Abstract
Pro-inflammatory cytokines produced in the tumor microenvironment facilitate tumor development and metastatic progression. In particular, TNF-α promotes cancer invasion and angiogenesis associated with epithelial-mesenchyme transition (EMT), however, the mechanisms underlying its induction of EMT in cancer cells remain unclear. Here we show that EMT and cancer stemness properties induced by chronic treatment with TNFα̣ are mediated by the upregulation of the transcriptional repressor Twist1. Exposure to TNF-α rapidly induced Twist1 mRNA and protein expression in normal breast epithelial and breast cancer cells. Both IKK-β and NF-κB p65 were required for TNF-α-induced expression of Twist1, suggesting the involvement of canonical NF-κB signaling. In support of this likelihood, we defined a functional NF-κB binding site in the Twist1 promoter and overexpression of p65 was sufficient to induce transcriptional upregulation of Twist1 along with EMT in mammary epithelial cells. Conversely, suppressing Twist1 expression abrogated p65-induced cell migration, invasion, EMT and stemness properties, establishing that Twist1 is required for NF-κB to induce these aggressive phenotypes in breast cancer cells. Taken together, our results establish a signaling axis through which the tumor microenvironment elicits Twist1 expression to promote cancer metastasis. We suggest that targeting NF-κB-mediated Twist1 upregulation may offer an effective a therapeutic strategy for breast cancer treatment.
Introduction
The transcriptional factor nuclear factor-kappa B (NFκB) was initially characterized as a central regulator in response to pathogens and viruses. Subsequently, studies found that NFκB is activated in a range of human cancers and to promote tumorigenesis via the regulation of target genes expression. In mammals, NF-κB binds to their target gene promoters as homo- or heterodimers composed of five subunits: RELA (p65), RELB, c-REL, NFκB1 (p105/p50), and NFκB2 (p100/p52). NFκB activation is exclusively regulated by two independent pathways. In the canonical pathway, NFκB activation is induced by various inflammatory stimuli, such as tumor necrosis factor-α (TNFα), interleukin-1 (IL-1), bacterial products e.g., lipopolysaccharide (LPS), chemical inducers e.g., phorbol-12-myristate-13-acetate (PMA), and reactive oxygen species (ROS) e.g., H2O2 through the IKKα/IKKβ/IKKγ complex. Upon stimulation, activated IKKβ phosphorylates the NFκB inhibitor, IκBα̣ at Ser32 and Ser36 and triggers its rapid degradation through the β-TrCP-mediated 26S proteasome proteolysis, resulting in the liberation of the NFκB. As a consequence, the NFκB heterodimer translocates to the nucleus, binds to its cognate DNA motifs in the promoters, and induces a myriad of gene expression involved in immune response (TNFα, IL-1, and COX2), cell proliferation (cyclin D1 and c-MYC), angiogenesis (VEGF, IL-6, and IL-8), cell survival (XIAP, BCL-xL, and c-IAP2), invasion (MMP-9), and EMT (Snail) (1) (2). In contrast, the noncanonical pathway is activated by different types of inflammatory stimuli via IKKα homodimers that modulates of B-cell development and adaptive immune response (3).
Epithelial-mesenchyme transition (EMT), a complex reprogramming process of epithelial cells, plays an indispensable role in tumor invasion and metastasis (4). The well-defined features of EMT include loss of epithelial markers (E-cadherin and α- and γ-catenin), gain of mesenchymal cell markers (fibronectin, vimentin, and N-cadherin), and the acquisition of migratory and invasive properties (5). Currently, studies show that EMT is controlled by a group of transcriptional repressors, such as Zeb-1/2, Twist1, Snail, and Slug. Upon activation, these repressors recruit histone deacetylases to the E-box elements of the E-cadherin promoter, resulting in transcriptional silence of E-cadherin expression (6). Twist1, known as a master regulator of morphogenesis, induces EMT to facilitate breast tumor metastasis (7). The role for Twist1 in EMT regulation has also been reported in many other cancer types, including those of the prostate (8) and uterus (9). In addition to that in patients with breast carcinoma, high expression of Twist1 also correlates with tumor invasion and metastasis in patients with esophageal squamous cell carcinomas (10), hepatocellular carcinoma (11), and gliomas (12).
Inflammation, hypoxia, and tumor-stroma interactions are the major activators of metastatic cascade. This tumor microenvironment, which consists of infiltrated immune cells and their secretary cytokines and/or chemokines, facilitates cancer cell motility, invasiveness, and metastatic potential (13) (14). To date, extensive studies have pointed to NFκB signaling as a critical inflammatory mediator in the response to invading pathogens. In addition, drugs and inhibitors aimed at targeting NFκB have shown promising clinical implications (15). Therefore, determining how NFκB mediates high malignancy to enhance cancer cell invasion, migration and subsequent metastasis may provide novel therapeutic value. Indeed, activation of the NFκB pathway is required for induction and maintenance of Ras- and TGFβ-dependent EMT (16). NFκB also binds to the promoter of the E-cadherin repressor, ZEB-1/2, resulting in regulation of the EMT phenotype (17). A recent study further suggested that inflammation-induced cell migration and invasion occur via NFκB-mediated stabilization of Snail (18). Despite the presence of antiapoptotic cross-talk between Twist1 and NFκB (19), the exact regulatory mechanism of NFκB in EMT regulation has yet to be determined. Here, we examine the role of NFκB activation in the EMT process and elucidate an important but underdeveloped pro-inflammation cytokine TNFα-mediated breast cancer metastasis through the initiation of EMT. We show that rapid activation of NFκB by TNFα upregulates Twist1 expression through nuclear translocation of p65, which in turn activates Twist1 gene expression, is an essential node for the chronic inflammation-induced EMT.
Experimental Procedures
Detailed information is included in Supplemental Information.
Cell Culture, Stable Transfectants and Transfection
MCF 10A, MCF-12A, MDA-MB-453, HBL-100, BT-549, and HEK-293 cells were obtained from American Type Culture Collection. GP293 cells were purchased from Clontech (Palo Alto, CA). IKKα-/-, IKKβ-/- and p65-/- MEFs were maintained as previous described (20) (21) (22). MCF 10A was cultured in DMEM/F12 medium supplemented with 5% horse serum, 10 μg/ml insulin, 20 ng/ml EGF, 100 ng/ml cholera toxin, and 500 ng/ml hydrocortisone. IKKβ stable transfectants in MDA-MB-435 cells were selected using blasticidin S as described previously (20). For transient transfection, cells were transiently transfected with DNA using an SN liposomes (23), lipofectamine™ 2000 (Invitrogen, Carlsbad, CA), or electroporation by a Nucleofector 1 device (Amaxa Biosystems, Koln, Germany) with electroporation buffer (137 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4, 6 mM glucose, and 20 mM HEPES, pH 7.0). For analysis of ligand-dependent Twist1 expression, cells were serum-starved overnight and harvested directly or after stimulation at different time points.
Mouse Model of Lung Metastasis
Tumor metastasis assays were performed using an intravenous breast cancer mouse model. The murine mammary tumor cell line 4T1-Luc was infected with lentiviral- based shRNA stable clones. Cells (1 × 105) were then injected into the lateral tail vein of BALB/c mice (The Jackson Laboratory; five mice per group). Two weeks later, Mice were injected intraperitoneally either with PBS or 10 mg/mice LPS in PBS. Lung metastasis was detected using an IVIS-100 imagining system (Xenogen). To measure lung metastases, animals were weighed before each experimental end point, and lung nodules were stained with India ink, excised, and counted immediately.
