Abstract
Although the term ‘epigenetics’ was coined nearly seventy years ago, its critical function in memory processing by the adult CNS has only recently been appreciated. The hypothesis that epigenetic mechanisms regulate memory and behavior was motivated by the need for stable molecular processes that evade turnover of the neuronal proteome. In this article, we discuss evidence that supports a role for neural epigenetic modifications in the formation, consolidation and storage of memory. In addition, we will review the evidence that epigenetic mechanisms regulate synaptic plasticity, a cellular correlate of memory. We will also examine how the concerted action of multiple epigenetic mechanisms with varying spatiotemporal profiles influence selective gene expression in response to behavioral experience. Finally, we will suggest key areas for future research that will help elucidate the complex, vital and still mysterious, role of epigenetic mechanisms in neural function and behavior.
Keywords: addiction, chromatin, DNA methylation, epigenetics, learning, memory
The renowned playwright, Tennessee Williams, once commented, “Life is all memory, except for the one present moment that goes by you so quickly you hardly catch it going”. This quote highlights the critical role of memory in the life and survival of the organism. Still, neuroscience is a long way from a precise understanding of the anatomical, cellular and molecular underpinnings of this process. By ‘memory’, we are not only referring to storage of factual information. Rather, learning and memory are defined more broadly as the acquisition and persistence of altered behavioral responses of the organism to an environmental stimulus [1]. While this definition is empirically biased, we feel it is appropriate in the context of the behavioral and molecular studies discussed below.
A newly formed memory must first be acquired (learned) and then be converted to a more persistent state in a process known as consolidation. Finally, stored memories are subject to retrieval upon re-exposure to the initial environmental stimulus [2]. It has been long appreciated that alterations in protein synthesis, gene expression and structural properties of neurons and synapses contribute to memory consolidation, storage and retrieval. Within the neuron, synaptic depolarization activates complex molecular signaling cascades that coalesce on specific gene loci, resulting in acute modulation of transcriptional efficacy. The resulting protein products are thought to produce stable alterations in cellular phenotype by influencing the structure and physiology of postsynaptic dendritic spines [3,4].
A central paradox in this explanation, however, becomes apparent when one considers the transient nature of novel memory-associated molecular products [5,6]. Although the memory trace can persist for years, these protein and RNA products are subject to half-lives on the order of minutes to hours [7]. Notably, the AMPA receptor, a vital regulator of synaptic strength and plasticity, is subject to a mere 30 h metabolic half-life [8]. Computational approaches therefore necessitate the existence of molecular markers that either evade macromolecular turnover or promote self-perpetuating signaling cascades. For instance, the calcium/calmodulin-dependent protein kinase and regulator of long-term memory and synaptic plasticity, CaMKII, undergoes activity-dependent autophosphorylation that persists in the absence of the initial activating stimulus (calcium influx into the cell) [9]. However, this example of short-term molecular memory is still subject to turnover and fails to account for long-term perpetuation of the memory trace.
The need for memory mechanisms that are impervious to molecular degradation has motivated a novel and burgeoning literature pertaining to epigenetic contributions to behavior. Traditionally, epigenetics has been defined as a set of stably heritable molecular phenotypes that do not affect the DNA sequence [10]. The term was precociously coined by developmental biologist, Conrad Waddington, who speculated that a layer of regulation upstream of the genome mediates gene–environment interactions which ultimately dictate the phenotype of the organism. Proposing a metaphorical ‘epigenetic landscape’, Waddington hypothesized these mechanisms help steer the increasing immutable cellular phenotype during development and differentiation. Epigenetics, therefore, would explain the diversity of cellular phenotypes produced and maintained in development despite nearly identical genomes. Displaying marked evolutionary insight, Waddington and his predecessors proposed that adaptive traits could appear in the absence of genetic mutation and that this phenotypic plasticity could become further rooted in genetic polymorphism [11].
In this article, we plan to summarize key findings in ‘neuroepigenetics’, specifically as it relates to lasting behavioral and cellular memory. We will begin with a brief overview of epigenetic molecular mechanisms and then discuss their relevance to brain physiology. We will then discuss how epigenetic mechanisms operate within the CNS during the formation and storage of memories. Next, we will review how specific epigenetic marks contribute to altered gene readout and discuss how these changes may be relevant for learning and memory. Finally, we will highlight key challenges in the field and offer speculations into future directions.
Molecular epigenetics
Traditionally, epigenetics has been defined as a set of heritable, stable molecular phenomena that modify gene expression and do not involve alterations to the DNA sequence of a cell. In light of the functionality of these same mechanisms in postmitotic cells including those in the brain, this definition has undergone considerable revision [12]. Here, we define molecular epigenetics or neuroepigenetic mechanisms as those which acutely or persistently modify transcription in cells, irrespective of their position in the cell cycle and which do not mutate the genome. This broader definition encompasses a number of distinct mechanisms, including those that have been co-opted by the nervous system for acute or persistent regulation of gene function in the absence of DNA sequence changes. Here, we will discuss the two canonical epigenetic mechanisms: post-translational modification of histone proteins and DNA methylation.
Chromatin: an epigenetic template
The diploid genome contained within a single cell spans 2 m of DNA. This requires several levels of organized compaction which allows the nucleus to envelope DNA and transcriptional machinery to access genes. This is achieved by chromatin, a complex structure consisting of DNA and closely apposed proteins known as histones. Four core proteins (H2A, H2B, H3 and H4) compose the histone octamer complex. Together with the 147 bp of surrounding DNA and linker DNA, this constitutes the nucleosome, the fundamental unit of chromatin. Functional epigenetic mechanisms acting on chromatin include posttranslational modifications of histones, covalent modification of DNA and chromatin remodeling.
Histone complexes comprise a globular domain and more loosely structured N-terminal tails [13]. These contain a large number of amino acid residues that are subject to covalent modifications including acetylation, phosphorylation, methylation, ubiquitination, sumoylation and ADP ribosylation. Histone writers include enzymes such as histone acetyltransferases (HATs) that catalyze addition of these moieties, although these vary in their target specificity. These modifications are subject to dynamic reversal by eraser enzyme complexes such as the distinct catalysts of histone deacetylation (histone deacetylases; HDACs) [14,15]. Histone modifications influence local gene expression through three mechanisms [16]. First, modifications affect the recruitment of transcription factors capable of activating or repressing gene transcription. Second, modifications to chromatin architecture directly influence binding of the machinery required for gene transcription. Finally, ATP-dependent complexes can be recruited to induce chromatin remodeling.
Histone acetylation, the best-documented transcriptionally permissive mark, occurs on the amino group nitrogen on lysine residues. Although initially positively charged due to protonation and closely juxtaposed with the anionic DNA backbone, the lysine residue becomes neutralized after acetylation because resonance stability of the resulting amide precludes protonation. This process leads to unraveling of nucleosome contacts and places chromatin in a permissive state. Moreover, readers containing bromodomains bind acetyl-lysine residues and can influence transcription directly or indirectly.
Histone phosphorylation predominantly occurs on serine and threonine residues and also influences chromatin tone by adding negative charge to the histone tail. This typically permissive mark is subject to dynamic regulation by kinases such as Aurora-B and IKKα and phosphatases such as protein phosphatase 1 (PP1) [17]. Histone phosphorylation plays a functional role in regulating immediate-early gene expression in vitro, and perhaps, also in chromosome dynamics during mitosis. Histone phosphorylation is especially intriguing in light of the broader function of phosphorylation in cellular signaling. This mechanism therefore may serve as a link between membrane-to-nuclear molecular cascades and chromatin dynamics.
Unlike acetylation and phosphorylation, histone methylation does not affect nucleosome charge and can play either a permissive or repressive function in gene expression. A number of histone methyltransferases (HMTs) and demethylases (HDMs) act as writers and erasers, respectively, and act on lysine and arginine substrates and regulate dynamic methylation [18]. Since multiple residues (e.g., – K4, K9, K27, K36 on histone H3) can each be modified with one, two or three methyl moieties, a high degree of combinatorial outcomes arise, thereby enabling complex modulation of transcriptional efficacy. Histone methylation marks are read directly by RNA polymerase II or by chromodomain-containing effectors. While H3K4 methylation is typically an activation mark, methylation at other residues is usually repressive. Likewise, the peptide modifications of histones including ubiquitination and sumoylation usually suppress transcription but appear to play ambiguous, context-dependent roles that are less well understood [19–21].
DNA methylation
Covalent modification of DNA canonically involves conversion of cytosines at CpG dinucleotides to 5-methylcytosine. These CpG sites occur at low frequency through the genome but also cluster into dense CpG islands, which are traditionally defined as DNA regions spanning at least 200 bp that possess an observed:expected CpG ratio of greater than 0.6. The latter are predominantly kept in a demethylated state and function in part to regulate local gene expression [22]. Although methylated promoter elements can either facilitate or inhibit expression, DNA methylation is usually associated with gene suppression and, as such, is thought to contribute to silencing of foreign genomic elements such as transposons and viral sequences [16,23]. Writers include de novo and maintenance isoforms of DNA methyltransferase (DNMT). Though previously assumed to be immutable due to the stability of the carbon–carbon bond, DNA methylation has been shown to undergo active reversal, although the identity and mechanism of action of these erasers is less well understood [24]. The methyl mark is read either through steric inhibition of transcription factor binding or through recruitment of reader proteins containing methyl-binding domains (MBDs). These either directly influence transcriptional efficiency or indirectly affect chromatin structure by recruiting HDACs [25].
Neural expression of the epigenetic machinery
The expression of several components of the epigenetic machinery discussed above in the nervous system drove interest into its then putative function in neurophysiology and behavior. For instance, a number of studies have documented a role for HDAC expression in neural development and function in the mature brain [26–28]. It is important to note that different homologs are expressed at different neurodevelopmental time points. For example, HDAC1 is largely confined to progenitor cells while HDAC2 is profoundly expressed postmitotically [29,30]. In addition, the histone methyltransferase complex GLP/G9a was shown to play a significant transcriptional role in forebrain cortical neurons [31].
More surprising was the finding of diverse DNMT expression in the developing and mature nervous system [32]. In this study, the reporter gene lacZ was fused to the promoters of the de novo methyltransferases DNMT3a or 3b to study temporal and spatial expression patterns. Immunohistochemistry revealed relatively acute embryonic expression of dnmt3b but broader expression of dnmt3a in the mature nervous system (including neurons and some glia) [32]. Intriguingly, although dnmt3a is expressed in neurons during the first three weeks of postnatal maturation, levels slowly decline in adulthood. This finding along with reports of attenuated DNA methylation in the aged brain suggests isoform-specific functional roles of DNMTs in development, maturation and aging in the CNS [33–35].
It is also surprising that DNMT1 was found to show robust expression into adulthood since this enzyme predominantly catalyzes maintenance of DNA methylation from hemimethylated DNA templates after cell division. Its role in the largely senescent brain is less clear, although studies indicate that it selectively silences reelin and gad1, genes associated with development/synaptic function and inhibitory interneuron physiology, respectively, in the cortex [36,37]. The fact that dnmt1 transcription negatively correlates with schizophrenia candidate gene expression implies a potential avenue of therapeutic intervention [38].
Although little is known about the function and molecular mechanisms underlying DNA demethylation, an important clue recently was uncovered in the hippocampus. A novel study utilizing a mammalian cDNA expression library and methylated reported constructs found that the growth arrest and DNA damage-inducible protein 45 (Gadd45) family contributes to active DNA demethylation [39], although this result is somewhat controversial. Another finding highlighted the expression of these genes in the dentate gyrus, a key anatomical region involved in downloading sensory information to the hippocampus [40]. In particular, the isoform Gadd45b exhibited robust upregulation in response to cell depolarization. Thus, Gadd45b is functionally associated with activity-regulated DNA demethylation and postnatal neurogenesis in the dentate gyrus.
Epigenetic contributions to neurophysiology
Neurons communicate by releasing chemical neurotransmitters which diffuse across synaptic clefts and activate receptors on the closely apposed neighboring cell varicosities. The target cells then undergo depolarization, the signature feature of neuronal activation, in the case of excitatory transmission. The sum of local electrical potentials passively affect somatic potential at the axon initial segment and, if threshold potential is reached, trigger an all-or-none burst of activity known as an action potential, the sine qua non of neural information processing [41]. The likelihood of spiking depends on the strength of postsynaptic responses to presynaptic activity. This parameter known as synaptic weight can undergo activity-dependent changes that persist for hours ex vivo. This phenomenon, long-term synaptic plasticity, is a well-characterized cellular correlate of long-term memory. A growing body of evidence implicates epigenetic mechanisms in synaptic function and plasticity and these are summarized in Table 1.
Table 1.
Summary of epigenetic regulation of synaptic function.
| Mechanism | Manipulation | Physiological phenotype |
|---|---|---|
| Histone acetylation | HDAC inhibition | Reduction in mEPSC frequency in mature neurons but enhances it in immature neurons Facilitates induction of LTF in Aplysia neurons Enhancement of LTP in the hippocampus and amygdala |
| CBP (histone acetyltransferase) inhibition or deletion |
Impairment of LTP in the hippocampus | |
| Histone methylation | Deletion of eed, a regulator of transcriptionally repressive histone methylation |
Augmention of LTP |
| Deletion of mll, a regulator of transcriptionally permissive histone methylation |
Impairment of LTP | |
| Histone ADP-ribosylation | Inhibition of PARP-1 | Impairment of LTP |
| DNA methylation | DNMT inhibition | Selective reduction in mEPSC frequency Reduction in spine density in nucleus accumbens |
| DNMT inhibition and genetic deletion of dnmt1 and dnmt3a |
Impairment of LTP | |
| Deletion or truncation of mecp2 | Reduction in paired-pulse ratio Increase in vesicle release probability Reduction in spontaneous neurotransmission Enhanced inhibitory tone |
|
| Genetic deletion of mbd1 | Impairment of LTP in the dentate gyrus | |
| Overexpression of DNMT3a | Increase spine density in nucleus accumbens | |
CBP: CREB-binding protein; DNMT: DNA methyltransferase; HDAC: Histone deacetylase; LTF: Long-term facilitation; LTP: Long-term potentiation; mEPSC: Miniature excitatory postsynaptic current.
Spontaneous neurotransmission
Although short-term synaptic function, unlike lasting plasticity, does not depend on gene expression changes, accumulating evidence points to a functional role of epigenetic signaling in baseline neurotransmission. An intriguing study highlights a functional role of DNA methylation in spontaneous miniature postsynaptic currents [42]. Pharmacological inhibition of DNMT activity in hippocampal cultures blocked excitatory (miniature excitatory postsynaptic currents; mEPSC) but not inhibitory (miniature inhibitory postsynaptic currents; mIPSC) neurotransmission, an effect that was sensitive to inhibition of transcription and presynaptic activity. One interpretation of these results is that glutamatergic pyramidal cells undergo activity-dependent changes in epigenetic signaling and gene expression and the resulting protein products modulate changes to cellular tone. The authors provide evidence for this model, showing that blockade of inhibitory transmission mimics the effect of DNMT inhibition on spontaneous activity. Further work is needed to support the notion that DNA methylation acts through modulation of homeostatic tone and to address the specificity of affected cell types.
