Abstract
Transposable elements (TEs) are highly abundant in the genome and capable of mobility, two properties that make them particularly prone to transfer horizontally between organisms. Although the impact of horizontal transfer (HT) of TEs is well recognized in prokaryotes, the frequency of this phenomenon and its contribution to genome evolution in eukaryotes remain poorly appreciated. Here, we provide evidence that a DNA transposon called SPIN has colonized the genome of 17 species of reptiles representing nearly every major lineage of squamates, including 14 families of lizards, snakes, and amphisbaenians. Slot blot analyses indicate that SPIN has amplified to high copy numbers in most of these species, ranging from 2,000–28,000 copies per haploid genome. In contrast, we could not detect the presence of SPIN in any of the turtles (seven species from seven families) and crocodiles (four species) examined. Genetic distances between SPIN sequences from species belonging to different squamate families are consistently very low (average = 0.1), considering the deep evolutionary divergence of the families investigated (most are >100 My diverged). Furthermore, these distances fall below interfamilial distances calculated for two genes known to have evolved under strong functional constraint in vertebrates (RAG1, average = 0.24 and C-mos, average = 0.27). These data, combined with phylogenetic analyses, indicate that the widespread distribution of SPIN among squamates is the result of at least 13 independent events of HTs. Molecular dating and paleobiogeographical data suggest that these transfers took place during the last 50 My on at least three different continents (North America, South America and, Africa). Together, these results triple the number of known SPIN transfer events among tetrapods, provide evidence for a previously hypothesized transoceanic movement of SPIN transposons during the Cenozoic, and further underscore the role of HT in the evolution of vertebrate genomes.
Keywords: horizontal transfer, transposable elements, reptiles
Introduction
Horizontal transfer (HT) is the passage of genetic material between reproductively isolated organisms. This mode of transmission is common in prokaryotes, where it often serves as a source of genomic innovation (Ochman et al. 2000; Koonin et al. 2001). HT is also increasingly recognized as an important evolutionary force shaping eukaryotic genomes. Most HT events characterized so far in eukaryotes correspond to prokaryote-to-eukaryote or organelle-to-nucleus gene transfers (see Marcet-Houben and Gabaldón 2009 and Danchin et al. 2010 for recent examples and Keeling and Palmer 2008 and Andersson 2009 for reviews). By comparison, relatively few cases of gene transfers have been described between multicellular eukaryotes (but see Gladyshev et al. 2008, Moran and Jarvik 2010 and Slot and Rokas 2011), and the majority of reported HTs between metazoans correspond to transfers of transposable elements (TEs; Schaack et al. 2010). TEs are segments of DNA that are able to move between different genomic loci, often duplicating themselves in the process (Craig et al. 2002). Two properties of these elements suggest that they may be more likely than genes to transfer horizontally between organisms: they are capable of mobility and they often represent the single most abundant component of eukaryotic genomes—for example, TEs make up ∼45% and ∼85% of the human and maize genomes, respectively (Lander et al. 2001; Schnable et al. 2009).
Over 200 solid cases of horizontal transfers of transposable elements (HTT) have been described so far in multicellular eukaryotes (Loreto et al. 2008; Schaack et al. 2010). This number may be viewed as surprisingly high given that only few large-scale searches have been conducted. In one of these searches, we computationally screened all eukaryotic genomes available in GenBank for the presence of four families of DNA-mediated (or class 2) transposons named SPACE INVADERS (or SPIN), OposCharlie1, hAT1, and ExtraTerrestrial (Pace et al. 2008; Gilbert et al. 2010). We found that these TEs were patchily distributed among metazoans and that they exhibited extremely high levels of nucleotide similarity between the different species in which they were found (up to 98%). Given the absence of purifying selection acting on the TE coding sequences following their genomic insertion and the deep evolutionary divergence times separating the various host species (80 to >500 My), we inferred that such levels of sequence similarity were incompatible with vertical inheritance of the TEs from a common metazoan ancestor. We concluded that the observed taxonomic distribution of these four families of TEs was the result of multiple events of HT. Estimates of the ages of these transposon families suggested that most of the transfers occurred within a relatively narrow evolutionary window, between 50 and 10 Ma (Gilbert et al. 2010). Together with other recent studies (Novick et al. 2010; Pagan et al. 2010; Thomas et al. 2010), these results indicate that the frequency and impact of HTT in metazoans may be underappreciated.
One limitation of the aforementioned studies is that they were based on computational analyses of relatively few genome sequences available in the databases. Although the number of eukaryotic genome sequences is increasing rapidly, it still accounts for a minuscule fraction of known extant species (less than 1,000 of >>1 million eukaryotes), and the distribution of sequenced genomes among taxa remains heavily biased toward species closely related to model organisms (Schaack et al. 2010). One way to improve our appreciation of the frequency of HTT and of its impact on eukaryotic genomes is to use a targeted and systematic experimental approach in order to extend the search for TEs known to have horizontally transferred to large taxonomic groups for which few or no complete genomes are available. Here, we initiate such a strategy by screening a comprehensive taxonomic sample of nonavian reptiles for the presence of SPIN transposons using a combination of polymerase chain reaction (PCR)/sequencing and slot blot hybridizations. Nonavian reptiles include four major lineages: the squamates (8,200 species; Hedges and Vidal 2009), the turtles (313 species; Shaffer 2009), the crocodiles (23 species; Brochu 2003), and the sphenodontians (one species; Hay et al. 2010). The rationale for targeting this taxonomic group was 2-fold. First, we knew that SPIN has invaded the green anole lizard Anolis carolinensis (Pace et al. 2008), the only nonavian reptile that has its genome completely sequenced, indicating that squamates are not refractory to HTT. Second, we have access to a diverse and representative collection of tissue samples through the UT Arlington Amphibian and Reptile Diversity Research Center.
Materials and Methods
DNA Extractions, PCR, Cloning, and Sequencing
The list of tissue samples used in this study is provided in supplementary table 1, Supplementary Material online. DNA was extracted using the DNeasy blood and tissue kit (Qiagen). The PCR screening for the presence of SPIN was conducted using two different sets of primers. The first one, designed to preferentially amplify small nonautonomous elements, included one primer anchored in the 5′ noncoding region of SPIN (NAF2 5′-CGA ACG ACC CTT TCA CAG G; position 41–59 of the SPIN superconsensus provided in Pace et al. (2008)) and one primer anchored in the 3′ noncoding region of SPIN (NAR2 5′-CAG TTC CTC ATG TTG TGG TGA C; position 2812–2833 of the SPIN superconsensus). The second set of primers was designed to amplify a ∼400-bp fragment of the SPIN transposase (SPINtpaseF2 5′-CAT GTT GCC TAC CTT ATC TGC; position 2005–2025 of SPIN superconsensus and SPINtpaseR2 5′-ACT TGA TAA CCA ACA AGC TGG; position 2379–2399 of SPIN superconsensus). PCR reactions were conducted using the following temperature cycling: initial denaturation at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 54 °C (for both primer sets) for 30 s, and elongation at 72 °C for 1 min, ending with a 10 min elongation step at 72 °C. Fragments from the PCR were visualized on a 1–2% agarose gel, cloned, and sequenced. Cloning was performed using the Strataclone PCR cloning kit (Stratagene) following manufacturer’s protocols and successfully transformed bacterial colonies were screened by PCR (same thermocycling program as above) using M13 primers (M13F 5′-GTA AAA CGA CGG CCA G; M13R 5′-CAG GAA ACA GCT ATG AC; Annealing temperature = 58 °C). Between five and ten amplicons from each cloning, products were directly sequenced using ABI’s BigDye sequencing mix (1.4 μl template PCR product, 0.4 μl BigDye, 2 μl manufacturer supplied buffer, 0.3 μl reverse primer, and 6 μl H2O). The thermocycler program was as follows: 2 min denaturation (96 °C) followed by 30 cycles alternating between 96 °C (30 s) and 60 °C (4 min), ending at 10 °C for 3 min. Sequencing reactions were ethanol precipitated and run on an ABI 3730 sequencer.
