Introduction
Depression and anxiety disorders have been linked to the dysfunction of serotonergic neurotransmission [1]. Five major classes of antidepressant drugs exist: monoamine oxidase inhibitors, selective serotonin reuptake inhibitors (SSRI), serotonin norepinephrine reuptake inhibitors, tricyclic compounds, and atypical antidepressant drugs [2,3]. Citalopram is a SSRI and its molecular target is the serotonin transporter (solute carrier family 6 member 4, SLC6A4) [4–6]; it inhibits serotonin reuptake from the synaptic cleft (see the selective serotonin reuptake inhibitors pharmacodynamics pathway at http://www.pharmgkb.org/do/serve?objId=PA161749006&objCls=Pathway, [7]).
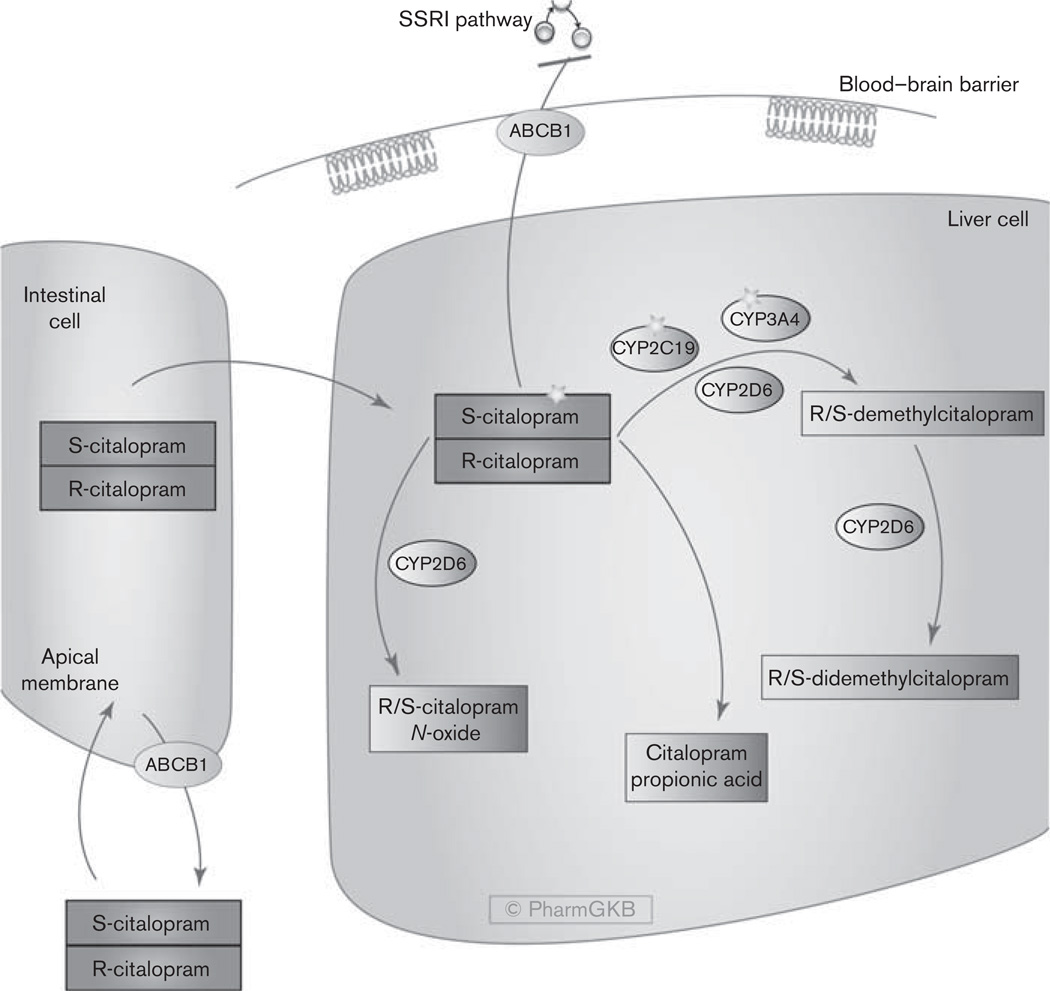
This summary briefly reviews the pharmacokinetics of citalopram (Fig. 1) and discusses the candidate pharmacogenes involved.