If DNA methylation affects neuronal function through epigenetic control of gene expression, it is likely that inhibition of molecules that bind to methylated DNA would mimic the effect of DNMT blockade. This was demonstrated by a number of studies examining neurotransmission deficits in mecp2 mutants [43]. This gene is mutated in Rett Syndrome, a debilitating neurodevelopmental disorder associated with learning and memory deficits from a young age. Deletion of mecp2 results in deficits in paired-pulse facilitation, a form of short-term plasticity; this was documented in hippocampal slices from null mutants but only from symptomatic animals [44]. A second study utilizing a truncated allele reported enhanced evoked activity – measured upon direct stimulation of presynaptic fibers – and deficits in short-term plasticity. These results imply enhanced vesicle release probability in mutants, suggesting epigenetic contribution to presynaptic function. The specific site of action is unclear, but one experiment suggests selective effects on release kinetics but not size or number of spontaneously recycling vesicles [42]. Intriguingly, pretreatment of cultured slices with S-adenosylmethionine (SAM), the methyl-donor and substrate for DNMT, reversed deficits in spontaneous neurotransmission in mecp2 null mutants [42]. This effect was only seen after prolonged incubation, suggesting the synaptic phenotype is dependent on long-term homeostatic gene expression changes that are epigenetically regulated. Consistent with these results, overexpression of MeCP2 led to enhanced short-term plasticity [45].
Rett Syndrome is also associated with aberrant neuronal maturation and morphology in the cortex [46,47]. Accordingly, reduced spontaneous activity from excitatory cortical pyramidal cells was reported in mecp2 mutants in the absence of alterations in action potential threshold [48]. Both frequency and amplitude of the mEPSCs was affected, suggesting MeCP2 contributes to both presynaptic vesicle release and postsynaptic sensitivity to glutamatergic input. Enhanced inhibitory synaptic charge in mutants further suggests epigenetic mechanisms regulate excitation/inhibition balance and homeostasis in cortical networks.
As stated earlier, substantial crosstalk between DNA methylation and histone modifications has been reported. MeCP2 in particular is known to interact directly with HDAC1 and 2 through its transcriptional repressor domain, suggesting that DNA methylation at certain loci indirectly silences gene expression through chromatin modification [25,43,49]. One would predict that HDAC inhibition would affect synaptic function similarly to mecp2 ablation. Indeed, a selective reduction in mEPSC frequency was observed upon broad HDAC inhibitor treatment [50]. This effect was sensitive to inhibition of transcription, suggesting mediation through gene expression changes – mostly like repression rather than induction. Mecp2 knockout neurons were impervious to HDAC inhibition, which is consistent with the notion of MeCP2 and HDAC acting as co-repressors. Surprisingly, HDAC inhibition in immature neurons enhanced mEPSC frequency, presynaptic vesicle mobilization and synaptogenesis [51]. However, this study confirmed the previously reported effect in mature neurons, suggesting that HDACs form a developmental switch that affects synapse function in accordance with cell maturation. It is notable that the effect in mature neurons was mediated selectively by HDAC2 rather than HDAC1. This implies isoform-specific functionality of HDACs in neuronal function.
Long-term synaptic plasticity
Although epigenetic processes modify baseline synaptic function and short-term plasticity, their contribution to lasting behavioral changes are more readily explained by long-term changes in synaptic weight. Indeed a number of studies implicate these mechanisms in long-term potentiation (LTP). Histone acetylation is by far the best characterized epigenetic mechanism in long-term plasticity. One of the first such insights came from an in vitro study of pharmacologically induced plasticity at sensorimotor synapses from the sea slug Aplysia [52]. Acute application of the neurotransmitter, serotonin, produces short-term facilitation whereas repeated application induces long-term facilitation (LTF). Intriguingly, HDAC inhibition in the presence of acute serotonin led to LTF. In addition, although repeated application of the peptide FMRFa normally induces long-term depression (LTD, a persistent reduction in synaptic strength), co-application of an HDAC inhibitor promoted LTF instead, even though the drug alone did not produce plasticity. These results provide evidence for a role of histone acetylation in a dynamic, bidirectional switch that controls the strength and direction of synaptic plasticity.
More recent studies confirm that histone acetylation acts as a regulator of long-term synaptic dynamics in the mammalian brain. Declarative memory is largely rooted in the mammalian hippocampus and depends vitally on long-term synaptic plasticity during memory consolidation. One of the earliest studies to suggest a link between LTP and histone acetylation showed robust enhancement of LTP induction and late phase magnitude upon bath application of trichostatin A (TSA) and sodium butyrate, two structurally distinct HDAC inhibitors [53]. Important control studies confirmed the dependence of this phenotype on transcription and a lack of effect on baseline neurotransmission, short-term plasticity and NMDA receptor function. There are four chemically and structurally distinct classes of HDACs and these drugs target at least ten isoforms constituting class I and II HDACs [29,34]. It is unclear which enzymes specifically contribute to synaptic plasticity. A recent study, however, illustrates that although both are class I molecules, HDAC2 but not HDAC1 selectively impairs LTP magnitude [26]. While LTP magnitude can be manipulated with HDAC blockade, the ease of LTP induction can also be modified. For instance, TSA treatment produces transcription-dependent, lasting LTP in response to a stimulus train that normally only results in early phase potentiation [54]. In addition, the structurally unrelated mammalian class III deacetylase SIRT1 was recently shown to positively regulate hippocampal LTP. This finding appears contrary to the above results, but SIRT1 appears to act via noncoding RNA regulation rather than direct influence of target effector genes.
The finding that canonical HDAC activity impairs synaptic plasticity suggests that opposing HAT activity augments it. This was most clearly demonstrated in the context of CREB-binding protein (CBP), a well characterized HAT. For instance, CBP haploinsufficiency in mice, a model for Rubinstein–Taybi syndrome (RTS), leads to selective deficits in late-phase, gene expression-dependent LTP and this effect is reversed upon treatment with a nonselective HDAC inhibitor [55]. Expression of a dominant negative truncated form of CBP also selectively impaired LTP but not baseline neurotransmission [56]. However, while this manipulation affected the ease of LTP induction, a more robust LTP induction paradigm produced normal plasticity, suggesting the mutation specifically blocked the ease of induction rather than capacity for LTP expression.
The ability of other histone modifications such as histone methylation to modulate synaptic plasticity and LTP is less well understood, in part because of the diverse array of enzymes that modulate this reaction. However, one study uncovered a function of Eed and Mll, members of the polycomb and trithorax group proteins, in hippocampal LTP [57]. These proteins function in complexes that in part affect repressive and permissive histone methylation marks, respectively. Haploinsufficiency of eed selectively enhanced LTP magnitude only after 30 minutes while mutation of mll produced a late-phase deficit. The temporal selectivity of these phenotypes is consistent with altered epigenetic regulation of effector gene expression. Likewise, ADP-ribosylation of histones, a much less well understood epigenetic mechanism, has also been implicated in synaptic plasticity. Inhibition of polyADP-ribose polymerase 1 (PARP-1) selectively impaired hippocampal LTP without affecting baseline function [58].
Studies have demonstrated that chromatin regulation of LTP is not specific to the hippocampus. For instance, a study shows that TSA treatment augments LTP in the amygdala [59]. However, this effect may be due more to acetylation of a transcription factor rather than histone tails. Further evidence comes from reports of epigenetic regulation of neuronal plasticity in the visual cortex. In one study, TSA treatment in vivo was shown to promote ocular dominance plasticity in a monocular deprivation paradigm [60]. Taken together, these results implicate chromatin modifications in neural plasticity across varying modalities.
The hypothesis that DNA methylation influences the behavior of the organism predicts similar regulation of synaptic plasticity. Both writers and readers of DNA methylation have been functionally linked to LTP. In hippocampal slice experiments, for example, bath application of two different DNMT inhibitors blocked both early and late phases of LTP [61]. Though epigenetic mechanisms influence cell phenotype by regulating transcription which normally only affects late LTP, it is not uncommon that manipulations of gene expression can also affect early phase LTP. An interesting follow-up result found that co-application of TSA reversed the effect of DNMT inhibition, verifying the importance of horizontal epigenetic crosstalk in synaptic plasticity [62]. Since these pharmacological agents vary in specificity and toxicity and are thought to require DNA synthesis for their action, a further study employed a genetic manipulation approach to verify these results [63]. When the maintenance and de novo isoforms, DNMT1 and 3a respectively, were both removed from forebrain neurons, a profound selective deficit in late-phase LTP was found in the absence of a baseline physiological phenotype. Only a minor reduction in hippocampal volume and normal cell count were reported in mutants, suggesting a selective effect on synaptic plasticity. In addition, enhanced LTD found in mutants implies that DNA methylation controls both direction and magnitude of late phase plasticity.
Readers of DNA methylation similarly contribute to dynamic regulation of synapse strength. Genetic deletion of the DNA-binding protein and silencer of gene expression, MBD1, impaired LTP in the dentate gyrus, a part of the hippocampus largely implicated in spatial memory [64]. Similarly, truncation or deletion of MeCP2 impairs LTP expression [44,65]. In addition, null mutants also exhibit attenuated LTD. These findings are consistent with the synaptic phenotype of the dnmt null mutants, again suggesting bidirectional regulation of long-term plasticity by DNA methylation.
Epigenetic regulation of memory
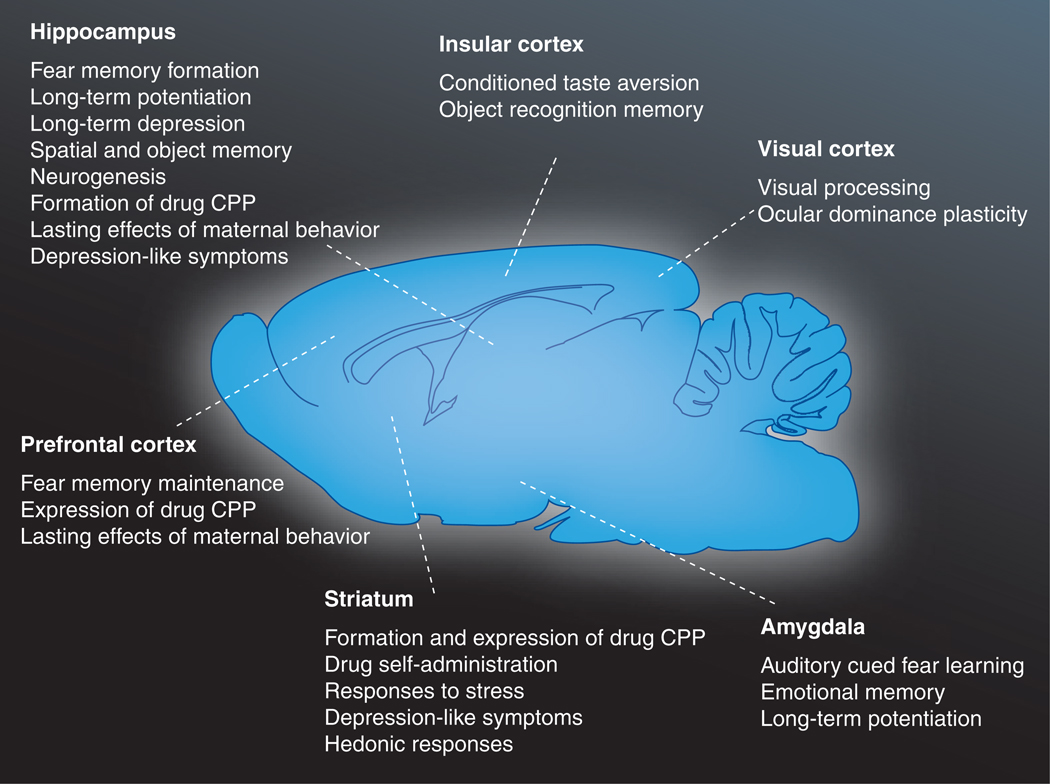
Epigenetic mechanisms in a several brain regions contribute to stable changes in behavior (see Figure 1 for summary), including learning and memory, drug addiction, depression and long-term responses to maternal care [66–71]. Here, we highlight the contributions of histone modifications and DNA methylation to memory dependent on several different areas. First, we will discuss hippocampus- and cortex-dependent phases of associative memory, as these anatomical regions are thought primarily to subserve the consolidation and storage of memory, respectively. Next, we will discuss the novel findings that chromatin dynamics regulate reward learning and addiction, mechanisms that rely on signaling in the striatum.
Figure 1. Functions known to be regulated by epigenetic mechanisms across brain regions.
Epigenetic mechanisms occur in a range of brain regions and regulate cell physiology and behavior associated with those anatomical areas (see text for details). It is important to note that certain memory tasks are associated with multiple regions, which may reflect temporally and spatially distinct phases of memory such as acquisition, consolidation and storage. Epigenetic mechanisms such as histone acetylation, in particular, appear to play a common role in many different memory modalities. However, other epigenetic marks such as DNA methylation have not received as much attention. Future work is needed to explore the diverse functionality of these mechanisms in multiple phases and categories of memory. Please note that these lists are not meant to be exhaustive and do not indicate that all of these functions are controlled by the same epigenetic marks in the same fashion.
CPP: Conditioned place preference.
Histone acetylation & hippocampus-dependent memory
A number of molecular memory mechanisms are phylogenetically conserved in mammals and epigenetic mechanisms have similarly been maintained throughout evolution [72]. For example, in accordance with the function of histone acetylation in synaptic plasticity in Aplysia, a recent finding points to similar regulation of memory in the invertebrate Chasmagnathus [73]. In measuring habituation in an associative contextual task, the authors confirmed enhanced memory retention in response to HDAC inhibition only when administered within either of two temporally distinct consolidation windows. In mammals, the hippocampus largely regulates contextual memory consolidation and, as such, as been the focus of studies of epigenetic regulation of spatial and contextual memory. Several results have highlighted the effect of HDAC inhibition on the enhancement of contextual memory in associative fear conditioning, spatial memory in the Morris water maze (MWM) and object recognition in wild-type rodents [53,54,74]. As with synaptic plasticity, these effects appear to be selectively regulated by specific HDAC isoforms. In particular, HDAC2 plays a critical function in these memory tasks, as over expression and deletion impaired or exaggerated hippocampus-dependent memory, respectively [26]. These studies illustrate congruent effects of HDAC manipulation on synaptic function and behavior. These findings are consistent with a model proposing that epigenetic regulation of histones affects memory through modulation of the neuron’s plasticity phenotype.