The identity of the species included in this study was verified by sequencing a portion of C-mos, a nuclear gene that has already been sequenced for many reptile species as part of several phylogenetic studies. PCR reactions for this gene were carried out using the primers and the protocol described in Saint et al. (1998). When C-mos was not available in public databases (five species, see supplementary table 1, Supplementary Material online), we sequenced a portion of the mitochondrial cytochrome oxidase subunit I (COI) using the following primers: (COItF 5′-CAC CAG ATA TAG CAT TTC CAC G and COItR 5′-GCT GGG GAT TTT ATG TTG ATT G for turtles and COIcF 5′-GCA CCC GAC ATA GCA TTY CC and COIcR 5′-TGG GTG GCC GAA GAA TCA G for crocodiles). PCR products were sequenced directly (without cloning), following the protocol described above. All C-mos and COI sequences produced in this study have been deposited in GenBank (accession numbers: JN090128–JN090158; see also supplementary data sets 1 and 2, Supplementary Material online), and SPIN sequences are provided in supplementary data sets 3–6, Supplementary Material online.
Slot Blot
For each species included on the slot blot, 1 μg of genomic DNA was denatured in a solution containing 0.4 M NaOH and 10 mM ethylenediaminetetraacetic acid at 90 °C for 10 min and deposited onto a Hybond-N+ membrane (Amersham) using a Minifold I Slot-Blot System (Whatman). The concentration of the DNA samples was measured using a NanoDrop (1000 Spectrophotometer) and by running an ethidium bromide–stained agarose gel. We noticed a few discrepancies between the DNA concentration measured with the NanoDrop and the relative intensity of the DNA samples on the agarose gel. In these instances, we chose to use the results of the ethidium bromide staining to adjust the amount of DNA deposited on the membrane and to ensure that these amounts were equal for all taxa. The membrane was then rinsed in 2× standard saline citrate (SSC), air dried for 3 min, and ultra violet cross-linked. In our previous reports based on computational scanning of SPIN in whole-genome sequences (Pace et al. 2008; Gilbert et al. 2010), we have observed that the copy numbers of both nonautonomous (NA) and autonomous (auto) SPIN can vary greatly between species. For example, there are 4,812 SPIN copies (4,807 NA; <5 auto) in the opossum (Monodelphis domestica) genome sequence and 33,123 SPIN copies (25,978 NA; 7,145 auto) in bushbaby (Otolmeur garnettii). In addition, we noticed that in some species such as the lizard A. carolinensis, SPIN elements may be highly fragmented. Thus, using a single short fragment of the SPIN autonomous sequence as a probe for the slot blot would only allow detecting a very minor portion of the total number of SPIN copies present in this genome. In order to enhance our ability to detect both autonomous and nonautonomous elements as well as fragmented copies, we used a mixture of three SPIN fragments as probe for hybridization. The first fragment is 260 bp long and corresponds to the very 5′ end of a SPIN copy, amplified in the genome of the tenrec (Echinops telfairi) using a primer anchored in the 5′ flanking region of this copy (5′dotblot2Fwd 5′-CTT GCC TAA TGT CTT AGA GCA G) as well as a primer anchored at position (221–244) of the SPIN superconsensus (5′dotblot2Rev 5′-TGT CAC ATA CAC CTG TAT TTG TAC). The second fragment is 304 bp long and corresponds to the very 3′ end of the same SPIN copy amplified in the tenrec genome using a primer anchored in the 3′ flanking region of this copy (3′dotblot54Rev 5′-GTT GGA AGA AAG GTC TAG GTC AG) and a primer anchored at position (2583–2603) of the SPIN superconsensus (3′dotblot55Fwd 5′-TGC TAC ACC ATG CTT CAA GAC). The third fragment corresponds to the portion of the transposase gene and was amplified in two different genomes (Ameiva undulata, Sceloporus adleri) using the primers SPINtpaseF2/R2 (see above for sequence). The PCR products of each of these fragments were cloned as described above. The products obtained from a second PCR performed on one clone for each of the fragments were then mixed and [α-32P]dCTP-labeled using the Random Primed DNA Labeling Kit (Roche). To verify that the transfer of DNA onto the nylon membrane was successful, we used as positive control a 242-bp fragment of the gene encoding the 28S ribosomal RNA, amplified in Cordylus tropidosternum with the following primer set (28SF 5′-AGG TGT CCT AAG GCG AGC; 28SR 5′-GAT AGG AAG AGC CGA CAT CG). This fragment was cloned and labeled as described above. The membrane was first hybridized with the 28S probe, washed and exposed during 24 h on X-ray film (Kodak). It was then stripped, exposed for 24 h to verify that no DNA was left on the membrane after stripping, and hybridized with the SPIN probe. After washing the excess of SPIN probe, the membrane was exposed for another 24 h on X-ray film. Hybridizations were performed in PerfectHyb Plus hybridization buffer (Sigma) at 65 °C. For each washing, the membrane was soaked twice 30 min into a solution of 1× SSC/0.1% sodium dodecyl sulfate.
To approximate the number of SPIN copies present in squamate genomes, three dilutions (2.5, 1, and 0.5 ng) of unlabeled probe were blotted onto the membrane. The number of SPIN fragments contained in each dilution (7.41 × 109, 3.095 × 109, and 1.5475 × 109, respectively) was calculated using the formula “copy number = (amount in ng × number/mole)/(length in bp × ng/g × g/mole of bp)” described and implemented on the website http://www.uri.edu/research/gsc/resources/cndna.html. The integrated density of the signal produced by each dilution on the dot blot was measured using ImageJ 1.43u (http://rsb.info.nih.gov/ij/). Based on the comparison of these three densities, we calculated that doubling the probe quantity led to a 1.25- to 1.3-fold increase of the signal. We therefore estimated the total copy numbers for each species by comparing the intensity of each signal to that of the signal obtained with the 1 ng dilution and assuming that a 1.27-fold change in density translates into a 2-fold change in copy number. In order to calculate the copy numbers per haploid genomes, we used C values taken from the Animal Genome Size Database (Gregory 2011) and assumed that 1 pg = 1 Gb. When multiple C values were given for one species, we used the average of these values. When a C value was not available for a species but was available for various other species of the same genus, we used the average of these values. When a C value was not available for any species belonging to the same genus as one of the species included in this study, we used an arbitrary C value of 2, corresponding to the average of all other squamate C values that we were able to find in the database. These genome sizes allowed us to estimate the number of haploid genomes present in 1 μg of DNA for each species using the formula given above.
Phylogenetic Analyses
All autonomous reptile SPIN sequences obtained in this study were aligned together with five SPIN sequences randomly extracted from the genome of the species where these elements have been previously described (Pace et al. 2008; Novick et al. 2010; Gilbert et al. 2010). The multiple alignment is provided in supplementary data set 6, Supplementary Material online. Because of the low copy number of autonomous SPIN elements in the lizard (A. carolinensis) and opossum (M. domestica) genome, only one sequence corresponding to the aligned region could be included for these two species. Similarly, all SPIN clones sequenced from Scincus scincus were identical, and we therefore included only one of them in the phylogenetic analysis. Sequences were aligned by hand using BioEdit 7.0.5.3 (Hall 2004), and regions absent in more than half of the sequences were removed. The model of nucleotide evolution best fitting this alignment (TPM3uf+G) was chosen based on the Akaike information criterion (AIC) in jModeltest 0.1.1 (Posada 2008). This model was then entered in PhyML 3.0 (Guindon and Gascuel 2003) to perform a bootstrap analysis involving 1,000 replicates in a maximum likelihood framework.
Distances between SPIN, C-mos, and RAG1 Sequences
Pairwise distances between the different reptile species included in this study were calculated for SPIN, C-mos, and RAG1 sequences. For SPIN, the distances were first calculated between majority rule SPIN consensus sequences derived for each species based on an alignment of individual copies (except in S. scincus for which we were able to isolate a single sequence). We also calculated the average distances between individual SPIN copies within each species where multiple SPIN copies were sequenced. For C-mos, distances were calculated between the sequences produced during this study (see above), and for RAG1, we used sequences available in GenBank for the species in which we found SPIN or for closely related species (See supplementary table 1, Supplementary Material online for accession numbers). The model of nucleotide evolution best fitting each alignment was chosen using the AIC in jModeltest (Posada 2008) and was entered in PAUP 4.0 (Swofford 2003) to calculate pairwise distances under maximum likelihood settings. The multiple alignments used to calculate these distances are provided in supplementary data sets 1–4, Supplementary Material online.
