Introduction
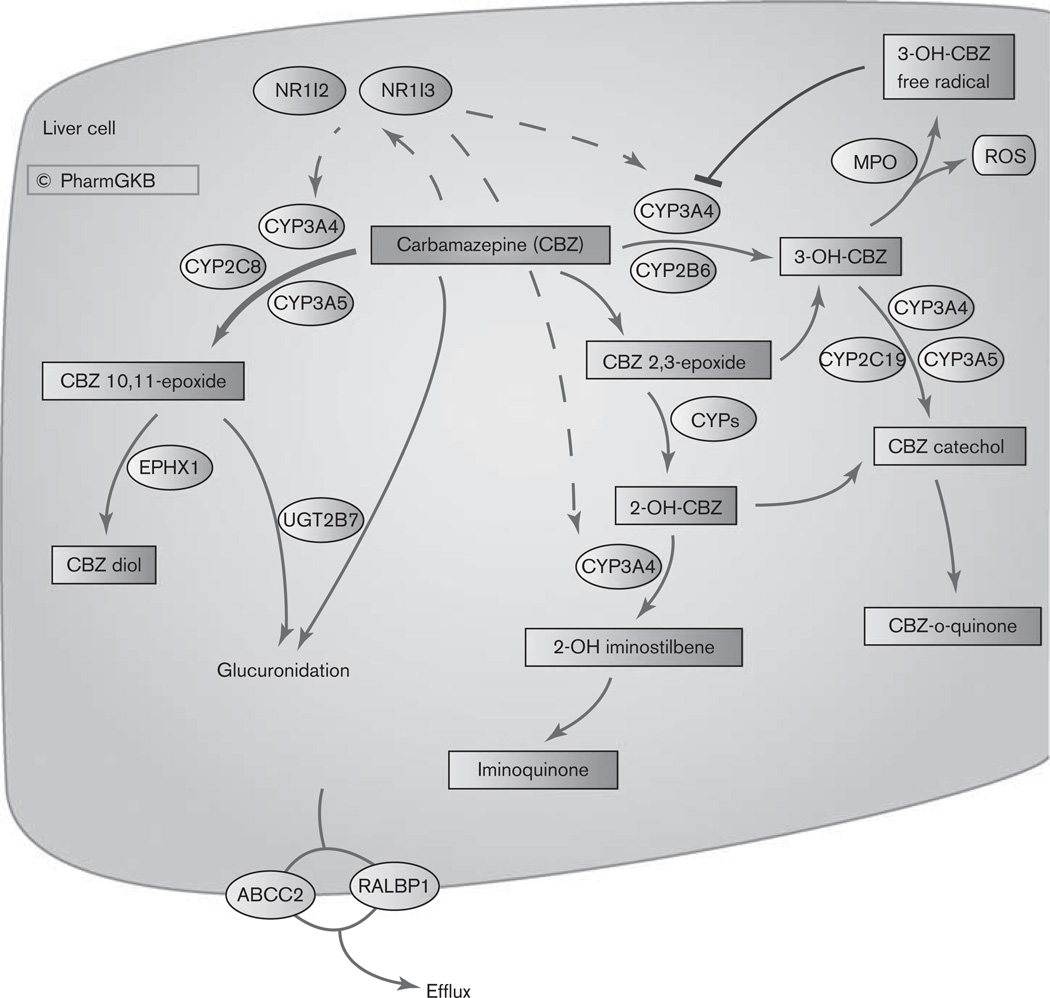
Carbamazepine (CBZ), a dibenzazepine, is a tricyclic compound used in the treatment of epilepsy, trigeminal neuralgia, and psychiatric mood disorders [1]. Serious adverse events have been reported for CBZ including Stevens–Johnson syndrome (SJS), toxic epidermal necrolysis (TEN), and Drug Reaction with Eosinophilia and Systemic Symptoms [2,3]. Other types of hypersensitivity reactions are also associated with CBZ including mild skin rashes, fever, eosinophilia, and cross-reactions to other anticonvulsants. Up to 80% of patients who have an idiopathic drug reaction to CBZ drugs will also have an adverse reaction to other anticonvulsants, further restricting treatment options [4]. In addition to adverse events, lack of efficacy can also be a problem, with as many as 30% of patients with epilepsy experiencing drug-resistance [5,6]. The mechanisms by which these events occur are not entirely clear although several candidate pharmacogenes have been associated with CBZ treatment responses. Current methods to individualize treatment involve therapeutic drug monitoring, the measurement of drug metabolites in patient samples posttreatment, and subsequent dose adjustment. Although this provides an accurate view of the drug-response phenotype, it still risks adverse events and cross-sensitivity. The ability to identify the patients that will benefit from CBZ, not suffer adverse events and define dose before treatment would be a highly valuable clinical tool. Here we present the current knowledge of CBZ pharmacogenomics (PGx) as a gene centered view of the pharmacokinetics of CBZ (Fig. 1) and collate the gene variants associated with CBZ responses.
