Abstract
Objective
Common genetic variants in a 58-kilobase region of chr 9p21, near the CDKN2A/CDKN2B tumor suppressor locus, are strongly associated with coronary artery disease. However, the underlying mechanism of action remains unknown.
Methods and Results
We previously reported a congenic mouse model harboring an atherosclerosis susceptibility locus and the region of homology with the human 9p21 locus. Microarray and transcript-specific expression analyses showed markedly decreased Cdkn2a expression, including both p16INK4a and p19ARF, but not Cdkn2b (p15INK4b), in macrophages derived from congenic mice compared to controls. Atherosclerosis studies in subcongenic strains revealed genetic complexity and narrowed one locus to a small interval including Cdkn2a/b. Bone marrow (BM) transplantation studies implicated myeloid lineage cells as the culprit cell type rather than resident vascular cells. To directly test the role of BM-derived Cdkn2a transcripts in atherogenesis and inflammatory cell proliferation, we performed a transplantation study utilizing Cdkn2a+/− cells in the Ldlr−/− mouse model. Cdkn2a-deficient BM recipients exhibited accelerated atherosclerosis, increased Ly6Chi pro-inflammatory monocytes and increased monocyte/macrophage proliferation compared to controls.
Conclusions
These data provide a plausible mechanism for accelerated atherogenesis in susceptible congenic mice, involving decreased expression of Cdkn2a and increased proliferation of monocyte/macrophages, with possible relevance to the 9p21 human locus.
Keywords: atherosclerosis, genetics, inflammation, leukocytes
Atherosclerotic vascular disease (ASVD) is the leading cause of death in Western societies. Complications of the disease are usually caused by rupture or erosion of an unstable atherosclerotic plaque, resulting in thrombus formation and arterial occlusion.1 The complex etiology involves both genetic and environmental factors. While genetic factors underlying traditional risk factors are well known,2 new data emerging from genome-wide association studies (GWASs) are revealing novel loci mediating disease susceptibility independent of traditional factors.3 In particular, a risk locus on chromosome (chr) 9p21.3 is strongly associated with ASVD (including myocardial infarction, stroke and aortic aneurysm) but independent of plasma cholesterol levels and hypertension.4–6 While the association with ASVD is robust and has been widely-replicated in different ethnic groups, the underlying pathogenic mechanism remains unknown.
The association at 9p21 appears to be the most robust common (minor allele frequency > 10%) genetic determinant for ASVD in the human genome. The SNPs most strongly associated with disease risk map to a 58-kb region containing a long, non-coding RNA (ANRIL) and lying ~100 kb centromeric to the CDKN2A/B locus encoding inhibitors of cellular proliferation. Multiple ANRIL splice variants exist, complicating genetic association studies.7 Decreased expression of the tumor suppressors p16INK4a, p19ARF, and p15INK4b has been observed among carriers of the ASVD risk allele in some studies8, 9 but not others.10–12 Targeted deletion of the homologous region in mice resulted in markedly decreased expression of Cdkn2a/b transcripts and increased proliferation of vascular smooth muscle cells (vSMCs) in vitro. However, no effect on atherosclerosis was observed in the Western diet-fed wild-type background, a highly athero-resistant model.13 Thus, the potential role of these transcripts and the pathogenic mechanism leading to increased atherosclerotic risk at the 9p21 locus remains unclear.
We have used a murine genetic approach to map atherosclerosis susceptibility loci, including a locus on mouse chr 4 that was confirmed in a congenic strain.14, 15 The effect on disease susceptibility was independent of plasma cholesterol levels, body weight and plasma glucose levels.15 We now report refined genetic mapping indicating that the introgressed interval contains at least two loci, one overlapping the Athsq1 QTL and one covering the region of homology with the human 9p21 ASVD locus. Gene expression studies revealed decreased mRNA levels of p16INK4a and p19ARF, but not p15INKb4, in macrophages derived from susceptible congenic mice. Mechanistically, we investigated the hypothesis that increased proliferation of macrophages (or mixed populations of monocyte/macrophages), due to reduced expression of Cdkn2a cell proliferation inhibitor transcripts in myeloid lineage cells, might be responsible, at least in part, for the accelerated atherosclerosis phenotype.
Methods
An expanded Methods section is available in the Online Supplemental Methods. B6.MOLF-Athsq1 congenic mice were bred as described.15 Cdkn2a−/− mice16 backcrossed into B617 were kindly provided by Dr. Sean Morrison (University of Michigan). Gene expression profiling or real-time PCR was carried out using peritoneal or splenic monocyte/macrophages. Raw profiling data are available in the NCBI GEO database (GSE 24342). Atherosclerotic lesions were quantified using morphometric analysis as described.14, 15 BM transplantation experiments were performed as described.18 Monocyte subsets were analyzed by standard flow cytometry analysis.19 Two-factor ANOVA and t tests were performed using STATVIEW 5.0 (Abacus Concepts, Inc). The threshold for significance was p = 0.05. Data shown are mean ± SEM.