Fig. 1.
Pharmacokinetics of the selective serotonin reuptake inhibitor citalopram (PA164713429; http://www.pharmgkb.org/do/serve?objId=PA164713429&objCls=Pathway). SSRI, selective serotonin reuptake inhibitor.
Metabolism of citalopram
After oral administration, citalopram is rapidly absorbed, with peak plasma levels observed approximately after 1–4 h and a plasma half-life of approximately 35 h after administration [8–10]. A substituted phthalane derivative with a tertiary amino acid side chain, citalopram is highly lipophilic and has one chiral center [11]. Its lipophilicity results in high bioavailability (approximately 80%) after oral administration [10]. Approximately 12–23% of orally dosed citalopram is excreted unchanged in the urine, and approximately 10% is excreted in the feces [12].
Citalopram and its N-demethylated metabolites exist as racemic compounds. In-vitro and in-vivo tests showed that the effects of citalopram and N-demethylcitalopram depend primarily on the S-enantiomers: S-citalopram and S-demethylcitalopram [13]. The in-vitro inhibition of serotonin uptake of S-citalopram and S-demethylcitalopram is 167 and 6.6 times more potent, respectively, than the R-enantiomers [13]. In radioligand-binding assays, demethylcitalopram showed a similar affinity for the human SLC6A4 as citalopram [14], but compared with citalopram, demethylcitalopram crosses the blood–brain barrier poorly [15].
S-citalopram is the active ingredient of escitalopram, another SSRI [16]. To a certain degree, R-citalopram counteracts the serotonin-enhancing action of the S-citalopram enantiomer [17,18]. As a result, escitalopram is a more potent antidepressant than citalopram, as the latter is a mixture of S-citalopram and R-citalopram [17].
Both enantiomers of citalopram are metabolized by the hepatic cytochrome P450 system, as depicted in Fig. 1. In-vitro studies showed that the formation of R/S-demethylcitalopram is catalyzed by the isoenzymes CYP2C19 and CYP3A4, with a minor role of CYP2D6 [19–21]. The subsequent N-demethylation to R/S-didemethylcitalopram is mediated by CYP2D6 [19,22,23]. The clearance of R/S-citalopram is stereoselective [10,24,25]. In in-vitro studies in human liver microsomes, CYP2C19, CYP3A4, and CYP2D6 seem to primarily metabolize the biologically active S-enantiomer [10,22]. The administration of the racemic compounds produces different steady-state concentrations of the R/S-stereoisomer. Furthermore, N-oxidation and deamination lead to R/S-citalopram N-oxide and citalopram propionic acid metabolites, respectively [26–28]. The N-oxidation step to R/S-citalopram N-oxide is also mediated by CYP2D6 [22]. A study using recombinant supersomes expressing human CYP2C19 showed that CYP2C19 forms the propionic acid metabolite of S-citalopram [29], in contrast to previous studies [22,28]. Monoamine oxidases type A and B and aldehyde oxidase probably form citalopram propionic acid from citalopram, N-demethylcitalopram and N-didemethylcitalopram [27,28]. As there is no clear consensus about the enzymes involved in this step, they are not illustrated in Fig. 1.
Citalopram and escitalopram are weak CYP2D6 inhibitors [30–32] and have weak or no effect on CYP1A2, CYP2C19, and CYP3A4 [30]. Demethylcitalopram is a one order of magnitude more potent inhibitor of CYP2D6 than citalopram and may mediate the mild interaction of the drug with other drugs metabolized by CYP2D6 [15].