Fig. 1.
Stylized liver cell depicting candidate genes involved in the pharmacokinetics of carbamazepine (CBZ). A fully interactive version is available online at http://www.pharmgkb.org/do/serve?objCls=Pathway&objId=PA165817070. 2-OH-CBZ, 2-hydroxy-CBZ; 3-OH-CBZ; 3-hydroxy-CBZ; MPO, myeloperoxidase; ROS, reactive oxygen species.
Pharmacokinetics
CBZ is almost completely metabolized in the liver with only approximately 5% of the drug excreted un-changed [7]. The major route of metabolism is conversion to CBZ 10,11-epoxide (CBZ-E) [1]. This reaction is primarily catalyzed by CYP3A4 although CYP2C8 also plays a role, and involvement of CYP3A5 has also been suggested (Fig. 1) [1,8]. Minor metabolic pathways include ring-hydroxylation to form 2-hydroxy-CBZ (2-OH-CBZ) and 3-hydroxy CBZ (3-OH-CBZ). The formation of each presumably proceeds by an epoxide intermediate (referred to as an arene oxide intermediate), with CYP2B6 and CYP3A4, being the major catalysts of 3-OH-CBZ formation [1] and multiple CYPs involved in 2-OH-CBZ formation [9]. Secondary metabolism of 2-OH-CBZ and 3-OH-CBZ by CYP3A4 represent two distinct potential bioactivation pathways. CYP3A4-dependent secondary oxidation of 2-OH-CBZ leads to the formation of thiol-reactive metabolites by an iminoquinone intermediate [10], whereas CYP3A4-dependent secondary oxidation of 3-OH-CBZ results in the formation of reactive metabolites capable of inactivating CYP3A4 [1] and forming covalent adducts [11]. 3-OH CBZ, and to a lesser extent 2-OH CBZ and CBZ, can be metabolized to form radicals by myeloperoxidase [12]. This releases reactive oxygen species and may lead to the formation of protein adducts. Covalent binding and protein adduct formation has also been observed for another antiepileptic drug, phenytoin, and is generally considered to be a necessary step in the pathogenesis of idiosyncratic reactions to this class of compounds [12].
CBZ stimulates the transcriptional upregulation of genes involved in its own metabolism, with autoinduction of CYP3A4 and CYP2B6, by nuclear receptors NR1I2 (PXR) and NR1I3 (CAR) [13–15]. Drug–drug interactions through CYP3A4 [16] and CYP2B6 [17] are well documented and can complicate the use of CBZ in polytherapy.
Some studies have suggested that glucuronidation is likely to play only a minor role in metabolism of CBZ and CBZ-E [7]. But other studies dispute the documenting involvement of UGT2B7 [18,19].
Transport
Variable transport of CBZ, particularly across the blood–brain barrier, may be responsible for variable CBZ response. Increased export from the brain has been discussed as a method of drug resistance with P-glycoprotein (PgP, coded by ABCB1) as the main focus [5]. Although studies in rats suggested PgP transport of CBZ [20], in-vitro assays and work in mice did not show evidence of CBZ transport by PgP [21,22] (not depicted in Fig. 1). RALBP1, also known as RLIP76, has been shown to transport CBZ and be involved in drug resistance [23]. Additional PGx evidence implicates ABCC2 as a potential pharmacogene for CBZ [24] however, cellular studies showed ABCC1, ABCC2, and ABCC5 did not transport CBZ in vitro [25]. See below for discussion of specific genomic variants in transporters and CBZ PGx.
Pharmacogenomics
Major histocompatibility locus variants
The most well-studied PGx variants with respect to CBZ are variants within major histocompatibility (MHC) locus in the human leukocyte antigen gene, HLA-B [26]. HLA-B codes for a protein that presents peptides to the immune system, identifying foreign or infected cells [27]. There are over 1500 alleles of HLA-B according to the IMGT/HLA Database [28]. Historically, these were identified by serotype phenotyping and although new allele subtypes are commonly identified by sequencing, the definitive genomic variants associated with most alleles are not well described. The extreme diversity of this locus in different ethnic groups means that different tag single nucleotide polymorphisms (SNPs) are associated with different serotypes in different populations. The serotype allele mostly associated with risk for the severe adverse drug reactions (ADRs), SJS, and TEN, in response to CBZ is HLA-B*1502 [29]. One mechanism that has been suggested for how CBZ hypersensitivity is triggered involves the proteasomal degradation and MHC-dependent presentation of CBZ metabolites [30]. The generation of free radicals is considered another possible mechanism. By forming adducts with CYP3A4 enzyme the radicals may also contribute to the cross-reactive hypersensitivity sometimes seen with other antiepileptic drugs [11]. An in-vitro study showed covalent binding of CBZ-modified peptides to the HLA-B*1502 protein that may lead to T-cell activation and SJS with this allele specifically [31].