As with synaptic plasticity, the best characterization of chromatin regulation in mammalian behavior comes from a number of studies investigating the consequences CBP mutations. Heterozygous ablation of cbp, for instance, selectively impaired long-term memory in fear conditioning and novel object recognition tasks [55]. In the former, the animal learns to associate a shock with a novel context (contextual component) and with an auditory tone (cued component). Mutants showed deficits in both of these tests, suggesting contributions from both hippocampus and amygdala, a part of the limbic system that regulates emotional behavior. In the novel object recognition task, the animal is initially exposed to two identical objects during training and then re-exposed to one familiar object and one novel object. Enhanced time interacting with the novel object is used to index memory. In both tasks, a deficit was found 24 h rather than 1 h after training. Intact short-term memory, which does not depend on transcription, is consistent with an epigenetic interpretation of these results. Still, since CBP plays direct roles in gene expression aside from its function as a HAT, an additional study created an inducible dominant negative transgenic lacking only the HAT domain of CBP [75]. Again, selective late-phase memory deficits were found in two object recognition tasks and the MWM. HDAC inhibition was found to reverse the recognition memory defect, consistent with the selective manipulation of histone acetylation. Surprisingly, no fear conditioning phenotype was found. These results suggest that while CBP regulates memory in a number of tasks, its epigenetic function may act more selectively in behavior.
The CREB dependence of CBP HAT function and temporal specificity in memory remains controversial. A recent study, for instance, shows that HDAC inhibition not only augments the consolidation of object recognition memory but also converts weak memory traces that normally dissipate into more robust, persistent memories [74]. These findings illustrate a role for histone acetylation in both the strength and induction threshold for lasting memory. HDAC inhibition further rescued memory deficits in mice expressing a form of CBP with a mutated CREB-binding domain. CBP acts as a transcriptional co-activator with CREB, a vital transcription factor in dynamic neuronal function [4,76]. Highlighting the dependence of the behavioral and synaptic function of CBP on intact CREB function, a study confirmed that creb mutants exhibiting fear memory and LTP deficits were resistant to rescue by HDAC inhibition [54]. The varying severity of mutations in these studies may help explain this disparity.
Manipulating the balance between acetylation and deacetylation therefore appears robustly to control the strength and uniquely the long-term duration of the memory trace. However, the absence of a short-term memory phenotype conflicts with another recent result. The authors produced a selective knockout of cbp in excitatory forebrain neurons and uncovered both short- and long-term deficits in contextual fear memory and object recognition [77]. More surprisingly, even chronic HDAC inhibitor treatment failed to rescue behavioral deficits in mutants despite a clear enhancement in wild-types. The authors suggest that residual HAT activity in the other cbp mutants may have been required for HDAC inhibition to function. These results are surprising since remaining HATs beside CBP were unable to compensate for the loss of CBP function, highlighting the critical and unique role of CBP in neuronal function. In particular, the HAT and CBP homolog p300/PCAF fails to compensate for CBP dysfunction. For instance, selective long-term deficits in spatial and contextual fear memory were found in mice with mutated p300 HAT domain [78]. These results mimic deficits seen with CBP mutants despite the high degree of sequence similarity between the two proteins. Differential association with transcriptional regulators and differences in target substrates may help explain these distinct roles [79,80]. Indeed, divergent functions were reported in motor learning, a cerebellum-dependent phenomenon [81]. It is possible that functions of CBP outside of the epigenetic regulation of chromatin may account for its additional role in short-term memory function. Since short-term memory depends more on cortical processing than hippocampal function, further studies are needed to address its anatomical resolution [82].
It is becoming increasingly evident that manipulation of histone acetylation patterns through HDAC inhibition may become a promising avenue of clinical therapeutics in neurodegenerative disease states. For example, behavioral rescue of memory impairments in Alzheimer’s disease and aging-associated cognitive dysfunction by HDAC inhibition have both been reported [83,84]. Epigenetic dysregulation and therapeutic implications in neurological disease states have been extensively reviewed elsewhere [21,34,85–90].
Histone acetylation & hippocampus-independent associative memory
Though mostly studied in hippocampus-dependent memory, histone acetylation in other brain regions similarly regulates memory processing. The cortex, for instance, regulates short-term working memory which is impaired in mice with genetic deletion of p300, although it is unclear if this effect is entirely due to its HAT function [91,92]. Long-term memory became impaired in mutants with age, but it is unclear whether this effect is a due to hippocampus- or c ortex-dependent mechanisms [92].
The cortex also functions in memory storage and epigenetic mechanisms may also regulate long-term cortical memory. A recent study, for instance, confirms a double dissociation between the hippocampus and insular cortex in related tasks measuring memory for object location and recognition, respectively [93]. HDAC inhibition improved memory only when infused into the respective brain region during the memory consolidation window. The study also highlights the influence of glucocorticoid receptor activation in this memory enhancing effect. Though glucocorticoid treatment augmented both forms of memory, no rescue of object location memory was found in cbp mutants, suggesting differential molecular functions of CBP in the hippocampus and insular cortex. Histone acetylation has also been implicated in insular cortex-dependent taste aversion and visual cortex-dependent recovery of visual acuity after monocular deprivation [94,95]. Further studies are needed to address the likely broader function of cortical histone modifications in postnatal behavior.
Additional histone modifications & associative memory
Histone phosphorylation is particularly intriguing because it provides a unique molecular signature that functions as a target of intracellular signaling cascades and a regulator of chromatin dynamics [21]. Recent studies have examined the function of the mitogen- and stress-activated protein kinase 1 (MSK1) in these signaling pathways and behavioral function. A germline knockout showed selective deficits in long-term spatial and contextual but not cued fear memory [96]. In accordance with findings from cbp mutants, HDAC inhibition failed to rescue this deficit in msk1 knockouts, suggesting a critical role for crosstalk between histone acetylation and phosphorylation or common upstream regulators of these modifications. Similarly, msk1/2 double knockouts displayed memory impairments in a forced swim memory paradigm, a task thought to be especially dependent on the dentate gyrus, a subfield of the hippocampus [97,98]. In addition to MSK, the α isoform of IκB kinase (IKK) complex, a known regulator of NF-κB disinhibition, was shown to regulate H3 phosphorylation in the hippocampus [99]. Inhibition of IKKα blocked contextual fear memory reconsolidation, a phenomenon in which memories become labile and subsequently strengthened following reactivation. Highlighting the evolutionary significance of this pathway, an earlier study identified a similar effect on memory consolidation in Chasmagnathus [100].
Taken together, these studies point to a critical function of histone kinases in memory formation. Future studies are needed to address the function of other kinases such as Aurora-B and Rsk-2 in memory-associated histone phosphorylation [101]. In addition, little is known about the role of dephosphorylation in memory. A recent study, however, shows PP1, a known memory suppressor, also acts on histones [102]. However, the pleitropic functionality of a number of these enzymes necessitates additional approaches to manipulate histone phosphorylation more selectively. For instance, PP1 inhibition produced alterations in histone acetylation and methylation in addition to phosphorylation. It is unclear whether these modifications results from epigenetic crosstalk specifically initiated by hyperphosphorylation or indirect effects of PP1 inhibition on other enzymes and transcription factors.
Recent studies have also uncovered a role for ADP-ribosylation of histones in behavioral memory. PARPs catalyze the addition of negatively charged ADP polymers which affect local chromatin architecture directly and the binding of transcription factors and chromatin remodeling complexes. PARP-1 was first discovered selectively to regulate long-term memory formation in an operant feeding paradigm in Aplysia [103]. Two recent studies confirm a conserved role of PARP-1 in mammalian memory formation. Intraventricular infusion or systemic injection of PARP-1 inhibitors impaired long-term performance in object recognition and place avoidance tasks [58,104]. Further investigations are needed into the role of additional histone modifications such as ubiquitination and sumoylation.
DNA methylation in memory formation & storage
Although most investigations into molecular epigenetic regulation of memory have focused on covalent histone modifications, a more recent and growing body of literature similarly implicates DNA methylation in mammalian performance in behavioral tasks. Observations from mutations in reader proteins provided early insights into a causative role of DNA methylation in memory. Mice lacking mbd1, for instance, showed deficits in spatial memory in the MWM [64]. Likewise, mecp2 truncation mutants showed poor performance in hippocampus-dependent spatial, contextual and social recognition memory paradigms [65]. These behavioral deficits mimic behavioral dysfunction in Rett syndrome patients, suggesting a conserved function of MeCP2 in mammals.
More direct evidence for a functional role of DNA methylation in behavior came from pharmacological and genetic manipulations of DNMTs. Infusions of the DNMT inhibitors zebularine or 5-azacytidine into the CA1 subfield of the hippocampus prior to or immediately after training impaired long-term performance in contextual fear conditioning [62,105,106]. Still, the lack of specificity, associated toxicity and requirement for DNA synthesis for the function of these agents called into question the conclusion that DNMTs specifically regulate long-term behavior [107]. A number of control experiments, however, have addressed these concerns. First, no lasting impairment in anterograde memory was found since repeated training produced robust memory in rats that had been treated with the inhibitor [106]. Second, infusion of a structurally unrelated competitive inhibitor, RG108, similarly impaired memory retention [105]. In addition, pretraining infusion of an HDAC inhibitor completely rescued the memory deficit [62]. This result provides evidence that histone acetylation and DNA methylation play complementary, permissive roles in memory and synaptic plasticity. These studies, however, failed to account for potentially off-target effects of DNMT inhibitors. This was addressed by genetic manipulation of dnmt genes. When both dnmt3a and dnmt1 were deleted in postnatal forebrain excitatory neurons, mutants displayed impaired long-term spatial and contextual memory [63]. No effects on hippocampal morphology or cell count were observed and only a minor reduction in volume was reported, suggesting a specific role in adult function. Taken together, these studies strongly implicate DNMT function in adult memory consolidation. Further investigations should use isoform-specific knockdown with RNA interference in a region-specific manner in adults to avoid completely any nonspecific effects that may have been missed in these studies.
The storage of memory is largely distinct from consolidation both anatomically and mechanistically. While these studies established the role of DNMTs in hippocampus-dependent memory formation, they fail to demonstrate the speculative ability of DNA methylation to regulate the long-term persistence of consolidated memory. It has been reported that remote contextual fear memory is largely dependent on the anterior cingulate cortex, a subregion of the dorsomedial prefrontal cortex [108]. A recent study capitalizing on these findings demonstrated a similar role of DNMTs in memory maintenance [109]. Intra-cortical infusion of distinct DNMT antagonists 29 days after training impaired memory retention. Importantly, the treatment failed to produce deficits if given 1 day after training, a period that precedes the time when memory is downloaded to the cortex. These results are the first to demonstrate that DNMTs regulate not only the initial formation of memory but also its perpetuation.
Aberrant DNA methylation has been reported in CNS disorders, but its potential manipulation for therapeutic use is still poorly understood. A small but growing set of studies, however, suggest functional changes in DNA methylation at pathologically dysregulated loci contribute to the Alzheimer’s disease phenotype and that these changes are promising targets of intervention. Indeed, manipulations of SAM, the substrate for DNA methylation reactions, epigenetically regulated expression of ps1 and bace in cell culture [110–112]. These enzymes catalyze processing of amyloid precursor protein (APP) and subsequent release of Ab, a hallmark pathological feature of Alzheimer’s disease. Enhancing DNA methylation attenuated Aβ load, suggesting molecules that enhance single-carbon transfer reactions may impede disease progression. A recent study confirms that folic acid treatment, a nonspecific way to enhance DNA methylation, in Alzheimer’s disease model mice partially augments the therapeutic effects of memantine in a memory task and suppresses neuronal toxicity [113]. In the same mouse model, B vitamin (including folate) deficiency was associated with site-specific demethylation near the ps1 transcriptional start site, suggesting that systemic manipulation of methyl-donor levels may promote cognitive rescue at least in part through epigenetic regulation of disease-associated loci.
While such interventions have not been attempted in patients, reduced DNA methylation in Alzheimer’s disease has been reported. Hypomethylation was confirmed in cortical postmortem tissue from patients and was exaggerated in neurons with characteristic pathological features [114]. It has yet to be determined which genes are most affected by aberrant DNA methylation, but it appears that aberrant epigenetic regulation of transcription in Alzheimer’s disease is highly locus-specific and associated with increased interindividual variability [115,116]. Consistent with the in vitro studies is the reportedly low systemic folate and high homocysteine, a metabolite of the DNA methylation reaction, in patients [117]. Collectively these results provide a case for DNA methylation as a target of therapeutic intervention in Alzheimer’s disease and perhaps more broadly in aging-associated cognitive loss. Future work may focus on developing means to enhance DNA methylation or perhaps inhibit demethylation selectively in the brain.
Epigenetic regulation of reward-related behaviors
Control of learned & unlearned responses to drugs of abuse
Exposure to drugs of abuse produces long-lasting structural and functional changes in brain reward circuits including the nucleus accumbens and ventral tegmental area [118–120]. These changes are believed to be mediated by equally long-lasting changes in expression of genes such as ΔFosB and creb [120]. A number of recent reports have also revealed that epigenetic mechanisms are critical for both learned and unlearned responses to drugs of abuse. The first of these studies focused on histone modifications and revealed that histone acetylation is a critical regulator of conditioned place preference (CPP) for cocaine. For example, treatment with an HDAC inhibitor prior to cocaine or morphine exposure results in an increased locomotor response to these drugs and boosts the formation of preferences for places associated with drug delivery [121–123]. Likewise, over-expression of HDAC4 in the nucleus accumbens results in a blunted conditioned place preference for cocaine and a decrease in the motivation to seek cocaine [121,124]. Similarly, overexpression of HDAC5 in the nucleus accumbens impairs the development of conditioned place preferences for cocaine, whereas treatment with TSA or genetic deletion of hdac5 results in enhanced drug CPP [123]. In addition, antagonism of a distinct class of HDACs (the sirtuins) within the nucleus accumbens impairs cocaine CPP and decreases lever press responses for intravenous cocaine [125]. Interestingly, histone acetylation can also play an important role in the reversal of drug-association memories, as HDAC inhibitors can facilitate the extinction of cocaine place preference [93].
In addition to histone acetylation, histone phosphorylation and methylation have both been implicated in behavioral responses to drugs of abuse. For example, viral knockdown of the histone dimethyltransferase G9a within the nucleus accumbens produces an increase in cocaine place preference, whereas the increased spine density caused by repeated cocaine administration was blocked by overexpression of G9a in the nucleus accumbens [126]. Moreover, specific ablation of methyltransferases, glp or g9a, in adult forebrain neurons (including the striatum) resulted in a number of deficits, including impaired sucrose preference, impaired fear conditioning and impaired exploratory behavior [31]. Conversely, increased histone phosphorylation is also necessary for a number of reward-related behaviors. Indeed, blocking the upstream signaling pathways that result in phosphorylation at serine 10 on H3 diminishes behavioral sensitization to both cocaine and morphine and blocks conditioned place preference for cocaine [127].