Immunohistochemistry of Human Breast Tumor Tissue Samples
Immunohistochemistry (IHC) was performed as described previously (20) (24) (25). Human tissue specimens were incubated with antibodies against IKKβ, p65, or Twist1 and a biotin-conjugated secondary antibody and then incubated with an avidin-biotin-peroxidase complex. Visualization was performed using amino-ethylcarbazole chromogen. The human breast tumor samples used in cell fractionation and Western blots were received from the breast tumor bank at The University of Texas MD Anderson Cancer Center. For statistical analysis, Fisher's exact test and Spearman rank correlation coefficient were used and a p-value less than 0.05 is considered statistically significant. According to histological scoring, the intensity of staining was ranked into four groups: high (score 3), medium (score 2), low (score 1), and negative (score 0).
Results
TNFα induces a rapid expression of Twist1
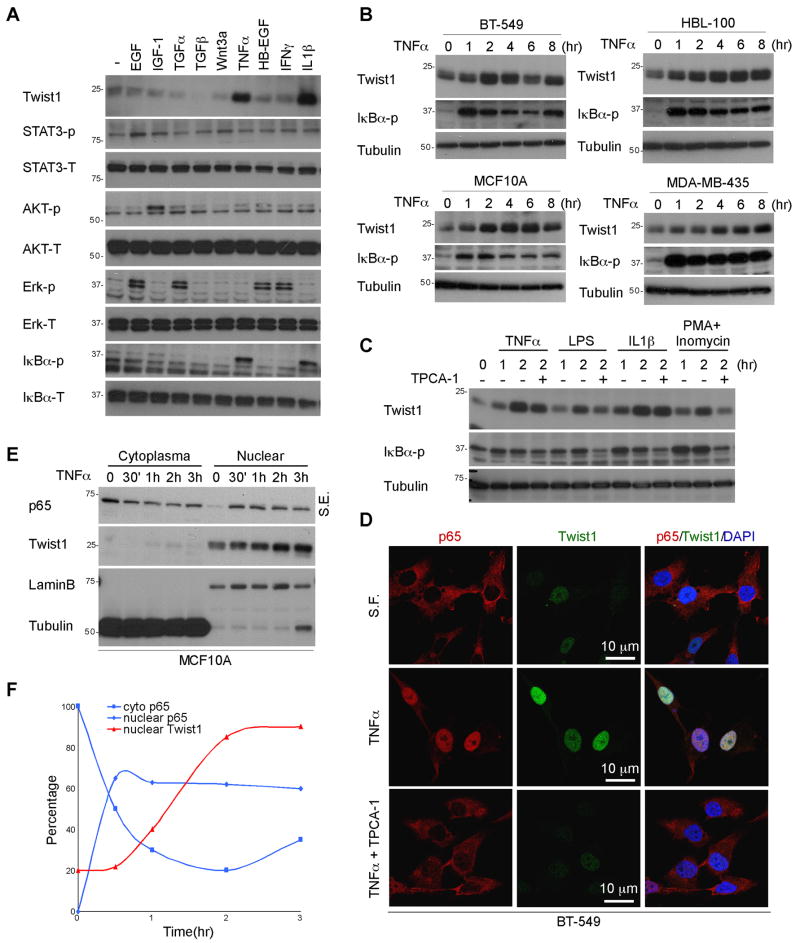
To study TNFα-mediated EMT regulation, mammary epithelial cells derived from normal tissue, MCF 10A and HBL-100 cells, were treated with TNFα in the presence or absence of TGFβ for several passages (Fig. S1A). As expected, we found that chronic exposure to TNFα enhanced TGFβ-induced EMT signaling as indicated by E-cadherin expression. However, continuous treatment with TNFα̣ to passage 4 alone (two days/passage) led to a loss of E-cadherin expression and promoted late EMT morphological changes compared to that of TGFβ treatment (Fig. S1A and S1B). To identify the genetic signatures that are involved in modulation of TNFα-mediated EMT, RT2 Profiler PCR array (SuperArray Bioscience Corporation, Frederick, MD) containing 84 well-characterized EMT mediators was performed. Between two tested cell lines, Twist1 mRNA was the only one found to be significantly upregulated upon TNFα stimulation (Fig. S1C). Various growth factors and cytokines including EGF, IGF-1, TGFα, TGFβ, Wnt3a, TNFα, IFNγ̣ HB-EGF, and IL-1β were tested to validate their ability to induce Twist1 expression. When MCF 10A cells were treated with various ligands for 2 hr, we found that TNFα rapidly induced Twist1 expression to a degree similar to that in cells treated with IL-1β (Fig. 1A). Next, we measured the time-dependent expression of Twist1 and found that it increased significantly after 1 hr of TNFα stimulation and reached at maximal level from between 2 to 4 hr (Fig. 1B). This regulation is present not only in mammary epithelial cells derived from normal tissue such as MCF 10A and HBL-100 but also in breast cancer cells (BT-549 and MDA-MB-435), suggesting that TNFα-induced Twist1 expression might be a general phenomenon (Fig. 1B). Next, to determine whether NFκB is responsible for the TNFα/IL-1β-induced Twist1 expression, several NFκB inducers such as LPS, PMA/Inomycin, and H2O2 as well as the IKKβ small molecule inhibitor TPCA-1 were used to test their effects on Twist1 expression. As shown in Figure 1C, Twist1 expression was upregulated in response to NFκB inducers with a similar degree of increase at 2 hr treatment. Similarly, Twist1 expression correlated with the activation status of NFκB (using phosphorylated IκBα as readout) in both MCF 10A and HBL-100 cells (Fig. 1C and S1D). Given TNFα activation induces p65 nuclear translocation, we examined endogenous p65 and Twist1 localization in BT-549 (Fig. 1D and Fig. S2) and MCF 10A (Fig. S1E) cells and found that both TNFα and IL1β induced nuclear translocation of p65 at 2 hr post treatment. Meanwhile, under the same exposure condition, we observed an increase in the level of nuclear Twist1 by confocal microscopy (Fig. 1D, center panel). To further confirm the upregulation of Twist1 and p65 nuclear translocation, nuclear and cytoplasmic fractions of MCF 10A cells were isolated at different time points upon treatment with TNFα (Fig. 1E). We observed TNFα̃ induced nuclear translocation of p65 at 30 min, whereas the nuclear expression of Twist1 began to increase at 1 hr after treatment (Fig. 1F). These results suggest that TNFα triggers a dynamic interaction between nuclear translocation of p65 and nuclear expression of Twist1.
Figure 1. Activation of NFκB induces Twist1 expression.
(A) Cells were serum-starved overnight and then treated with 30 ng/ml EGF, 25 ng/ml IGF-1, 1 μg/ml TGFα, 100 nM TGFβ, 30 ng/ml Wnt3a, 10 ng/ml TNFα, 10 ng/ml HB-EGF, and 50 ng/ml IFNγ̣for 2 hr. Protein expression were analyzed by Western blot.
(B) BT-549, HBL-100, MCF 10A and MDA-MB-435 cells were serum-starved overnight and then treated with 10 ng/ml TNFα for the indicated periods.
(C) Serum-starved MCF 10A cells were treated with various NFκB activators, TNFα̣ 10 ng/ml), LPS (1 ng/ml), IL1β (10 ng/ml) and PMA/Inomycin (PMA, 10 nM and Inomycin, 100 nM) at indicated time point. TPCA-1 was applied 30 min before the experiment.