HLA*1502 allele is strongly associated with CBZ-induced SJS/TEN in Taiwanese, Chinese, Indians, and Chinese–Americans but not in Caucasians or Japanese individuals [29,32–38]. This has led the clinical labeling from the Food and Drug Administration to recommend testing only in individuals with ancestry genetically at risk populations. The tag SNPs for HLA-B*1502 in Han Chinese HapMap samples are SNPs rs3909184 and rs2844682 [39].
HLA-A*3101 has been associated with CBZ-induced ADRs in Asians [32]. Recently, two independent genome-wide association studies showed association of HLA-A* 3101 with CBZ-induced ADRs in Caucasians [40] and Asians [41]. The tag SNP in linkage with HLA-A*3101 in the Asian population was rs1633021 [41] and in the Caucasian population was rs1061235 [40].
As a result of high degree of linkage across the MHC region, tagging SNPs may tag for a functional variant in another gene. The variants rs3909184 (within FLOT1 gene), rs2844682 (MUC21), rs1059510 (HLA-E), rs1264511 (intergenic), rs3130690 (intergenic), rs2848716 (intergenic), rs750332 (BAT2), rs2227956 (HSPA1A, HSPA1L, LSM2), rs1043620 (HSPA1A, HSPA1L, LSM2), rs506770 (HSPA1A, HSPA1L), rs2395402 (LEMD2) rs986475 (LST1, LTB, NCR3), rs2894342 (MLN) and rs1800629 (TNF:(−308)G > A) have been associated with CBZ-induced ADRs (including SJS, TEN, maculopapular eruption, and hypersensitivity syndrome) [32,42,43] (see variant annotations at http://www.pharmgkb.org/do/serve?objId=PA448785&objCls=Drug#tabview=tab2 for more details). In addition, the HLA-B*0702 allele was shown to protect against severe CBZ hypersensitivity (mostly Drug Reaction with Eosinophilia and Systemic Symptoms) in a small study of Caucasians [36].
As not all individuals with the HLA*1502 allele experience ADRs, it is still unclear which particular SNPs are causative and which are just tagging SNPs, or which other mechanisms (e.g. possible haplotype combinations), may be protective and prevent occurrence of ADRs in HLA*1502 carriers. The definition of which SNPs are causative for the CBZ-induced ADRs, as opposed to linked to the serological phenotype, will aid in better identifying those patients at risk for ADR particularly in those without Asian ancestry.
Metabolizing enzyme variants
Variants in CBZ metabolizing enzymes have been shown to affect CBZ pharmacokinetics although studies have been few and without replication. The reduced function protein CYP3A4*16 (rs12721627) shows decreased clearance in in-vitro systems [44,45] therefore potentially requiring altered dosing in individuals with this variant (found at a frequency of 1–5% in populations from Japan, Korea, and Mexico). Clearance of CBZ may be altered in vivo by CYP3A5 variants [46,47] (for a description of CYP3A5*3 see http://www.pharmgkb.org/search/annotatedGene/cyp3a5/variant.jsp). A small study on Korean individuals with epilepsy found that CYP3A5 nonexpressors (CYP3A5*3, rs776746) had higher clearance of CBZ and higher plasma levels than CYP3A5 expressors, a finding that seems incongruent but could be explained by autoinduction of CYP3A genes [46]. However, a larger study of Japanese epilepsy patients did not find a difference although this study included patients on comedications that may have further induced CYP3A4 [47]. Variants in EPHX1 have also been associated with altered CBZ metabolism [48]. A haplotype of rs1051740 (EPHX1:Y113H) and rs2234922 (EPHX1:H139R) showed increased plasma CBZ-diol/CBZ-E ratios in vivo in Japanese epilepsy patients [48]. Studies of polymorphisms in metabolizing drugs and effect on CBZ-induced ADRs have been mostly negative with one study associating a SNP in the 3′UTR of CYP2B6 (rs1042389) with maculopapular eruption and hypersensitivity syndrome but this was not significant after the Bonferroni correction [32].
