Abstract
The current paradigm of human genetics research is to analyze variation of a single data type (i.e., DNA sequence or RNA levels) to detect genes and pathways that underlie complex traits such as disease state or drug response. While these studies have detected thousands of variations that associate with hundreds of complex phenotypes, much of the estimated heritability, or trait variability due to genetic factors, remain unexplained. We may be able to account for a portion of the missing heritability if we incorporate a systems biology approach into these analyses. Rapid technological advances will make it possible for scientists to explore this hypothesis via the generation of high-throughput omics data – transcriptomic, proteomic and methylomic to name a few. Analyzing this ‘meta-dimensional’ data will require clever statistical techniques that allow for the integration of qualitative and quantitative predictor variables. For this article, we examine two major categories of approaches for integrated data analysis, give examples of their use in experimental and in silico datasets, and assess the limitations of each method.
Keywords: computational methods, data integration, pharmacogenomics, systems biology
One of the primary goals of current human genetics research is to elucidate the genetic architecture of complex heritable traits, such as drug response. Technological advancements have been crucial in driving the direction of these studies. For example, affordable high-throughput SNP genotyping has made the genome-wide association study a popular analysis strategy for finding genetic variants that associate with a specific phenotype. While genome-wide association studies have been successful at finding SNPs that point to novel biological underpinnings of disease [101], almost all have very small effect sizes and cumulatively account for only a small proportion of the estimated heritability of the trait. This phenomenon has led many scientists to hypothesize where the remainder of the heritability might lie. One idea is that traditional analytical methods that examine the effect of one variant at a time are not robust to the complexity of the genetic architecture of these traits [1]. For example, if the true genetic model involves SNPs in different genes with little or no effect when they occur individually but a very large effect when they occur together, traditional single locus analysis would likely fail. This concept of complex genetic etiology also encompasses the possibility that the true model involves variation at different levels of biological regulation, or a ‘meta-dimensional’ model. For example, if a susceptibility model includes both the hypermethylation of a gene and a particular allelic variant at a SNP to pass the threshold for the trait to occur, detection would require an analytical method that can integrate gene expression and genotype data.
Recent advancements in high-throughput techniques for measuring quantitative variables across their respective ‘omes’, such as gene expression [2, 3], protein expression [4], and methylation patterns [5], allow for meta-dimensional analyses to be carried out. There are several key reasons performing a meta-dimensional analysis would allow for a more thorough and informative interrogation of risk etiology than a single data type analysis. Several reviews go over these arguments in detail [6–9] with the main ideas being:
-
▪
Multiple datatypes may compensate for missing or unreliable information in any one data type;
-
▪
Different sources of data that point to the same gene or pathway are less likely to be false positives and could indicate functionality;
-
▪
The full biological model may only be detected if different levels of regulation are considered in an analysis.
One limitation to meta-dimensional analyses for human genetics studies is that data acquisition and quantification of RNA and protein levels is not as straightforward as for germline DNA sequence. Confident genotype calls can be made from relatively small quantities of DNA acquired from a variety of tissue types that have been stored properly for an extended period of time [10–12]. Conversely, gene- and protein-expression levels are dynamic and, in part, dependent on the tissue from which they were extracted [13, 14]. These sources of variability make the process of collecting samples to generate data more complex. However, this complexity is also a strength of meta-dimensional analyses in that the dynamic nature of specific datatypes reflects the true nature of biology and may allow for discoveries that would be missed by only examining DNA sequence variation.
One potential and rapidly expanding source of meta-dimensional data for pharmacogenomics research is the biobank [15, 16]. Biobanks are repositories for biological specimens that will be stored for an extended period of time for future clinical or research use. Currently, there are enormous efforts underway to create population-based biobanks with specimens linked to clinical and environmental data [17–19]. Careful collection, handling and storage of blood and other tissues in these banks [20–22] would allow scientists to gather information from different levels of biological regulation on numerous subjects without the added cost and effort of standard recruitment.