(D) MCF 10A were treated with TNFα for 2 hr. After fixation, the cellular location of endogenous p65 (red) and Twist1 (green) were analyzed by confocal microscopy. Cell nuclei were stained with DAPI (blue). S.F., Serum-free.
(E) MCF 10A cells were treated with TNFα at different time point. Cytosolic and nuclear p65 and Twist1 protein were separated using hypotonic buffer. Tubulin and lamin B indicate cytosolic and nuclear fraction, respectively. S.E., short exposure; L.E., long exposure.
(F) Densitometric analysis of the Western blot.
IKKβ is also required for TNFα-induced Twist1 expression
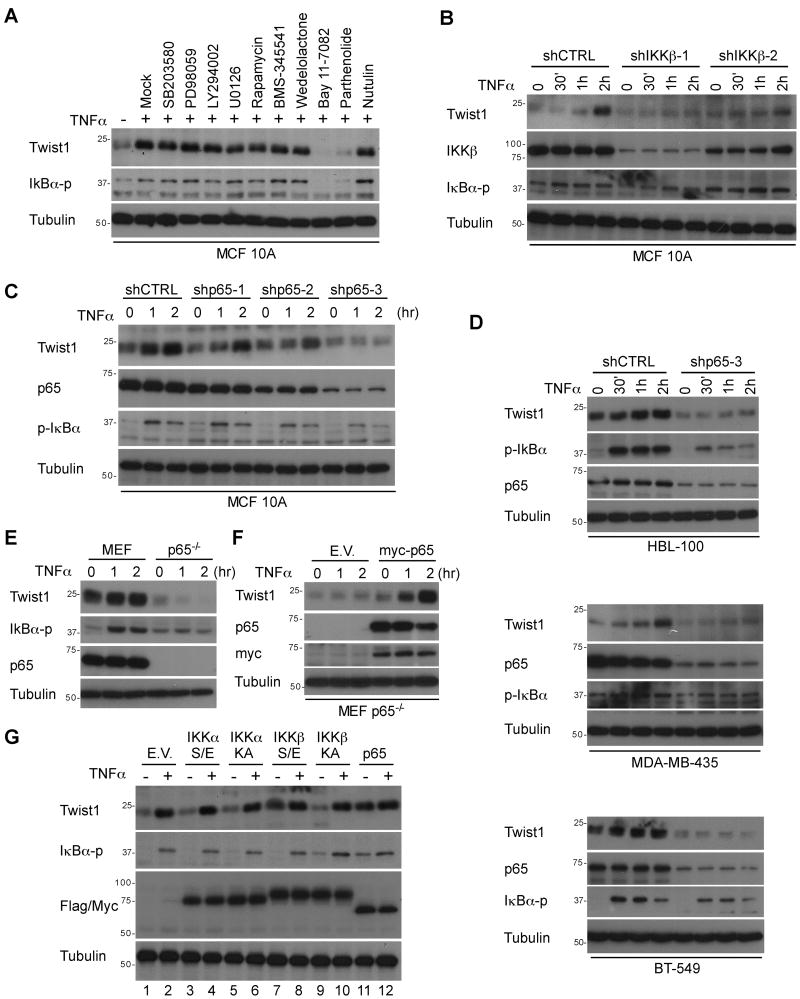
Because TNFα can induce activation of various signaling pathways, we wanted to determine which signaling cascade is responsible for TNFα-mediated Twist1 expression. To do so, MCF 10A cells were serum-starved overnight and pretreated with various inhibitors prior to TNFα stimulation. We found that upregulation of Twist1 by TNFα was not affected by MAPK/ERK, mTOR, p38, or JNK kinases inhibitors. In contrast, IKKβ inhibitors, BAY 11-7082 and parthenonlide, both abrogated TNFα-induced Twist1 expression (Fig. 2A). To diminish the off-target effect of these chemical inhibitors and further validate the role of IKKβ in TNFα induced Twist1 expression, we introduced a lentiviral-based IKKβ shRNA into MCF 10A cells. Consistently, silencing IKKβ expression level also attenuated TNFα-induced Twist1 expression (Fig. 2B). Interestingly, we showed that activation of IKKα via RANKL treatment (Fig. S3A) or silencing IKKα expression (Fig. S3B) had no effect on TNFα-induced Twist1 expression. We also performed experiments using previously established IKKβ and IKKα knockout Mouse Embryonic Fibroblasts (MEFs) (20). As shown in Figure S3C, we detected TNFα-induced Twist1 expression in wild-type MEFs but not in IKKβ-deficient MEFs. Re-expression of wild-type IKKβ but not an IKKβ kinase-dead mutant (KA) restored TNFα-induced Twist1 expression (Fig. S3D), suggesting that the kinase activity of IKKβ is required. Similarly, in low IKKβ-expressing MDA-MB-453 cells, Twist1 expression was not affected by TNFα, however, reintroducion of IKKβ by stable transfection elevated the TNFα-induced Twist1 expression (20)(Fig. S3E). Altogether, we conclude that the canonical IKKβ-dependent NFκB signaling is required for TNFα-induced Twist1 expression.
Figure 2. NFκB is required for TNFα-mediated Twist1 expression.
(A) MCF 10A cells were treated with SB203580, PD98059, LY294002, U0126, Rapamycin, BMS-345541, Wedelolactone, 40 μmol/L Bay 11-7082, 80 μmol/L Parthenolide, and Nutulin for 30 min.
(B) Two individual shIKKβ stable clones of MCF 10A cells were serum-starved overnight and treated with TNFα at various time points.
(C) ShCTRL and shp65 stable clone of MCF 10A cells were serum-starved overnight and treated with TNFα at various time points.
(D) Control or shp65-3 was expressed in HBL-100, BT-549 and MDA-MB-453 cells followed by treatment with TNFα or a vehicle for up to 2 hr. The protein expression of Twist1, p65, and p-IκBα were analyzed using Western blot.
(E) MEF and p65-/- MEF cells were serum-starved overnight and then treated with 10 ng/ml TNFα or a vehicle.
(F) Myc-p65 was transient transfected into p65-/- MEFs to restore p65 expression. TNFα-mediated Twist1 expression was analyzed using Western blot.
(G) Flag-tagged IKK or Myc-tagged p65 was transiently expressed in HBL-100 cells. The protein expression of Twist1 and p-IκBα were examined using Western blot.
TNFα-mediated Twist1 expression is dependent on p65 activation by IKKβ
Since activation of NFκB cascade usually results in nuclear translocation and activation of p65, we hypothesize that p65 might be involved in TNFα-induced Twist1 expression. To elucidate the causal relationship between p65 and Twist1, p65 was stably knocked down using three independent shRNAs in MCF 10A cells. We found that knock-down of endogenous p65 expression attenuated TNFα-induced Twist1 expression (Fig. 2C). Moreover, stable clones harboring high levels of p65 expression showed a higher Twist1 expression in respond to TNFα treatment. These results also ruled out the off-target effects due to shRNA-mediated gene silencing (Fig. 2C). Consistently, knock-down of p65 expression also inhibited TNFα-induced Twist1 expression in BT-549, HBL-100 and MDA-MB-435 cells (Fig. 2D). In addition, TNFα rapidly induced Twist1 expression in wild-type (p65+/+) MEFs but not in p65-deficient (p65-/-) MEFs (Fig. 2E). Restoration of myc-tagged p65 in p65-/- MEFs rescued TNFα-induced Twist1 expression, further supporting that p65 is required for TNFα-mediated Twist1 expression (Fig. 2F). To further confirm this finding, we expressed constitutively active (S/E) or kinase dead (KD) IKKα, IKKβ, or p65 in HBL-100 cells and then treated with TNFα. Expression of both constitutively active IKKβ (Fig. 2G, lane 7) and p65 (Fig. 2G, lane 11) was sufficient to induce Twist1 expression to a degree similar to that of TNFα treatment. To establish a clinical relevance of inhibition of NFκB-mediated Twist1 expression, both MCF 10A and HBL-100 cells were pretreated with nonsteroidial anti-inflammatory drugs (NSAIDs) and subjected to TNFα stimulation. When these cells were pretreated with another commonly used NFκB inhibitor Sanguinarine and tosyl phenylalanyl chloromethyl ketine 1 (TPCK-1), TNFα-induced Twist1 expression was abolished (Fig. S3F and Fig. S3G). Therefore, targeting NFκB-mediated Twist1 expression implicates a novel aspect for breast cancer therapy.