Transporter variants
Study on the PGx of CBZ transport and resistance are similarly conflicting and in need of replication in larger cohorts. The well-known ABCB1 variant 3435C > T, rs1045642 CC genotype was associated with drug-resistant epilepsy in a cohort of 315 British patients although the drugs used in this study were not specified [49] (for a full description of ABCB1:3435C > T see http://www.pharmgkb.org/search/annotatedGene/abcb1/variant.jsp). Several studies since then have found no association of this variant (see meta-analysis by Bournissen et al. [50]) and these too did not separate patients by treatment. A study of 464 Chinese epilepsy patients associated ABCB1 variants rs3789243 and rs2032582 with CBZ resistance [51], but a study of 228 North Indian epileptics did not replicate this association [52]. Although it could be that different haplotype structures or racial background may have influenced these results, a subanalysis of the Bournissen meta-analysis, which looked at European cohorts and Asian cohorts separately also found no evidence of association of ABCB1:3435C > T with drug resistance [50].
Initial studies of RALBP1 expression pointed toward a role in drug-resistant epilepsy [23] however, two studies of RALBP1 variants in British cohorts failed to find association for all treatments [53,54]. Although there was a weak association in the small subset of patients (n = 81) on CBZ only for rs329017 in which the P values were not significant but the researchers felt warranted further study [54]. One association that has been replicated is with the ABCC2 SNP c.1249 G > A (p.V417I, rs2273697). This variant was associated with neurological ADRs in 146 Korean individuals with epilepsy receiving CBZ and validated in an independent cohort of the same ethnicity [24]. An additional SNP in ABCC2 (−24C > T, rs717620) has been associated with lack of response to CBZ in young Caucasian epilepsy patients [6].
Pharmacodynamic variants
Although not depicted in the figure, the targets of CBZ in the brain sodium channels SCN1A, SCN1B, SCN2A, and SCN3A have pharmacogenomic consequences. The variant SCN1A IVS5N + 5 G > A (rs3812718, also reported as SCN1A IVS4–91 G > A) has been associated with high-dose requirements in patients with epilepsy [55,56]. Variants in SCN2A and SCN3A may contribute to CBZ resistance [57,58] in individuals with epilepsy. In-vitro evidence from mice also suggests SCN1B as a potential pharmacogene for CBZ that may warrant further study [59].
Conclusions
The HLA alleles (HLA-B*1502 and HLA-A*3101) are the most important pharmacogenomic variants for carbamazepine to date. Although it is encouraging that labeling changes have been made for CBZ that have been shown to prevent severe side effects [60], we still need to understand the mechanism by which these events occur and how ethnicity influences this so as to develop more reliable tests based on causative variants that can be applied in all individuals regardless of race or ancestry.
Although preliminary data has been collected to show influence of genomic variation on CBZ metabolism, studies have been small and not validated. Studies appear to be heterogeneous with respect to ethnicity and assessment of ADRs. Study on defining the PGx of drug resistance has been complicated by common cotreatment with several antiepileptic drugs. There is a need for larger studies that have sufficient numbers in each of the documented treatment groups with well-defined phenotypes. In addition, studies including DNA sequencing, micro-RNA, or epigenetic analyses are lacking. Thus, more work is needed to translate observed differences in metabolism and pharmacokinetics into using genomic variation for predictive dosing.
Acknowledgements
The authors thank Fen Liu for assistance with the graphics. This study is supported by the NIH/NIGMS (R24 GM61374 R01 GM58883 and U01 HD044239).
Footnotes
Conflicts of interest
There are no conflicts of interest.
References