Analyses of Selection
For each squamate species where we found SPIN elements, we tested whether the pattern of mutations observed between each copy and the consensus (an estimate of the ancestral founder element) was significantly different from what is expected if the sequence is evolving neutrally using the codon-based Z-test in MEGA 4.036 with the Nei–Gojobori method and the Jukes–Cantor correction (500 bootstrap replicates). The results of the test showed that the number of nonsynonymous substitutions accumulated on SPIN copies since their insertion in the various squamates was not significantly different from the number of synonymous substitutions. This result indicates that the SPIN copies are evolving neutrally after insertion in the genome.
Paleobiogeographic Data
In order to infer the most likely distribution of squamate species at the time of the SPIN transfers, we gathered fossil and molecular dating evidence from the literature (fig. 1). Currently, Amphisbaena alba is widely distributed in South America, and the ancestors of the genus Amphisbaena are thought to have originated on this continent >80 Ma (Macey et al. 2004). The family Teiidae (whiptails) likely originated in South America during the Paleocene (>55 Ma), and the presence of A. undulata in Central America and southern Mexico is probably the result of recent dispersal (Giugliano et al. 2007). According to Fuller et al. (1998), varanid lizards arose in Asia around 65 Ma and migrated to Africa, where Varanus exanthematicus is found today, during the late Tertiary period. The oldest African fossils attributed to the varanid family are found in the early Miocene of Kenya (∼18 Ma; Clos 1995). The current distribution of the anguid lizard Mesaspis moreletii covers most highlands of Nuclear Central America including southern Mexico. The species is part of the Gerrhonotinae, a subfamily that arose in North America at least 50 Ma (Macey et al. 1999). Beaded lizards (Helodermatidae), today distributed in the southwestern United States, Mexico and Guatemala, originated in North America about 35 Ma (Douglas et al. 2010). Furthermore, the fossil record indicates that the ancestors of this family occurred in North America during the early Cretaceous (>100 Ma; Cifelli and Nydam 1995; Nydam 2000). The snakes Hypsiglena torquata (Dipsadidae) and Nerodia erythrogaster (Natricidae) are found today in the southwestern United States and Mexico and southeastern United States, respectively. The two species belong to the superfamily Dipsadoidea, which likely originated in Asia during the Tertiary. Biogeographical scenarios are, however, equivocal regarding the precise date at which the ancestors of the Dipsadidae and Natricidae first migrated to the New World (Pinou et al. 2004). The Dipsadoidea may have entered the New World either at the same time as the Colubridae (16–10 Ma; Vidal et al. 2000) or at the same time as the Boidae (48 Ma; Estes and Hutchinson 1980). The three viperid snakes (Agkistrodon contortrix, Crotalus atrox, and Sistrurus catenatus) are widely distributed in North America. It is thought that viperid snakes migrated from Asia to the New World through the Bering Land Bridge and the earliest North American fossil attributed to the family is from the Early Miocene (22 Ma old; Douglas et al. 2006). Among iguanids, anole lizards and the subfamily Phrynosomatinae (to which S. adleri belongs) are endemic to Central-South America and North America, respectively. Although the biogeographic history of the family Iguanidae is complex, both fossil evidence and molecular dating suggest that the ancestors of these two taxa arose in the New World more than 50 Ma (Noonan and Chippindale 2006; Smith 2009). Agama agama is currently restricted to Africa, but it is unclear whether African agamids originated in Africa or Asia and the age of their ancestor has not been precisely determined within the Cenozoic (Macey et al. 2000; Okajima and Kumazawa 2010). Scincus scincus is currently found in Africa, and it is thought that the family Scincidae (skinks) originated in Africa during the Cenozoic more than 23 Ma (Greer 1970; Whiting et al. 2003; Carranza et al. 2008). Lepidophyma flavimaculatum and the 30 other species of the Xantusidae family (night lizards) are all endemic to North and Central America and Cuba, and molecular dating indicates that they arose on this region around 60 Ma (Vicario et al. 2003). Hemidactylus geckos most likely originated in the Old World, but it is unclear where and when exactly (Carranza and Arnold 2006). Cordylid or spinytail lizards (Cordylidae) are all distributed in sub-saharan Africa and their origin dates back to more than 35 Ma on this continent (Stanley et al. 2011). In addition, the sister family of Cordylidae (Gerrhosauridae, plated lizards) is also native from Africa, and the oldest fossils attributable to the (Cordylidae + Gerrhosauridae) lineage are from the upper cretaceous (>65 Ma) of Madagascar (Krause et al. 2003).
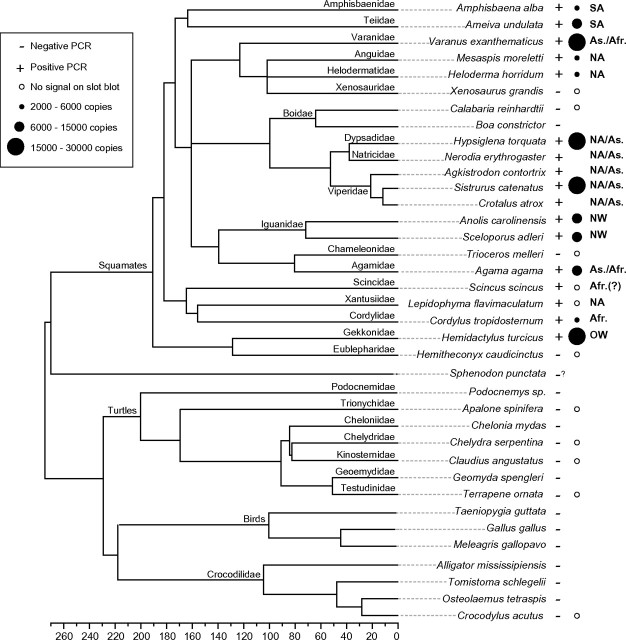
FIG. 1.
Phylogenetic tree of nonavian reptile taxa screened in this study and overview of the results. The phylogenetic relationships and divergence times are taken from Hedges and Vidal (2009), Shaffer (2009), and Roos et al. (2007). The tree summarizes the results of the slot blot and PCR screening. SPIN copy numbers are derived from supplementary figure 1, Supplementary Material online (see also Materials and Methods and supplementary table 1, Supplementary Material online). Also listed is the likely geographic distribution of the taxa during the last ∼50 My (the time during which SPIN were transferred horizontally to the various squamate lineages). The references used to infer these ancestral distributions are cited in the Materials and Methods. SA = South America; NA = North America; As. = Asia; Afr. = Africa; NW = New World; OW = Old World; slash bar = or/and (e.g., NA/SA indicates that the taxa were present either in North America, either in South America, or on both continent at the time of the transfer). Museum voucher numbers of the tissues, when available, are listed in supplementary table 1, Supplementary Material online. The “–?” symbol next to tuatara indicates that we have not found any SPIN sequence in the BAC inserts available in NCBI but that given that these sequences only represent a small portion of the tuatara genome (see Results), we cannot conclude on whether SPIN is present or absent in this genome.
Results
Taxonomic Sample and PCR Screening
Our search for the presence of SPIN transposons in reptiles began with an initial PCR screen of 46 squamate species (snake, lizards, and amphisbaenians), 6 species of crocodiles and 23 species of turtles. The two other groups of reptiles (birds and tuatara) were not included in this study, but it is worth mentioning that our Blast searches using the SPIN superconsensus as a query on the chicken, zebra finch, and turkey genome (the three birds species for which whole-genome sequences are available at present, Hillier et al. 2004; Dalloul et al. 2010; Warren et al. 2010) did not yield any significant hit, suggesting that SPIN is absent from these species. We also extended these searches to the sequence of ten tuatara bacterial artificial chromosome (BAC) clones produced by the NIH Intramural Sequencing Center and representing a total of 1.6 Mb. This search did not retrieve any positive hit either but given that these BAC sequences represent only a small fraction of the tuatara genome (C value = 5 pg, Olmo 1981), we cannot exclude that SPIN is not present in this genome.