TNFα-induced Twist1 expression is transcriptionally regulated by p65
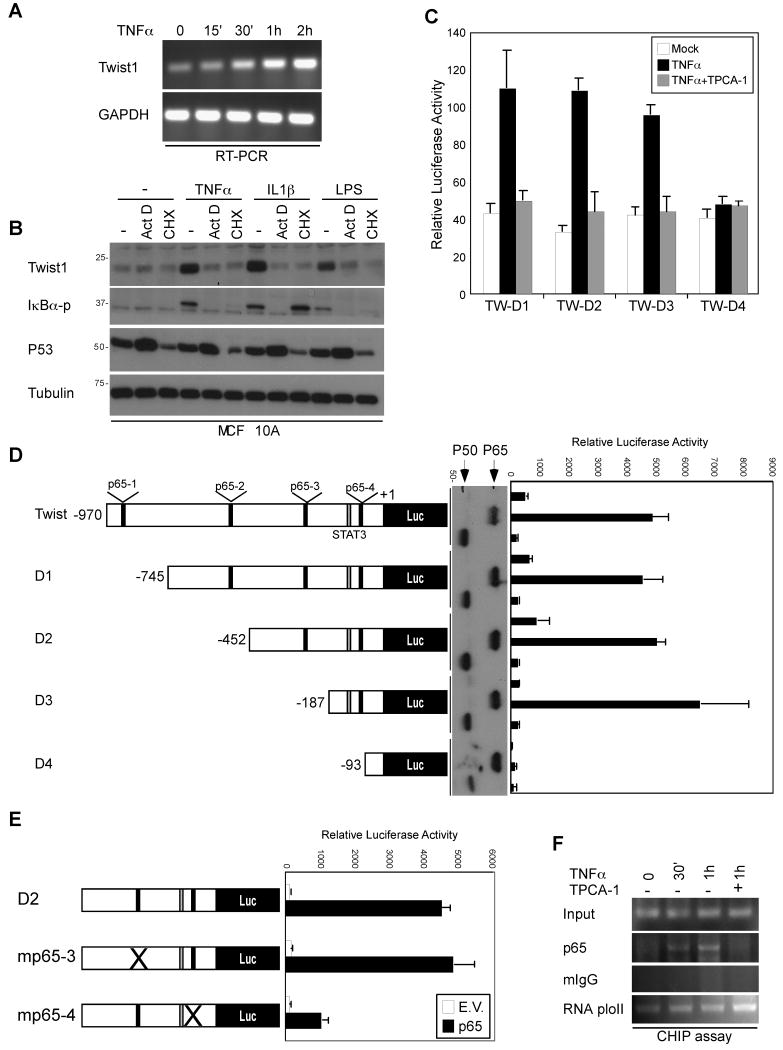
Because the TNFα-induced Twist1 expression requires p65, it would be of interest to determine whether TNFα-induced Twist1 expression is transcriptionally regulated. Indeed, TNFα elevated Twist1 mRNA expression at 1 hr of treatment in MCF 10A and HBL-100 cells (Fig. 3A and data not shown). Consistent with this finding, Twist1 expression induced by TNFα, LPS or IL1β was abrogated when cells were pretreated with a transcription inhibitor actinomycin D (Act D) or a protein synthesis inhibitor cycloheximide (CHX) (Fig. 3B). Since Twist1 undergoes protein degradation via 26S proteasome machinery (26), we also tested whether the activation of NFκB affects Twist1 protein stability. As shown in Figures S4C and S4D, the Twist1 protein half-life was not influenced by TNFα treatment or co-expression of p65, suggesting that TNFα induces Twist1 expression exclusively via transcriptional regulation.
Figure 3. p65 transcriptionally regulates Twist1 expression.
(A) mRNAs isolated from TNFα treated MCF 10A cells were subjected to RT-PCR using primer sets specific against Twist1 and GAPDH.
(B) MCF 10A cells were pretreated with 500 ng/ml Actinomycin D (Act D) and 10 μg/ml cycloheximide (CHX) for 1 hr, stimulated with various agents for 2 hr, and subjected to Western blot with the indicated antibodies.
(C) HEK-293 cells transfected with the indicated Twist1 promoter were treated in the presence or absence of 10 ng TNFα and TPCA-1 for 2 hr. The luciferase activity was measured and normalized according to Renilla luciferase activity.
(D) A series of deletion mutants of the Twist1 promoter were introduced to HEK-293 cells together with or without p65 and p50 (expression showed in the middle panel).
(E) Identification of p65 binding site on Twist1 promoter. Wild-type and p65-binding-element-mutated Twist1 promoter luciferase were transiently expressed in HEK-293 cells. The relative luciferase activity is present as the means ± standard error from three independent experiments.
(F) Chromatin immunoprecipitation (ChIP) of p65 in respond to TNFα treatment.
p65 binds directly to the Twist1 promoter to regulate its expression
The p65 protein is a multifunctional transcription factor that elicits its physiologic function by regulating target gene expression upon NFκB activation. To investigate the molecular mechanism by which TNFα induces Twist1 expression, we employed three bioinformatics programs to identify the putative binding sites for p65 on the Twist1 promoter. We found that the Twist1 promoter sequence from -970 to +1 contains four p65 binding sites and two of which are highly consensus among three predications (Fig. S4A), suggesting that p65 might regulate Twist1 expression by directly binding to its promoter. Using a luciferase reporter construct, Twist1-Luc responded to TNFα stimulation in HEK-293 (Fig. 3C) and MCF10A cells (Fig. S4E). In contrast, treatment with TPCA-1 (an IKKβ̣ inhibitor) abrogated TNFα-mediated Twist1 promoter activities. Moreover, co-expression of p65 and Twist1-Luc significantly enhanced the reporter activity but not IKKα or dominant negative IKKβ (Fig. S4B).
Furthermore, to locate the authentic p65 binding sites, a nested deletion of Twist1-Luc (D1, D2, D3, and D4) was generated. Among the five constructs, a p65-4 element alone on the Twist1 promoter maintained high reporter activity by p65 induction, indicating that the critical p65 DNA-binding elements are located in the 120-bp region of the promoter (Fig. 3D). To pinpoint the exact binding motifs, we introduce point mutations into the p65-3 and p65-4 elements of Twist1 D4-Luc (Fig. 3E, left panel). Ablation of the p65-4 binding site on the Twist1 promoter abrogated p65-mediated Twist1 expression (Fig. 3E). We also transient transfected Twist1 D4-Luc into a stable clone of MCF 10A expressing p65 shRNA, and show that cells harboring high level of p65 posses higher reporter activity, confirming that endogenous p65 is critical for Twist1 expression (Fig. S4F and S4G).