Genetic variants in genes involved in the metabolism of citalopram
Citalopram is an ABCB1 substrate and is actively transported from the brain. The efficacy of citalopram in people possessing the 2677TT (rs2032583) genotype of ABCB1 is likely to be diminished [33,34], although another study did not confirm this result [35]. Kinetic studies using ABCC1-enriched membrane vesicles revealed that citalopram is a substrate of ABCC1 [36]. The c.4002G > A (rs2230671) polymorphism in the ABCC1 gene showed a significant association with remission state at 8 weeks with the A allele being strongly associated with the remitted group and fewer adverse effects from citalopram use [36]. Individuals carrying the A allele of the c.4002G > A single nucleotide polymorphism had significantly increased ABCC1 mRNA levels in peripheral blood cells [36]. Brain immunostaining studies suggest that ABCC1 is expressed at the basolateral membrane of the blood–brain barrier; a higher ABCC1 expression might increase the level of citalopram in the brain [36].
The plasma concentration of citalopram is affected by CYP2C19 variants: poor metabolizers of CYP2C19 have a reduced clearance of citalopram and escitalopram [37–45]. Patients with the CYP2C19*17 (rs12248560) allele have a lower serum concentration of S-citalopram and citalopram [38,46,47]. The CYP2D6 poor metabolizer genotype in combination with CYP2C19 poor metabolizer genotype can increase the half-life of citalopram; in one patient, this resulted in severe adverse effects [39]. In volunteers who received an oral dose of 20 mg of citalopram, the influence of the CYP2D6*1/*4 genotype on the biotransformation of citalopram was very low in CYP2C19 extensive metabolizers, whereas its influence was more apparent in CYP2C19*2 (rs4244285) allele carriers [48]. The Sequenced Treatment Alternatives to Relieve Depression (STAR*D) study [49,50] provides the largest cohort assembled to date of DNA of patients with major depressive disorder treated with citalopram and followed prospectively for up to 12 weeks. This cohort has provided information about the effect of genetic variations on the response to citalopram and consequent remission from major depressive disorder as well as treatment-emergent adverse effects [51]. Surprisingly, and in contrast to smaller studies, polymorphisms in the pharmacokinetic genes CYP2D6, CYP2C19, CYP3A4, CYP3A5, and ABCB1 were not associated with antidepressant response in an initial study in the STAR*D cohort [35]. A recent study analyzed the relationship between genotype-based categories derived from genotyping the CYP2C19 and CPP2D6 genes and the clinical endpoints of drug tolerance and remission of depressive symptoms in white non-Hispanic patients of the STAR*D sample [52]. The CYP2C19*2 allele was associated with lower odds of tolerance, but CYP2D6 genotype-based categories were not found to be significantly associated with tolerance [52]. In a subset of patients who were able to tolerate the medication, carriers of two loss-of-function CYP2C19 alleles had higher odds of remission, whereas carriers of the increased activity allele CYP2C19*17 showed a trend of association of lower remission [52].
Thus, the pharmacokinetics of citalopram is affected by CYP2D6 and CYP2C19 genotypes, but the clinically relevant effect greatly varies between studies [35,42,52] (Table 1) and no predictive algorithm has been demonstrated.
Table 1.