Our initial screen yielded positive PCR products for most squamate species (32 of 46) and negative amplifications for all crocodiles and turtles (not shown). Based on these results, we narrowed down our taxonomic sampling to retain only the species for which we could obtain sufficient good quality DNA in order to repeat PCRs, cloning, and sequencing and to perform a slot blot analysis. In addition, we selected taxa so as to maximize the coverage of the phylogenetic diversity of nonavian reptiles, and we preferentially chose those species for which the gene C-mos was available in GenBank in order to facilitate the validation of species ID (see Materials and Methods). In the end, we thoroughly screened 20 species representing 18 of the 58 families of squamates (and 14 of the ∼32 non-snake families, Hedges and Vidal 2009), 4 species representing the two families of crocodiles (Brochu 2003), and 7 species representing half of the 14 families of turtles (Shaffer 2009). The results of this screen are summarized in figure 1. In summary, we were able to amplify and sequence multiple copies of nonautonomous and/or autonomous SPIN in 16 of 20 squamate species. PCRs at annealing temperatures lower than the one used in squamates yielded DNA bands on agarose gel electrophoresis in some species of turtles and crocodiles, but none of them were confirmed as SPIN by DNA sequencing.
Ruling Out Contamination
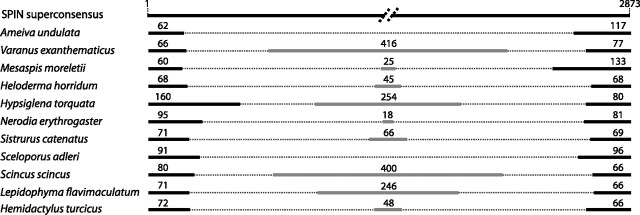
SPIN sequences previously characterized in other metazoans are typically more than 95% identical between the different species in which they occur and most do not form monospecific phylogenetic groups (Pace et al. 2008; Gilbert et al. 2010). This means that contamination of genomic DNA would be very difficult to detect in a PCR screen like the one deployed here. Several lines of evidence, however, suggest that the results presented herein are not due to cross DNA or tissue sample contaminations. First, we verified the species ID of all tissue samples used by sequencing the gene C-mos or COI. For each of the C-mos and COI sequence obtained, the best Blast hit in GenBank was the expected species, or a species of the same family in cases where the gene of the expected species was not available in Genbank (note: all sequences generated in this study have now been deposited in Genbank). Furthermore, all the validated sequence reads were clean, without double peaks that would have indicated that some of our DNA samples were heavily contaminated. Third, of the 13 squamate species that yielded positive SPIN PCR/sequence and that were included on the slot blot (see below and suppplementary fig. 1, Supplementary Material online), 10 produced a strong signal, unlikely to result from minor DNA contaminations (see text below for the three species that did not produced a strong signal). Fourth, the structure of each nonautonomous element was species specific (fig. 2 and supplementary data set 5, Supplementary Material online), that is, we never sequenced the same subfamily of nonautonomous elements in two different species. Nonautonomous class 2 elements are generally amplified from deletion derivatives of autonomous elements that most likely result from aborted double-stranded gap repair following excision (Engels et al. 1990). On occasion, a non-TE “filler” DNA segment may be inserted during the gap repair process, producing a more complex nonautonomous element with an extraneous internal region (Rubin and Levy 1997; Yan et al. 1999). All but two subfamilies of nonautonomous elements sequenced in squamate species correspond to such complex nonautonomous elements (fig. 2). This type of rearranged elements appears to be particularly common in the lizard A. carolinensis, the only squamate for which whole-genome sequences are available so far in GenBank (Novick et al. 2010). Interestingly, the non-TE fragments differed in all species both in terms of length and primary sequence. This observation coupled to the fact that the positions of the deletion breakpoints often vary between species (fig. 2) suggests that the different nonautonomous elements originated independently in each squamate lineage where SPIN was identified and therefore provides strong evidence against contamination. We reextracted DNA and resequenced multiple SPIN sequences in the two species that produced no signal on the slot blot (L. flavimaculatum, S. scincus). In both cases, we obtained nonautonomous elements structurally identical to those we had sequenced previously, suggesting that SPIN are indeed present in these two species, though most likely at copy numbers that are too low to be detected by slot blot hybridization. Finally, it is worth mentioning that for one of the squamate family included in this study (Viperidae), the presence of SPIN was also verified through another independent investigation based on low-coverage (4.5%) high-throughput genome sequencing of the copperhead (A. contortix) (Castoe et al. 2011). Searching the sequence data generated by these investigators, we identified hundreds of SPIN sequences (autonomous and nonautonomous, together occupying ∼80 kb of DNA) that are more than 90% identical to the SPIN superconsensus reconstructed in Pace et al. (2008). Among these, we found two SPIN fragments identical to the transposase region isolated herein by PCR. Note these two fragments are also 97% identical to the SPIN consensus of another pitviper examined, the massassauga rattlesnake (S. catenatus), and all of these sequences cluster phylogenetically (see below) with the SPIN copies sequenced from the two pitvipers included in our screening (S. catenatus and C. atrox).
FIG. 2.
Schematic representation of the structure of nonautonomous SPIN elements sequenced in various species of squamates. For each nonautonomous element, the 5′ and 3′ regions that are homologous to the autonomous element are represented in black (the length of each sequence is indicated on top of them in bp). The internal non-SPIN region is represented in gray. All internal non-SPIN regions differ in terms of primary sequence (no homology between them) and length (indicated on top of each sequence in bp).
Evidence for Multiple Horizontal Transfers of SPIN in Squamates
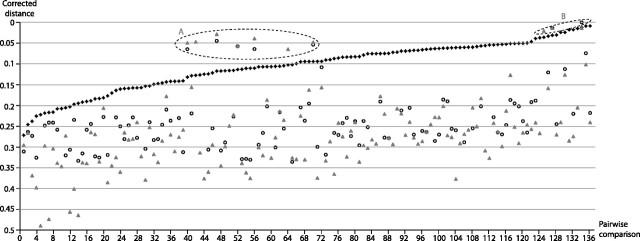
For most squamate species in which we detected SPIN, we were able to sequence between three and five segments of transposase coding regions (supplementary table 1, Supplementary Material online). All clones (n = 8) sequenced in S.scincus were identical, which together with the absence of signal on the slot blot for this species suggests that there might be only one or a few fragmented remnants of autonomous SPIN in this species, as in the genome of A. carolinensis (Pace et al. 2008). Nucleotide genetic distances between individual SPIN copies within species were extremely low (supplementary table 1, Supplementary Material online; average = 0.034; standard deviation [SD] = 0.02; range = 0.08–0.01). To assess SPIN distances between species, we reconstructed a majority-rule SPIN consensus for each species and computed pairwise distances between all consensus sequences (fig. 3; supplementary table 2, Supplementary Material online). All these comparisons showed distances that are also extremely low (average = 0.1; SD = 0.06; range = 0.008–0.27). These distances fall within the range of those computed previously for the same region of the autonomous SPIN sequence among metazoan species in which SPIN is known to have been introduced horizontally (Pace et al. 2008; Gilbert et al. 2010; Novick et al. 2010). As in these previous studies, we found no evidence of purifying selection acting on the coding sequence of SPIN in squamates (see Materials and Methods). This suggests that, like in other metazoans, SPIN elements are not evolving under functional constraint after insertion in squamate genomes. Almost all (113 of 120) SPIN pairwise distances computed herein involve species that diverged from each other more than 100 Ma (supplementary table 2, Supplementary Material online). Given these deep divergence times and the absence of purifying selection acting on SPIN sequences, the extremely low pairwise SPIN distances seem incompatible with a scenario invoking vertical inheritance of these transposons from the ancestor of squamates. Indeed, for most pairwise comparisons (114 of 120), the distances calculated for SPIN are much lower than those computed for RAG1 (supplementary table 2, Supplementary Material online; average = 0.24; SD = 0.07; range = 0.01–0.33) and C-mos (supplementary table 2, Supplementary Material online; average = 0.27; SD = 0.09; range = 0.01–0.49), two essential genes that must have evolved under strong functional constraint in virtually all vertebrate lineages (fig. 3). The average difference between pairwise distances calculated for these genes and for SPIN are 0.14 for RAG1 (SD = 0.06; range = 0.01–0.23) and 0.18 for C-mos (SD = 0.06; range = 0.02–0.31). Together, these data strongly suggest that the presence of SPIN in most of the major squamate lineages examined in this study is the result of independent HT events that occurred after these lineages diverged from each other.