To further examine the binding of p65 to the Twist1 promoter in vivo, a chromatin immunoprecipitation (ChIP) assay was performed using stable MCF 10A-p65 cells. Upon TNFα stimulation, nuclear p65 bound to the human Twist1 gene promoter at 30 min. In contrast, IgG did not associate with the Twist1 promoter at a detectable level. The binding of p65 to the Twist1 promoter was released by treatment with TPCA-1 (Fig. 3F). Moreover, gel sift assay was also conducted to confirm that p65 is bound to the Twist1 promoter in vitro (data not shown). Collectively, these results suggest that p65 regulates Twist1 transcription by directly binding to the Twist1 promoter in a TNFα-dependent fashion.
p65-mediated Twist1 expression results in EMT
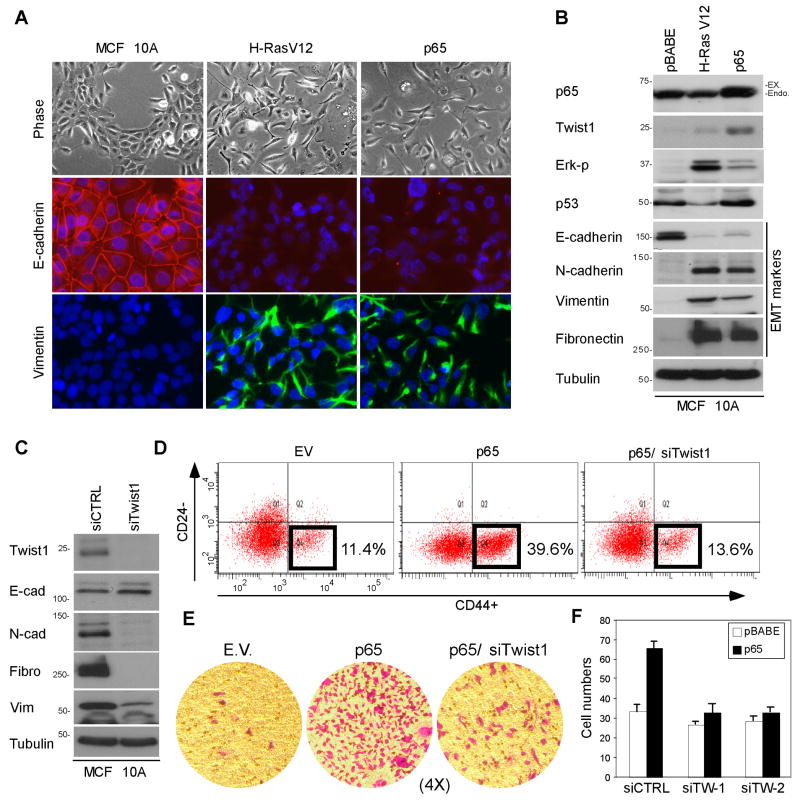
To determine the functional consequences of p65 activation in breast cancer cells, ectopic expression of p65 in MCF 10A cells was accomplished using a retroviral infection. H-RasV12, which is known to induce EMT in various types of cells (27), was used as a positive control. Compared with empty vector-infected cells (pBABE), p65-expressing cells exhibited spindle-like morphology, loss of cell contact, and formation of vimentin fibers reminiscent of EMT (Fig. 4A, phase contrast micrograph). The EMT-like phenotypic changes were confirmed by detecting expression of characteristic molecular markers using immunofluorescence (Fig. 4A, immunostaining) and Western blot (Fig. 4B). In p65-expressing cells, the expression of mesenchymal markers fibronectin, N-cadherin, and vimentin was significantly upregulated, whereas that of the epithelial marker E-cadherin was downregulated. We observed similar results using MCF-12A cells infected with a p65-expressing retrovirus (Fig. S5A, S5B and S5C). MCF 10A-p65 cells demonstrated increased cellular migration and invasion abilities as measured by a wound-healing assay and Boyden chamber assay in media lacking EGF, respectively (Fig. 4E and 4F). Interestingly, we observed a significant upregulation of Twist1 expression in p65-overexpressing MCF 10A and MCF-12A cells. The increase in Twist1 expression was further enhanced by treatment with TNFα (Fig. S5D).
Figure 4. p65 overexpression upregulates Twist1 expression and induces changes in epithelial cell morphology.
(A) Phase-contrast and immunofluorescent micrographs showing the morphological appearance of MCF 10A cells infected with pBABE (empty vector) as compared with that of cells infected with pBABE-H-Ras V12 and pBABE-p65.
(B) Western blot analysis of the protein expression for p65, Twist1, and EMT markers in MCF 10A stable clones shown in Figure 4A.
(C) Western blot analysis of mesenchymal markers in MCF 10A-p65 cells with Twist1 siRNA.
(D) Abrogation of p65-mediated cancer stem cell population by Twist1 suppression.
(E) Reduction of p65-mediated cell invasion by Twist1 suppression.
(F) Reduction of p65-mediated cell migration by Twist1 suppression.
To test whether upregulation of Twist1 expression is required for p65-induced EMT, Twist1 expression was knocked down in the MCF 10A-p65 stable cells. This knockdown inhibited cell migration, invasion, and formation of EMT phenotype (Fig. 4C), suggesting that Twist1 is required for p65-mediated EMT phenotypic changes. It has been documented that Twist1 modulates breast cancer stem cells via transcriptional suppression of CD24 expression (28). Therefore, we asked whether the p65-Twist1 axis regulates breast cancer cells side population. Indeed, p65 overexpression could induce CD24-/CD44+ population in two different breast epithelial cell lines (MCF 10A in Fig. 4D and MCF-12A in Fig. S5C) and that downregulation of Twist1 expression by siRNA partially reversed the stem cell molecular signature by reducing p65 induced cancer stem cell population (Fig. 4D, right panel). In addition, we found that Twist1 is required for p65-mediated mammosphere formation (Fig. S5E). Together with cell migration and invasion assays (Fig. 4E and 4F), these results identified a prerequisite role for Twist1 in p65-mediated breast tumor progression.