- 1.Pearce RE, Lu W, Wang Y, Uetrecht JP, Correia MA, Leeder JS. Pathways of carbamazepine bioactivation in vitro. III. The role of human cytochrome P450 enzymes in the formation of 2,3-dihydroxycarbamazepine. Drug Metab Dispos. 2008;36:1637–1649. doi: 10.1124/dmd.107.019562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hung SI, Chung WH, Liu ZS, Chen CH, Hsih MS, Hui RC, et al. Common risk allele in aromatic antiepileptic-drug induced Stevens-Johnson syndrome and toxic epidermal necrolysis in Han Chinese. Pharmacogenomics. 2010;11:349–356. doi: 10.2217/pgs.09.162. [DOI] [PubMed] [Google Scholar]
- 3.Ganeva M, Gancheva T, Lazarova R, Troeva J, Baldaranov I, Vassilev I, et al. Carbamazepine-induced drug reaction with eosinophilia and systemic symptoms (DRESS) syndrome: report of four cases and brief review. Int J Dermatol. 2008;47:853–860. doi: 10.1111/j.1365-4632.2008.03637.x. [DOI] [PubMed] [Google Scholar]
- 4.Shear NH, Spielberg SP. Anticonvulsant hypersensitivity syndrome. In vitro assessment of risk. J Clin Invest. 1988;82:1826–1832. doi: 10.1172/JCI113798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sisodiya SM, Goldstein DB. Drug resistance in epilepsy: more twists in the tale. Epilepsia. 2007;48:2369–2370. doi: 10.1111/j.1528-1167.2007.01260_1.x. [DOI] [PubMed] [Google Scholar]
- 6.Ufer M, Mosyagin I, Muhle H, Jacobsen T, Haenisch S, Hasler R, et al. Non-response to antiepileptic pharmacotherapy is associated with the ABCC2-24C > T polymorphism in young and adult patients with epilepsy. Pharmacogenet Genomics. 2009;19:353–362. doi: 10.1097/fpc.0b013e328329940b. [DOI] [PubMed] [Google Scholar]
- 7.Kim KA, Oh SO, Park PW, Park JY. Effect of probenecid on the pharmacokinetics of carbamazepine in healthy subjects. Eur J Clin Pharmacol. 2005;61:275–280. doi: 10.1007/s00228-005-0940-7. [DOI] [PubMed] [Google Scholar]
- 8.Kerr BM, Thummel KE, Wurden CJ, Klein SM, Kroetz DL, Gonzalez FJ, et al. Human liver carbamazepine metabolism. Role of CYP3A4 and CYP2C8 in 10,11-epoxide formation. Biochem Pharmacol. 1994;47:1969–1979. doi: 10.1016/0006-2952(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 9.Pearce RE, Vakkalagadda GR, Leeder JS. Pathways of carbamazepine bioactivation in vitro I. Characterization of human cytochromes P450 responsible for the formation of 2- and 3-hydroxylated metabolites. Drug Metab Dispos. 2002;30:1170–1179. doi: 10.1124/dmd.30.11.1170. [DOI] [PubMed] [Google Scholar]
- 10.Pearce RE, Uetrecht JP, Leeder JS. Pathways of carbamazepine bioactivation in vitro: II. The role of human cytochrome P450 enzymes in the formation of 2-hydroxyiminostilbene. Drug Metab Dispos. 2005;33:1819–1826. doi: 10.1124/dmd.105.004861. [DOI] [PubMed] [Google Scholar]
- 11.Kang P, Liao M, Wester MR, Leeder JS, Pearce RE, Correia MA. CYP3A4-Mediated carbamazepine (CBZ) metabolism: formation of a covalent CBZ-CYP3A4 adduct and alteration of the enzyme kinetic profile. Drug Metab Dispos. 2008;36:490–499. doi: 10.1124/dmd.107.016501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu W, Uetrecht JP. Peroxidase-mediated bioactivation of hydroxylated metabolites of carbamazepine and phenytoin. Drug Metab Dispos. 2008;36:1624–1636. doi: 10.1124/dmd.107.019554. [DOI] [PubMed] [Google Scholar]
- 13.Pippenger CE. Clinically significant carbamazepine drug interactions: an overview. Epilepsia. 1987;28(Suppl 3):S71–S76. doi: 10.1111/j.1528-1157.1987.tb05781.x. [DOI] [PubMed] [Google Scholar]
- 14.Oscarson M, Zanger UM, Rifki OF, Klein K, Eichelbaum M, Meyer UA. Transcriptional profiling of genes induced in the livers of patients treated with carbamazepine. Clin Pharmacol Ther. 2006;80:440–456. doi: 10.1016/j.clpt.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 15.Mo SL, Liu YH, Duan W, Wei MQ, Kanwar JR, Zhou SF. Substrate specificity, regulation, and polymorphism of human cytochrome P450 2B6. Curr Drug Metab. 2009;10:730–753. doi: 10.2174/138920009789895534. [DOI] [PubMed] [Google Scholar]
- 16.Dresser GK, Spence JD, Bailey DG. Pharmacokinetic-pharmacodynamic consequences and clinical relevance of cytochrome P450 3A4 inhibition. Clin Pharmacokinet. 2000;38:41–57. doi: 10.2165/00003088-200038010-00003. [DOI] [PubMed] [Google Scholar]