FIG. 3.
Graph illustrating the pairwise corrected distances of SPIN (black lozenges), RAG1 (gray triangles), and C-mos (open circles) between the squamate lineages included in this study. The distances are derived from all possible pairwise comparisons (n = 136; labeled on the x axis) between the 17 species of squamates in which SPIN was found. Distances as well as divergence times for each pairwise comparison are listed in supplementary table 2, Supplementary Material online. Before plotting the distance values, they were sorted by ascending order of SPIN distances. Distances falling into A and B dotted circles correspond to pairwise comparisons for which RAG1 and C-mos distances are lower than SPIN distances (see Results).
SPIN Phylogeny
The general topology of the SPIN phylogeny (fig. 4) is unresolved and star-like, indicating that, following insertion, most transposon copies accumulate discrete substitutions at the neutral rate of the species (consistent with the pattern observed for other DNA transposon families, e.g., see Robertson 2002). In several instances (e.g., Hemidactylus turcicus, L. flavimaculatum, Heloderma horridum, Tenrec ecaudatus, and Otolemur garnettii) and as previously observed (Pace et al. 2008; Gilbert et al. 2010), SPIN copies do not form monospecific clusters, suggesting that the genome of multiple species were invaded by very similar or identical founder elements. In some species (e.g., A. agama, A. undulata, and N. erythrogaster, S. alderi), however, SPIN copies cluster into strongly supported monospecific groups separated from the other SPIN copies by relatively long branches (fig. 4). This pattern indicates that the founder element invading these species is phylogenetically distinct from the elements identified in other metazoan genomes. There are two instances in the phylogeny where SPIN copies from different host species cluster together in a strongly supported clade (color branches in fig. 4). The first cluster includes copies from an amphisbaenian (A. alba), a marsupial (M. domestica), and a hemipteran insect (Rhodnius prolixus). This grouping is not only completely incongruent with the species phylogeny, further supporting HT, it may also be indicative of direct transfers between these species given their geographical and ecological overlap (see Discussion). The second cluster groups SPIN copies from the three viperid snakes examined in this study (bootstrap = 65). Given the taxonomic proximity of these snakes, this grouping most likely reflects that SPIN was horizontally introduced in a common ancestor of these species (see below).
FIG. 4.
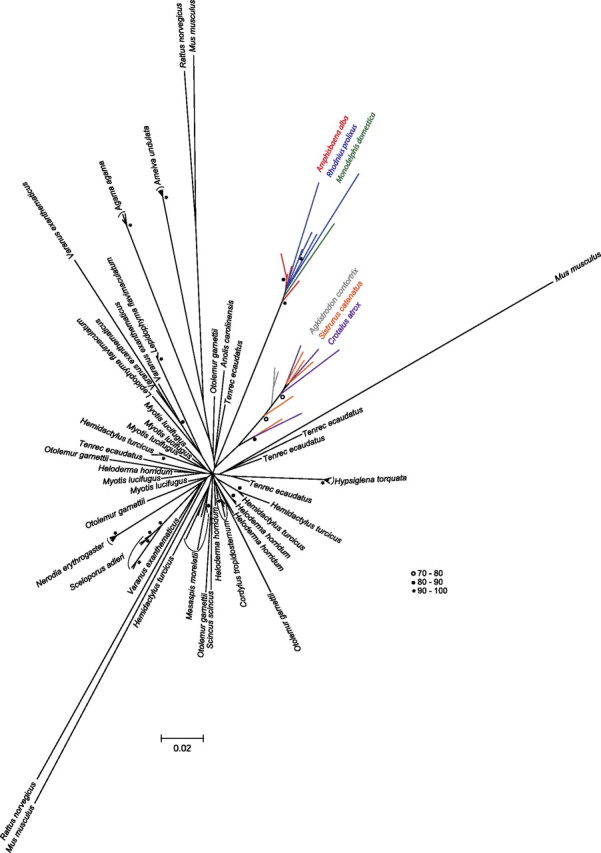
Phylogenetic analysis of SPIN sequences. The tree is based on an alignment of a ∼400-bp region of the SPIN transposase. It includes all autonomous squamate SPIN copies sequenced in this study and five copies randomly extracted from the genomes of the species in which SPIN has been previously characterized. The genome of Anolis carolinensis and Monodelphis domestica contain only one autonomous SPIN copy that includes the transposase region used in this analysis. The phylogenetic analysis was carried out in a maximum likelihood framework using PhyML 3.0. Bootstrap values (BP) were obtained after analysis of 1,000 replicates of the original matrix and are shown as open circles (70 < BP < 80), black circles (81 < BP < 90), or asterisks (90 < BP < 100).
SPIN versus RAG1 and C-mos Distances: Exceptions to the Trend
For all pairwise comparisons where SPIN distances are lower than the distances for RAG1 and C-mos (126 of 136 pairwise comparisons), the introduction of SPIN via HT must have occurred at a time when RAG1 and C-mos were already very divergent from each other (considering purifying selection), that is, well after the speciation events separating each pair of species compared. For ten pairs of species, however, the distances calculated between SPIN consensus sequences are greater than the distances calculated for the two genes (see A and B dotted circles on fig. 3 and supplementary table 2, Supplementary Material online). All ten comparisons involve snake species that have diverged from each other less than 55 Ma and are at least 50 My less divergent than most other species pairs. Two hypotheses may explain the inverse relationship between SPIN and RAG1/C-mos distances for these ten pairs of species: 1) SPIN was horizontally transferred in the ancestor of the five species involved in these comparisons and was subsequently transmitted vertically and accumulated mutations faster (at the neutral rate of each species) than RAG1 and C-mos that are evolving under functional constraints and 2) SPIN was introduced horizontally in each species but the time at which these HT events occurred was very close to the time at which the species diverged, such that at the time of the transfer, RAG1 and C-mos were still very similar between each two lineages of snakes. Some of these comparisons involve three viperid snakes (A. contortrix, C. atrox, and S. catenatus; dotted circle B on fig. 3) that diverged less than 22 Ma (Douglas et al. 2006). The SPIN copies from these three snakes cluster together in the phylogeny (fig. 4), consistent with the idea that a distinct variant of SPIN was horizontally transferred to a common ancestor of these three species. The phylogeny is, however, inconclusive regarding the relationship of the other snake SPIN copies (sequenced from N. erythrogaster and H. torquata), and it is therefore unclear whether these two species each acquired the element via HT or whether there was a single transfer event in their common ancestor.
Discussion
In this study, we have shown that SPIN transposons have infiltrated the germ lines of many species of lizards, snakes, and at least one amphisbaenian spreading over most of the diversity of extant squamates (fig. 1). This distribution implies that SPIN was transferred independently at least 13 times during the evolution of squamates, which triples the number of HT events of SPIN previously reported among diverse metazoans (Pace et al. 2008; Gilbert et al. 2010; Novick et al. 2010). This estimate is conservative as it assumes a single HT event in the common ancestor of all snake species tested positive for SPIN. Yet, the patchy distribution of SPIN among snakes (SPIN is absent in the two species of Boidae included in this study) is suggestive of multiple transfers. In any case, these data considerably extend our appreciation of the frequency of HTT in nonavian reptiles, which was previously based on only one sequenced genome, that of the green anole lizard (Gilbert et al. 2010; Novick et al. 2010; Thomas et al. 2010).
The activity of SPIN in squamates has resulted in the spread and accumulation of large numbers of elements in a lineage-specific fashion (between 2,000 and 28,000 per haploid genome as revealed by slot blot analysis; fig. 1; supplementary fig. 1 and table 1, Supplementary Material online). Further studies will be necessary to assess the evolutionary impact of these invasions on the structure and function of squamate genomes, but it is tantalizing to speculate that they have provided a rich source of raw material for genetic innovation, as observed for other DNA transposons (Feschotte and Pritham 2007). Some recent studies indeed suggest that TEs have been instrumental in the size expansion of the squamate Hox gene clusters and that they may have had a major impact on the evolution of body plan in this group (Di-Poï et al. 2009, 2010). Whether SPIN invasions and other HTT events could have been involved in the spectacular diversification of squamates during the Tertiary leading to the formation of more than 8,000 extant species (Hedges and Vidal 2009) is a fascinating question that warrants further investigation.