Increasing evidence indicates that DNA methylation also plays a critical role in the regulation of drug-related behavior. For example, knockdown of the methyl binding protein MeCP2 in the dorsal striatum prevents the escalated increase in cocaine intake observed in animals under extended access conditions and flattens the dose-response curve for cocaine [128]. Furthermore, knockdown of MeCP2 in the nucleus accumbens boosts the locomotor activating effects of amphetamine but decreases amphetamine-induced CPP [129]. Likewise, mice with a hypomorphic mutation in mecp2 (resulting in deletion of the C-terminus) also possess exaggerated locomotor responses to amphetamine and impaired amphetamine CPP [129]. Importantly, mecp2 mutant mice also fail to show increased spine density in the NAc in response to chronic amphetamine treatment [129]. Taken together, these results suggest that DNA methylation within the striatum may regulate responses to drugs of abuse. Consistent with this hypothesis, a recent study revealed that inhibition of DNA methyltransferase activity using zebularine significantly impaired the induction of locomotor sensitization following seven days of cocaine treatment [130]. Moreover, site-specific inhibition of DNMT activity with RG-108 boosts the development of cocaine CPP, whereas overexpression of the DNMT3a isoform within the nucleus accumbens impairs cocaine place preference while mimicking cocaine’s effects on spine density [119]. Furthermore, DNA methylation within the hippocampus and prelimbic cortex is also necessary for the formation and expression of cocaine place preferences, respectively, indicating that epigenetic changes in brain regions outside of the striatum are also key regulators of drug memories [126].
Control of responses to stress, depression & natural rewards
In addition to drug-related behaviors, burgeoning literature has also revealed that epigenetic patterns within the striatum are important for responses to stressful situations and depression. Thus, mice lacking hdac5 exhibit exaggerated responses to social defeat stress, including an avoidance of novel mice and stress-induced anhedonia [123]. In contrast, treatment with two different HDAC inhibitors in another report essentially rescued the deficits produced by chronic social defeat stress on social interaction, anhedonia and immobility in the forced swim test [125]. These behavioral effects are very similar to those produced by antidepressants such as fluoxetine and indicate that HDAC inhibitors may be useful in treatment as antidepressant drugs. Consistent with the above findings, DNA methylation also regulates stress responses. Thus, overexpression of DNMT3a produced prodepressive responses on acute defeat stress and forced swim tests, whereas DNMT inhibition reversed social avoidance phenotype following chronic social defeat in a manner similar to the antidepressant fluoxetine [119].
These data suggest that epigenetic regulation of gene expression patterns may be a central component in long-term responses to many different rewarding and stressful experiences. However, very little is known about whether learned experiences surrounding natural rewards like food, water and social interaction are capable of producing epigenetic alterations in the same way that drugs of abuse can. Indeed, although natural reward learning can activate the same biochemical pathways that lead to the epigenetic modifications described above [127,131,132], it is unclear if drugs of abuse and stress are uniquely capable of altering histone marks or DNA methylation patterns. This question is especially relevant given that these modifications presumably evolved to support the formation or storage of associations between actions or stimuli and rewards in the environment. However, it is also possible that drugs of abuse usurp these mechanisms in a way that allows drug memories to become especially potent and induce drug relapse after extended drug abstinence. Therefore, future studies will be required to enable comparisons between the epigenetic modifications induced by learning for natural rewards and learning for drug rewards.
Epigenetic signatures in neural function: upstream mechanisms & effectors
The above studies raise several important questions regarding epigenetic control of neural function and behavior. Are manipulations of epigenetic readers and erasers associated with changes in epigenetic marks? If so, do these span the genome broadly or affect selective gene expression? To what extent do these marks act cooperatively? Do they synergize to affect transcription or act in an additive manner? Do epigenetic signatures in memory formation differ from those associated with nonspecific depolarization? Are different marks associated with different brain regions or specific cell types? These questions have motivated numerous investigations into activity- and experience-dependent epigenetic mechanisms in the brain. Although further research into these problems is necessary, our current understanding suggests that anatomical-, locus- and experience-specific marks functionally regulate synaptic physiology and memory formation. These mechanisms and downstream effectors are outlined in Table 2.
Table 2.
Summary of epigenetic processes in memory and memory-associated transcription.
| Mechanism | Findings |
|---|---|
| Histone acetylation | HDAC inhibition promotes associative memory consolidation and H3 acetylation in Chasmagnathus HDAC activity is negatively associated with hippocampus (especially HDAC2) and cortex-dependent memory in mammals and may operate through induction of Nr4a1 and Nr4a2 transcription factors Contextual fear conditioning promotes H3 acetylation while latent inhibition training induces H4 acetylation Forced swim training induces H3K14 acetylation hdac2 deletion enhanced H4 and H2B acetylation as well as numerous targets of CREB-dependent transcription CBP inhibition produces memory deficits that vary by the severity and selectivity of the manipulation Overexpression of HDACs in the nucleus accumbens impairs conditioned place preference for cocaine; HDAC inhibitors increase cocaine place preference |
| Histone phosphorylation |
msk1 deletion impaired spatial and contextual memory and deletion of msk1 and 2 impaired forced swim memory. These tasks are associated with H3S10 phosphorylation and H3K14 acetylation |
| Histone methylation | Contextual fear conditioning produces enhancements in H3K4me3 and H3K9me2 marks and this appears to regulate genes including zif268 and bdnf Overexpression of the histone methyltransferase G9a in the nucleus accumbens impairs conditioned place preference for cocaine; viral knockdown of G9a increases cocaine place preference |
| Histone ADP-ribosylation | PARP-1 inhibition blocked memory consolidation in Aplysia and rodents and memory formation led to ADP ribosylation of H1 |
| DNA methylation | Deletion of mbd1 and mecp2 impair long-term memory in rodents Inhibition in the hippocampus or genetic ablation of DNMTs selectively impairs long-term memory consolidation DNMT inhibition in the cortex impairs remote contextual fear memory storage DNMT inhibition or knockout in the nucleus accumbens boosts development of conditioned place preference for cocaine, whereas overexpression of DNMT3a in the nucleus accumbens reduces cocaine place preference DNMT inhibition in the hippocampus prevents development of conditioned placed preference for cocaine, whereas DNMT inhibition in the prefrontal cortex prevents expression of cocaine place preference after learning Fear conditioning produces dynamic alterations in DNA methylation at loci including reelin pp1β, bdnf and zif268 in the hippocampus during memory consolidation Remote memory storage is associated with lasting hypermethylation and downregulation of calcineurin in the anterior cingulate cortex |
CBP: CREB-binding protein; DNMT: DNA methyltransferase; HDAC: Histone deacetylase.
Insights from invertebrates
A pioneering study on epigenetic regulation of synaptic plasticity in cultured Aplysia neurons confirmed specific histone acetylation patterns in response to activity. Serotonin application promoted acetylation of histone H3 and H4 on lysines 14 and 8, respectively, in the promoter of c/ebp, an immediate-early gene positively associated with memory function [52]. This effect was correlated with release of CREB2, a repressive transcription factor. Consequently, FMRFa application induced HDAC5 recruitment, histone deacetylation and CREB2 binding. These findings suggest bidirectional regulation of synaptic plasticity is functionally dependent on epigenetic regulation of memory-associated genes. Likewise, in Chasmagnathus, HDAC inhibition in vivo and training both increased acetyl H3 signal in the central brain [73]. Critically, HDAC inhibition facilitated acetylation in trained crabs too. Unlike the Aplysia study, this result shows a global increase in acetylation. This may represent a net effect of different site-specific modifications. In addition, in the mollusk Helix lucorum, taste aversion training augmented H3 acetylation specifically in command neurons and this effect was sensitive to inhibition of the signaling protein MEK [133]. Finally, ADP-ribosylation of the linker histone H1 and activation of PARP-1 were associated with long-term facilitation and associative memory in Aplysia [103].
Chromatin marks in mammalian memory
These and other epigenetic signatures appear to be conserved in the context of memory and synaptic plasticity in the mammalian brain. For instance, novel object recognition training induced PARP-1 activity in the hippocampus and cerebral cortex [104]. Similar results were found with high-frequency stimulation of hippocampal slices and these results were associated with increased polyADP-ribose (PAR) polymer formation [58]. Histone variant H1 is a target of polymerization and subsequent clearance. Accordingly, PARP-1 function led to recruitment of transcription factors NF-κB and CREB and RNA polymerase II to immediate-early gene promoters. This may functionally depend on histone acetylation, as PARP-1 was shown in vitro to promote neurotrophin-mediated histone H4 acetylation [134]. Intriguingly, PARP-1 itself is a target of PAR modification and interacts with phosphorylated ERK, a key upstream regulatory factor activated by MEK, in neuronal plasticity function, in a positive feedback mechanism. This mechanism could account for sustained activity of PARP-1 and possibly implicate it in lasting memory storage in addition to its f unction in consolidation.
Neuronal activation in vivo has been functionally associated with transient and lasting histone acetylation. Acute electroconvulsive shock, for instance, produced short-term H4 hyperacetylation in the promoter of c-fos and immediate early gene, while chronic shock led to sustained hypoacetylation [135]. Similar dissociative patterns in creb and bdnf were found, suggesting that the pattern of cellular input induces site- and duration-specific modifications in the hippocampus that correlate with transcription rate. In a model of status epilepticus, a prolonged seizure state, H4 hypoacetylation and downregulation was associated with glur2, which encodes an AMPA receptor subunit [136]. Likewise, H4 acetylation and H3 phosphorylation and upregulation of c-fos and c-jun were also reported [137]. These effects were dependent on the HAT function of CBP, highlighting its crucial in vivo role in transcriptome plasticity.
Given that activity-dependent epigenetic modifications in the hippocampus are input-specific, it is not surprising that memory function is similarly regulated. Contextual fear training selectively enhanced H3 acetylation while latent inhibition, a variant in which prior exposure to the context causes the animal to diassociate the context and shock, affected only H4 acetylation [53]. This result argues that epigenetic modulation of gene expression is tuned to behavior-specific patterns of activity. In addition, upstream signaling is also regulated in a selective fashion. Contextual training depends on the NMDA receptor and downstream MAP kinase activation, which results in H3 acetylation. A different mechanism may underlie H4 acetylation and this is less well understood. Intriguingly, a selective mark, H4K12 acetylation, is attenuated after fear memory training in aged mice [84]. This study suggests aging-related cognitive decline is associated with a specifically disrupted histone acetylation/ deacetylation balance. As with hippocampus-dependent memory, gustatory memory depends on MAPK activation and histone and cytosolic protein acetylation [94].
Likewise, histone acetylation marks at important plasticity genes appear to play a critical role in responses to drugs of abuse. For example, studies using chromatin immunoprecipitation have revealed that acute cocaine experience produces a robust but transient acetylation at histone H4 and phosphoacetylation of H3 at the promoter region for the c-Fos gene [121]. Likewise, chronic cocaine administration results in increased acetylation of H3 at the bdnf and cdk5 genes in the striatum [121]. Subsequent studies using microarray technology revealed that cocaine experience produces a number of changes in histone modifications that are linked to altered transcription of hundreds of gene targets [123]. Interestingly, many of these changes were controlled by a specific striatally-enriched HDAC (HDAC5), which is dynamically shuttled in and out of the cell nucleus following cocaine exposure.
Mechanisms of the establishment and role of histone acetylation are becoming clearer, but some results remain ambiguous. cbp haploinsufficiency resulted in H2B hypoacetylation while forebrain deletion of cbp produced H3 and H2B hypoacetylation and both were reversible by HDAC inhibition [55,77]. Early-response gene induction was normal in the former case while reductions in transcripts pertaining to the calcium-dependent CaM kinase signaling and glutamate receptor signaling proteins were downregulated in the latter. These results may reflect differential effects by cell type or variable extent of CBP dysfunction. It is unclear how much is truly due to impaired HAT activity. In the amygdala, fear conditioning spurs HAT activity and interaction between NF-κB and both CBP and HDAC3 [59]. Still, the relative contributions of acetylation of histone and non-histone proteins as well as target gene loci remain uncertain. It is also unclear whether MAPK signaling also mediates experience-dependent epigenetic effects in amygdala.
Studies of locus-specific histone modifications uncover a number of target genes implicated in memory. HDAC2 removal was linked to global increases in H4K12, H4K5 and H2B acetylation and enhanced expression of CREB-CBP pathway target genes such as zif268/egr-1 and bdnf and other genes implicated in synaptic remodeling and plasticity [26,138–140]. Also of note, HDAC2 enrichment was found at the promoter of pkmζ, a critical gene in memory maintenance [141]. Memory enhancement by HDAC inhibiton is thought to act through multiple gene targets such as these, but a recent study posits a crucial, specific dependence of this effect on CREB-mediated induction of the transcription factors, Nr4a1/nurr77/NGFI-B and Nr4a2 [54].
Most of these studies employ training paradigms that incorporate emotionally arousing experiences. Glucocorticoids, which are known to mediate such memories, were recently shown to regulate the memory-enhancing effects of HDAC inhibition [93]. Corticosterone enhanced H3K14 acetylation in a manner dependent on protein kinase A (PKA), a cystosolic mediator of membrane-to-nucleus signaling. Since glucocorticoids influence stress responses, it is not surprising that stress-related memory formation is associated with similar epigenetic marks. Forced swimming training induced the permissive marks, H3S10 phosphorylation and H3K14 acetylation in the dentate gyrus and these are mediated by the NMDA receptor, MEK and MSK1/2 [97]. In addition, both the glucocorticoid and NMDA receptors are necessary for memory- and novelty-associated phosphoacetylation and c-fos induction [98]. Downstream effectors mediating epigenetic modifications include MSK itself and Elk-1 binding to serum response elements and recruitment of CBP and p300. In addition, immobilization stress epigenetically suppresses transcript-specific bdnf expression [142]. As with stress-related memory, learning in MWM and fear conditioning is associated with ERK-dependent phosphoacetylation in the hippocampus [98,143]. MSK1 appears to play a critical role in this epigenetic tag during fear memory consolidation, as msk1 null mutants showed impaired learning-induced CREB phosphorylation and phosphoacetylation [96]. Although NMDAR and glucocorticoid receptor-mediated signaling play a significant role in memory-associated chromatin modifications, the in vivo function of neuromodulators is less well understood. However, dopamine, acetylcholine and glutamate have been shown to regulate phosphoacetylation in vitro [144].
A comprehensive appreciation of chromatin dynamics in memory necessitates a discussion of histone methylation. The roles of readers, writers and erasers of histone methylation marks are poorly understood, but one study links contextual fear memory to dynamic modulation of the permissive H3K4me3 and suppressive H3K9me2 marks in the hippocampus [145]. Target genes include zif268 and bdnf. Similarly, a recent report also reveal dynamic modulation of histone methylation in the striatum response to cocaine experience and these changes in methylation were associated with altered expression of a number of genes, including cdk5, Nf-κB, arc and bdnf exon VI [126]. Intriguingly, HDAC inhibition suppresses H3K9 methylation, suggesting histone acetylation acts upstream of methylation [146]. The growing evidence for co-occurring histone modifications and extensive epigenetic crosstalk engendered the controversial ‘histone code’ hypothesis, in which combinatorial marks putatively produce a concerted effect on local transcription [21,147,148]. Whether a specific histone mark results from recruitment of writers and erasers in response to other marks or common upstream enzymes process multiple marks simultaneously is unclear. However, one study suggests PP1 functions as such a suppressor of multiple permissive signatures including H3S10 phosphorylation, H3 and H4 acetylation and H3K36 methylation that bidirectionally affect genes such as creb and Nf-κB [102]. In line with these observations, PP1 inhibition enhanced object recognition memory and associated epigenetic modifications. Comprehensively, these studies show that multiple signaling cascades converge onto effectors that orchestrate an array of epigenetic marks. To a degree, the anatomical and genetic location of these effects reflects the organism’s environment and behavior.