- 17.Zhu M, Kaul S, Nandy P, Grasela DM, Pfister M. Model-based approach to characterize efavirenz autoinduction and concurrent enzyme induction with carbamazepine. Antimicrob Agents Chemother. 2009;53:2346–2353. doi: 10.1128/AAC.01120-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Staines AG, Coughtrie MW, Burchell B. N-glucuronidation of carbamazepine in human tissues is mediated by UGT2B7. J Pharmacol Exp Ther. 2004;311:1131–1137. doi: 10.1124/jpet.104.073114. [DOI] [PubMed] [Google Scholar]
- 19.Hara Y, Nakajima M, Miyamoto K, Yokoi T. Morphine glucuronosyltransferase activity in human liver microsomes is inhibited by a variety of drugs that are co-administered with morphine. Drug Metab Pharmacokinet. 2007;22:103–112. doi: 10.2133/dmpk.22.103. [DOI] [PubMed] [Google Scholar]
- 20.Potschka H, Fedrowitz M, Loscher W. P-glycoprotein and multidrug resistance-associated protein are involved in the regulation of extracellular levels of the major antiepileptic drug carbamazepine in the brain. Neuroreport. 2001;12:3557–3560. doi: 10.1097/00001756-200111160-00037. [DOI] [PubMed] [Google Scholar]
- 21.Owen A, Pirmohamed M, Tettey JN, Morgan P, Chadwick D, Park BK. Carbamazepine is not a substrate for P-glycoprotein. Br J Clin Pharmacol. 2001;51:345–349. doi: 10.1046/j.1365-2125.2001.01359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luna-Tortos C, Fedrowitz M, Loscher W. Several major antiepileptic drugs are substrates for human P-glycoprotein. Neuropharmacology. 2008;55:1364–1375. doi: 10.1016/j.neuropharm.2008.08.032. [DOI] [PubMed] [Google Scholar]
- 23.Awasthi S, Hallene KL, Fazio V, Singhal SS, Cucullo L, Awasthi YC, et al. RLIP76, a non-ABC transporter, and drug resistance in epilepsy. BMC Neurosci. 2005;6:61. doi: 10.1186/1471-2202-6-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim WJ, Lee JH, Yi J, Cho YJ, Heo K, Lee SH, et al. A nonsynonymous variation in MRP2/ABCC2 is associated with neurological adverse drug reactions of carbamazepine in patients with epilepsy. Pharmacogenet Genomics. 2010;20:249–256. doi: 10.1097/FPC.0b013e328338073a. [DOI] [PubMed] [Google Scholar]
- 25.Luna-Tortos C, Fedrowitz M, Loscher W. Evaluation of transport of common antiepileptic drugs by human multidrug resistance-associated proteins (MRP1, 2 and 5) that are overexpressed in pharmacoresistant epilepsy. Neuropharmacology. 2010;58:1019–1032. doi: 10.1016/j.neuropharm.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 26.Gatanaga H, Honda H, Oka S. Pharmacogenetic information derived from analysis of HLA alleles. Pharmacogenomics. 2008;9:207–214. doi: 10.2217/14622416.9.2.207. [DOI] [PubMed] [Google Scholar]
- 27.Orr HT, Lopez de Castro JA, Lancet D, Strominger JL. Complete amino acid sequence of a papain-solubilized human histocompatibility antigen, HLA-B7-2. Sequence determination and search for homologies. Biochemistry. 1979;18:5711–5720. doi: 10.1021/bi00592a030. [DOI] [PubMed] [Google Scholar]
- 28.Robinson J, Mistry K, McWilliam H, Lopez R, Parham P, Marsh SG. The IMGT/HLA database. Nucleic Acids Res. 2011;39(Database issue):D1171–D1176. doi: 10.1093/nar/gkq998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung WH, Hung SI, Hong HS, Hsih MS, Yang LC, Ho HC, et al. Medical genetics: a marker for Stevens-Johnson syndrome. Nature. 2004;428:486. doi: 10.1038/428486a. [DOI] [PubMed] [Google Scholar]
- 30.Gueant JL, Gueant-Rodriguez RM, Gastin IA, Cornejo-Garcia JA, Viola M, Barbaud A, et al. Pharmacogenetic determinants of immediate and delayed reactions of drug hypersensitivity. Curr Pharm Des. 2008;14:2770–2777. doi: 10.2174/138161208786369795. [DOI] [PubMed] [Google Scholar]
- 31.Yang CW, Hung SI, Juo CG, Lin YP, Fang WH, Lu IH, et al. HLA-B*1502-bound peptides: implications for the pathogenesis of carbamazepine-induced Stevens-Johnson syndrome. J Allergy Clin Immunol. 2007;120:870–877. doi: 10.1016/j.jaci.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 32.Hung SI, Chung WH, Jee SH, Chen WC, Chang YT, Lee WR, et al. Genetic susceptibility to carbamazepine-induced cutaneous adverse drug reactions. Pharmacogenet Genomics. 2006;16:297–306. doi: 10.1097/01.fpc.0000199500.46842.4a. [DOI] [PubMed] [Google Scholar]