An interesting aspect of SPIN HTs previously noticed is that the transfers apparently took place on at least three different continents (Africa, Eurasia, and South America) within a relatively narrow evolutionary window (15–46 Ma) (Pace et al. 2008; Gilbert et al. 2009, 2010). One way to estimate the time at which HTT took place is to calculate the pairwise divergence between all individual TE copies and an ancestral founder copy, which can be approximated by the consensus sequence of the TE family. Because TEs typically evolve neutrally after insertion in the genome (e.g., Lampe et al. 2003 and see Results), the time of the transfers can be approximated by dividing these pairwise divergences by the nuclear neutral substitution rate of the various species lineages in which HTT has occurred (Khan et al. 2006; Pace et al. 2008). The average pairwise corrected divergences between each SPIN copy sequenced in this study and the SPIN consensus reconstructed for each squamate species ranges between 0.002 and 0.05 (average = 0.019; SD = 0.014). Because there is no reliable estimate of neutral substitution rate for nonavian reptiles, we used the two most extreme neutral rates known so far for amniotes (2 × 10−9 and 4.5 × 10−9 substitutions per site per year [subst./site/year]) as a range to estimate the timing of SPIN HTs in squamates. These rates were taken from a study of multiple introns in birds (2 × 10−9–3.9 × 10−9 subst./site/year; Axelsson et al. 2004) and on two studies of ancestral orthologous repeats shared between several placental mammals (2.2 × 10−9 –4.5 × 10−9 subst./site/year; Waterston et al. 2002; Pace et al. 2008). Applying these rates to the squamate SPIN pairwise distances yields invasion times ranging from 667 thousand years (using the smallest average divergence [0.002] and the fastest rate [4.5 × 10−9 subst./site/year]) and 25 My (using the largest average divergence [0.05] and the slowest rate [2 × 10−9 subst./site/year]). These rough estimates indicate that the multiple horizontal introductions of SPIN in squamates all occurred within the same evolutionary timeframe (and some perhaps more recently) as those previously reported in other tetrapods (Pace et al. 2008; Gilbert et al. 2010).
These estimates should be used cautiously, however, not only because of uncertainties and variations in neutral molecular clocks but also because we analyzed only few SPIN copies for each species and a relatively small segment (∼400 bp) of coding sequence. For the sake of the discussion, we assume that all SPIN transfers in squamates took place within the past 25 My, and it is important to note that even if the SPIN invasion times were twice older than our oldest estimate (i.e., as old as 50 My), the following biogeographical inferences would all remain valid. Robust fossil evidence from this period and molecular dating suggests that the ancestors of Amphisbaena (Amphisbaenidae), those of Mesaspis (Anguidae), Heloderma (Helodermatidae) and Lepidophyma (Xantusiidae), and those of Cordylus (Cordylidae) were distributed on the same three continents occupied today by their extant descendants, namely South America, North America, and Africa, respectively (fig. 1 and see Materials and Methods). Together, these observations confirm the hypothesis of at least one transoceanic transfer of SPIN elements (See Gilbert et al. 2009 for discussion), and they extend the geographic range of SPIN HTs to North America. Based on the paleobiogeographical data available for the Varanidae, Agamidae, snakes, and Hemidactylus over the same period (see Materials and Methods), it is possible that SPIN HT also took place in Asia (fig. 1). Further sampling of additional SPIN sequences and of other species will be required to infer more precise estimates of the time and geographical span of the transfers.
This study, together with earlier reports of HTT on a wide geographical and taxonomic scale (e.g., Pace et al. 2008; Gilbert et al. 2010), continues to raise the question of the mechanisms underpinning such transfers. A number of possible vectors and scenarios have been proposed in the literature (e.g., Houck et al. 1991, Piskurek and Okada 2007; reviewed in Loreto et al. 2008 and Schaack et al. 2010) but none have been fully verified in nature. In a recent study, we proposed that host–parasite interactions might have facilitated HTT (Gilbert et al. 2010). This hypothesis was based on the observation that four families of DNA transposons (including SPIN; see Introduction) known to have transferred horizontally in multiple species of tetrapods are also found in the kissing bug (R. prolixus; Triatominae), a South American hemipteran insect feeding on the blood of a wide range of vertebrate hosts. Strikingly, two of the transposons (SPIN and OposCharlie1) identified in R. prolixus are almost identical (>98% identity) and cluster phylogenetically with those of the opossum, one of the bug’s preferred hosts in nature. This data are consistent with a transfer between host and parasite, an act that could have been facilitated by frequent physical contacts between the bug’s saliva and the blood of its hosts when the bug is feeding. The phylogenetic analysis presented herein (fig. 4), although based only on a portion of the SPIN autonomous sequence, recovers a strongly supported grouping of R. prolixus and opossum SPIN sequences. Interestingly, this clade also includes all SPIN copies isolated from A. alba. Amphisbaenians are enigmatic worm-like fossorial squamates found in tropical and semitropical regions of the world (Macey et al. 2004). Amphisbaena alba is found throughout most of South America, east of the Andes (Colli and Zamboni 1999), a geographic range that largely overlaps with that of R. prolixus and triatomine bugs in general (Lent and Wygodzinsky 1979). There is no published observation of R. prolixus feeding specifically on A. alba, but numerous triatomine species have been reported to feed on various squamate species (Lent and Wygodzinsky 1979). In addition, triatomines are commonly found under fallen logs, among exposed roots and under loose bark, as well as in the burrows of some mammalian species (Lent and Wygodzinsky 1979). Given the geographical and potential ecological overlap of triatomine bugs and A. alba in South America, the hypothesis of a transfer of SPIN between these two species seems plausible. These observations further indicate that triatomine bugs might have played a central role in the spread of SPIN across diverse South American tetrapods.
Another intriguing trend emerging from this work lies in our inability to detect SPIN elements in any of the crocodiles and turtles examined, while we found evidence for their repeated invasions in nearly all major squamate lineages and in several mammals as reported previously (Pace et al. 2008). Though our taxonomic sample includes only 7 of 313 and 4 of 23 described species of turtles and crocodiles, respectively, it does capture a substantial portion of their evolutionary history (fig. 1). Therefore, if HT of SPIN occurred at all in these taxa, its frequency is likely to be much lower than in squamates and mammals. The reasons for this difference remain unclear, but it might reflect a lack of (or reduced) interaction between the putative vectors of SPIN HT and crocodiles and turtles and/or a lower tolerance or better genomic defense of crocodile and turtle genomes against the amplification of some TEs. We note, however, that retroelements and other DNA transposons have been identified in crocodiles and turtles (Ray et al. 2005; Shedlock et al. 2007; Kordis 2009). Another simple nonmutualistic explanation for the absence of SPIN HTs in crocodiles and turtles is that these taxa have diversified in a much fewer number of lineages than squamates during the tertiary, leading to a much lower number of extant species (8,200 squamates vs. 313 turtles and 23 crocodiles; Brochu 2003; Hedges and Vidal 2009; Shaffer 2009). Based on these numbers only, the odds of successful HT may have simply been lower in turtles and crocodiles than in squamates. A more systematic screen of crocodiles, turtles, and other tetrapods as well as some of their parasites should further our understanding of the taxonomic and geographic spread of these transposons and of the mechanisms and factors promoting HTT and lead to a greater appreciation of the impact of HTT on the evolution of eukaryotic genomes.
Supplemental Material
Supplementary figure S1, tables S1 and S2, and data sets 1–6 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We are grateful to Andrea Acevedo, Christian Cox, Oscar Flores-Villela, Thomas Eimermacher, Carl Franklin, Alan Kardon (San Antonio Zoo), Jesse Meik, Terry Robinson, Anne Ropiquet, Jeff Streicher, Walter Schargel, and Luis Sigler (Dallas World Aquarium) for the generous gift of tissue samples. In particular, Carl Franklin was pivotal in providing testudine and crocodilian samples and data. We thank Sarah Schaack and members of the Genome Biology Group at the University of Texas at Arlington for useful suggestions during the preparation of the manuscript. This work was supported by grant R01GM77582 to C.F. from the National Institutes of Health.