Results
Genetic complexity of an atherosclerosis susceptibility locus containing Athsq1 as well as a 5.4-9 Mb interval containing Cdkn2a/b
A series of subcongenic strains was generated to narrow a previously-reported15 54-Mb interval on chr 4 harboring an atherosclerosis susceptibility gene(s). Mice were bred to homozygosity, fed 9-wk WTD and analyzed for atherosclerotic lesion development. Mice carrying a 17- or 11-Mb proximal subinterval, but not the 4-Mb proximal tip, exhibited approximately 2-fold greater mean lesion area than non-congenic controls (p<0.0001) (Figure 1A–B). The effect was observed in both males and females. The negative lesion phenotype of the 4-Mb subcongenic was confirmed in a second cohort (Supplemental Figure I). These data narrow the proximal interval to a 5.4-9 Mb region containing 8–21 known protein coding genes including Cdkn2a/b (Figure 1C). Of note, this locus did not segregate with versican accumulation (data not shown), a phenotype originally observed in the full congenic strain.15 An additional distal locus was identified in a subcongenic strain carrying a non-overlapping donor interval (Supplemental Figure IB–C). The distal locus overlaps the linkage peak from the original Athsq1 mapping cross, but detailed mapping and phenotypic assessment have not been carried out. Together, the data indicate genetic complexity of the full 54-Mb interval, with refined mapping of the proximal locus leading to identification of a critical region containing 8–21 genes.
Figure 1. Refined mapping of an atherosclerosis susceptibility locus to a 5.4-9-Mb region including the region of homology with a human ASVD risk interval on 9p21.
A, Physical map of mouse chr 4 and MOLF donor intervals (white boxes) carried by congenic strains in the B6-Ldlr−/− background. Mb, megabase; b/b, homozygosity for B6 alleles; m/m, homozygosity for MOLF alleles. The rectangle indicates the narrowed interval defined by data in (b). B, Mean lesion areas in control (b/b) and congenic strains fed 9-wk WTD. Two-factor ANOVA performed with square root transformation. Horizontal bars indicate group means for males (circles) and females (triangles). NS, not significant. C, Microsatellite markers delineating proximal (D4Mit349/D4Mit27) and distal (D4Mit350/D4Mit154) recombination breakpoints of the refined risk interval. The interval includes the region of homology with a human risk interval on 9p21.
Having previously demonstrated a role for BM-derived cells in determining the pro-atherosclerotic phenotype of Athsq1 congenics,15 we now performed a reverse BM transplantation to determine whether vSMCs or endothelial cells contributed to the phenotype as well. B6-Ldlr−/− BM was injected into lethally-irradiated 17-Mb subcongenics and non-congenic controls. Following repopulation of the BM, mice were fed 11-wk WTD and analyzed for atherosclerotic lesion development. No difference in mean lesion area was observed between the groups (Supplemental Figure II). Similar results were obtained for males and females. Importantly, these data indicate that resident cells of the vessel wall are not involved in the accelerated atherosclerosis phenotype associated with the proximal 17-Mb locus. Thus, we focused further mechanistic studies of this strain in BM-derived monocyte/macrophages.
Microarray and transcript-specific gene expression analyses reveal decreased macrophage expression of p16INK4a and p19ARF, but not p15INK4b, in Athsq1 congenic mice
As an independent approach to narrow the list of candidate genes (>600 genes in the full 54-Mb congenic interval and ~160 genes in the 17-Mb interval), we performed gene expression analysis using microarrays. Elicited peritoneal macrophages derived from full (54-Mb) congenics and non-congenic controls were collected after 6-wk WTD feeding. Differentially-expressed genes were defined as exhibiting ≥20% difference in expression level with a significance threshold corrected for multiple testing.20 Three to six of the differentially expressed transcripts reside within the narrowed 5.4-9-Mb proximal interval and could be considered as causal candidate genes (Table 1). Strikingly, the most significant difference was a ~6-fold decrease in Cdkn2a levels in congenic compared to control macrophages. Cdkn2a encodes two transcripts involved in cell proliferation regulation: p16INK4a and p19ARF. 21 The transcripts utilize different promoters and alternative reading frames resulting in proteins with no amino acid homology.22 Because of the Cdkn2a exonic stucture, the microarray could not discern p16INK4a vs p19ARF transcripts. Transcript-specific rtPCR confirmed the results in the 17-Mb subcongenic strain and showed reduced expression of both p16INK4a and p19ARF, but not the adjacent p15INK4b transcript, in macrophages from the atherosclerosis-prone 54- and 17-Mb congenics compared to non-congenic controls (Figure 2A). Similar results were observed for both transcripts in resident peritoneal macrophages, and for p16INK4a in splenic monocyte/macrophages, from 54- and 17-Mb congenic mice compared to controls (Figure 2B–C). Of note, gene expression in peripheral blood monocytes was low (with Ct values >38) and we could not reliably detect a difference between congenics and controls. Thus, an atherosclerosis-prone murine strain exhibits decreased expression of the p16INK4a and p19ARF tumor suppressor genes in macrophages and mixed monocyte/macrophage populations.
Table 1. Cdkn2a is the most differentially expressed macrophage-derived gene residing in the 5.4-9-Mb mapped susceptibility interval.
Fold-change listed for all transcripts encoded within the 5.4–9-Mb interval and exhibiting a ≥20% difference in expression level in peritoneal macrophages derived from Athsq1 54-Mb full congenic or non-congenic (b/b) mice fed 6-wk WTD. N=10 mice/group. Note that Ptplad2 is listed twice (+1.24, −1.30 fold changes), with known splice variants exhibiting different expression patterns.