DNA methylation marks in memory
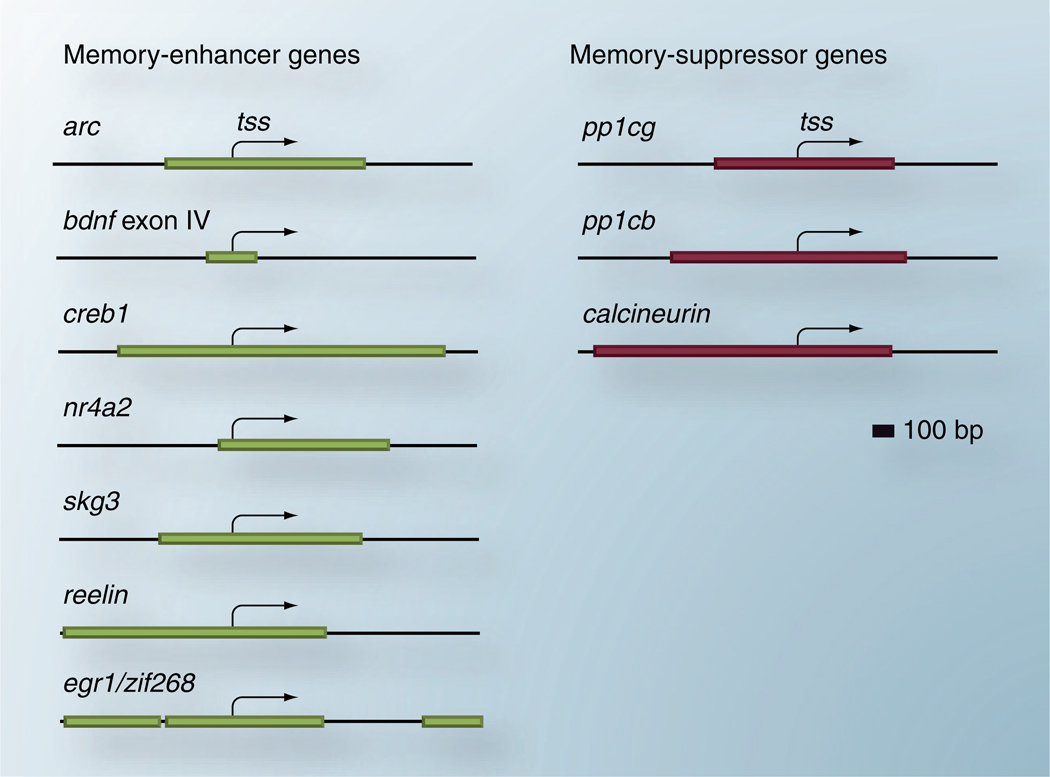
A smaller but growing body of evidence points to active DNA methylation alterations in response to neuronal activity and behavior. As mentioned above, DNMT inhibitors have pronounced effects on memory formation and storage as well as behavioral responses to drugs of abuse. However, the ability of DNA methylation to affect these behaviors implies that the plasticity genes which are important regulators of these experiences contain active sites for methylation and/or demethylation of DNA. Indeed, this appears to be the case. In fact, a large number of genes which have previously been shown to positively and negatively modulate behavioral memory contain dense CpG islands surrounding their promoter regions (Figure 2). Thus, these genes are potential targets for changes in the machinery that underlies DNA methylation in neurons.
Figure 2. CpG islands span memory-associated genes.
The majority of mammalian gene promoters are associated with CpG islands. Here, we show a large number of memory-associated transcription start sites are embedded within CpG islands, suggesting potential regulation by DNA methylation. Genes both positively (left) and negatively (right) linked to memory and synaptic plasticity are associated with these dense, CpG-rich regions.
Early evidence from cell culture models showed neuronal depolarization produces bdnf exon IV promoter demethylation associated with release of MeCP2 [149]. This may act through upstream activators such as those discussed above, as PKC activation in hippocampal slices similarly produced bdnf exon I demethylation [61]. In contrast to activity-regulated demethylation, direct inhibition of DNMT also led to reelin and site-specific bdnf demethylation in vitro and in vivo [42,61,105]. The requirement for DNA synthesis of such inhibitors suggests active demethylation and methylation may occur at a baseline rate in a base excision-repair fashion at neuron-specific loci [150–152]. Indeed, the putative mediator of active DNA demethylation, Gadd45b, is necessary for bdnf exon IX demethylation [40]. In addition, altered DNMT expression may contribute to demethylation in neurons. Expression of DNMT3a and DNMT1 was suppressed by depolarization in a cortical neuron culture and accompanied bdnf upregulation [153]. Still, this conflicts with the finding that PKC activation and fear memory formation upregulated DNMT3a and DNMT3b [61,106]. This suggests unique activity patterns differentially regulate DNA methylation machinery. It is also possible in the latter studies that enhanced DNMT expression actually promoted demethylation directly through DNMT action [152]. However, the fact that acute DNMT inhibition in the hippocampus in naive animals produces region-specific demethylation events in bdnf challenges this interpretation [105]. It is also now clear that changes in the expression of DNMTs also occurs in the striatum in response to cocaine administration [119,130], suggesting that this may be a general mechanism for alterations in DNA methylation profiles throughout the brain.
DNA methylation may also control neuronal tone by regulating ion channel expression. For instance, DNMT blockade and a chronic intermittent ethanol paradigm led to demethylation and upregulation of NR2B, an NMDA receptor subunit [154–157]. Similar effects were found upon activity suppression in cortical neurons, suggesting DNA methylation mediates lasting homeostatic plasticity [158]. Many of these induced demethylation events are associated with release of MeCP2 and other repressive factors [149,158]. Likewise, an induced plasticity state by HDAC inhibition was associated with permissive epigenetic marks in bdnf and reduced HDAC1, MeCP2 and MBD1 binding [159].
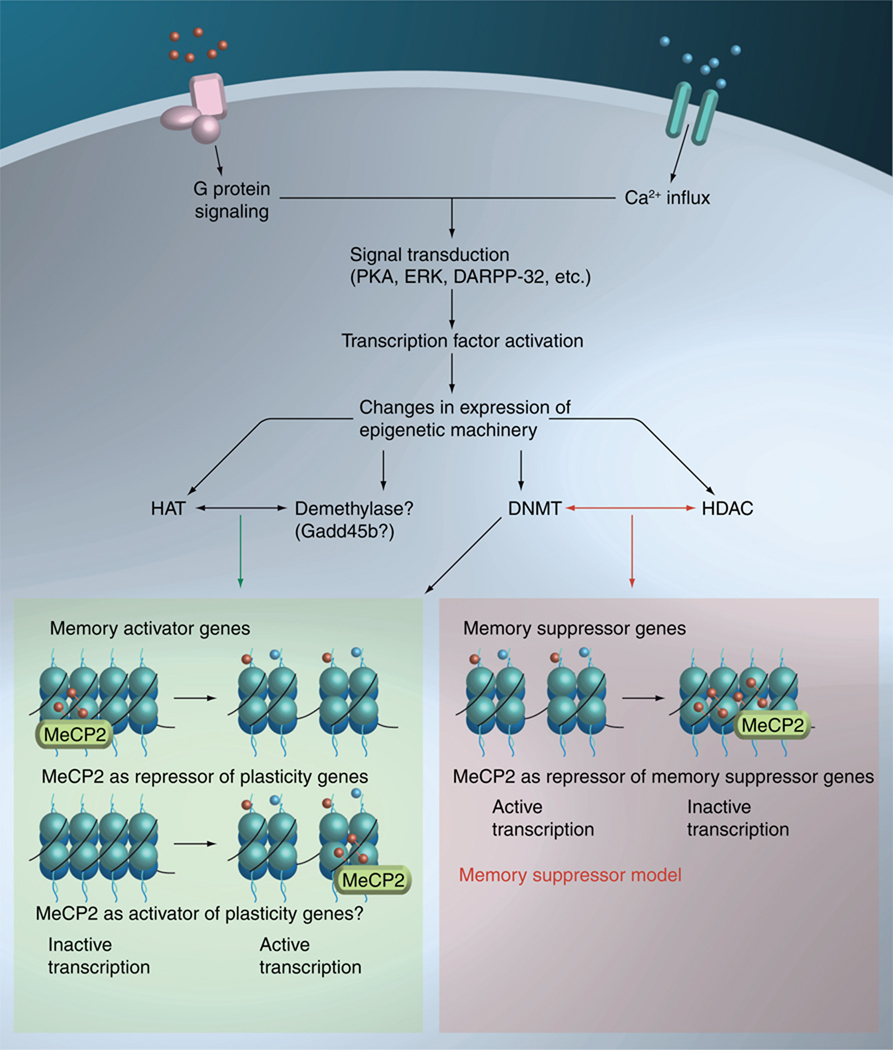
How then might changes in DNA methylation directly lead to memory formation and stabilization? Recent data support a model in which dynamic DNA methylation interacts with other epigenetic changes to program memory-associated behavior (Figure 3). For example, contextual fear conditioning enhanced and reduced methylation at the promoters of pp1β and reelin, respectively, with commensurate decrease and increase in transcription [106]. This pattern of epigenetic modulation is consistent with the reported roles of pp1β and reelin as memory-suppressive and memory-permissive genes, respectively [139,160,161]. This suggests memory consolidation is associated with bidirectional, site-specific DNA methylation dynamics. DNMT blockade immediately after training drove further demethylation of both genes and notably doubled pp1β transcripts. The latter finding fits with the surprising fact that DNMT inhibition, which presumably would enhance gene expression broadly, blocks hippocampus-dependent memory. DNA methylation may therefore mediate behavioral function more by silencing memory suppressors than by affecting memory activators [162]. However, while promoter-specific DNA methylation events were found in bdnf after fear conditioning, DNMT inhibition prior to training paradoxically inhibited exon IV demethylation [105]. This implies DNMT is functioning in active demethylation itself or that this effect is an indirect consequence of DNMT activity at other suppressor gene loci. PP1 may play a crucial role, but other suppressors may be acting similarly. An additional, although not mutually exclusive possibility, is that histone modifications may be necessary for DNA methylation changes. Indeed, HDAC inhibition reversed DNMT inhibitor-induced memory deficits and DNMT inhibition in slices blocked active H3 acetylation [61,62]. In line with the suppressor hypothesis, remote memory storage in the cortex was shown to be sensitive to DNMT inhibition before retrieval of 1 month-old memories [109]. In particular, this treatment disrupted lasting memory-specific hypermethylation of the promoter of calcineurin, a phosphatase and memory suppressor gene and led to its upregulation [139]. This study is the first to demonstrate a functional role of DNA methylation in cortical memory storage, although it is unclear whether the true alteration in cellular phenotype acts by promoting synaptic plasticity, permanently preventing further plasticity or by affecting the global responsiveness of the cell [71]. Further, it is unclear how specifically, if at all, these events affect cortical pyramidal cells, the principle computational units of the cortex, in comparison to other cells. Of note, excessive cortical DNMT expression associated with psychosis acts in part by silencing gad67 and reelin expression in inhibitory interneurons [163]. DNMT may also function to enhance network activity in memory consolidation by indirectly modulating inhibitory tone, but this has not been shown.
Figure 3. Memory activator versus suppressor model.
The synaptic membrane harbors key signal transduction mediators acting through ion influx and G-protein-coupled receptors. Signaling proceeds to the nucleus through kinase and phosphatase cascades that impinge on transcription factor modulation. Acute effects include altered expression of proteins constituting the epigenetic machinery that regulates histone acetylation, DNA methylation and other effects. Canonically, histone acetylation and phosphorylation (denoted by circles on histone tails) and DNA demethylation induce transcription while histone deacetylation and DNA methylation (denoted by circles on DNA strand) silence genes. Acting at memory activator loci (left panel), DNA demethylation and histone acetylation/phosphorylation would result in transcription of important plasticity genes. However, it is also possible that DNA methylation at these loci enhances transcription by MeCP2 recruitment and subsequent, noncanonical activation. Acting at memory suppressor loci (right panel), such as those encoding phosphatases, the DNA methylation would repress these genes and promote cellular activity and plasticity by disinhibiting the neuron. This model helps explain why DNA methylation is memory-permissive (i.e., why DNMT inhibition impairs memory formation and maintenance). It is surprising that DNA methylation and histone acetylation are both memory-permissive even though they canonically affect gene expression through opposite directions. Differences in locus-specificity as suggested by this model could help explain this perplexing finding. It is also noteworthy that although these models may help to explain memory formation, entirely different models may be necessary to explain prolonged memory stabilization across time, wherein further activation of plasticity genes may be counterproductive to memory storage.
DNMT: DNA methyltransferase; HAT: Histone acetyltransferases; HDAC: Histone deacetylases.
Alternatively, DNA methylation in the context of memory may act as a transcriptional enhancer (Figure 3). For instance, ZIF268 expression is enhanced during memory consolidation despite an increase in DNA methylation [145]. Still, the enhanced H3K4 trimethylation in the same region may have overcome or tuned the potential influence of DNA methylation on expression. Alternatively, the recruited MeCP2 may be acting as a transcriptional activator in a gene- and behavior-specific manner. Of note, a neuronal genome-wide analysis revealed that most MeCP2-bound promoters are actively expressed, not repressed [164]. These observations imply the existence of a complex layer of dynamic and persisting epigenetic regulation of gene expression that is specific to the pattern of behavior of the organism. A more inclusive activator and suppressor model, therefore, may be appropriate in defining how DNA methylation affects the neural transcriptome.
It remains perplexing that most of these mechanisms observed in the context of cell activation are relatively transient, in contrast to the canonical interpretation of epigenetics as a set of stable mechanisms associated with cellular differentiation. Insights from these studies on brain tissue show instead that epigenetic marks are written and erased dynamically in response to sensory input. In associative memory, this finding is consistent with the well-established role of the hippocampus in the conversion of short-term to long-term memory. This consolidation process lasts for a matter of hours, similar to the time-scale, for instance, of DNA methylation changes in the hippocampus after contextual fear conditioning. Indeed, reelin and pp1β methylation and expression return to baseline within 24 h of training [106]. In addition, DNMT blockade 6 h after training, a period that lies outside of the consolidation window, failed to block memory. This suggests DNA methylation has evolved as an acute rather than stable transcriptional regulator of experience-sensitive gene expression in the hippocampus. This interpretation conforms to interpretation that computational units in the hippocampus must be primed for acquisition of information after the consolidation of one episodic event is complete. In theory, stable modification of the epigenome during consolidation would eventually cause epigenetic saturation and inability to react to future events. In combination with the above results, it appears an unidentified DNA demethylase complex or protein is necessary to explain events such as rapid reelin demethylation or the return of pp1β methylation to baseline after fear conditioning. Gadd45b and other putative regulators of this process have been identified in the hippocampus and future work is needed to delineate their function in memory and associated epigenetics.