References
- Andersson JO. Gene transfer and diversification of microbial eukaryotes. Annu Rev Microbiol. 2009;63:177–193. doi: 10.1146/annurev.micro.091208.073203. [DOI] [PubMed] [Google Scholar]
- Axelsson E, Smith NG, Sundstrom H, Berlin S, Ellegren H. Male-biased mutation rate and divergence in autosomal, z-linked and w-linked introns of chicken and turkey. Mol Biol Evol. 2004;21:1538–1547. doi: 10.1093/molbev/msh157. [DOI] [PubMed] [Google Scholar]
- Brochu CA. Phylogenetic approaches toward crocodylian history. Annu Rev Earth Planet Sci. 2003;31:357–397. [Google Scholar]
- Carranza S, Arnold EN. Systematics, biogeography, and evolution of Hemidactylus geckos (Reptilia: Gekkonidae) elucidated using mitochondrial DNA sequences. Mol Phylogenet Evol. 2006;38:531–545. doi: 10.1016/j.ympev.2005.07.012. [DOI] [PubMed] [Google Scholar]
- Carranza S, Arnold EN, Geniez P, Roca J, Mateo JA. Radiation, multiple dispersal and parallelism in the skinks, Chalcides and Sphenops (Squamata: Scincidae), with comments on Scincus and Scincopus and the age of the Sahara Desert. Mol Phylogenet Evol. 2008;46:1071–1094. doi: 10.1016/j.ympev.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Castoe TA, Hall KT, Guibotsy Mboulas ML, et al. (13 co-authors). Forthcoming. Discovery of highly divergent repeat landscapes in snake genomes using high throughput sequencing. Genome Biol. advance online. 2011 doi: 10.1093/gbe/evr043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifelli RL, Nydam RL. Primitive, helodermatid-like platynotan from the early cretaceous of Utah. Herpetologica. 1995;51:286–291. [Google Scholar]
- Clos LM. A new species of Varanus (Reptilia, Sauria) from the Miocene of Kenya. J Vertebr Paleontol. 1995;15:254–267. [Google Scholar]
- Colli GR, Zamboni DS. Ecology of the worm-lizard Amphisbaena alba in the Cerrado of central Brazil. Copeia. 1999;3:733–742. [Google Scholar]
- Craig NL, Craigie R, Gellert M, Lambowitz AM. Mobile DNA II. Washington (DC): American Society for Microbiology Press; 2002. [Google Scholar]
- Dalloul RA, Long JA, Zimin AV, et al. (71 co-authors) Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biol. 2010;8:e1000475. doi: 10.1371/journal.pbio.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danchin EGJ, Rosso MN, Vieira P, de Almeida-Engler J, Coutinho PM, Henrissat B, Abada P. Multiple lateral gene transfers and duplications have promoted plant parasitism ability in nematodes. Proc Natl Acad Sci U S A. 2010;107:17651–17656. doi: 10.1073/pnas.1008486107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di-Poï N, Montoya-Burgos JI, Duboule D. Atypical relaxation of structural constraints in Hox gene clusters of the green anole lizard. Genome Res. 2009;19:602–610. doi: 10.1101/gr.087932.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di-Poï N, Montoya-Burgos JI, Miller H, Pourquié O, Milinkovitch MC, Duboule D. Changes in Hox genes’ structure and function during the evolution of the squamate body plan. Nature. 2010;464:99–103. doi: 10.1038/nature08789. [DOI] [PubMed] [Google Scholar]
- Douglas ME, Douglas MR, Schuett GW, Beck DD, Sullivan BK. Conservation phylogenetics of helodermatid lizards using multiple molecular markers and a supertree approach. Mol Phylogenet Evol. 2010;55:153–167. doi: 10.1016/j.ympev.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Douglas ME, Douglas MR, Schuett GW, Porras LW. Evolution of rattlesnakes (Viperidae; Crotalus) in the warm deserts of western North America shaped by Neogene vicariance and Quaternary climate change. Mol Ecol. 2006;15:3353–3374. doi: 10.1111/j.1365-294X.2006.03007.x. [DOI] [PubMed] [Google Scholar]
- Engels WR, Johnsonschlitz DM, Eggleston WB, Sved J. High-frequency P-element loss in Drosophila is homolog dependent. Cell. 1990;62:515–525. doi: 10.1016/0092-8674(90)90016-8. [DOI] [PubMed] [Google Scholar]
- Estes R, Hutchinson JH. Eocene lower vertebrates from Ellesmere Islands, Canadian Arctic Archipelago. Paleogeogr Palaeoclimatol Palaeoecol. 1980;30:325–347. [Google Scholar]
- Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller S, Baverstock P, King D. Biogeographic origins of goannas (Varanidae): a molecular perspective. Mol Phylogenet Evol. 1998;9:294–307. doi: 10.1006/mpev.1997.0476. [DOI] [PubMed] [Google Scholar]
- Gilbert C, Pace JK, Feschotte C. Horizontal SPINning of transposons. Commun Integr Biol. 2009;2:117–119. doi: 10.4161/cib.7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Schaack S, Pace JK, 2nd, Brindley PJ, Feschotte C. A role for host-parasite interactions in the horizontal transfer of transposons across phyla. Nature. 2010;464:1347–1350. doi: 10.1038/nature08939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giugliano LG, Garcia Collevatti R, Rinaldi Colli G. Molecular dating and phylogenetic relationships among Teiidae (Squamata) inferred by molecular and morphological data. Mol Phylogenet Evol. 2007;45:168–179. doi: 10.1016/j.ympev.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Gladyshev EA, Meselson M, Arkhipova IR. Massive horizontal gene transfer in bdelloid rotifers. Science. 2008;320:1210–1213. doi: 10.1126/science.1156407. [DOI] [PubMed] [Google Scholar]
- Greer AE. A subfamilial classification of scincid lizards. Bull Mus Comp Zool. 1970;139:151–183. [Google Scholar]
- Gregory TR. Animal Genome Size Database. 2011. [Internet]. [cited 2011 jan]. Available from: http://www.genomesize.com. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hall T. BioEdit version 5.0.6. 2004. [Internet]. [cited 2010 dec]. Available from: http://www.mbio.ncsu.edu/bioedit/bioedit.html. [Google Scholar]
- Hay JM, Sarre SD, Lambert DM, Allendorf FW, Daugherty CH. Genetic diversity and taxonomy: a reassessment of species designation in tuatara (Sphenodon: Reptilia) Conserv Genet. 2010;11:1063–1081. [Google Scholar]
- Hedges SB, Vidal N. Lizards, snakes and amphisbaenians (Squamata) In: Blair Hedges S, Kumar S, editors. The timetree of life. New York (NY): Oxford University Press; 2009. pp. 383–389. [Google Scholar]
- Hillier LW, Miller W, Birney E, et al. (175 co-authors) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Houck MA, Clark JB, Peterson KR, Kidwell MG. Possible horizontal transfer of Drosophila genes by the mite Proctolaelaps regalis. Science. 1991;253:1125–1128. doi: 10.1126/science.1653453. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Palmer JD. Horizontal transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Aravind L. Horizontal gene transfer in prokaryotes: quantification and classification. Annu Rev Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordis D. Transposable elements in reptilian and avian (sauropsida) genomes. Cytogenet Genome Res. 2009;127:94–111. doi: 10.1159/000294999. [DOI] [PubMed] [Google Scholar]
- Krause DW, Evans SE, Gao KQ. First definitive record of Mesozoic lizards from Madagascar. J Vertebr Paleontol. 2003;23:842–856. [Google Scholar]
- Lampe DJ, Witherspoon DJ, Soto-Adames FN, Robertson HM. Recent horizontal transfer of mellifera subfamily mariner transposons into insect lineages representing four different orders shows that selection acts only during horizontal transfer. Mol Biol Evol. 2003;20:554–562. doi: 10.1093/molbev/msg069. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, et al. (255 co-authors) Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lent H, Wygodzinsky P. Revision of the Triatominae (Hemiptera, Reduviidae), and their significance as vectors of Chagas disease. Bull Am Mus Nat Hist. 1979;163:123–520. [Google Scholar]