Pharmacogenomic associations of genetic variants in pharmacokinetic genes involved in the metabolism of citalopram
| Gene | Variant | Phenotype | References |
|---|---|---|---|
| ABCB1 | rs2032583 | Diminished efficacy of citalopram | [33,34] |
| No effect on efficacy of citalopram | [35] | ||
| ABCC1 | rs2230671 | Associated with remission state at 8-week citalopram treatment | [36] |
| CYP2C19 | *2 (rs4244285) | Associated with lower odds of tolerance | [52] |
| No association with antidepressant response | [35] | ||
| CYP2C19 | *17 (rs12248560) | Trend of association of lower remission | [52] |
| No association with antidepressant response | [35] |
Conclusion
Serum drug levels have not consistently been associated with citalopram response [53], directing the pharmacogenomic interest more toward pharmacodynamic genes. Numerous studies with the goal of identifying genetic markers that might help to predict variation in response to treatment with citalopram have investigated the effect of polymorphisms in pharmacodynamic genes, mostly involved in the serotonin signaling pathway (see SSRI pathway [7], http://www.pharmgkb.org/do/serve?objId=PA161749006&objCls=Pathway). To date, genome-wide association studies have found no association of variants in pharmacokinetic genes with citalopram [54] or ecitalopram [55] response or remission. Instead, these studies found that variants in yet unexplored pathways showed the highest association signal [54–56]. Thus, although knowledge of the pharmacokinetics of citalopram may be important for avoiding drug–drug interactions, it may have a minimal role to play in the development of predictive profiles for SSRI response. Future studies involving polygenic single nucleotide polymorphism score analysis or meta-analysis of multiple genome-wide association study datasets, may be more successful in defining the impact of pharmacokinetic polymorphisms as a subset of the variation that influences citalopram response.
Acknowledgements
The authors thank Feng Liu for assistance with the graphics. This study is supported by the National Institutes of Health/National Institute of General Medical Sciences (R24GM61374).
Footnotes
Conflicts of interest
There are no conflicts of interest.
References
- 1.Pollock BG. Citalopram: a comprehensive review. Expert Opin Pharmacother. 2001;2:681–698. doi: 10.1517/14656566.2.4.681. [DOI] [PubMed] [Google Scholar]
- 2.Briley M, Moret C. Neurobiological mechanisms involved in antidepressant therapies. Clin Neuropharmacol. 1993;16:387–400. doi: 10.1097/00002826-199310000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Stahl SM, Grady MM, Moret C, Briley M. SNRIs: their pharmacology, clinical efficacy, and tolerability in comparison with other classes of antidepressants. CNS Spectr. 2005;10:732–747. doi: 10.1017/s1092852900019726. [DOI] [PubMed] [Google Scholar]
- 4.Plenge P, Gether U, Rasmussen SG. Allosteric effects of R- and S-citalopram on the human 5-HT transporter: evidence for distinct high- and low-affinity binding sites. Eur J Pharmacol. 2007;567:1–9. doi: 10.1016/j.ejphar.2007.03.055. [DOI] [PubMed] [Google Scholar]
- 5.Owens MJ, Knight DL, Nemeroff CB. Second-generation SSRIs: human monoamine transporter binding profile of escitalopram and R-fluoxetine. Biol Psychiatry. 2001;50:345–350. doi: 10.1016/s0006-3223(01)01145-3. [DOI] [PubMed] [Google Scholar]