Inflammation-induced Twist1 upregulation increases metastatic potential
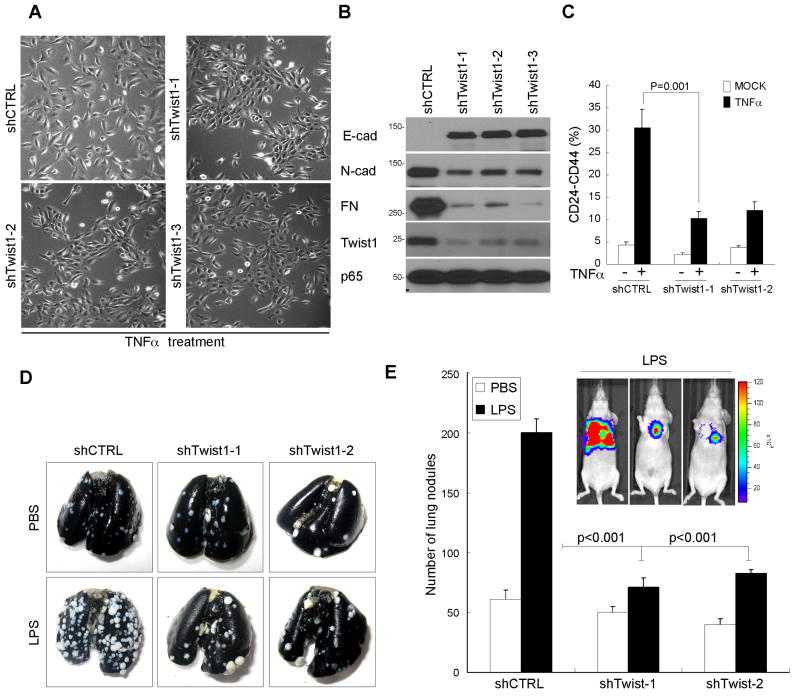
To test whether constitutive Twist1 expression contributes to TNFα-induced EMT, the endogenous Twist1 was knocked down in MCF 10A cells using a lentivirus-based shRNA (shTwist1). We found that three independent shRNA constructs efficient knocked down endogenous Twist1 expression, as confirmed by Western blot (Fig. 5B). Inhibition of Twist1 expression in MCF 10A cells significantly reduced TNFα-mediated EMT at passage 3, whereas cells infected with shRNA against luciferase (shCTRL) exhibited EMT (Fig. 5A). Moreover, Twist1 knockdown resulted in increased E-cadherin and reduced fibronectin expression. Thus, suppression of Twist1 expression in MCF 10A cells partially reversed TNFα-induced EMT (see above). Consistent with these phenotypic changes, TNFα-induced breast cancer stem cell population was abolished by Twist1 inhibition (Fig. 5C). We then confirmed our finding using a xenograft lung metastasis model in which administration of the inflammation inducer LPS enhance lung metastasis in mice. Our in vivo metastasis assay showed that knockdown of Twist1 expression in 4T1-Luc cells antagonized LPS-induced metastasis by measuring the number of lung nodules formed in mice (Fig. 5D and 5E). Although Twist1 had little effect on intrinsic metastatic potential, it significantly impacted inflammation-induced metastasis (82% lower in lung nodules versus shCTRL in LPS treated mice). Thus, these results suggest that inflammation induced upregulation of Twist1 expression plays an essential role in breast cancer metastasis.
Figure 5. Twist1 is required for inflammation-induced metastasis.
(A) Phase-contrast images of EMT morphotypic changes in MCF 10A shCTRL and shTwist1 stable cells treated with TNFα.
(B) Western blot of the EMT markers from cells in (A)
(C) Abrogation of TNFα-mediated cancer stem cell population by Twist1 suppression.
(D) Representative photograph of metastatic lung nodules. 4T1-Luc cells with control and two shTwist1 stable clones were injected into BALB/c mice via the tail vein. The mice then received intraperitoneal injection of saline or 10 μg LPS. Seven days later, the mice were sacrificed and the entire lungs were stained with India ink and resected.
(E) Quantification of the lung nodules in Figure 5C. The error bars represent standard deviation for n=5. *indicates P < 0.0002).
IKKβ/nuclear p65 associates positively with Twist1 in cancer cell lines and primary breast carcinomas
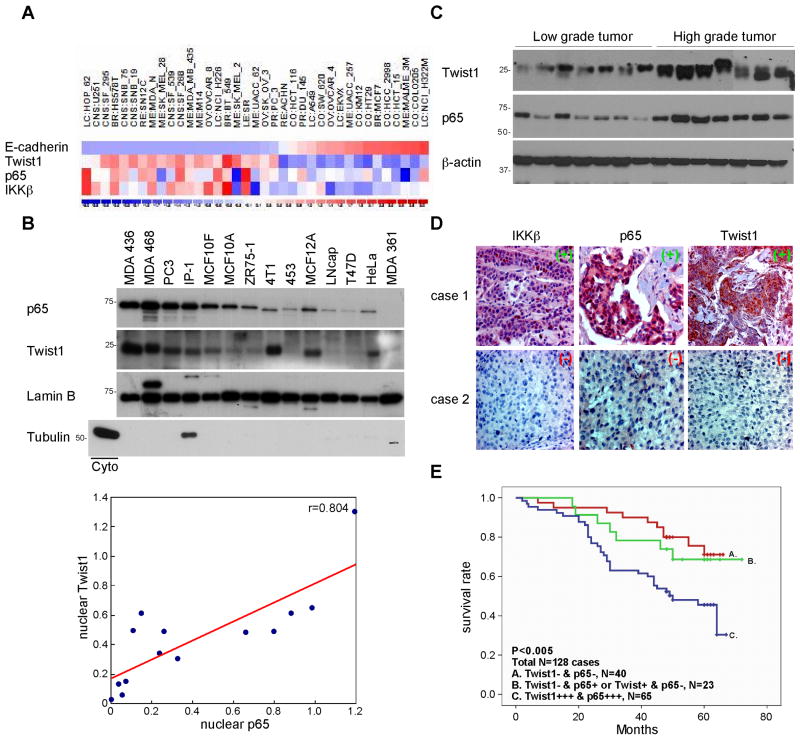
To elucidate the clinical relevance of NFκB activation and Twist1 expression, the association of their cDNA expression was examined by reanalyzing NCI-60 microarray databases from a total of 60 various cancer cell lines. A strong correlation was found between Twist1, p65, and IKKβ expression (data not shown). To determine the significance of p65/Twist1 in the EMT, we selected 37 cell lines from the NCI-60 panel and found that expression of the Twist1 was inversely correlated with that of E-cadherin (correlation coefficient r <−0.8) (Figs. 6A, S6A, and S6B), indicating the functional significance of the Twist1 in these cell lines. As shown in Figure 6A, the expression of Twist1 was significantly correlated with that of p65 (r = 0.529) (Fig. S6D) and IKKβ (r = 0.630) (Fig. S6C).
Figure 6. Clinical association of IKKβ, p65, and Twist1 expression with survival of breast cancer patients.
(A) Heat map generated using 37 cell lines from the NCI-60 panel showing the levels of expression of E-cadherin, Twist1, p65 and IKKβ.
(B) Western blot analysis of p65 and Twist1 expression in nuclear fraction isolated from 14 cell lines.
(C) Western blot analysis of p65 and Twist1 expression in high- and low-grade human breast tumor samples.
(D) IHC staining of human breast cancer samples showing the expression of IKKβ, p65 and Twist1.
(E) Kaplan-Meier overall survival curves of p65 and Twist1.
We next asked whether overexpression of Twist1 in the breast cancer cells might be a result from NFκB activation. Because nuclear p65 reflects the active state of NFκB (18) and the functional Twist1 is known to localize in the nucleus, we measured the expression of p65 and Twist1 in nuclear extracts from 14 different cancer cell lines (Fig. 6B). As expected, the nuclear fraction of p65 level was highly correlated with the nuclear Twist1 (r = 0.804, P = 0.0013) (Fig. 6D). These results are also consistent with earlier finding that nuclear Twist1 expression is associated with p65 nuclear translocation (Figs. 1D, 1E, and 1F). To determine the clinical correlation of p65 and Twist1 protein expression in human breast cancer, we examine their expression in 14 freshly isolated low- and high-grade breast tumor samples. Based on our data, p65 and Twist1 expression levels are elevated in high-grade tumors indicating that coexpression of p65 and Twist1 enhances the aggressive phenotype of breast cancer cells (Fig. 6C).