- 33.Wu XT, Hu FY, An DM, Yan B, Jiang X, Kwan P, et al. Association between carbamazepine-induced cutaneous adverse drug reactions and the HLA-B* 1502 allele among patients in central China. Epilepsy Behav. 2010;19:405–408. doi: 10.1016/j.yebeh.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 34.Mehta TY, Prajapati LM, Mittal B, Joshi CG, Sheth JJ, Patel DB, et al. Association of HLA-B*1502 allele and carbamazepine-induced Stevens-Johnson syndrome among Indians. Indian J Dermatol Venereol Leprol. 2009;75:579–582. doi: 10.4103/0378-6323.57718. [DOI] [PubMed] [Google Scholar]
- 35.Lonjou C, Thomas L, Borot N, Ledger N, de Toma C, LeLouet H, et al. A marker for Stevens-Johnson syndrome: ethnicity matters. Pharmacogenomics J. 2006;6:265–268. doi: 10.1038/sj.tpj.6500356. [DOI] [PubMed] [Google Scholar]
- 36.Alfirevic A, Jorgensen AL, Williamson PR, Chadwick DW, Park BK, Pirmohamed M. HLA-B locus in Caucasian patients with carbamazepine hypersensitivity. Pharmacogenomics. 2006;7:813–818. doi: 10.2217/14622416.7.6.813. [DOI] [PubMed] [Google Scholar]
- 37.Locharernkul C, Loplumlert J, Limotai C, Korkij W, Desudchit T, Tongkobpetch S, et al. Carbamazepine and phenytoin induced Stevens-Johnson syndrome is associated with HLA-B*1502 allele in Thai population. Epilepsia. 2008;49:2087–2091. doi: 10.1111/j.1528-1167.2008.01719.x. [DOI] [PubMed] [Google Scholar]
- 38.Kaniwa N, Saito Y, Aihara M, Matsunaga K, Tohkin M, Kurose K, et al. HLA-B locus in Japanese patients with anti-epileptics and allopurinol-related Stevens-Johnson syndrome and toxic epidermal necrolysis. Pharmacogenomics. 2008;9:1617–1622. doi: 10.2217/14622416.9.11.1617. [DOI] [PubMed] [Google Scholar]
- 39.de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38:1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCormack M, Alfirevic A, Bourgeois S, Farrell JJ, Kasperaviciute D, Carrington M, et al. HLA-A*3101 and carbamazepine-induced hypersensitivity reactions in Europeans. N Engl J Med. 2011;364:1134–1143. doi: 10.1056/NEJMoa1013297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ozeki T, Mushiroda T, Yowang A, Takahashi A, Kubo M, Shirakata Y, et al. Genome-wide association study identifies HLA-A*3101 allele as a genetic risk factor for carbamazepine-induced cutaneous adverse drug reactions in Japanese population. Hum Mol Genet. 2011;20:1034–1041. doi: 10.1093/hmg/ddq537. [DOI] [PubMed] [Google Scholar]
- 42.Alfirevic A, Mills T, Harrington P, Pinel T, Sherwood J, Jawaid A, et al. Serious carbamazepine-induced hypersensitivity reactions associated with the HSP70 gene cluster. Pharmacogenet Genomics. 2006;16:287–296. doi: 10.1097/01.fpc.0000189800.88596.7a. [DOI] [PubMed] [Google Scholar]
- 43.Pirmohamed M, Lin K, Chadwick D, Park BK. TNFalpha promoter region gene polymorphisms in carbamazepine-hypersensitive patients. Neurology. 2001;56:890–896. doi: 10.1212/wnl.56.7.890. [DOI] [PubMed] [Google Scholar]
- 44.Maekawa K, Yoshimura T, Saito Y, Fujimura Y, Aohara F, Emoto C, et al. Functional characterization of CYP3A4.16: catalytic activities toward midazolam and carbamazepine. Xenobiotica. 2009;39:140–147. doi: 10.1080/00498250802617746. [DOI] [PubMed] [Google Scholar]
- 45.Maekawa K, Harakawa N, Yoshimura T, Kim SR, Fujimura Y, Aohara F, et al. CYP3A4*16 and CYP3A4*18 alleles found in East Asians exhibit differential catalytic activities for seven CYP3A4 substrate drugs. Drug Metab Dispos. 2010;38:2100–2104. doi: 10.1124/dmd.110.034140. [DOI] [PubMed] [Google Scholar]
- 46.Park PW, Seo YH, Ahn JY, Kim KA, Park JY. Effect of CYP3A5*3 genotype on serum carbamazepine concentrations at steady-state in Korean epileptic patients. J Clin Pharm Ther. 2009;34:569–574. doi: 10.1111/j.1365-2710.2009.01057.x. [DOI] [PubMed] [Google Scholar]