| Probe ID | Map Location (Mb) |
Gene symbol |
Gene name | Fold change (relative to b/b) |
P-value |
|---|---|---|---|---|---|
| 100093087_TGI_at | 88.3 | Klhl9 | kelch-like 9 | 1.29 | 0.0001 |
| 100103804_TGI_at | 88.0 | Ptplad2 | protein tyrosine phosphatase-like A domain containing 2 | 1.24 | 0.0065 |
| 100102382_TGI_at | 88.8 | Mtap | methylthioadenosine phosphorylase | −1.26 | 7.77E-05 |
| 100116677_TGI_at | 88.0 | Ptplad2 | protein tyrosine phosphatase-like A domain containing 2 | −1.30 | 0.0071 |
| 100097525_TGI_at | 94.7 | Jun | Jun oncogene | −1.39 | 0.0028 |
| 100113480_TGI_at | 93.0 | Tusc1 | tumor suppressor candidate 1 | −1.88 | 5.36E-07 |
| 100099086_TGI_at | 88.9 | Cdkn2a | cyclin-dependent kinase inhibitor 2A | −5.98 | 1.25E-06 |
Figure 2. Decreased expression of p16INK4a and p19ARF, but not p15INK4b, cell proliferation inhibitor transcripts in BM-derived cells from chr 4 congenic mice compared to controls.
rtPCR results in cells derived from B6-Ldlr−/− (b/b) and congenic (54- or 17-Mb) mice. A, concanavalin A-elicited peritoneal macrophages, 6-wk WTD, n=10 mice/group; B, resident peritoneal macrophages, 9-wk WTD, n=8–12 mice/group; C, splenic CD11b+ monocyte/macrophages, 15-wk WTD. n=7–9 mice/group. ANOVA (A) or t-test (B, C) performed with log transformation. *p≤0.05, **p≤0.005, †p≤0.0005 compared to b/b controls.
BM-specific Cdkn2a-deficiency results in accelerated atherosclerosis, increased Ly6Chi monocytes, and increased BrdU incorporation into monocytes and macrophages
To directly test the hypothesis that decreased Cdkn2a expression in BM-derived cells is pro-atherogenic, we performed a BM transplantation study using a previously described Cdkn2a-deficient mouse. The targeted mutation replaces exons 2/3 with a neo cassette, knocking out both p16INK4a and p19ARF expression16, and has been crossed into a uniform B6 background.17 In an effort to simulate the modest reduction of p16INK4a and p19ARF expression observed in congenics compared to controls, we used donor B6-Ldlr+/−mice heterozygous for the Cdkn2a null allele. B6-Ldlr−/− recipients were injected with B6-Ldlr+/− or B6-Ldlr+/−, Cdkn2a+/− BM and fed 10-wk WTD following repopulation of the BM. No differences were observed in body weight, plasma total cholesterol, HDL or triglycerides (Supplemental Table I). Resident peritoneal macrophages exhibited a 2.5-fold decrease in expression of p16INK4a but not p19ARF or p18INK4c (another INK4-class gene encoded by a distant region of mouse chr 4) in B6-Ldlr+/−, Cdkn2a+/− recipients compared to controls (Figure 3A). Similarly, splenic CD11b+ monocyte/macrophages exhibited ~4-fold decreases in both p16INK4a and p19ARF but not p18INK4c (Figure 3B). Importantly, mean atherosclerotic lesion area was increased in B6-Ldlr+/−, Cdkn2a+/− recipients compared to controls (p=0.04, two-factor ANOVA) (Figure 3C), with non-significant single-sex increases of 34% and 17% for males and females, respectively. Thus, heterozygous BM-specific deficiency of Cdkn2a is sufficient to confer a modestly accelerated atherosclerosis phenotype in mice.
Figure 3. BM-specific Cdkn2a deficiency is sufficient to promote atherosclerosis in B6-Ldlr−/− mice.
A–B, Transcript-specific rtPCR results for Cdkn2a (p16INK4a and p19ARF) and p18INK4c (another INK4-class gene encoded by a distant region of mouse chr 4). N=7 mice/group. Unpaired t-test performed with log transformation. C, Mean lesion areas from B6-Ldlr−/− mice transplanted with B6-Ldr+/− or B6-Ldr+/−, Cdkn2a+/− BM and fed 10-wk WTD. Two-factor ANOVA performed with square root transformation. Horizontal bars indicate group means for males (circles) and females (triangles). *p≤0.05.
To test potential atherogenic mechanisms consistent with decreased p16INK4a and/or p19ARF expression in monocyte/macrophages, we assessed apoptosis and measures of monocytosis in the BM transplanted mice. TUNEL staining of atherosclerotic lesions revealed few positive cells and no difference between B6-Ldlr+/−, Cdkn2a+/− recipients compared to controls (data not shown). No differences were observed in circulating white blood cell counts or total monocytes in blood derived from B6-Ldlr+/−, Cdkn2a+/− recipients compared to controls (data not shown). However, flow cytometry analysis of blood monocytes revealed increased Ly6Chi monocytes after chow (0-wk timepoint) and WTD feeding (Figure 4A–B). Increased percentages of Ly6Chi and decreased percentages of Ly6Clo subsets (Figure 4B) resulted in a significant increase in the ratio of Ly6Chi:Ly6Clo monocytes in the circulation of B6-Ldlr+/−, Cdkn2a+/− recipients compared to controls (Supplemental Figure 3A). Note that the 0- and 2-wk timepoints are pre-lesional indicating that increased Ly6Chi monocytes are not secondary to accelerated atherosclerosis.15 Similar results were observed for the 17-Mb congenic strain compared to non-congenic controls (Supplemental Figure IIIB). Moreover, an increased ratio of Ly6Chi:Ly6Clo monocytes was observed in splenic monocytes derived from B6-Ldlr−/−, Cdkn2a+/− mice compared to B6-Ldlr−/− controls (p=0.003) (Supplemental Figure IVA–B).
Figure 4. Increased inflammatory Ly6Chi monocytes, mediated by increased cell proliferation, in the circulation of B6-Ldlr−/− mice transplanted with Ldlr+/−, Cdkn2a+/− BM compared to controls.