Clinical significance of activation of the IKKβ/p65/Twist1 axis in a cohort of primary breast carcinomas
To further examine our findings in human primary tumors, we studied the expression of IKKβ, p65, and Twist1 in 115 human primary breast tumor specimens using immunohistochemistry (IHC) analysis. Twist1 was detected in 67 (51%) of the 82 specimens with high p65 expression but in only 10 (7.6%) of the 49 specimens with low p65 expression, indicating that p65 expression associates with high levels of Twist1 expression (p < 0.0001) (Fig. 6E). Consistent with this finding, we found that IKKβ expression associates with Twist1 (p < 0.023) (Table 1) and p65 expression (p < 0.017) (Table S1) expression. Next, we analyzed their expression with other clinical record and found strong activation of IKKβ/p65/Twist1 axis in patients with lymph node metastasis patient (Table S2). We also analyzed the expression of p65 and Twist1 in breast tumor tissues and correlated the findings with patient survival data. The Kaplan-Meier overall survival curves showed that high p65 and Twist1 expression levels were associated with poor survival (Fig. S6E and S6F). However, the combination of p65 and Twist1 expression was a better predictor of survival than was either factor alone (p < 0.02 versus p < 0.005) (Fig. 6F). Taken together, the IHC staining data further strengthened the notion that activation of the IKKβ complex induces nuclear translocation of p65 and subsequently upregulation of Twist1 expression, which contributes to the promotion of EMT phenotype and is associated with poor clinical outcome of breast cancer patients.
Table 1. Relationships between expression of Twist1, NFκB/p65 and IKKβ in surgical specimens of breast cancer.
| Expression of Twist1 | |||||
|---|---|---|---|---|---|
| -/+ | ++/+++ | Total | P value | ||
| NFκB | -/+ | 39 (29.8%) | 10 (7.6%) | 49 (37.4%) | |
| ++/+++ | 15 (11.5%) | 67 (51.1%) | 82 (62.6%) | ||
| Total | 54 (41.2%) | 77 (58.8%) | 131 (100%) | P<0.0001 | |
| IKKβ | -/+ | 39 (30.5%) | 42 (32.8%) | 81 (63.3%) | |
| ++/+++ | 13 (10.2%) | 34 (26.6%) | 47 (36.7%) | ||
| Total | 52 (40.6%) | 76 (59.4%) | 128 (100%) | P<0.023 | |
Positive correlation between Twist1, NFκB/p65 and IKKβ calculated using the Pearson Chi-Square analysis.
Discussion
Chronic inflammation-induced metastasis has long been considered as a major challenge in cancer therapy and is a primary cause of mortality in many cancers. Understanding the underlying mechanism governing the metastatic nature is therefore critical and may uncover therapeutic interventions. In the present study, we investigated an important but underdeveloped signaling axis that controls inflammatory cytokine-and promotes EMT. Despite the essential role of TGFβ-dependent Smad regulation in EMT, we discovered a novel aspect by which p65 transactivation of Twist1 expression is required for TNFα-induced EMT. On the basis of our findings, we propose a model in which elevated TNFα̣ from macrophages or the tumor microenvironment, upregulates the canonical NFκB signaling through the activation of IKKβ but not IKKα. The liberated cytoplasmic p65 then translocates to the nucleus, recognizes a cognate sequence on the Twist1 promoter, induces Twist1 expression, and promotes tumor metastasis (Fig. S6G).
IKKβ is a component of the classic IKK complex, which is composed of three subunits: two catalytic kinases (IKKα and IKKβ) and a regulatory scaffold partner (IKKγ̣. Upon stimulation by either TNFα or IL-1β, activated IKKβ phosphorylates the NFκB inhibitor IκBα and disrupts the nuclear retention of NFκB. In fact, IKKβ does more than simply induce IκBα degradation for its tumorigenesis activity. For example, IKKβ directly phosphorylates p65 to promote its interaction with transcriptional co-activators and enhance its transactivation (29). Moreover, IKKβ-induced TSC1 phosphorylation inhibits its association with GTPase-activating protein (TSC2), alters mTOR activity, allows VEGF-A expression, and promotes tumorigenesis (20). In the present study, we found that both IKKβ and p65 are mutually exclusive important in TNFα-mediated Twist1 regulation. Both stable knockdown and overexpression of IKKβ affects Twist1 expression. Given that constitutive active IKKβ induces EMT in EpRas cells (16), the involvement of IKKβ in EMT supports our hypothesis. Here, we identified a mechanism for IKKβ-mediated tumor metastasis via upregulation of Twist1 expression. In addition to the requirement of IKKβ for TNFα-mediated Twist1 expression, constitutively activated IKKβ promotes Twist1 expression, which may in turn contribute to the EMT phenotype.
Twist1 is a bHLH transcription factor that has been known as an essential player in the aggressive phenotype of EMT (7). Given that EMT is usually accompanied by an increase in stem cell-like properties to facilitate metastatic colonization as well as drug resistance (30) (31), researchers recently demonstrated that Twist1 induces cancer stem-cell ability by inhibiting CD24 gene expression (28). Surprisingly, we found that p65-induced EMT is also accompanied by the acquisition of cancer stem cell properties. In addition, downregulation of Twist1 expression suppressed p65-mediated malignancy, including EMT and stemness, suggesting that Twist1 is a central modulator downstream from NFκB. By in vivo metastasis experimental model, suppression of Twist1 expression reduced LPS-meditated lung metastasis. Therefore, the current study strongly supports the notion that p65 and Twist1 oncoproteins interact to regulate the expression of a series target genes involved in aggressive cancer behavior. This regulation may likely contribute to inflammation-induced breast cancer metastasis.
Despite frequent reports of Twist1 overexpression in human cancers, transcriptional regulation of the human Twist1 genes remains largely unknown. Previously, we showed that EGFR cooperates with Signal Transducer and Activator of Transcription 3 (STAT3) to induce EMT in breast cancer cells via upregulation of Twist1 gene expression (24). In addition, STAT3 has been shown to transcriptionally activate Twist1 expression, resulting in AKT2-mediated oncogenic properties (32). A recent study demonstrated that knockdown of STAT3 expression in murine 4T1 mammary tumor cells led to altered expression of Twist1 (32). Moreover, regulation of the murine Twist genes has involved NFκB (33) and Wnt1/TCF/h-catenin pathways (34). However, the NFκB and TCF/h-catenin response elements found in the mouse Twist1 gene promoters are not present in the human Twist1 gene. Herein, we provide the first evidence to show that TNFα stimulates p65 to bind to the human Twist1 promoter and regulate its transcription. Using TF Search and TESS transcription factor search tools together with biochemical analysis, we identified a p65-binding site on the Twist1 promoter in response to TNFα treatment. Because the murine Twist1 promoter also contains the p65 consensus site, this novel axis is reminiscent of an evolutionarily conserved mechanism.
Given that Twist1 undergoes caspase-mediated cleavage and proteasome-mediated degradation under apoptotic stimuli (26), investigation of the Twist1 protein stability in response to NFκB activation is conceivable. To date, p65 has been shown to enhance Snail protein stability by recruiting COP9 signalosome 2 (CSN2) complexes to inhibit β-TRCP-mediated degradation (18). In contrast, our result exclude the possibility that p65 affects Twist1 protein stability, albeit over a short period. We report herein that expression of human Twist1 gene is directly upregulated by p65-mediated transcriptional activation in response to chronic inflammation.
Several lines of evidence demonstrate that TNFα-mediated Twist1 expression in breast cancer cells contributes to their aggressive phenotype. We demonstrated in this study that TNFα and various NFκB activators induce Twist1 expression in both normal breast epithelial and breast cancer cells; 2) both canonical modules of NFκB signaling, IKKβ̣ and p65, are required for TNFα-mediated Twist1 expression; 3) Twist1 expression is required and correlates with p65-mediated cancer progression; 4) downregulation of Twist1 expression reduces TNFα-mediated EMT and tumor metastasis. The fact that Twist1 promoter also contains a functional p65 binding motif, therefore, we propose that, breast cancer cell metastasis induced by pro-inflammatory cytokine TNFα is coordinated by a canonical NFκB signaling involved in Twist1 activation. The in-depth analysis of this novel axis may improve understanding of breast cancer signaling and therefore introduce a therapeutic strategy for targeting breast cancer malignancy.
