piece of evidence played an important role in the decision to develop the OVA1 IVDMIA.22
Figure 2.
Evidence of the portability of a biomarker’s ability to extend its discriminatory power to samples from independent and geographically distant clinical sites. (a) Plot of an unsupervised cluster analysis of samples from a prospective study at a single site. The plot shows the samples in the first two-dimensional principal component analysis (PCA) of data from seven ovarian cancer biomarkers (not including CA-125). (b) Using the same PCA projection coefficients from A, samples from five additional geographically distant clinical sites are plotted in the same two-dimensional PCA component space. The persistence of clustering patterns and separation between benign and malignant ovarian tumors from site to site is an important piece of evidence to support further IVDMIA development. Four of the seven biomarkers were late added to CA-125 to form the OVA1® (Vermillion, Inc., Austin, TX) biomarker panel.
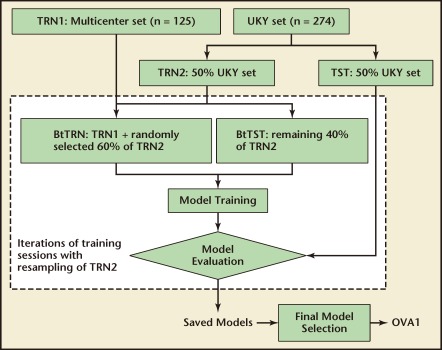
The training samples of OVA1 were from two prospectively collected sample sets. The first set included 274 consecutive samples from the University of Kentucky (UKY) Medical Center (167 benign, 29 low malignant potential [LMP] tumor, 63 epithelial ovarian cancer [EOC], 3 other ovarian cancer, and 12 other cancer). The second set consisted of 125 samples from a multi-center, prospective study (33 EOC and 92 benign). Both sample sets had similar inclusion and exclusion criteria that required the subjects to be women aged 18 years or older diagnosed with an ovarian tumor and a subsequently confirmed malignancy status by surgery. The use of these samples in multivariate model derivation involved extensive statistical resampling (bootstrap) to select and test models that are likely to have a robust performance and generalize well in patients from different clinical sites. Figure 3 shows the usage of training samples in OVA1 model derivation. Iterations of training sessions in which the composition of training samples were altered each time by bootstrap resampling generated many multivariate models. The selection of the final OVA1 model was based on model performance on training samples, in-training test samples, and a set of set-aside samples that was a randomly selected half of the UKY samples and never used in model training.
Figure 3.
Data usage diagram for the derivation of OVA1® (Vermillion, Inc, Austin, TX) multivariate model. During model derivation, 50% of the University of Kentucky (UKY) samples were randomly selected as a set-aside test set. The remaining 50% of the UKY samples became TRN2. In each of the iterative training sessions, TRN2 was resampled to add to TRN1 (the multicenter samples) to form an actual training set btTRN. The remaining samples of TRN2 served as btTST for in-training testing. The set-aside set was not directly involved in model derivation. It contributed to the selection of a pool of candidate models and the final choice of the OVA1 model.
Conclusions
Advances in genomic and proteomic technologies and the push for personalized medicine have driven the development and application of biomarkers for risk assessment, early detection, diagnosis, prognosis, treatment selection, and monitoring. However, the need for evidence-based medicine also demands that such applications are based on scientific data from statistically sound clinical studies. IVDMIA combines multiple biomarkers into a single-valued index and therefore makes it possible to validate its clinical utilities using well-established procedures and protocols similar to that for traditional IVD tests.
The collective use of multiple biomarkers offers some level of flexibility during IVDMIA development to shape its performance characteristics for a specific clinical application. It is therefore of paramount importance to have a clearly defined intended use prior to committing into a full-fledged IVDMIA development program. In this article, OVA1 was used as an example to illustrate several key elements in the development of IVDMIA. The current intended use of OVA1 is supported by a large-scale, prospective, multicenter clinical study. The performance characteristics of OVA1, to a large degree, were by design optimized for its predefined intended use and limited by the discriminatory powers of its component biomarkers. It is expected that with ongoing effort to introduce additional or replacement biomarkers, the performance of future successors of OVA1 will be further improved.
Main Points.
The advantages of an in vitro diagnostic multivariate index assay (IVDMIA) in comparison to a single biomarker assay are based on the premise that the single-valued index, with its aggregated information from complementary biomarkers, will outperform each of its component biomarkers used individually.
The ability of multivariate models to capture complex patterns in high-dimensional data also means that non-disease-related artifacts that happen to confound the samples used to train the models will also be captured.
The inclusion of biomarkers in an IVDMIA requires that they are complementary, and that they collectively outperform a single marker with respect to the test’s intended use.
OVA1® (Vermillion, Inc., Austin, TX) combines results from five tests—CA-125 II, prealbumin, apolipoprotein A-1, β2-microglobulin, and transferrin—into a single-valued index between 0 and 10; a higher value corresponds to a higher risk of malignancy.
The addition of OVA1 to clinical assessment brings significant improvement in sensitivity. This is, however, at the cost of a reduced specificity. During the construction of the OVA1 multivariate model and the choice of cutoff values, a conscious decision was made to emphasize the need for a high sensitivity. This decision took into consideration the need to mitigate the safety concern of OVA1 with respect to its predefined intended use. Because OVA1 is to be used prior to the decision to refer to a specialist, a high sensitivity minimizes the risk of false-negative results for patients who actually have malignant diseases.
Footnotes
Dr. Zhang is the inventor of the OVA1 algorithm, and as such is entitled to royalty payments from the sale of OVA1® through a license agreement between Johns Hopkins University and Vermillion, Inc.