A, Flow cytometry analysis of blood monocytes from Ldlr+/− or Ldlr+/−, Cdkn2a+/− BM recipients fed chow or WTD. Monocytes were gated as CD45+CD115+ Ly6Chi or CD45+CD115+Ly6Clo. B, Quantification of Ly6Chi/ Ly6Clo cells among total CD45+CD115+ cells. C, Analysis of proliferating CD45+CD115+ monocytes at the 10-wk timepoint. Cells were gated as Ly6Chi BrdU+. D, Quantification of BrdU+ cells among total CD45+CD115+ monocytes. N=11 mice/group. *p≤0.05, **p≤0.005, †p≤0.0005.
To test for a direct effect of heterozygous Cdkn2a deficiency on monocyte proliferation, we injected mice with BrdU before sacrifice at the 10-wk timepoint. B6-Ldlr+/−, Cdkn2a+/− recipients exhibited an increased percentage of BrdU+ monocytes compared to controls, indicating increased monocyte proliferation (Figure 4C–D). Moreover, the increase in BrdU incorporation occurs mainly in the Ly6Chi subset (Figure 4C, left panel).
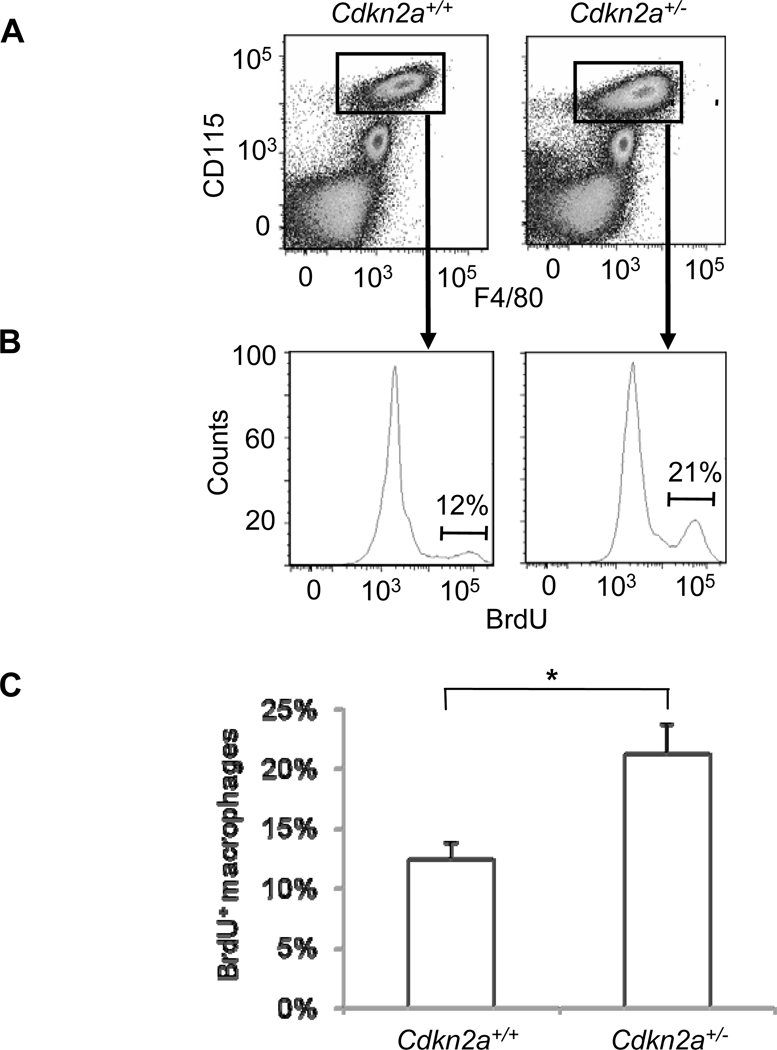
To test for an effect of BM-derived Cdkn2a deficiency on tissue macrophage proliferation, we assayed elicited peritoneal macrophages by flow cytometry following intraperitoneal BrdU injection. While there was no difference in the total number of CD45+CD115+F4/80+ macrophages (Figure 5A), there was a significant increase in the percentage of BrdU+ macrophages from B6-Ldlr−/−, Cdkn2a+/− mice compared to B6-Ldlr−/− controls (p<0.04) (Figure 5B–C). We also tested for increased BrdU incorporation within lesions of a small cohort of B6-Ldlr−/−, Cdkn2a+/− mice compared to controls using immunohistochemistry. While ample numbers of cells (mean=42–47 cells) stained positively following a 7-day pulse, the within-group variation was high and power calculations indicated that we would need to study 50 mice/group to have an 80% chance of detecting a significant difference (p<0.05) comparable in magnitude to the difference in lesion area (i.e. ~30%).
Figure 5. Increased proliferation of tissue macrophages derived from B6-Ldlr−/− mice transplanted with Ldlr+/−, Cdkn2a+/− BM compared to controls.
A, Flow cytometry analysis of concanavalin A-elicited peritoneal macrophages from B6-Ldlr−/−or B6-Ldlr−/−, Cdkn2a+/− mice fed 4–5 wks WTD. Macrophages were gated as CD45+CD115+F4/80+. B, Analysis of proliferating CD45+CD115+F4/80+ macrophages at the 4–5 wk timepoint. Cells were gated as BrdU+. C, Quantification of BrdU+ cells among total CD45+CD115+F4/80+ macrophages. N=6–7 mice/group. *p≤0.05.
Together, these data provide direct evidence for a suppressive effect of BM-derived p16INK4a and/or p19ARF expression on inflammatory monocyte/macrophage proliferation.