In contrast to the temporally pliable epigenetic regulation of gene expression following memory acquisition, the lasting storage of memory demands self-perpetuating or otherwise stable molecular mechanisms that evade protein turnover. Stable epigenetic marks in the prefrontal cortex corresponding to sensory input and memory have been shown, but it is unclear what governs this dissociation between these persistent DNA methylation marks and more malleable ones the hippocampus. One possibility is that cortical cells are developmentally tuned to undergo stable epigenetic changes by utilizing unique molecular mechanisms that ‘lock’ the epigenome in response to specific behaviors. In this model, hippocampal cells may lack or suppress these stabilizing signals. Alternatively, specific patterns of activity in the different brain regions that correspond to sensory input may modulate this switch. This network scale interpretation ignores any inherent, developmental differences between the cells in the hippocampus and cortex and reflects the differences in organization between these areas. In any scenario, it appears likely that a locking switch exists, but its identity is presently unclear. It is also unclear to what extent lasting epigenetic modifications mediate other functions besides remote memory storage. Most hippocampal studies have only found acute modifications, although H3K9 dimethylation is reduced in the hippocampus 1 day after training [145]. However, this effect is not specific for memory formation. It remains to be seen to what extent, if any, persistent epigenetics controls neural function in other behavioral paradigms and other brain regions.
Conclusion & future perspective
Epigenetic mechanisms in the brain are alluring candidates for regulation of behavioral function. Phenomena such as posttranslational modifications of histones and covalent modification of DNA have emerged as necessary regulators of synaptic physiology and memory, although as noted above these molecular changes vary considerably in anatomical locus and temporal dynamics. To an extent, these differences reflect the recent or remote patterns of environmental cues and behavioral events, suggesting epigenetics has evolved to encode information from an organism’s experiences. These mechanisms affect behavioral phenotype indirectly by regulating cellular phenotype through dynamic or persistent transcriptional control.
Despite our growing understanding of these mechanisms, however, future studies are needed to address a number of challenges. For instance, the breadth of gene targets affected by these mechanisms has yet to be uncovered by previous analyses, marking the need for high-throughput studies such as epigenome-wide screens. In addition, it is unclear which specific cells experience the epigenetic modifications reported above. Thus, cell type-specific studies will be critical for our understanding of how unique circuits within the same brain region may differentially control behavioral function via epigenetic modifications. Further, the role of DNA methylation in the cortex in persisting memory storage has only been appreciated recently. Although this result is very exciting, the specific role of this modification in maintaining the memory engram will need to be explored in future studies. Moreover, the role that additional mechanisms such as histone modifications play in this long-term maintenance of memory has yet to be examined.
So-called noncanonical epigenetic mechanisms such as RNA interference have also been found in brain function, although their specific role in memory and behavior is poorly understood at the present time. However, a recent study demonstrated that a novel class of noncoding RNA, enhancer RNAs, responds to the H3K4me1 signal and may regulate local gene expression in neurons [165]. Indeed, growing evidence suggests a key role for noncoding miRNAs and siRNAs in memory and synaptic function [166–168]. Notably, SIRT1 regulates memory and synaptic plasticity through a specific miRNA, suggesting crosstalk between histone acetylation and noncanonical epigenetics may be necessary for either to alter neural function [169]. This is a promising area for future research. Finally, studies have shown that epigenetic mechanisms regulate postnatal neurogenesis in the dentate gyrus [170]. In light of the reported but controversial contribution of neurogenesis to memory, it remains to be shown whether epigenetic regulation of hippocampus-dependent memory relies on neurogenesis [129]. Thus, future studies will be necessary to examine this link and to determine whether the role of epigenetic marks in neurogenesis differs from their role in learning and memory. These and other exciting avenues of research will enhance our understanding of the breadth of influence of epigenetics in the adult nervous system and of the molecular underpinnings of memory formation and behavior.
Executive summary.
-
□
Lasting changes in behavioral responses to environmental cues, an empirical definition of memory, require persistent molecular correlates in the brain and this necessity drove much of the initial interest in epigenetic mechanisms as regulators of memory function.
Molecular epigenetics
-
□
Dynamic and stable covalent alterations of histone and DNA itself, the core constituents of chromatin, constitute the two canonical epigenetic mechanisms.
-
□
Isoform-specific expression of the epigenetic machinery, consisting of readers, writers and erasers of epigenetic marks, has been confirmed in the developing, nascent and mature brain.
Epigenetic contributions to neurophysiology
-
□
A functional role of DNA methylation and histone acetylation in spontaneous neurotransmission and short-term synaptic plasticity has been confirmed with pharmacological and genetic approaches in vitro.
-
□
Lasting changes in synaptic efficacy, a cellular correlate of long-term memory, are sensitive to alterations in histone methylation, acetylation, ADP-ribosylation and DNA methylation. This result is consistent with the well-established finding that late-phase synaptic plasticity depends on stable changes in gene expression.
Epigenetic regulation of memory
-
□
Studies of invertebrates and mammals confirm that the action of epigenetic readers, writers and erasers in a range of brain regions subserves long-term memory in paradigms associated with those anatomical locations.
-
□
The influence of epigenetics on memory also acts in a spatiotemporally specific manner. Notably, dynamic DNA methylation changes in the hippocampus underlie consolidation whereas persistent patterns in the cortex propogate long-term storage.
Epigenetic regulation of reward-related behaviors
-
□
As with associative, spatial and recognition memory, epigenetic mechanisms regulate stereotyped behavioral responses in rodents to drugs of abuse, stress and natural rewards.
Epigenetic signatures in neural function: upstream mechanisms & effectors
-
□
Consistent with the reported role of the epigenetic machinery in memory and synaptic function, epigenetic modifications to chromatin are associated with specific patterns of activity in response to the organism’s experience and behavior.
-
□
A number of membrane-to-nucleus signaling cascades translate neuronal activity patterns into alterations in transcriptional efficacy which subserve the dynamic cellular phenotype.
-
□
Combinations of co-occurring permissive and repressive signatures are associated with acute and persistent regulation of effector gene transcription, suggesting an ‘epigenetic code’ modulates discrete patterns of gene-expression changes in line with specific environmental cues and behavior.
Future perspective
-
□
Many challenges in addressing epigenetic contributions to memory remain. Further investigation into the breadth and combinatorial pattern of epigenetic signatures with powerful tools such as genome-wide association approaches is needed. In addition, studies will need to address how epigenetic marks alter the physiology of specific cells and network function associated with memory formation and storage. A number of noncanonical epigenetic mechanisms in brain function have also been identified but still poorly understood.
Acknowledgments
Financial & competing interests disclosure
Jeremy J Day is supported by a grant from the National Institute on Drug Abuse (1F32DA029419).
Footnotes
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Sweatt JD. Mechanisms of Memory. Burlington, MA, USA: Elsevier; 2010. [Google Scholar]
- 2.Abel T, Lattal KM. Molecular mechanisms of memory acquisition, consolidation and retrieval. Curr. Opin. Neurobiol. 2001;11(2):180–187. doi: 10.1016/s0959-4388(00)00194-x. [DOI] [PubMed] [Google Scholar]
- 3.Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiol. Rev. 2009;89(1):121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stevens CF. CREB and memory consolidation. Neuron. 1994;13(4):769–770. doi: 10.1016/0896-6273(94)90244-5. [DOI] [PubMed] [Google Scholar]
- 5.Bailey CH, Kandel ER, Si K. The persistence of long-term memory: a molecular approach to self-sustaining changes in learning-induced synaptic growth. Neuron. 2004;44(1):49–57. doi: 10.1016/j.neuron.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 6.Roberson ED, Sweatt JD. A biochemical blueprint for long-term memory. Learn Mem. 1999;6(4):381–388. [PMC free article] [PubMed] [Google Scholar]
- 7.Price JC, Guan S, Burlingame A, Prusiner SB, Ghaemmaghami S. Analysis of proteome dynamics in the mouse brain. Proc. Natl Acad. Sci. USA. 2010;107(32):14508–14513. doi: 10.1073/pnas.1006551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Archibald K, Perry MJ, Molnar E, Henley JM. Surface expression and metabolic half-life of AMPA receptors in cultured rat cerebellar granule cells. Neuropharmacology. 1998;37(10–11):1345–1353. doi: 10.1016/s0028-3908(98)00135-x. [DOI] [PubMed] [Google Scholar]
- 9.Silva AJ, Stevens CF, Tonegawa S, Wang Y. Deficient hippocampal long-term potentiation in α-calcium-calmodulin kinase II mutant mice. Science. 1992;257(5067):201–206. doi: 10.1126/science.1378648. [DOI] [PubMed] [Google Scholar]
- 10.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23(7):781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price TD, Qvarnstrom A, Irwin DE. The role of phenotypic plasticity in driving genetic evolution. Proc. Biol. Sci. 2003;270(1523):1433–1440. doi: 10.1098/rspb.2003.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bird A. Perceptions of epigenetics. Nature. 2007;447(7143):396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 13.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Brownell JE, Zhou J, Ranalli T, et al. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84(6):843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 15.Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272(5260):408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 16.Allis CD, Reinberg D. Epigenetics. Cold Spring Harbor, USA: Cold Spring Harbor Laboratory Press; 2007. [Google Scholar]
- 17.Nowak SJ, Corces VG. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 2004;20(4):214–220. doi: 10.1016/j.tig.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 18.Peters AH, Schubeler D. Methylation of histones: playing memory with DNA. Curr. Opin. Cell Biol. 2005;17(2):230–238. doi: 10.1016/j.ceb.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 2008;20(3):341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc. Natl Acad. Sci. USA. 2003;100(23):13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graff J, Mansuy IM. Epigenetic codes in cognition and behaviour. Behav. Brain Res. 2008;192(1):70–87. doi: 10.1016/j.bbr.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 22.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 23. Chahrour M, Jung SY, Shaw C, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320(5880):1224–1229. doi: 10.1126/science.1153252.▪▪ Demonstrates that MeCP2, a key protein that binds to methylated CpG sites of DNA, can activate transcription through interactions with the transcription factor CREB, and repress transcription by associating with histone deacetylases (HDACs). This study reported that the vast majority of genes altered by MeCP2 overexpression exhibited increased expression levels, suggesting that it positively regulates gene expression. It had previously been assumed that DNA methylation in the CNS was generally a transcriptionally repressive mark.