- 6.Horschitz S, Hummerich R, Schloss P. Down-regulation of the rat serotonin transporter upon exposure to a selective serotonin reuptake inhibitor. Neuroreport. 2001;12:2181–2184. doi: 10.1097/00001756-200107200-00027. [DOI] [PubMed] [Google Scholar]
- 7.Sangkuhl K, Klein TE, Altman RB. Selective serotonin reuptake inhibitors pathway. Pharmacogenet Genomics. 2009;19:907–909. doi: 10.1097/FPC.0b013e32833132cb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Overo KF. Preliminary studies of the kinetics of citalopram in man. Eur J Clin Pharmacol. 1978;14:69–73. doi: 10.1007/BF00560260. [DOI] [PubMed] [Google Scholar]
- 9.Kragh-Sorensen P, Overo KF, Petersen OL, Jensen K, Parnas W. The kinetics of citalopram: single and multiple dose studies in man. Acta Pharmacol Toxicol (Copenh) 1981;48:53–60. doi: 10.1111/j.1600-0773.1981.tb01587.x. [DOI] [PubMed] [Google Scholar]
- 10.Rocha A, Marques MP, Coelho EB, Lanchote VL. Enantioselective analysis of citalopram and demethylcitalopram in human and rat plasma by chiral LC-MS/MS: application to pharmacokinetics. Chirality. 2007;19:793–801. doi: 10.1002/chir.20452. [DOI] [PubMed] [Google Scholar]
- 11.Willetts J, Lippa A, Beer B. Clinical development of citalopram. J Clin Psychopharmacol. 1999;19:36S–46S. doi: 10.1097/00004714-199910001-00004. [DOI] [PubMed] [Google Scholar]
- 12.Baumann P. Pharmacology and pharmacokinetics of citalopram and other SSRIs. Int Clin Psychopharmacol. 1996;11(Suppl 1):5–11. doi: 10.1097/00004850-199603001-00002. [DOI] [PubMed] [Google Scholar]
- 13.Hyttel J, Bogeso KP, Perregaard J, Sanchez C. The pharmacological effect of citalopram residues in the (S)-(+)-enantiomer. J Neural Transm Gen Sect. 1992;88:157–160. doi: 10.1007/BF01244820. [DOI] [PubMed] [Google Scholar]
- 14.Tatsumi M, Groshan K, Blakely RD, Richelson E. Pharmacological profile of antidepressants and related compounds at human monoamine transporters. Eur J Pharmacol. 1997;340:249–258. doi: 10.1016/s0014-2999(97)01393-9. [DOI] [PubMed] [Google Scholar]
- 15.Caccia S. Metabolism of the newer antidepressants: an overview of the pharmacological and pharmacokinetic implications. Clin Pharmacokinet. 1998;34:281–302. doi: 10.2165/00003088-199834040-00002. [DOI] [PubMed] [Google Scholar]
- 16.Huska MT, Catalano G, Catalano MC. Serotonin syndrome associated with the use of escitalopram. CNS Spectr. 2007;12:270–274. doi: 10.1017/s1092852900021027. [DOI] [PubMed] [Google Scholar]
- 17.Sanchez C. The pharmacology of citalopram enantiomers: the antagonism by R-citalopram on the effect of S-citalopram. Basic Clin Pharmacol Toxicol. 2006;99:91–95. doi: 10.1111/j.1742-7843.2006.pto_295.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhong H, Hansen KB, Boyle NJ, Han K, Muske G, Huang X, et al. An allosteric binding site at the human serotonin transporter mediates the inhibition of escitalopram by R-citalopram: kinetic binding studies with the ALI/VFL-SI/TT mutant. Neurosci Lett. 2009;462:207–212. doi: 10.1016/j.neulet.2009.07.030. [DOI] [PubMed] [Google Scholar]
- 19.Rochat B, Amey M, Gillet M, Meyer UA, Baumann P. Identification of three cytochrome P450 isozymes involved in N-demethylation of citalopram enantiomers in human liver microsomes. Pharmacogenetics. 1997;7:1–10. doi: 10.1097/00008571-199702000-00001. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi K, Chiba K, Yagi T, Shimada N, Taniguchi T, Horie T, et al. Identification of cytochrome P450 isoforms involved in citalopram N-demethylation by human liver microsomes. J Pharmacol Exp Ther. 1997;280:927–933. [PubMed] [Google Scholar]