Discussion
Prior to the identification of 9p21 as a risk locus for human ASVD, we used a murine genetic approach to identify the homologous region of mouse chr 4 as a modifier of atherosclerosis susceptibility. In the current study, we provide refined genetic mapping and transcriptional evidence that Cdkn2a mediates at least some of the altered atherosclerosis susceptibility of this region through altered expression in macrophages. In support of this model, heterozygous deficiency of Cdkn2a transcripts in BM-derived cells was found to be sufficient to confer accelerated atherogenesis in the B6-Ldlr−/− background. The moderate (~2–4-fold) reduction in p16INK4a and p19ARF expression associated with increased atherosclerosis in these murine models is consistent with the reduction of p16INK4a and p19ARF expression in T cells of carriers of the 9p21 risk allele.8 Moreover, our study shows for the first time that the underlying pathogenic mechanism may involve increased proliferation/expansion of the Ly6Chi inflammatory monocyte population in the circulation as well as increased proliferation of tissue macrophages. This provides a plausible mechanism to account, in part, for accelerated atherogenesis in the chr 4 congenic mouse, with possible relevance to the mechanism of increased atherogenesis in humans bearing the 9p21 risk allele.
Consistent with our BM transplantation study, whole-body deletion of p19ARF was recently shown to be pro-atherosclerotic in the B6-Apoe−/− background.23 The mechanism of action suggested in the Apoe−/− model was decreased apoptosis, although the culprit cell type was not identified. We did not observe differences in plaque apoptosis in either the Athsq1 congenic model or mice carrying BM-deficiency of Cdkn2a compared to respective controls. The discrepancy between the two studies could be due to different experimental designs (BM vs whole body deficiency), different targeted alleles (p16INK4a/p19ARF vs p19ARF alone), different mouse models (Ldlr−/− vs Apoe−/−), or a non-target effect of a carrier gene in one of the mutant models. However, while decreased apoptosis may accelerate early lesion formation, increased apoptosis likely contributes to lesion progression to advanced plaques with clinically significant consequences24 such as MI associated with 9p21.
Chr 4 mouse deletion mutants with decreased expression of all three Cdkn2a/b transcripts exhibited increased proliferation of vSMCs 25. However, vSMC proliferation does not readily explain the human athero-thrombotic phenotype since this generally leads to cap thickening and plaque stabilization.26 The results of a reverse BM transplantation experiment reported herein were inconsistent with a role for vSMCs in the atherosclerosis phenotype of Athsq1 17-Mb subcongenic mice (Supplementary Figure 2). In line with this, Folkersen et al27 failed to demonstrate an association between vascular gene expression of CDKN2A/B and 9p21 risk genotype.
Evidence for an association between elevated WBC counts and ASVD risk has been documented in more than a dozen prospective epidemiological studies.28 There is also substantial evidence in animal models for a causal relationship between monocytosis and atherogenesis.29–32 In mouse models of diet-induced atherosclerosis, the subset of Ly6Chi expressing monocytes expands with hypercholesterolemia and selectively enters sites of inflammation including atherosclerotic lesions, compared to the Ly6Clo subset.33, 34 In addition, single gene mutations resulting in reduced blood monocytes also reduced atherosclerosis independent of plasma cholesterol levels.35–38 While the relationship between monocytosis and atherosclerosis has been well-documented, detailed mechanisms responsible for monocytosis are lacking. Consistent with a recent finding,19 our study suggests that the control of myeloid cell proliferation during hypercholesterolemia is an important factor determining magnitude of leukocytosis and the atherogenic response.
A variety of mechanisms for CDKN2A/B and/or ANRIL involvement in the 9p21 ASVD locus have been proposed. Regulation of CDKN2A/B gene expression has been suggested to occur via cis or trans mechansim(s) involving either the structurally overlapping ANRIL,9, 39 or other regulatory motifs residing within the 58-kb risk locus.11, 40 It is also possible that ANRIL may mediate effects at 9p21 without involvement of CDKN2A/B, through regulation of gene expression at an unlinked locus.41 The mouse genome does not contain a contiguous sequence with homology to ANRIL. However, markedly decreased expression of Cdkn2a/b transcripts in the 70-kb deletion mutant suggests the existence of a cis-acting enhancer.13 In our Athsq1 congenic mouse, there is a loss of coordinate regulation with decreased expression of Cdkn2a (p16INK4a and p19ARF) transcripts but not Cdkn2b (p15INK4b). Thus, in our model, the causal variant is likely to be cis-acting and specific to the Cdkn2a locus.
The 9p21 locus has been associated with multiple vascular phenotypes. These include myocardial infarction,5 abdominal aortic and intracranial aneurysms,42 ischemic stroke,43, 44 and peripheral artery disease.45 Interestingly, monocyte/macrophage recruitment is an important process in cerebral aneurysm formation and Ccl2 deficiency has been shown to inhibit macrophage accumulation in aneurysmal walls and significantly decrease aneurysm formation in an experimentally induced mouse model.46 In another study, increased immunostaining of CD68 antigen was observed in intracranial aneurysms compared to control tissue.47 In ischemic stroke, macrophage accumulation has been associated with severity of brain injury.48 Thus, alterations in monocyte/macrophage proliferation could potentially provide a common underlying mechanism for vascular phenotypes associated with the 9p21 locus.
Supplementary Material
Acknowledgments
Sources of Funding
This work was supported by National Institutes of Health (NIH) research grants RO1 HL102206 (CLW), HL54591 (ART) and AG024379 (NES), and a Burroughs-Wellcome Clinical Scientist Award in Translational Research (NES).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
Oscar Puig works at Merck Research Laboratories.