- 47.Seo T, Nakada N, Ueda N, Hagiwara T, Hashimoto N, Nakagawa K, et al. Effect of CYP3A5*3 on carbamazepine pharmacokinetics in Japanese patients with epilepsy. Clin Pharmacol Ther. 2006;79:509–510. doi: 10.1016/j.clpt.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 48.Nakajima Y, Saito Y, Shiseki K, Fukushima-Uesaka H, Hasegawa R, Ozawa S, et al. Haplotype structures of EPHX1 and their effects on the metabolism of carbamazepine-10,11-epoxide in Japanese epileptic patients. Eur J Clin Pharmacol. 2005;61:25–34. doi: 10.1007/s00228-004-0878-1. [DOI] [PubMed] [Google Scholar]
- 49.Siddiqui A, Kerb R, Weale ME, Brinkmann U, Smith A, Goldstein DB, et al. Association of multidrug resistance in epilepsy with a polymorphism in the drug-transporter gene ABCB1. N Engl J Med. 2003;348:1442–1448. doi: 10.1056/NEJMoa021986. [DOI] [PubMed] [Google Scholar]
- 50.Bournissen FG, Moretti ME, Juurlink DN, Koren G, Walker M, Finkelstein Y. Polymorphism of the MDR1/ABCB1 C3435T drug-transporter and resistance to anticonvulsant drugs: a meta-analysis. Epilepsia. 2009;50:898–903. doi: 10.1111/j.1528-1167.2008.01858.x. [DOI] [PubMed] [Google Scholar]
- 51.Kwan P, Wong V, Ng PW, Lui CH, Sin NC, Poon WS, et al. Gene-wide tagging study of association between ABCB1 polymorphisms and multidrug resistance in epilepsy in Han Chinese. Pharmacogenomics. 2009;10:723–732. doi: 10.2217/pgs.09.32. [DOI] [PubMed] [Google Scholar]
- 52.Grover S, Bala K, Sharma S, Gourie-Devi M, Baghel R, Kaur H, et al. Absence of a general association between ABCB1 genetic variants and response to antiepileptic drugs in epilepsy patients. Biochimie. 2010;92:1207–1212. doi: 10.1016/j.biochi.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 53.Soranzo N, Kelly L, Martinian L, Burley MW, Thom M, Sali A, et al. Lack of support for a role for RLIP76 (RALBP1) in response to treatment or predisposition to epilepsy. Epilepsia. 2007;48:674–683. doi: 10.1111/j.1528-1167.2007.00926.x. [DOI] [PubMed] [Google Scholar]
- 54.Leschziner GD, Jorgensen AL, Andrew T, Williamson PR, Marson AG, Coffey AJ, et al. The association between polymorphisms in RLIP76 and drug response in epilepsy. Pharmacogenomics. 2007;8:1715–1722. doi: 10.2217/14622416.8.12.1715. [DOI] [PubMed] [Google Scholar]
- 55.Tate SK, Depondt C, Sisodiya SM, Cavalleri GL, Schorge S, Soranzo N, et al. Genetic predictors of the maximum doses patients receive during clinical use of the anti-epileptic drugs carbamazepine and phenytoin. Proc Natl Acad Sci U S A. 2005;102:5507–5512. doi: 10.1073/pnas.0407346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heinzen EL, Yoon W, Tate SK, Sen A, Wood NW, Sisodiya SM, et al. Nova2 interacts with a cis-acting polymorphism to influence the proportions of drug-responsive splice variants of SCN1A. Am J Hum Genet. 2007;80:876–883. doi: 10.1086/516650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kwan P, Poon WS, Ng HK, Kang DE, Wong V, Ng PW, et al. Multidrug resistance in epilepsy and polymorphisms in the voltage-gated sodium channel genes SCN1A, SCN2A, and SCN3A: correlation among phenotype, genotype, and mRNA expression. Pharmacogenet Genomics. 2008;18:989–998. doi: 10.1097/FPC.0b013e3283117d67. [DOI] [PubMed] [Google Scholar]
- 58.Holland KD, Kearney JA, Glauser TA, Buck G, Keddache M, Blankston JR, et al. Mutation of sodium channel SCN3A in a patient with cryptogenic pediatric partial epilepsy. Neurosci Lett. 2008;433:65–70. doi: 10.1016/j.neulet.2007.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uebachs M, Opitz T, Royeck M, Dickhof G, Horstmann MT, Isom LL, et al. Efficacy loss of the anticonvulsant carbamazepine in mice lacking sodium channel beta subunits via paradoxical effects on persistent sodium currents. J Neurosci. 2010;30:8489–8501. doi: 10.1523/JNEUROSCI.1534-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen P, Lin JJ, Lu CS, Ong CT, Hsieh PF, Yang CC, et al. Carbamazepine-induced toxic effects and HLA-B*1502 screening in Taiwan. N Engl J Med. 2011;364:1126–1133. doi: 10.1056/NEJMoa1009717. [DOI] [PubMed] [Google Scholar]