- 24.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 2010;11(9):607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones PL, Veenstra GJ, Wade PA, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998;19(2):187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 26.Guan JS, Haggarty SJ, Giacometti E, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459(7243):55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris MJ, Karra AS, Monteggia LM. Histone deacetylases govern cellular mechanisms underlying behavioral and synaptic plasticity in the developing and adult brain. Behav. Pharmacol. 2010;21(5–6):409–419. doi: 10.1097/FBP.0b013e32833c20c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janssen C, Schmalbach S, Boeselt S, Sarlette A, Dengler R, Petri S. Differential histone deacetylase mRNA expression patterns in amyotrophic lateral sclerosis! J. Neuropathol. Exp. Neurol. 2010;69(6):573–581. doi: 10.1097/NEN.0b013e3181ddd404. [DOI] [PubMed] [Google Scholar]
- 29.Riccio A. Dynamic epigenetic regulation in neurons: enzymes, stimuli and signaling pathways. Nat. Neurosci. 2010;13(11):1330–1337. doi: 10.1038/nn.2671. [DOI] [PubMed] [Google Scholar]
- 30.MacDonald JL, Roskams AJ. Histone deacetylases 1 and 2 are expressed at distinct stages of neuro-glial development. Dev. Dyn. 2008;237(8):2256–2267. doi: 10.1002/dvdy.21626. [DOI] [PubMed] [Google Scholar]
- 31.Schaefer A, Sampath SC, Intrator A, et al. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron. 2009;64(5):678–691. doi: 10.1016/j.neuron.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J. Neurosci. Res. 2005;79(6):734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- 33.Thompson RF, Atzmon G, Gheorghe C, et al. Tissue-specific dysregulation of DNA methylation in aging. Aging Cell. 9(4):506–518. doi: 10.1111/j.1474-9726.2010.00577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Penner MR, Roth TL, Barnes CA, Sweatt JD. An epigenetic hypothesis of aging-related cognitive dysfunction. Front. Aging Neurosci. 2010;2:9. doi: 10.3389/fnagi.2010.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Penner MR, Roth TL, Chawla MK, et al. Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiol. Aging. 2010 doi: 10.1016/j.neurobiolaging.2010.01.009. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noh JS, Sharma RP, Veldic M, et al. DNA methyltransferase 1 regulates reelin mRNA expression in mouse primary cortical cultures. Proc. Natl Acad. Sci. USA. 2005;102(5):1749–1754. doi: 10.1073/pnas.0409648102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma RP, Gavin DP, Grayson DR. CpG methylation in neurons: message, memory, or mask? Neuropsychopharmacology. 2010;35(10):2009–2020. doi: 10.1038/npp.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levenson JM. DNA (cytosine-5) methyltransferase inhibitors: a potential therapeutic agent for schizophrenia. Mol. Pharmacol. 2007;71(3):635–637. doi: 10.1124/mol.106.033266. [DOI] [PubMed] [Google Scholar]
- 39.Barreto G, Schafer A, Marhold J, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445(7128):671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 40. Ma DK, Jang MH, Guo JU, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323(5917):1074–1077. doi: 10.1126/science.1166859.▪Reveals a novel potential mechanism for the demethylation of methylated cytosines in DNA. Methylated cytosine is converted to thymine and then removed through base excision repair mechanisms, under control of growth arrest and DNA damage protein 45b (Gadd45b)
- 41.Kandel JHS, Jessell TM. Principles of Neural Science. 4th Edition. McGraw-Hill Medical; 2000. [Google Scholar]
- 42.Nelson ED, Kavalali ET, Monteggia LM. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J. Neurosci. 2008;28(2):395–406. doi: 10.1523/JNEUROSCI.3796-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monteggia LM, Kavalali ET. Rett syndrome and the impact of MeCP2 associated transcriptional mechanisms on neurotransmission. Biol. Psychiatry. 2009;65(3):204–210. doi: 10.1016/j.biopsych.2008.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asaka Y, Jugloff DG, Zhang L, Eubanks JH, Fitzsimonds RM. Hippocampal synaptic plasticity is impaired in the Mecp2-null mouse model of Rett syndrome. Neurobiol. Dis. 2006;21(1):217–227. doi: 10.1016/j.nbd.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 45.Collins AL, Levenson JM, Vilaythong AP, et al. Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Hum. Mol. Genet. 2004;13(21):2679–2689. doi: 10.1093/hmg/ddh282. [DOI] [PubMed] [Google Scholar]
- 46.Belichenko PV, Oldfors A, Hagberg B, Dahlstrom A. Rett syndrome: 3-D confocal microscopy of cortical pyramidal dendrites and afferents. Neuroreport. 1994;5(12):1509–1513. [PubMed] [Google Scholar]
- 47.Kaufmann WE, MacDonald SM, Altamura CR. Dendritic cytoskeletal protein expression in mental retardation: an immunohistochemical study of the neocortex in Rett syndrome. Cereb. Cortex. 2000;10(10):992–1004. doi: 10.1093/cercor/10.10.992. [DOI] [PubMed] [Google Scholar]
- 48.Dani VS, Chang Q, Maffei A, Turrigiano GG, Jaenisch R, Nelson SB. Reduced cortical activity due to a shift in the balance between excitation and inhibition in a mouse model of Rett syndrome. Proc. Natl Acad. Sci. USA. 2005;102(35):12560–12565. doi: 10.1073/pnas.0506071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki M, Yamada T, Kihara-Negishi F, Sakurai T, Oikawa T. Direct association between PU.1 and MeCP2 that recruits mSin3A–HDAC complex for PU.1-mediated transcriptional repression. Oncogene. 2003;22(54):8688–8698. doi: 10.1038/sj.onc.1207182. [DOI] [PubMed] [Google Scholar]
- 50.Nelson ED, Kavalali ET, Monteggia LM. MeCP2-dependent transcriptional repression regulates excitatory neurotransmission. Curr. Biol. 2006;16(7):710–716. doi: 10.1016/j.cub.2006.02.062. [DOI] [PubMed] [Google Scholar]
- 51.Akhtar MW, Raingo J, Nelson ED, et al. Histone deacetylases 1 and 2 form a developmental switch that controls excitatory synapse maturation and function. J. Neurosci. 2009;29(25):8288–8297. doi: 10.1523/JNEUROSCI.0097-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guan Z, Giustetto M, Lomvardas S, et al. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell. 2002;111(4):483–493. doi: 10.1016/s0092-8674(02)01074-7. [DOI] [PubMed] [Google Scholar]
- 53.Levenson JM, O’Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J. Biol. Chem. 2004;279(39):40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- 54.Vecsey CG, Hawk JD, Lattal KM, et al. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J. Neurosci. 2007;27(23):6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alarcon JM, Malleret G, Touzani K, et al. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42(6):947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 56.Wood MA, Kaplan MP, Park A, et al. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn Mem. 2005;12(2):111–119. doi: 10.1101/lm.86605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim SY, Levenson JM, Korsmeyer S, Sweatt JD, Schumacher A. Developmental regulation of Eed complex composition governs a switch in global histone modification in brain. J. Biol. Chem. 2007;282(13):9962–9972. doi: 10.1074/jbc.M608722200. [DOI] [PubMed] [Google Scholar]
- 58.Fontan-Lozano A, Suarez-Pereira I, Horrillo A, del-Pozo-Martin Y, Hmadcha A, Carrion AM. Histone H1 poly[ADP]-ribosylation regulates the chromatin alterations required for learning consolidation. J. Neurosci. 2010;30(40):13305–13313. doi: 10.1523/JNEUROSCI.3010-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yeh SH, Lin CH, Gean PW. Acetylation of nuclear factor-κB in rat amygdala improves long-term but not short-term retention of fear memory. Mol. Pharmacol. 2004;65(5):1286–1292. doi: 10.1124/mol.65.5.1286. [DOI] [PubMed] [Google Scholar]
- 60.Putignano E, Lonetti G, Cancedda L, et al. Developmental downregulation of histone posttranslational modifications regulates visual cortical plasticity. Neuron. 2007;53(5):747–759. doi: 10.1016/j.neuron.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 61.Levenson JM, Roth TL, Lubin FD, et al. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J. Biol. Chem. 2006;281(23):15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- 62.Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol. Learn Mem. 2008;89(4):599–603. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feng J, Zhou Y, Campbell SL, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010;13(4):423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao X, Ueba T, Christie BR, et al. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc. Natl Acad. Sci. USA. 2003;100(11):6777–6782. doi: 10.1073/pnas.1131928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moretti P, Levenson JM, Battaglia F, et al. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J. Neurosci. 2006;26(1):319–327. doi: 10.1523/JNEUROSCI.2623-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat. Rev. Neurosci. 2007;8(5):355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- 67.Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat. Neurosci. 2006;9(4):519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 68. Weaver IC, Cervoni N, Champagne FA, et al. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7(8):847–854. doi: 10.1038/nn1276.▪Reveals that poor maternal behavior induces potentially life-long changes in epigenetic status at genes associated with stress responsivity. These changes are reversed by cross-fostering or infusion of histone deacetylases inhibitors
- 69.Weaver IC, Diorio J, Seckl JR, Szyf M, Meaney MJ. Early environmental regulation of hippocampal glucocorticoid receptor gene expression: characterization of intracellular mediators and potential genomic target sites. Ann. NY Acad. Sci. 2004;1024:182–212. doi: 10.1196/annals.1321.099. [DOI] [PubMed] [Google Scholar]
- 70.Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol. Psychiatry. 2009;65(9):760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Day JJ, Sweatt JD. DNA methylation and memory formation. Nat. Neurosci. 2010;13(11):1319–1323. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Levenson JM, Sweatt JD. Epigenetic mechanisms: a common theme in vertebrate and invertebrate memory formation. Cell Mol. Life Sci. 2006;63(9):1009–1016. doi: 10.1007/s00018-006-6026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Federman N, Fustinana MS, Romano A. Histone acetylation is recruited in consolidation as a molecular feature of stronger memories. Learn Mem. 2009;16(10):600–606. doi: 10.1101/lm.1537009. [DOI] [PubMed] [Google Scholar]
- 74.Stefanko DP, Barrett RM, Ly AR, Reolon GK, Wood MA. Modulation of long-term memory for object recognition via HDAC inhibition. Proc. Natl Acad. Sci. USA. 2009;106(23):9447–9452. doi: 10.1073/pnas.0903964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42(6):961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Josselyn SA, Nguyen PV. CREB, synapses and memory disorders: past progress and future challenges. Curr. Drug Targets CNS Neurol. Disord. 2005;4(5):481–497. doi: 10.2174/156800705774322058. [DOI] [PubMed] [Google Scholar]
- 77.Chen G, Zou X, Watanabe H, van Deursen JM, Shen J. CREB binding protein is required for both short-term and long-term memory formation. J. Neurosci. 2010;30(39):13066–13077. doi: 10.1523/JNEUROSCI.2378-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oliveira AM, Wood MA, McDonough CB, Abel T. Transgenic mice expressing an inhibitory truncated form of p300 exhibit long-term memory deficits. Learn Mem. 2007;14(9):564–572. doi: 10.1101/lm.656907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kalkhoven E. CBP and p300: HATs for different occasions. Biochem. Pharmacol. 2004;68(6):1145–1155. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 80.Ramos YF, Hestand MS, Verlaan M, et al. Genome-wide assessment of differential roles for p300 and CBP in transcription regulation. Nucleic Acids Res. 38(16):5396–5408. doi: 10.1093/nar/gkq184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Oliveira AM, Abel T, Brindle PK, Wood MA. Differential role for CBP and p300 CREB-binding domain in motor skill learning. Behav. Neurosci. 2006;120(3):724–729. doi: 10.1037/0735-7044.120.3.724. [DOI] [PubMed] [Google Scholar]
- 82.Burke SN, Barnes CA. Neural plasticity in the ageing brain. Nat. Rev. Neurosci. 2006;7(1):30–40. doi: 10.1038/nrn1809. [DOI] [PubMed] [Google Scholar]
- 83.Kilgore M, Miller CA, Fass DM, et al. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer’s disease. Neuropsychopharmacology. 2010;35(4):870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Peleg S, Sananbenesi F, Zovoilis A, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328(5979):753–756. doi: 10.1126/science.1186088.▪▪Demonstrates that the aging process is associated with an inability to produce a specific histone modification (acetylation at H4K12) and learning deficits. Interestingly, treatment with an histone deacetylases inhibitor restored this modification and rescued memory deficits, suggesting potential as a therapeutic target in disorders of learning and memory
- 85.Graff J, Mansuy IM. Epigenetic dysregulation in cognitive disorders. Eur. J. Neurosci. 2009;30(1):1–8. doi: 10.1111/j.1460-9568.2009.06787.x. [DOI] [PubMed] [Google Scholar]
- 86.Fischer A, Sananbenesi F, Mungenast A, Tsai LH. Targeting the correct HDAC(s) to treat cognitive disorders. Trends Pharmacol. Sci. 2010;31(12):605–617. doi: 10.1016/j.tips.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 87.Selvi BR, Cassel JC, Kundu TK, Boutillier AL. Tuning acetylation levels with HAT activators: Therapeutic strategy in neurodegenerative diseases. Biochim. Biophys. Acta. 2010;1799(10–12):840–853. doi: 10.1016/j.bbagrm.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 88.D’Mello SR. Histone deacetylases as targets for the treatment of human neurodegenerative diseases. Drug News Perspect. 2009;22(9):513–524. doi: 10.1358/dnp.2009.9.1428871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chuang DM, Leng Y, Marinova Z, Kim HJ, Chiu CT. Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends Neurosci. 2009;32(11):591–601. doi: 10.1016/j.tins.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mai A, Rotili D, Valente S, Kazantsev AG. Histone deacetylase inhibitors and neurodegenerative disorders: holding the promise. Curr. Pharm. Des. 2009;15(34):3940–3957. doi: 10.2174/138161209789649349. [DOI] [PubMed] [Google Scholar]
- 91.Duclot F, Jacquet C, Gongora C, Maurice T. Alteration of working memory but not in anxiety or stress response in p300/CBP associated factor (PCAF) histone acetylase knockout mice bred on a C57BL/6 background. Neurosci. Lett. 2010;475(3):179–183. doi: 10.1016/j.neulet.2010.03.077. [DOI] [PubMed] [Google Scholar]
- 92.Maurice T, Duclot F, Meunier J, et al. Altered memory capacities and response to stress in p300/CBP-associated factor (PCAF) histone acetylase knockout mice. Neuropsychopharmacology. 2008;33(7):1584–1602. doi: 10.1038/sj.npp.1301551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Roozendaal B, Hernandez A, Cabrera SM, et al. Membrane-associated glucocorticoid activity is necessary for modulation of long-term memory via chromatin modification. J. Neurosci. 2010;30(14):5037–5046. doi: 10.1523/JNEUROSCI.5717-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Swank MW, Sweatt JD. Increased histone acetyltransferase and lysine acetyltransferase activity and biphasic activation of the ERK/ RSK cascade in insular cortex during novel taste learning. J. Neurosci. 2001;21(10):3383–3391. doi: 10.1523/JNEUROSCI.21-10-03383.2001.▪Among the first reports to demonstrate epigenetic changes in the adult CNS during learning, specifically the induction of histone acetylation following taste learning. This report also implicates the MAPK signaling pathway in the initiation of epigenetic changes. The MAPK pathway plays a critical role in learning and memory in numerous brain areas
- 95.Silingardi D, Scali M, Belluomini G, Pizzorusso T. Epigenetic treatments of adult rats promote recovery from visual acuity deficits induced by long-term monocular deprivation. Eur. J. Neurosci. 2010;31(12):2185–2192. doi: 10.1111/j.1460-9568.2010.07261.x. [DOI] [PubMed] [Google Scholar]
- 96.Chwang WB, Arthur JS, Schumacher A, Sweatt JD. The nuclear kinase mitogen- and stress-activated protein kinase 1 regulates hippocampal chromatin remodeling in memory formation. J. Neurosci. 2007;27(46):12732–12742. doi: 10.1523/JNEUROSCI.2522-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chandramohan Y, Droste SK, Arthur JS, Reul JM. The forced swimming-induced behavioural immobility response involves histone H3 phospho-acetylation and c-Fos induction in dentate gyrus granule neurons via activation of the N-methyl-D-aspartate/ extracellular signal-regulated kinase/ mitogen- and stress-activated kinase signalling pathway. Eur. J. Neurosci. 2008;27(10):2701–2713. doi: 10.1111/j.1460-9568.2008.06230.x. [DOI] [PubMed] [Google Scholar]
- 98.Reul JM, Hesketh SA, Collins A, Mecinas MG. Epigenetic mechanisms in the dentate gyrus act as a molecular switch in hippocampus-associated memory formation. Epigenetics. 2009;4(7):434–439. doi: 10.4161/epi.4.7.9806. [DOI] [PubMed] [Google Scholar]