References
- 1.Lusis AJ. Atherosclerosis. Nature. 2000;407:233–241. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McPherson R. Chr 9p21 and coronary artery disease. N Engl J Med. 2010;362:1736–1737. doi: 10.1056/NEJMcibr1002359. [DOI] [PubMed] [Google Scholar]
- 3.Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, Absher D, Aherrahrou Z, Allayee H, Altshuler D, Anand SS, Andersen K, Anderson JL, Ardissino D, Ball SG, Balmforth AJ, Barnes TA, Becker DM, Becker LC, Berger K, Bis JC, Boekholdt SM, Boerwinkle E, Braund PS, Brown MJ, Burnett MS, Buysschaert I, Carlquist JF, Chen L, Cichon S, Codd V, Davies RW, Dedoussis G, Dehghan A, Demissie S, Devaney JM, Diemert P, Do R, Doering A, Eifert S, Mokhtari NE, Ellis SG, Elosua R, Engert JC, Epstein SE, de Faire U, Fischer M, Folsom AR, Freyer J, Gigante B, Girelli D, Gretarsdottir S, Gudnason V, Gulcher JR, Halperin E, Hammond N, Hazen SL, Hofman A, Horne BD, Illig T, Iribarren C, Jones GT, Jukema JW, Kaiser MA, Kaplan LM, Kastelein JJ, Khaw KT, Knowles JW, Kolovou G, Kong A, Laaksonen R, Lambrechts D, Leander K, Lettre G, Li M, Lieb W, Loley C, Lotery AJ, Mannucci PM, Maouche S, Martinelli N, McKeown PP, Meisinger C, Meitinger T, Melander O, Merlini PA, Mooser V, Morgan T, Muhleisen TW, Muhlestein JB, Munzel T, Musunuru K, Nahrstaedt J, Nelson CP, Nothen MM, Olivieri O, Patel RS, Patterson CC, Peters A, Peyvandi F, Qu L, Quyyumi AA, Rader DJ, Rallidis LS, Rice C, Rosendaal FR, Rubin D, Salomaa V, Sampietro ML, Sandhu MS, Schadt E, Schafer A, Schillert A, Schreiber S, Schrezenmeir J, Schwartz SM, Siscovick DS, Sivananthan M, Sivapalaratnam S, Smith A, Smith TB, Snoep JD, Soranzo N, Spertus JA, Stark K, Stirrups K, Stoll M, Tang WH, Tennstedt S, Thorgeirsson G, Thorleifsson G, Tomaszewski M, Uitterlinden AG, van Rij AM, Voight BF, Wareham NJ, Wells GA, Wichmann HE, Wild PS, Willenborg C, Witteman JC, Wright BJ, Ye S, Zeller T, Ziegler A, Cambien F, Goodall AH, Cupples LA, Quertermous T, Marz W, Hengstenberg C, Blankenberg S, Ouwehand WH, Hall AS, Deloukas P, Thompson JR, Stefansson K, Roberts R, Thorsteinsdottir U, O'Donnell CJ, McPherson R, Erdmann J, Samani NJ. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, Konig IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, Gudbjartsson DF, Magnusson KP, Andersen K, Levey AI, Backman VM, Matthiasdottir S, Jonsdottir T, Palsson S, Einarsdottir H, Gunnarsdottir S, Gylfason A, Vaccarino V, Hooper WC, Reilly MP, Granger CB, Austin H, Rader DJ, Shah SH, Quyyumi AA, Gulcher JR, Thorgeirsson G, Thorsteinsdottir U, Kong A, Stefansson K. A common variant on chr 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 6.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC. A common allele on chr 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunnington MS, Keavney B. Genetic mechanisms mediating atherosclerosis susceptibility at the chr 9p21 locus. Curr Atheroscler Rep. 2011;13:193–201. doi: 10.1007/s11883-011-0178-z. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Sanoff HK, Cho H, Burd CE, Torrice C, Mohlke KL, Ibrahim JG, Thomas NE, Sharpless NE. Ink4/arf transcript expression is associated with chr 9p21 variants linked to atherosclerosis. PLoS One. 2009;4:e5027. doi: 10.1371/journal.pone.0005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an ink4/arf-associated non-coding rna correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holdt LM, Beutner F, Scholz M, Gielen S, Gabel G, Bergert H, Schuler G, Thiery J, Teupser D. Anril expression is associated with atherosclerosis risk at chr 9p21. Arterioscler Thromb Vasc Biol. 2010;30:620–627. doi: 10.1161/ATVBAHA.109.196832. [DOI] [PubMed] [Google Scholar]
- 11.Jarinova O, Stewart AF, Roberts R, Wells G, Lau P, Naing T, Buerki C, McLean BW, Cook RC, Parker JS, McPherson R. Functional analysis of the chr 9p21.3 coronary artery disease risk locus. Arterioscler Thromb Vasc Biol. 2009;29:1671–1677. doi: 10.1161/ATVBAHA.109.189522. [DOI] [PubMed] [Google Scholar]
- 12.Cunnington MS, Santibanez Koref M, Mayosi BM, Burn J, Keavney B. Chr 9p21 snps associated with multiple disease phenotypes correlate with anril expression. PLoS Genet. 2010;6:e1000899. doi: 10.1371/journal.pgen.1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, Blow MJ, Cohen JC, Rubin EM, Pennacchio LA. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;18:409–412. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Welch CL, Bretschger S, Latib N, Bezouevski M, Guo Y, Pleskac N, Liang CP, Barlow C, Dansky H, Breslow JL, Tall AR. Localization of atherosclerosis susceptibility loci to chrs 4 and 6 using the ldlr knockout mouse model. Proc Natl Acad Sci U S A. 2001;98:7946–7951. doi: 10.1073/pnas.141239098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seidelmann SB, Kuo C, Pleskac N, Molina J, Sayers S, Li R, Zhou J, Johnson P, Braun K, Chan C, Teupser D, Breslow JL, Wight TN, Tall AR, Welch CL. Athsq1 is an atherosclerosis modifier locus with dramatic effects on lesion area and prominent accumulation of versican. Arterioscler Thromb Vasc Biol. 2008;28:2180–2186. doi: 10.1161/ATVBAHA.108.176800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the ink4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 17.Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16ink4a and p19arf senescence pathways. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han S, Liang CP, Westerterp M, Senokuchi T, Welch CL, Wang Q, Matsumoto M, Accili D, Tall AR. Hepatic insulin signaling regulates vldl secretion and atherogenesis in mice. J Clin Invest. 2009;119:1029–1041. doi: 10.1172/JCI36523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yvan-Charvet L, Pagler T, Gautier EL, Avagyan S, Siry RL, Han S, Welch CL, Wang N, Randolph GJ, Snoeck HW, Tall AR. Atp-binding cassette transporters and hdl suppress hematopoietic stem cell proliferation. Science. 2010;328:1689–1693. doi: 10.1126/science.1189731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puig O, Wang I-M, Cheng P, Zhou P, Roy S, Cully D, Peters M, Benita Y, Thompson J, Cai T-Q. Transcriptome profiling and network analysis of genetically hypertensive mice identifies potential pharmacological targets of hypertension. Physiol. Genomics. 2010;42A:24–32. doi: 10.1152/physiolgenomics.00010.2010. [DOI] [PubMed] [Google Scholar]
- 21.Sherr CJ. Principles of tumor suppression. Cell. 2004;116:235–246. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- 22.Kim WY, Sharpless NE. The regulation of ink4/arf in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez-Navarro H, Abu Nabah YN, Vinue A, Andres-Manzano MJ, Collado M, Serrano M, Andres V. P19(arf) deficiency reduces macrophage and vascular smooth muscle cell apoptosis and aggravates atherosclerosis. J Am Coll Cardiol. 2010;55:2258–2268. doi: 10.1016/j.jacc.2010.01.026. [DOI] [PubMed] [Google Scholar]
- 24.Tabas I. Macrophage death and defective inflammation resolution in atherosclerosis. Nat Rev Immunol. 2009;10:36–46. doi: 10.1038/nri2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clarke MC, Littlewood TD, Figg N, Maguire JJ, Davenport AP, Goddard M, Bennett MR. Chronic apoptosis of vascular smooth muscle cells accelerates atherosclerosis and promotes calcification and medial degeneration. Circ Res. 2008;102:1529–1538. doi: 10.1161/CIRCRESAHA.108.175976. [DOI] [PubMed] [Google Scholar]
- 27.Folkersen L, Kyriakou T, Goel A, Peden J, Malarstig A, Paulsson-Berne G, Hamsten A, Hugh W, Franco-Cereceda A, Gabrielsen A, Eriksson P. Relationship between cad risk genotype in the chr 9p21 locus and gene expression. Identification of eight new anril splice variants. PLoS One. 2009;4:e7677. doi: 10.1371/journal.pone.0007677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coller BS. Leukocytosis and ischemic vascular disease morbidity and mortality: Is it time to intervene? Arterioscler Thromb Vasc Biol. 2005;25:658–670. doi: 10.1161/01.ATV.0000156877.94472.a5. [DOI] [PubMed] [Google Scholar]
- 29.Ortlepp JR, Metrikat J, Albrecht M, Maya-Pelzer P. Relationship between physical fitness and lifestyle behaviour in healthy young men. Eur J Cardiovasc Prev Rehabil. 2004;11:192–200. doi: 10.1097/01.hjr.0000131578.48136.85. [DOI] [PubMed] [Google Scholar]
- 30.Bovill EG, Bild DE, Heiss G, Kuller LH, Lee MH, Rock R, Wahl PW. White blood cell counts in persons aged 65 years or more from the cardiovascular health study. Correlations with baseline clinical and demographic characteristics. Am J Epidemiol. 1996;143:1107–1115. doi: 10.1093/oxfordjournals.aje.a008687. [DOI] [PubMed] [Google Scholar]
- 31.Feldman DL, Mogelesky TC, Liptak BF, Gerrity RG. Leukocytosis in rabbits with diet-induced atherosclerosis. Arterioscler Thromb. 1991;11:985–994. doi: 10.1161/01.atv.11.4.985. [DOI] [PubMed] [Google Scholar]
- 32.Averill LE, Meagher RC, Gerrity RG. Enhanced monocyte progenitor cell proliferation in bone marrow of hyperlipemic swine. Am J Pathol. 1989;135:369–377. [PMC free article] [PubMed] [Google Scholar]