- 21.von Moltke LL, Greenblatt DJ, Grassi JM, Granda BW, Venkatakrishnan K, Duan SX, et al. Citalopram and desmethylcitalopram in vitro: human cytochromes mediating transformation, and cytochrome inhibitory effects. Biol Psychiatry. 1999;46:839–849. doi: 10.1016/s0006-3223(98)00353-9. [DOI] [PubMed] [Google Scholar]
- 22.Olesen OV, Linnet K. Studies on the stereoselective metabolism of citalopram by human liver microsomes and cDNA-expressed cytochrome P450 enzymes. Pharmacology. 1999;59:298–309. doi: 10.1159/000028333. [DOI] [PubMed] [Google Scholar]
- 23.Matsui E, Hoshino M, Matsui A, Okahira A. Simultaneous determination of citalopram and its metabolites by high-performance liquid chromatography with column switching and fluorescence detection by direct plasma injection. J Chromatogr B Biomed Appl. 1995;668:299–307. doi: 10.1016/0378-4347(95)00073-r. [DOI] [PubMed] [Google Scholar]
- 24.Bondolfi G, Chautems C, Rochat B, Bertschy G, Baumann P. Non-response to citalopram in depressive patients: pharmacokinetic and clinical consequences of a fluvoxamine augmentation. Psychopharmacology (Berl) 1996;128:421–425. doi: 10.1007/s002130050152. [DOI] [PubMed] [Google Scholar]
- 25.Sidhu J, Priskorn M, Poulsen M, Segonzac A, Grollier G, Larsen F. Steady-state pharmacokinetics of the enantiomers of citalopram and its metabolites in humans. Chirality. 1997;9:686–692. doi: 10.1002/(SICI)1520-636X(1997)9:7<686::AID-CHIR9>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 26.Berzas-Nevado JJ, Villasenor-Llerena MJ, Guiberteau-Cabanillas C, Rodriguez-Robledo V. Enantiomeric screening of racemic citalopram and metabolites in human urine by entangled polymer solution capillary electrophoresis: an innovatory robustness/ruggedness study. Electrophoresis. 2006;27:905–917. doi: 10.1002/elps.200500413. [DOI] [PubMed] [Google Scholar]
- 27.Nikisch G, Mathe AA, Czernik A, Eap CB, Jimenez-Vasquez P, Brawand-Amey M, Baumann P. Stereoselective metabolism of citalopram in plasma and cerebrospinal fluid of depressive patients: relationship with 5-HIAA in CSF and clinical response. J Clin Psychopharmacol. 2004;24:283–290. doi: 10.1097/01.jcp.0000125680.89843.a6. [DOI] [PubMed] [Google Scholar]
- 28.Rochat B, Kosel M, Boss G, Testa B, Gillet M, Baumann P. Stereoselective biotransformation of the selective serotonin reuptake inhibitor citalopram and its demethylated metabolites by monoamine oxidases in human liver. Biochem Pharmacol. 1998;56:15–23. doi: 10.1016/s0006-2952(98)00008-2. [DOI] [PubMed] [Google Scholar]
- 29.Rudberg I, Reubsaet JL, Hermann M, Refsum H, Molden E. Identification of a novel CYP2C19-mediated metabolic pathway of S-citalopram in vitro. Drug Metab Dispos. 2009;37:2340–2348. doi: 10.1124/dmd.109.029355. [DOI] [PubMed] [Google Scholar]
- 30.Spina E, Santoro V, D’Arrigo C. Clinically relevant pharmacokinetic drug interactions with second-generation antidepressants: an update. Clin Ther. 2008;30:1206–1227. doi: 10.1016/s0149-2918(08)80047-1. [DOI] [PubMed] [Google Scholar]
- 31.Noehr-Jensen L, Zwisler ST, Larsen F, Sindrup SH, Damkier P, Brosen K. Escitalopram is a weak inhibitor of the CYP2D6-catalyzed O-demethylation of (+)-tramadol but does not reduce the hypoalgesic effect in experimental pain. Clin Pharmacol Ther. 2009;86:626–633. doi: 10.1038/clpt.2009.154. [DOI] [PubMed] [Google Scholar]