- Loreto EL, Carareto CM, Capy P. Revisiting horizontal transfer of transposable elements in Drosophila. Heredity. 2008;100:545–554. doi: 10.1038/sj.hdy.6801094. [DOI] [PubMed] [Google Scholar]
- Macey JR, Papenfuss TJ, Kuehl JV, Fourcade HM, Boore JL. Phylogenetic relationships among amphisbaenian reptiles based on complete mitochondrial genomic sequences. Mol Phylogenet Evol. 2004;33:22–31. doi: 10.1016/j.ympev.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Macey JR, Schulte JA, 2nd, Larson A, Ananjeva NB, Wang Y, Pethiyagoda R, Rastegar-Pouyani N, Papenfuss TJ. Evaluating trans-tethys migration: an example using acrodont lizard phylogenetics. Syst Biol. 2000;49:233–256. doi: 10.1093/sysbio/49.2.233. [DOI] [PubMed] [Google Scholar]
- Macey JR, Schulte JA, 2nd, Larson A, Tuniyev BS, Orlov N, Papenfuss TJ. Molecular phylogenetics, tRNA evolution, and historical biogeography in Anguid lizards and related taxonomic families. Mol Phylogenet Evol. 1999;12:250–272. doi: 10.1006/mpev.1999.0615. [DOI] [PubMed] [Google Scholar]
- Marcet-Houben M, Gabaldón T. Acquisition of prokaryotic genes by fungal genomes. Trends Genet. 2009;26:5–8. doi: 10.1016/j.tig.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Moran NA, Jarvik T. Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science. 2010;328:624–627. doi: 10.1126/science.1187113. [DOI] [PubMed] [Google Scholar]
- Noonan BP, Chippindale PT. Vicariant origin of malagasy reptiles supports late cretaceous antarctic land bridge. Am Nat. 2006;168:730–741. doi: 10.1086/509052. [DOI] [PubMed] [Google Scholar]
- Novick P, Smith J, Ray D, Boissinot S. Independent and parallel lateral transfer of DNA transposons in tetrapod genomes. Gene. 2010;449:85–94. doi: 10.1016/j.gene.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Nydam RL. A new taxon of helodermatid-like lizard from the Albian-Cenomanian of Utah. J Vertebr Paleontol. 2000;20:285–294. [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Okajima Y, Kumazawa Y. Mitochondrial genomes of acrodont lizards: timing of gene rearrangements and phylogenetic and biogeographic implications. BMC Evol Biol. 2010;10:141. doi: 10.1186/1471-2148-10-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmo E. Evolution of genome size and DNA-base composition in reptiles. Genetica. 1981;57:39–50. [Google Scholar]
- Pace JK, 2nd, Gilbert C, Clark MS, Feschotte C. Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Proc Natl Acad Sci U S A. 2008;105:17023–17028. doi: 10.1073/pnas.0806548105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagan HJ, Smith JD, Hubley RM, Ray DA. PiggyBac-ing on a primate genome: novel elements, recent activity and horizontal transfer. Genome Biol Evol. 2010;2:293–303. doi: 10.1093/gbe/evq021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinou T, Vicario S, Marschner M, Caccone A. Relict snakes of North America and their relationships within Caenophidia, using likelihood-based Bayesian methods on mitochondrial sequences. Mol Phylogenet Evol. 2004;32:563–574. doi: 10.1016/j.ympev.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Piskurek O, Okada N. Poxviruses as possible vectors for horizontal transfer of retroposons from reptiles to mammals. Proc Natl Acad Sci U S A. 2007;104:12046–12051. doi: 10.1073/pnas.0700531104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Ray DA, Hedges DJ, Herke SW, Fowlkes JD, Barnes EW, LaVie DK, Goodwin LM, Densmore LD, Batzer MA. Chompy: an infestation of MITE-like repetitive elements in the crocodilian genome. Gene. 2005;362:1–10. doi: 10.1016/j.gene.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Robertson HM. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington (DC): ASM; 2002. pp. 1093–1110. [Google Scholar]
- Roos J, Aggarwal RK, Janke A. Extended mitogenomic phylogenetic analyses yield new insight into crocodylian evolution and their survival of the Cretaceous–Tertiary boundary. Mol Phylogenet Evol. 2007;45:663–673. doi: 10.1016/j.ympev.2007.06.018. [DOI] [PubMed] [Google Scholar]
- Rubin E, Levy AA. Abortive gap repair: underlying mechanism for Ds element formation. Mol Cell Biol. 1997;17:6294–6302. doi: 10.1128/mcb.17.11.6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint KM, Austin CC, Donnellan SC, Hutchinson MN. C-mos, a nuclear marker useful for squamate phylogenetic analysis. Mol Phylogenet Evol. 1998;10:259–263. doi: 10.1006/mpev.1998.0515. [DOI] [PubMed] [Google Scholar]
- Schaack S, Gilbert C, Feschotte C. Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol Evol. 2010;25:537–546. doi: 10.1016/j.tree.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, et al. (157 co-authors) The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- Shaffer HB. Turtles (Testudines) In: Blair Hedges S, Kumar S, editors. The timetree of life. New York (NY): Oxford University Press; 2009. pp. 398–401. [Google Scholar]
- Shedlock AM, Botka CW, Zhao S, Shetty J, Zhang T, Liu JS, Deschavanne PJ, Edwards SV. Phylogenomics of nonavian reptiles and the structure of the ancestral amniote genome. Proc Natl Acad Sci U S A. 2007;104:2767–2772. doi: 10.1073/pnas.0606204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot JC, Rokas A. Horizontal transfer of a large and highly toxic secondary metabolic gene cluster between fungi. Curr Biol. 2011;21:134–139. doi: 10.1016/j.cub.2010.12.020. [DOI] [PubMed] [Google Scholar]
- Smith KT. Eocene lizards of the clade Geiseltaliellus from Messel and Geiseltal, Germany, and the early radiation of Iguanidae (Reptilia: Squamata) Bull Peabody Mus Nat Hist. 2009;50:219–306. [Google Scholar]
- Stanley EL, Bauer AM, Jackman TR, Branch WR, Mouton Ple F. Between a rock and a hard polytomy: rapid radiation in the rupicolous girdled lizards (Squamata: Cordylidae) Mol Phylogenet Evol. 2011;58:53–70. doi: 10.1016/j.ympev.2010.08.024. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony (* and other methods), version 4. Sunderland (MA): Sinauer; 2003. [Google Scholar]
- Thomas J, Schaack S, Pritham EJ. Pervasive horizontal transfer of rolling-circle transposons among animals. Genome Biol Evol. 2010;2:656–664. doi: 10.1093/gbe/evq050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicario S, Caccone A, Gauthier J. Xantusiid “night” lizards: a puzzling phylogenetic problem revisited using likelihood-based Bayesian methods on mtDNA sequences. Mol Phylogenet Evol. 2003;26:243–261. doi: 10.1016/s1055-7903(02)00313-5. [DOI] [PubMed] [Google Scholar]
- Vidal N, Kindl SG, Wong A, Hedges SB. Phylogenetic relationships of xenodontine snakes inferred from 12S and 16S ribosomal RNA sequences. Mol Phylogenet Evol. 2000;14:389–402. doi: 10.1006/mpev.1999.0717. [DOI] [PubMed] [Google Scholar]
- Warren WC, Clayton DF, Ellegren H, et al. (82 co-authors) The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, et al. (222 co-authors) Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Whiting AS, Bauer AM, Sites FW., Jr Phylogenetic relationships and limb loss in sub-Saharan African scincine lizards (Squamata: Scincidae) Mol Phylogenet Evol. 2003;29:582–598. doi: 10.1016/s1055-7903(03)00142-8. [DOI] [PubMed] [Google Scholar]
- Yan X, Martinez-Fèrez IM, Kavchok S, Dooner HK. Origination of Ds elements from Ac elements in maize: evidence for rare repair synthesis at the site of Ac excision. Genetics. 1999;152:1733–1740. doi: 10.1093/genetics/152.4.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.