- 33.Swirski FK, Libby P, Aikawa E, Alcaide P, Luscinskas FW, Weissleder R, Pittet MJ. Ly-6chi monocytes dominate hypercholesterolemia-associated monocytosis and give rise to macrophages in atheromata. J Clin Invest. 2007;117:195–205. doi: 10.1172/JCI29950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tacke F, Alvarez D, Kaplan TJ, Jakubzick C, Spanbroek R, Llodra J, Garin A, Liu J, Mack M, van Rooijen N, Lira SA, Habenicht AJ, Randolph GJ. Monocyte subsets differentially employ ccr2, ccr5, and cx3cr1 to accumulate within atherosclerotic plaques. J Clin Invest. 2007;117:185–194. doi: 10.1172/JCI28549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajavashisth T, Qiao JH, Tripathi S, Tripathi J, Mishra N, Hua M, Wang XP, Loussararian A, Clinton S, Libby P, Lusis A. Heterozygous osteopetrotic (op) mutation reduces atherosclerosis in ldl receptor- deficient mice. J Clin Invest. 1998;101:2702–2710. doi: 10.1172/JCI119891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith JD, Trogan E, Ginsberg M, Grigaux C, Tian J, Miyata M. Decreased atherosclerosis in mice deficient in both macrophage colony-stimulating factor (op) and apolipoprotein e. Proc Natl Acad Sci U S A. 1995;92:8264–8268. doi: 10.1073/pnas.92.18.8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Combadiere C, Potteaux S, Rodero M, Simon T, Pezard A, Esposito B, Merval R, Proudfoot A, Tedgui A, Mallat Z. Combined inhibition of ccl2, cx3cr1, and ccr5 abrogates ly6c(hi) and ly6c(lo) monocytosis and almost abolishes atherosclerosis in hypercholesterolemic mice. Circulation. 2008;117:1649–1657. doi: 10.1161/CIRCULATIONAHA.107.745091. [DOI] [PubMed] [Google Scholar]
- 38.Saederup N, Chan L, Lira SA, Charo IF. Fractalkine deficiency markedly reduces macrophage accumulation and atherosclerotic lesion formation in ccr2−/− mice: Evidence for independent chemokine functions in atherogenesis. Circulation. 2008;117:1642–1648. doi: 10.1161/CIRCULATIONAHA.107.743872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding rna anril and methylated histone h3 lysine 27 by polycomb cbx7 in transcriptional silencing of ink4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, Ren B, Fu XD, Topol EJ, Rosenfeld MG, Frazer KA. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature. 2011;470:264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sato K, Nakagawa H, Tajima A, Yoshida K, Inoue I. Anril is implicated in the regulation of nucleus and potential transcriptional target of e2f1. Oncol Rep. 2010;24:701–707. doi: 10.3892/or_00000910. [DOI] [PubMed] [Google Scholar]
- 42.Helgadottir A, Thorleifsson G, Magnusson KP, Gretarsdottir S, Steinthorsdottir V, Manolescu A, Jones GT, Rinkel GJ, Blankensteijn JD, Ronkainen A, Jaaskelainen JE, Kyo Y, Lenk GM, Sakalihasan N, Kostulas K, Gottsater A, Flex A, Stefansson H, Hansen T, Andersen G, Weinsheimer S, Borch-Johnsen K, Jorgensen T, Shah SH, Quyyumi AA, Granger CB, Reilly MP, Austin H, Levey AI, Vaccarino V, Palsdottir E, Walters GB, Jonsdottir T, Snorradottir S, Magnusdottir D, Gudmundsson G, Ferrell RE, Sveinbjornsdottir S, Hernesniemi J, Niemela M, Limet R, Andersen K, Sigurdsson G, Benediktsson R, Verhoeven EL, Teijink JA, Grobbee DE, Rader DJ, Collier DA, Pedersen O, Pola R, Hillert J, Lindblad B, Valdimarsson EM, Magnadottir HB, Wijmenga C, Tromp G, Baas AF, Ruigrok YM, van Rij AM, Kuivaniemi H, Powell JT, Matthiasson SE, Gulcher JR, Thorgeirsson G, Kong A, Thorsteinsdottir U, Stefansson K. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40:217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 43.Matarin M, Brown WM, Singleton A, Hardy JA, Meschia JF. Whole genome analyses suggest ischemic stroke and heart disease share an association with polymorphisms on chr 9p21. Stroke. 2008;39:1586–1589. doi: 10.1161/STROKEAHA.107.502963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gschwendtner A, Bevan S, Cole JW, Plourde A, Matarin M, Ross-Adams H, Meitinger T, Wichmann E, Mitchell BD, Furie K, Slowik A, Rich SS, Syme PD, MacLeod MJ, Meschia JF, Rosand J, Kittner SJ, Markus HS, Muller-Myhsok B, Dichgans M. Sequence variants on chr 9p21.3 confer risk for atherosclerotic stroke. Ann Neurol. 2009;65:531–539. doi: 10.1002/ana.21590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cluett C, McDermott MM, Guralnik J, Ferrucci L, Bandinelli S, Miljkovic I, Zmuda JM, Li R, Tranah G, Harris T, Rice N, Henley W, Frayling TM, Murray A, Melzer D. The 9p21 myocardial infarction risk allele increases risk of peripheral artery disease in older people. Circ Cardiovasc Genet. 2009;2:347–353. doi: 10.1161/CIRCGENETICS.108.825935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aoki T, Kataoka H, Ishibashi R, Nozaki K, Egashira K, Hashimoto N. Impact of monocyte chemoattractant protein-1 deficiency on cerebral aneurysm formation. Stroke. 2009;40:942–951. doi: 10.1161/STROKEAHA.108.532556. [DOI] [PubMed] [Google Scholar]
- 47.Krischek B, Kasuya H, Tajima A, Akagawa H, Sasaki T, Yoneyama T, Ujiie H, Kubo O, Bonin M, Takakura K, Hori T, Inoue I. Network-based gene expression analysis of intracranial aneurysm tissue reveals role of antigen presenting cells. Neuroscience. 2008;154:1398–1407. doi: 10.1016/j.neuroscience.2008.04.049. [DOI] [PubMed] [Google Scholar]
- 48.Chauveau F, Cho TH, Berthezene Y, Nighoghossian N, Wiart M. Imaging inflammation in stroke using magnetic resonance imaging. Int J Clin Pharmacol Ther. 2010;48:718–728. doi: 10.5414/cpp48718. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.