- 32.Jeppesen U, Gram LF, Vistisen K, Loft S, Poulsen HE, Brosen K. Dose-dependent inhibition of CYP1A2, CYP2C19 and CYP2D6 by citalopram, fluoxetine, fluvoxamine and paroxetine. Eur J Clin Pharmacol. 1996;51:73–78. doi: 10.1007/s002280050163. [DOI] [PubMed] [Google Scholar]
- 33.Uhr M, Tontsch A, Namendorf C, Ripke S, Lucae S, Ising M, et al. Polymorphisms in the drug transporter gene ABCB1 predict antidepressant treatment response in depression. Neuron. 2008;57:203–209. doi: 10.1016/j.neuron.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 34.Nikisch G, Eap CB, Baumann P. Citalopram enantiomers in plasma and cerebrospinal fluid of ABCB1 genotyped depressive patients and clinical response: a pilot study. Pharmacol Res. 2008;58:344–347. doi: 10.1016/j.phrs.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Peters EJ, Slager SL, Kraft JB, Jenkins GD, Reinalda MS, McGrath PJ, Hamilton SP. Pharmacokinetic genes do not influence response or tolerance to citalopram in the STAR*D sample. PLoS One. 2008;3:e1872. doi: 10.1371/journal.pone.0001872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee SH, Lee MS, Lee JH, Kim SW, Kang RH, Choi MJ, et al. MRP1 polymorphisms associated with citalopram response in patients with major depression. J Clin Psychopharmacol. 30:116–125. doi: 10.1097/JCP.0b013e3181d2ef42. [DOI] [PubMed] [Google Scholar]
- 37.Rudberg I, Hendset M, Uthus LH, Molden E, Refsum H. Heterozygous mutation in CYP2C19 significantly increases the concentration/dose ratio of racemic citalopram and escitalopram (S-citalopram) Ther Drug Monit. 2006;28:102–105. doi: 10.1097/01.ftd.0000189899.23931.76. [DOI] [PubMed] [Google Scholar]
- 38.Rudberg I, Mohebi B, Hermann M, Refsum H, Molden E. Impact of the ultrarapid CYP2C19*17 allele on serum concentration of escitalopram in psychiatric patients. Clin Pharmacol Ther. 2008;83:322–327. doi: 10.1038/sj.clpt.6100291. [DOI] [PubMed] [Google Scholar]
- 39.Herrlin K, Yasui-Furukori N, Tybring G, Widen J, Gustafsson LL, Bertilsson L. Metabolism of citalopram enantiomers in CYP2C19/CYP2D6 phenotyped panels of healthy Swedes. Br J Clin Pharmacol. 2003;56:415–421. doi: 10.1046/j.1365-2125.2003.01874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sindrup SH, Brosen K, Hansen MG, Aaes-Jorgensen T, Overo KF, Gram LF. Pharmacokinetics of citalopram in relation to the sparteine and the mephenytoin oxidation polymorphisms. Ther Drug Monit. 1993;15:11–17. doi: 10.1097/00007691-199302000-00002. [DOI] [PubMed] [Google Scholar]
- 41.Yu BN, Chen GL, He N, Ouyang DS, Chen XP, Liu ZQ, Zhou HH. Pharmacokinetics of citalopram in relation to genetic polymorphism of CYP2C19. Drug Metab Dispos. 2003;31:1255–1259. doi: 10.1124/dmd.31.10.1255. [DOI] [PubMed] [Google Scholar]
- 42.Yin OQ, Wing YK, Cheung Y, Wang ZJ, Lam SL, Chiu HF, Chow MS. Phenotype-genotype relationship and clinical effects of citalopram in Chinese patients. J Clin Psychopharmacol. 2006;26:367–372. doi: 10.1097/01.jcp.0000227355.54074.14. [DOI] [PubMed] [Google Scholar]
- 43.Noehr-Jensen L, Zwisler ST, Larsen F, Sindrup SH, Damkier P, Nielsen F, Brosen K. Impact of CYP2C19 phenotypes on escitalopram metabolism and an evaluation of pupillometry as a serotonergic biomarker. Eur J Clin Pharmacol. 2009;65:887–894. doi: 10.1007/s00228-009-0657-0. [DOI] [PubMed] [Google Scholar]