References
- 1.US Department of Health and Human Services, authors. Draft Guidance for Industry, Clinical Laboratories, and Staff: In Vitro Diagnostic Multivariate Index Assays. [Accessed April 2, 2012]. http://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/ucm071455.pdf.
- 2.Catalona WJ, Smith DS, Wolfert RL, et al. Evaluation of percentage of free serum prostate-specific antigen to improve specificity of prostate cancer screening. JAMA. 1995;274:1214–1220. [PubMed] [Google Scholar]
- 3.Ransohoff DF. Bias as a threat to the validity of cancer molecular-marker research. Nat Rev Cancer. 2005;5:142–149. doi: 10.1038/nrc1550. [DOI] [PubMed] [Google Scholar]
- 4.Ransohoff DF. How to improve reliability and efficiency of research about molecular markers: roles of phases, guidelines, and study design. J Clin Epidemiol. 2007;60:1205–1219. doi: 10.1016/j.jclinepi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 5.Kozak KR, Amneus MW, Pusey SM, et al. Identification of biomarkers for ovarian cancer using strong anion-exchange ProteinChips: potential use in diagnosis and prognosis. Proc Natl Acad Sci U S A. 2003;100:12343–12348. doi: 10.1073/pnas.2033602100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petricoin EF, Ardekani AM, Hitt BA, et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 7.Zhou M, Guan W, Walker LD, et al. Rapid mass spectrometric metabolic profiling of blood sera detects ovarian cancer with high accuracy. Cancer Epidemiol Biomarkers Prev. 2010;19:2262–2271. doi: 10.1158/1055-9965.EPI-10-0126. [DOI] [PubMed] [Google Scholar]
- 8.Pepe MS, Feng Z. Improving biomarker identification with better designs and reporting. Clin Chem. 2011;57:1093–1095. doi: 10.1373/clinchem.2011.164657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueland FR, Desimone CP, Seamon LG, et al. Effectiveness of a multivariate index assay in the preoperative assessment of ovarian tumors. Obstet Gynecol. 2011;117:1289–1297. doi: 10.1097/AOG.0b013e31821b5118. [DOI] [PubMed] [Google Scholar]
- 10.Ware Miller R, Smith A, DeSimone CP, et al. Performance of the American College of Obstetricians and Gynecologists’ ovarian tumor referral guidelines with a multivariate index assay. Obstet Gynecol. 2011;117:1298–1306. doi: 10.1097/AOG.0b013e31821b1d80. [DOI] [PubMed] [Google Scholar]
- 11.Engelen MJA, Kos HE, Willemse PH, et al. Surgery by consultant gynecologic oncologists improves survival in patients with ovarian carcinoma. Cancer. 2006;106:589–598. doi: 10.1002/cncr.21616. [DOI] [PubMed] [Google Scholar]
- 12.Hillner BE, Smith TJ, Desch CE. Hospital and physician volume or specialization and outcomes in cancer treatment: importance in quality of cancer care. J Clin Oncol. 2000;18:2327–2340. doi: 10.1200/JCO.2000.18.11.2327. [DOI] [PubMed] [Google Scholar]
- 13.Le T, Giede C, Salem S, et al. Initial evaluation and referral guidelines for management of pelvic/ovarian masses [article in English, French] J Obstet Gynecol Can. 2009;31:668–680. doi: 10.1016/s1701-2163(16)34254-2. [DOI] [PubMed] [Google Scholar]
- 14.Myers ER, Bastian LA, Havrilesky LJ, et al. Management of adnexal mass. Evid Rep Technol Assess (Full Rep) 2006;130):1–145. [PMC free article] [PubMed] [Google Scholar]
- 15.American College of Obstetricians and Gynecologists Committee on Gynecologic Practice, authors. Committee Opinion No. 477: the role of the obstetrician-gynecologist in the early detection of epithelial ovarian cancer. Obstet Gynecol. 2011;117:742–746. doi: 10.1097/AOG.0b013e31821477db. [DOI] [PubMed] [Google Scholar]
- 16.van der Zee AG, Engelen MJ, Schaapveld M, et al. Primary surgery by a gynecological oncologist improves the prognosis in patients with ovarian carcinoma. Ned Tijdschr Geneeskd. 2009;153:15–19. [PubMed] [Google Scholar]
- 17.Carney ME, Lancaster JM, Ford C, et al. A population-based study of patterns of care for ovarian cancer: who is seen by a gynecologic oncologist and who is not? Gynecol Oncol. 2002;84:36–42. doi: 10.1006/gyno.2001.6460. [DOI] [PubMed] [Google Scholar]
- 18.Earle CC, Schrag D, Neville BA, et al. Effect of surgeon specialty on processes of care and outcomes for ovarian cancer patients. J Natl Cancer Inst. 2006;98:172–180. doi: 10.1093/jnci/djj019. [DOI] [PubMed] [Google Scholar]
- 19.Cramer DW , Jr, Bast RC , Jr, Berg CD, et al. Ovarian cancer biomarker performance in Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial specimens. Cancer Prev Res (Phila) 2011;4:365–374. doi: 10.1158/1940-6207.CAPR-10-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rai AJ, Zhang Z, Rosenzweig J, et al. Proteomic approaches to tumor marker discovery. Arch Pathol Lab Med. 2002;126:1518–1526. doi: 10.5858/2002-126-1518-PATTMD. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Z, Bast RC, Yu Y, et al. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res. 2004;64:5882–5890. doi: 10.1158/0008-5472.CAN-04-0746. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Z, Chan DW. The road from discovery to clinical diagnostics: lessons learned from the first FDA-cleared in vitro diagnostic multivariate index assay of proteomic biomarkers. Cancer Epidemiol Biomarkers Prev. 2010;19:2995–2999. doi: 10.1158/1055-9965.EPI-10-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]