- 99.Lubin FD, Sweatt JD. The IκB kinase regulates chromatin structure during reconsolidation of conditioned fear memories. Neuron. 2007;55(6):942–957. doi: 10.1016/j.neuron.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Merlo E, Freudenthal R, Romano A. The IκB kinase inhibitor sulfasalazine impairs long-term memory in the crab Chasmagnathus. Neuroscience. 2002;112(1):161–172. doi: 10.1016/s0306-4522(02)00049-0. [DOI] [PubMed] [Google Scholar]
- 101.Zocchi L, Sassone-Corsi P. Joining the dots: from chromatin remodeling to neuronal plasticity. Curr. Opin. Neurobiol. 2010;20(4):432–440. doi: 10.1016/j.conb.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Koshibu K, Graff J, Beullens M, et al. Protein phosphatase 1 regulates the histone code for long-term memory. J. Neurosci. 2009;29(41):13079–13089. doi: 10.1523/JNEUROSCI.3610-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cohen-Armon M, Visochek L, Katzoff A, et al. Long-term memory requires polyADP-ribosylation. Science. 2004;304(5678):1820–1822. doi: 10.1126/science.1096775. [DOI] [PubMed] [Google Scholar]
- 104.Goldberg S, Visochek L, Giladi E, Gozes I, Cohen-Armon M. PolyADP-ribosylation is required for long-term memory formation in mammals. J. Neurochem. 2009;111(1):72–79. doi: 10.1111/j.1471-4159.2009.06296.x. [DOI] [PubMed] [Google Scholar]
- 105.Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J. Neurosci. 2008;28(42):10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53(6):857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 107.Juttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc. Natl Acad. Sci. USA. 1994;91(25):11797–11801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Frankland PW, Bontempi B, Talton LE, Kaczmarek L, Silva AJ. The involvement of the anterior cingulate cortex in remote contextual fear memory. Science. 2004;304(5672):881–883. doi: 10.1126/science.1094804. [DOI] [PubMed] [Google Scholar]
- 109. Miller CA, Gavin CF, White JA, et al. Cortical DNA methylation maintains remote memory. Nat. Neurosci. 2010;13(6):664–666. doi: 10.1038/nn.2560.▪▪ Bisulfite sequencing and subcloning were used to reveal that learning induces long-lasting (at least 30 days) changes in DNA methylation at specific genes. Moreover, inhibiting DNA methylation with site-specific infusions of a DNA methyltransferase (DNMT) inhibitor impaired remote memory maintenance, suggesting that ongoing DNA methylation is required to support long-term memories
- 110.Fuso A, Nicolia V, Cavallaro RA, et al. B-vitamin deprivation induces hyperhomocysteinemia and brain S-adenosylhomocysteine, depletes brain S-adenosylmethionine, and enhances PS1 and BACE expression and amyloid-β deposition in mice. Mol. Cell Neurosci. 2008;37(4):731–746. doi: 10.1016/j.mcn.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 111.Fuso A, Seminara L, Cavallaro RA, D’Anselmi F, Scarpa S. S-adenosylmethionine/homocysteine cycle alterations modify DNA methylation status with consequent deregulation of PS1 and BACE and β-amyloid production. Mol. Cell Neurosci. 2005;28(1):195–204. doi: 10.1016/j.mcn.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 112.Scarpa S, Fuso A, D’Anselmi F, Cavallaro RA. Presenilin 1 gene silencing by S-adenosylmethionine: a treatment for Alzheimer disease? FEBS Lett. 2003;541(1–3):145–148. doi: 10.1016/s0014-5793(03)00277-1. [DOI] [PubMed] [Google Scholar]
- 113.Chen TF, Huang RF, Lin SE, Lu JF, Tang MC, Chiu MJ. Folic Acid potentiates the effect of memantine on spatial learning and neuronal protection in an Alzheimer’s disease transgenic model. J. Alzheimers Dis. 2010;20(2):607–615. doi: 10.3233/JAD-2010-1396. [DOI] [PubMed] [Google Scholar]
- 114.Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic changes in Alzheimer’s disease: decrements in DNA methylation. Neurobiol. Aging. 2010;31(12):2025–2037. doi: 10.1016/j.neurobiolaging.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Silva PN, Gigek CO, Leal MF, et al. Promoter methylation analysis of SIRT3, SMARCA5, HTERT and CDH1 genes in aging and Alzheimer’s disease. J. Alzheimers Dis. 2008;13(2):173–176. doi: 10.3233/jad-2008-13207. [DOI] [PubMed] [Google Scholar]
- 116.Wang SC, Oelze B, Schumacher A. Age-specific epigenetic drift in late-onset Alzheimer’s disease. PLoS One. 2008;3(7):e2698. doi: 10.1371/journal.pone.0002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Scarpa S, Cavallaro RA, D’Anselmi F, Fuso A. Gene silencing through methylation: an epigenetic intervention on Alzheimer disease. J. Alzheimers Dis. 2006;9(4):407–414. doi: 10.3233/jad-2006-9406. [DOI] [PubMed] [Google Scholar]
- 118.Robinson TE, Kolb B. Persistent structural modifications in nucleus accumbens and prefrontal cortex neurons produced by previous experience with amphetamine. J. Neurosci. 1997;17(21):8491–8497. doi: 10.1523/JNEUROSCI.17-21-08491.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.LaPlant Q, Vialou V, Covington HE, 3rd, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat. Neurosci. 2010;13(9):1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nestler EJ. Molecular basis of long-term plasticity underlying addiction. Nat. Rev. Neurosci. 2001;2(2):119–128. doi: 10.1038/35053570. [DOI] [PubMed] [Google Scholar]
- 121. Kumar A, Choi KH, Renthal W, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48(2):303–314. doi: 10.1016/j.neuron.2005.09.023.▪Exposure to drugs of abuse produces long-term plasticity and prolonged changes in gene expression in brain reward regions. This report employed chromatin immunoprecipitation techniques to show that exposure to cocaine produces rapid changes in histone acetylation and phosphorylation at specific gene targets, including cFos, ΔFosB and BDNF thus revealing an epigenetic mechanism by which drug administration can control gene readout
- 122.Sanchis-Segura C, Lopez-Atalaya JP, Barco A. Selective boosting of transcriptional and behavioral responses to drugs of abuse by histone deacetylase inhibition. Neuropsychopharmacology. 2009;34(13):2642–2654. doi: 10.1038/npp.2009.125. [DOI] [PubMed] [Google Scholar]
- 123.Renthal W, Maze I, Krishnan V, et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56(3):517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 124.Wang L, Lv Z, Hu Z, et al. Chronic cocaine-induced H3 acetylation and transcriptional activation of CaMKIIα in the nucleus accumbens is critical for motivation for drug reinforcement. Neuropsychopharmacology. 2010;35(4):913–928. doi: 10.1038/npp.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Renthal W, Kumar A, Xiao G, et al. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62(3):335–348. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Maze I, Covington HE, 3rd, Dietz DM, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327(5962):213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Stipanovich A, Valjent E, Matamales M, et al. A phosphatase cascade by which rewarding stimuli control nucleosomal response. Nature. 2008;453(7197):879–884. doi: 10.1038/nature06994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Im HI, Hollander JA, Bali P, Kenny PJ. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA. Nat. Neurosci. 2010;13(9):1120–1127. doi: 10.1038/nn.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Deng W, Aimone JB, Gage FH. New neurons and new memories: how does adult hippocampal neurogenesis affect learning and memory? Nat. Rev. Neurosci. 2010;11(5):339–350. doi: 10.1038/nrn2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Anier K, Malinovskaja K, Aonurm-Helm A, Zharkovsky A, Kalda A. DNA methylation regulates cocaine-induced behavioral sensitization in mice. Neuropsychopharmacology. 2010;35(12):2450–2461. doi: 10.1038/npp.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shifett MW, Martini RP, Mauna JC, Foster RL, Peet E, Thiels E. Cue-elicited reward-seeking requires extracellular signal-regulated kinase activation in the nucleus accumbens. J. Neurosci. 2008;28(6):1434–1443. doi: 10.1523/JNEUROSCI.2383-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shifett MW, Mauna JC, Chipman AM, Peet E, Thiels E. Appetitive Pavlovian conditioned stimuli increase CREB phosphorylation in the nucleus accumbens. Neurobiol. Learn Mem. 2009;92(3):451–454. doi: 10.1016/j.nlm.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Danilova AB, Kharchenko OA, Shevchenko KG, Grinkevich LN. Histone H3 acetylation is asymmetrically induced upon learning in identified neurons of the food aversion network in the mollusk Helix lucorum. Front. Behav. Neurosci. 2010;4:180. doi: 10.3389/fnbeh.2010.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cohen-Armon M, Visochek L, Rozensal D, et al. DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Mol. Cell. 2007;25(2):297–308. doi: 10.1016/j.molcel.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 135.Tsankova NM, Kumar A, Nestler EJ. Histone modifications at gene promoter regions in rat hippocampus after acute and chronic electroconvulsive seizures. J. Neurosci. 2004;24(24):5603–5610. doi: 10.1523/JNEUROSCI.0589-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Huang Y, Doherty JJ, Dingledine R. Altered histone acetylation at glutamate receptor 2 and brain-derived neurotrophic factor genes is an early event triggered by status epilepticus. J. Neurosci. 2002;22(19):8422–8428. doi: 10.1523/JNEUROSCI.22-19-08422.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sng JC, Taniura H, Yoneda Y. Histone modifications in kainate-induced status epilepticus. Eur. J. Neurosci. 2006;23(5):1269–1282. doi: 10.1111/j.1460-9568.2006.04641.x. [DOI] [PubMed] [Google Scholar]
- 138.Malkani S, Wallace KJ, Donley MP, Rosen JB. An egr-1 (zif268) antisense oligodeoxynucleotide infused into the amygdala disrupts fear conditioning. Learn Mem. 2004;11(5):617–624. doi: 10.1101/lm.73104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Baumgartel K, Genoux D, Welzl H, et al. Control of the establishment of aversive memory by calcineurin and Zif. Nat. Neurosci. 2008;11(5):572–578. doi: 10.1038/nn.2113. [DOI] [PubMed] [Google Scholar]
- 140.Monfils MH, Cowansage KK, LeDoux JE. Brain-derived neurotrophic factor: linking fear learning to memory consolidation. Mol. Pharmacol. 2007;72(2):235–237. doi: 10.1124/mol.107.038232. [DOI] [PubMed] [Google Scholar]
- 141.Sacktor TC. PKMzeta, LTP maintenance, and the dynamic molecular biology of memory storage. Prog. Brain Res. 2008;169:27–40. doi: 10.1016/S0079-6123(07)00002-7. [DOI] [PubMed] [Google Scholar]
- 142.Fuchikami M, Morinobu S, Kurata A, Yamamoto S, Yamawaki S. Single immobilization stress differentially alters the expression profile of transcripts of the brain-derived neurotrophic factor (BDNF) gene and histone acetylation at its promoters in the rat hippocampus. Int. J. Neuropsychopharmacol. 2009;12(1):73–82. doi: 10.1017/S1461145708008997. [DOI] [PubMed] [Google Scholar]
- 143.Chwang WB, O’Riordan KJ, Levenson JM, Sweatt JD. ERK/MAPK regulates hippocampal histone phosphorylation following contextual fear conditioning. Learn Mem. 2006;13(3):322–328. doi: 10.1101/lm.152906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Crosio C, Heitz E, Allis CD, Borrelli E, Sassone-Corsi P. Chromatin remodeling and neuronal response: multiple signaling pathways induce specific histone H3 modifications and early gene expression in hippocampal neurons. J. Cell Sci. 2003;116(Pt 24):4905–4914. doi: 10.1242/jcs.00804. [DOI] [PubMed] [Google Scholar]
- 145.Gupta S, Kim SY, Artis S, et al. Histone methylation regulates memory formation. J. Neurosci. 2010;30(10):3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.An W. Histone acetylation and methylation: combinatorial players for transcriptional regulation. Subcell Biochem. 2007;41:351–369. [PubMed] [Google Scholar]
- 147.Oliver SS, Denu JM. Dynamic interplay between histone H3 modifications and protein interpreters: emerging evidence for a ‘histone language’. Chembiochem. 2010;12(2):299–307. doi: 10.1002/cbic.201000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Meaney MJ, Ferguson-Smith AC. Epigenetic regulation of the neural transcriptome: the meaning of the marks. Nat. Neurosci. 2010;13(11):1313–1318. doi: 10.1038/nn1110-1313. [DOI] [PubMed] [Google Scholar]
- 149.Martinowich K, Hattori D, Wu H, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302(5646):890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 150.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135(7):1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kangaspeska S, Stride B, Metivier R, et al. Transient cyclical methylation of promoter DNA. Nature. 2008;452(7183):112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 152.Metivier R, Gallais R, Tiffoche C, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452(7183):45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 153.Sharma RP, Tun N, Grayson DR. Depolarization induces downregulation of DNMT1 and DNMT3a in primary cortical cultures. Epigenetics. 2008;3(2):74–80. doi: 10.4161/epi.3.2.6103. [DOI] [PubMed] [Google Scholar]
- 154.Marutha Ravindran CR, Ticku MK. Effect of 5-azacytidine on the methylation aspects of NMDA receptor NR2B gene in the cultured cortical neurons of mice. Neurochem. Res. 2009;34(2):342–350. doi: 10.1007/s11064-008-9783-9. [DOI] [PubMed] [Google Scholar]
- 155.Marutha Ravindran CR, Ticku MK. Changes in methylation pattern of NMDA receptor NR2B gene in cortical neurons after chronic ethanol treatment in mice. Brain Res. Mol. Brain Res. 2004;121(1–2):19–27. doi: 10.1016/j.molbrainres.2003.10.025. [DOI] [PubMed] [Google Scholar]
- 156.Marutha RCR, Ticku MK. Role of CpG islands in the up-regulation of NMDA receptor NR2B gene expression following chronic ethanol treatment of cultured cortical neurons of mice. Neurochem. Int. 2005;46(4):313–327. doi: 10.1016/j.neuint.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 157.Qiang M, Denny A, Chen J, Ticku MK, Yan B, Henderson G. The site specific demethylation in the 5′-regulatory area of NMDA receptor 2B subunit gene associated with CIE-induced up-regulation of transcription. PLoS One. 2010;5(1):e8798. doi: 10.1371/journal.pone.0008798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lee S, Kim W, Ham BJ, Chen W, Bear MF, Yoon BJ. Activity-dependent NR2B expression is mediated by MeCP2-dependent epigenetic regulation. Biochem. Biophys. Res. Commun. 2008;377(3):930–934. doi: 10.1016/j.bbrc.2008.10.082. [DOI] [PubMed] [Google Scholar]
- 159.Tian F, Hu XZ, Wu X, et al. Dynamic chromatin remodeling events in hippocampal neurons are associated with NMDA receptor-mediated activation of Bdnf gene promoter 1. J. Neurochem. 2009;109(5):1375–1388. doi: 10.1111/j.1471-4159.2009.06058.x. [DOI] [PubMed] [Google Scholar]
- 160.Levenson JM, Qiu S, Weeber EJ. The role of reelin in adult synaptic function and the genetic and epigenetic regulation of the reelin gene. Biochim. Biophys. Acta. 2008;1779(8):422–431. doi: 10.1016/j.bbagrm.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 161.Koshibu K, Graff J, Mansuy IM. Nuclear protein phosphatase-1: an epigenetic regulator of fear memory and amygdala long-term potentiation. Neuroscience. 2011;173:30–36. doi: 10.1016/j.neuroscience.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 162.Abel T, Martin KC, Bartsch D, Kandel ER. Memory suppressor genes: inhibitory constraints on the storage of long-term memory. Science. 1998;279(5349):338–341. doi: 10.1126/science.279.5349.338. [DOI] [PubMed] [Google Scholar]
- 163.Costa E, Dong E, Grayson DR, Guidotti A, Ruzicka W, Veldic M. Reviewing the role of DNA (cytosine-5) methyltransferase overexpression in the cortical GABAergic dysfunction associated with psychosis vulnerability. Epigenetics. 2007;2(1):29–36. doi: 10.4161/epi.2.1.4063. [DOI] [PubMed] [Google Scholar]
- 164.Jiang Y, Langley B, Lubin FD, et al. Epigenetics in the nervous system. J. Neurosci. 2008;28(46):11753–11759. doi: 10.1523/JNEUROSCI.3797-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kim TK, Hemberg M, Gray JM, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Smalheiser NR, Lugli G, Thimmapuram J, Cook EH, Larson J. Endogenous siRNAs and noncoding RNA-derived small RNAs are expressed in adult mouse hippocampus and are up-regulated in olfactory discrimination training. RNA. 2011;17(1):166–181. doi: 10.1261/rna.2123811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Edbauer D, Neilson JR, Foster KA, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR. Neuron. 2010;65(3):373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Rajasethupathy P, Fiumara F, Sheridan R, et al. Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron. 2009;63(6):803–817. doi: 10.1016/j.neuron.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gao J, Wang WY, Mao YW, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR. Nature. 2010;466(7310):1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ma DK, Marchetto MC, Guo JU, Ming GL, Gage FH, Song H. Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat. Neurosci. 2010;13(11):1338–1344. doi: 10.1038/nn.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]