- 44.Jin Y, Pollock BG, Frank E, Cassano GB, Rucci P, Muller DJ, et al. Effect of age, weight, and CYP2C19 genotype on escitalopram exposure. J Clin Pharmacol. 2010;50:62–72. doi: 10.1177/0091270009337946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsai MH, Lin KM, Hsiao MC, Shen WW, Lu ML, Tang HS, et al. Genetic polymorphisms of cytochrome P450 enzymes influence metabolism of the antidepressant escitalopram and treatment response. Pharmacogenomics. 11:537–546. doi: 10.2217/pgs.09.168. [DOI] [PubMed] [Google Scholar]
- 46.Ohlsson Rosenborg S, Mwinyi J, Andersson M, Baldwin RM, Pedersen RS, Sim SC, et al. Kinetics of omeprazole and escitalopram in relation to the CYP2C19*17 allele in healthy subjects. Eur J Clin Pharmacol. 2008;64:1175–1179. doi: 10.1007/s00228-008-0529-z. [DOI] [PubMed] [Google Scholar]
- 47.de Vos A, Van der Weide J, Loovers HM. Association between CYP2C19*17 and metabolism of amitriptyline, citalopram and clomipramine in Dutch hospitalized patients. Pharmacogenomics J. 2010 doi: 10.1038/tpj.2010.39. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 48.Fudio S, Borobia AM, Pinana E, Ramirez E, Tabares B, Guerra P, et al. Evaluation of the influence of sex and CYP2C19 and CYP2D6 polymorphisms in the disposition of citalopram. Eur J Pharmacol. 2010;626:200–204. doi: 10.1016/j.ejphar.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 49.Fava M, Rush AJ, Trivedi MH, Nierenberg AA, Thase ME, Sackeim HA, et al. Background and rationale for the sequenced treatment alternatives to relieve depression (STAR*D) study. Psychiatr Clin North Am. 2003;26:457–494. doi: 10.1016/s0193-953x(02)00107-7. [DOI] [PubMed] [Google Scholar]
- 50.Trivedi MH, Rush AJ, Wisniewski SR, Nierenberg AA, Warden D, Ritz L, et al. Evaluation of outcomes with citalopram for depression using measurement-based care in STAR*D: implications for clinical practice. Am J Psychiatry. 2006;163:28–40. doi: 10.1176/appi.ajp.163.1.28. [DOI] [PubMed] [Google Scholar]
- 51.Laje G, Perlis RH, Rush AJ, McMahon FJ. Pharmacogenetics studies in STAR*D: strengths, limitations, and results. Psychiatr Serv. 2009;60:1446–1457. doi: 10.1176/appi.ps.60.11.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mrazek DA, Biernacka JM, O’Kane DJ, Black JL, Cunningham JM, Drews MS, et al. CYP2C19 variation and citalopram response. Pharmacogenet Genomics. 2011;21:1–9. doi: 10.1097/fpc.0b013e328340bc5a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rasmussen BB, Brosen K. Is therapeutic drug monitoring a case for optimizing clinical outcome and avoiding interactions of the selective serotonin reuptake inhibitors? Ther Drug Monit. 2000;22:143–154. doi: 10.1097/00007691-200004000-00001. [DOI] [PubMed] [Google Scholar]
- 54.Garriock HA, Kraft JB, Shyn SI, Peters EJ, Yokoyama JS, Jenkins GD, et al. A genome-wide association study of citalopram response in major depressive disorder. Biol Psychiatry. 2010;67:133–138. doi: 10.1016/j.biopsych.2009.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uher R, Perroud N, Ng MY, Hauser J, Henigsberg N, Maier W, et al. Genome-wide pharmacogenetics of antidepressant response in the GENDEP project. Am J Psychiatry. 2010;167:555–564. doi: 10.1176/appi.ajp.2009.09070932. [DOI] [PubMed] [Google Scholar]
- 56.Perroud N, Uher R, Ng MY, Guipponi M, Hauser J, Henigsberg N, et al. Genome-wide association study of increasing suicidal ideation during antidepressant treatment in the GENDEP project. Pharmacogenomics J. 2010 doi: 10.1038/tpj.2010.70. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]