Supplementary Material
Acknowledgments
We would like to thank Don Norwood at Scientific Publication at MD Anderson Cancer Center for editing and Dr. Stephanie A. Miller for critical reading of the manuscript. This work was partially supported by several National Institutes of Health grants—PO1 grant CA09903, RO1 grant CA109311, Breast Cancer SPORE P50 CA116199, Breast Cancer Research Foundation (to G.N.H), National Breast Cancer Foundation, Inc. (to M.-C.H), Sister Institution Fund of China Medical University and Hospital and MD Anderson Cancer Center, Cancer Research Center of Excellence and Department of Defense Postdoctoral Fellowship (W81XXWH-10-1-0598 to C.-W. L). In memoriam, Mrs. Serena Lin-Guo for her courageous fight against cancer.
References
- 1.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–59. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 2.Luo JL, Kamata H, Karin M. IKK/NF-kappaB signaling: balancing life and death--a new approach to cancer therapy. J Clin Invest. 2005;115:2625–32. doi: 10.1172/JCI26322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaisho T, Takeda K, Tsujimura T, Kawai T, Nomura F, Terada N, et al. IkappaB kinase alpha is essential for mature B cell development and function. J Exp Med. 2001;193:417–26. doi: 10.1084/jem.193.4.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–54. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 5.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–58. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–28. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 7.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Kwok WK, Ling MT, Lee TW, Lau TC, Zhou C, Zhang X, et al. Up-regulation of TWIST in prostate cancer and its implication as a therapeutic target. Cancer Res. 2005;65:5153–62. doi: 10.1158/0008-5472.CAN-04-3785. [DOI] [PubMed] [Google Scholar]
- 9.Kyo S, Sakaguchi J, Ohno S, Mizumoto Y, Maida Y, Hashimoto M, et al. High Twist expression is involved in infiltrative endometrial cancer and affects patient survival. Hum Pathol. 2006;37:431–8. doi: 10.1016/j.humpath.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 10.Yuen HF, Chan YP, Wong ML, Kwok WK, Chan KK, Lee PY, et al. Upregulation of Twist in oesophageal squamous cell carcinoma is associated with neoplastic transformation and distant metastasis. J Clin Pathol. 2007;60:510–4. doi: 10.1136/jcp.2006.039099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuo N, Shiraha H, Fujikawa T, Takaoka N, Ueda N, Tanaka S, et al. Twist expression promotes migration and invasion in hepatocellular carcinoma. BMC Cancer. 2009;9:240. doi: 10.1186/1471-2407-9-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elias MC, Tozer KR, Silber JR, Mikheeva S, Deng M, Morrison RS, et al. TWIST is expressed in human gliomas and promotes invasion. Neoplasia. 2005;7:824–37. doi: 10.1593/neo.04352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee DF, Hung MC. Advances in targeting IKK and IKK-related kinases for cancer therapy. Clin Cancer Res. 2008;14:5656–62. doi: 10.1158/1078-0432.CCR-08-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Y, Zhou BP. TNF-alpha/NF-kappaB/Snail pathway in cancer cell migration and invasion. Br J Cancer. 102:639–44. doi: 10.1038/sj.bjc.6605530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 16.Huber MA, Azoitei N, Baumann B, Grunert S, Sommer A, Pehamberger H, et al. NF-kappaB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J Clin Invest. 2004;114:569–81. doi: 10.1172/JCI21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chua HL, Bhat-Nakshatri P, Clare SE, Morimiya A, Badve S, Nakshatri H. NF-kappaB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: potential involvement of ZEB-1 and ZEB-2. Oncogene. 2007;26:711–24. doi: 10.1038/sj.onc.1209808. [DOI] [PubMed] [Google Scholar]
- 18.Wu Y, Deng J, Rychahou PG, Qiu S, Evers BM, Zhou BP. Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion. Cancer Cell. 2009;15:416–28. doi: 10.1016/j.ccr.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pham CG, Bubici C, Zazzeroni F, Knabb JR, Papa S, Kuntzen C, et al. Upregulation of Twist-1 by NF-kappaB blocks cytotoxicity induced by chemotherapeutic drugs. Mol Cell Biol. 2007;27:3920–35. doi: 10.1128/MCB.01219-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee DF, Kuo HP, Chen CT, Hsu JM, Chou CK, Wei Y, et al. IKK beta suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway. Cell. 2007;130:440–55. doi: 10.1016/j.cell.2007.05.058. [DOI] [PubMed] [Google Scholar]
- 21.Huang WC, Ju TK, Hung MC, Chen CC. Phosphorylation of CBP by IKKalpha promotes cell growth by switching the binding preference of CBP from p53 to NF-kappaB. Mol Cell. 2007;26:75–87. doi: 10.1016/j.molcel.2007.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng J, Miller SA, Wang HY, Xia W, Wen Y, Zhou BP, et al. beta-catenin interacts with and inhibits NF-kappa B in human colon and breast cancer. Cancer Cell. 2002;2:323–34. doi: 10.1016/s1535-6108(02)00154-x. [DOI] [PubMed] [Google Scholar]
- 23.Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, et al. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–37. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 24.Lo HW, Hsu SC, Xia W, Cao X, Shih JY, Wei Y, et al. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 2007;67:9066–76. doi: 10.1158/0008-5472.CAN-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, et al. EZH2 Promotes Expansion of Breast Tumor Initiating Cells through Activation of RAF1-beta- Catenin Signaling Cancer Cell. 19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Demontis S, Rigo C, Piccinin S, Mizzau M, Sonego M, Fabris M, et al. Twist is substrate for caspase cleavage and proteasome-mediated degradation. Cell Death Differ. 2006;13:335–45. doi: 10.1038/sj.cdd.4401744. [DOI] [PubMed] [Google Scholar]
- 27.Janda E, Lehmann K, Killisch I, Jechlinger M, Herzig M, Downward J, et al. Ras and TGF[beta] cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. J Cell Biol. 2002;156:299–313. doi: 10.1083/jcb.200109037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vesuna F, Lisok A, Kimble B, Raman V. Twist modulates breast cancer stem cells by transcriptional regulation of CD24 expression. Neoplasia. 2009;11:1318–28. doi: 10.1593/neo.91084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakurai H, Chiba H, Miyoshi H, Sugita T, Toriumi W. IkappaB kinases phosphorylate NF-kappaB p65 subunit on serine 536 in the transactivation domain. J Biol Chem. 1999;274:30353–6. doi: 10.1074/jbc.274.43.30353. [DOI] [PubMed] [Google Scholar]
- 30.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Cheng GZ, Zhang WZ, Sun M, Wang Q, Coppola D, Mansour M, et al. Twist is transcriptionally induced by activation of STAT3 and mediates STAT3 oncogenic function. J Biol Chem. 2008;283:14665–73. doi: 10.1074/jbc.M707429200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sosic D, Richardson JA, Yu K, Ornitz DM, Olson EN. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-kappaB activity. Cell. 2003;112:169–80. doi: 10.1016/s0092-8674(03)00002-3. [DOI] [PubMed] [Google Scholar]
- 34.Howe LR, Watanabe O, Leonard J, Brown AM. Twist is up-regulated in response to Wnt1 and inhibits mouse mammary cell differentiation. Cancer Res. 2003;63:1906–13. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.