Another possible source for multiple high-throughput datatypes are immortalized lymphoblastoid cell lines (LCLs) [23]. Genotype, gene expression and drug-response phenotype data on a number of established LCLs, such as those derived from the individuals that participated in the International HapMap Project, have already been generated and been made publicly available [24–27]. In addition, many of these cell lines are commercially available through the Coriell Institute for Medical Research making it possible for scientists to perform in-house functional experiments [102]. While several interesting findings have come about from LCLs, such as mapping the determinants of human gene-expression variation within and between human populations [28–35], there are some critical limitations to this model. A number of reviews have examined in detail the strengths and weaknesses of using immortalized LCLs for human genetics research [36–41]. For example, one strength of this model is that pharmacological experiments using highly toxic drugs may be done using cell lines that would be difficult or unethical using human subjects [38]. Conversely, a major weakness to this approach is that LCLs have been shown to consistently express only approximately 50% of known genes [37]. If the true disease etiology consists of altered expression levels for genes that are not expressed in LCLs, these variables would be missed.
Data integration is a broad topic and the user-specific definition will be crucial in deciding which analytical method to use. The key focus of this article is integration of different types of high-throughput data for detecting biological models that associate with a complex human phenotype. Thus far, two main categories of analytical techniques have been used to integrate different datatypes:
-
▪
Determine the correlation of the independent variables with each other and with the trait of interest to map disease loci (multistage approach);
-
▪
Combine all of the data initially and then allowing the computational method to find meta-dimensional models (simultaneous analysis).
Table 1 lists the studies that have integrated high-throughput qualitative (genotype) and quantitative (gene or protein expression) datatypes specifically to detect biological variation that underlies complex human traits. By far, the multistage approach has dominated the field of integrated analyses. The goals of this article are to assess the benefits and limitations of the techniques that have been used for the multistage approach and to describe promising computational methods for simultaneous analysis of meta-dimensional data.
Table 1.
Description of published meta-dimensional analyses for complex human traits.
| Study (year) |
Type | Data source and type | Phenotype | Statistical method |
Results | Ref. |
|---|---|---|---|---|---|---|
| Huang et al. (2007) | MS | LCL genotype and gene-expression data | Etoposide-induced cytotoxicity | Triangle method | 63 SNPs that associate with the phenotype through expression of 45 genes | [42] |
| Huang et al. (2007) | MS | LCL genotype and gene-expression data | Cisplatin-induced cytotoxicity | Triangle method | 17 SNPs that associate with the phenotype through expression of 26 genes | [43] |
| Emilsson et al. (2008) | MS | Genotype and gene-expression data from blood and adipose tissue | Obesity-related traits | Pathway analysis | Transcriptional network SNPs associated with obesity traits | [50] |
| Huang et al. (2008) | MS | LCL genotype and gene-expression data + validation LCLs | Daunorubicin-induced cytotoxicity | Triangle method | 63 SNPs that associate with the phenotype through expression of 61 genes; two CEU SNPs validated in independent LCL sample | [44] |
| Huang et al. (2008) | MS | LCL genotype and gene-expression data | Carboplatin-induced cytotoxicity | Triangle method | 46 SNPs that associate with the phenotype through expression of 38 genes | [45] |
| Choy et al. (2008) | MS | LCL genotype and gene-expression data | Cell growth response to 5-fluorouracil, methotrexate, 6-mercaptopurine, SAHA and simvastatin | Variation of triangle method | Three SNPs nominally associated for methotrexate, 5-fluorouracil and simvastatin | [40] |
| Hartford et al. (2009) | MS | LCL genotype and gene-expression data + validation LCLs | Population-specific cytarabine arabinoside cytotoxicity | Triangle method | 26 and 33 SNPs that associate with phenotype through expression of 12 and 36 genes for CEU and YRI, respectively; two genes validated in independent LCL sample | [46] |
| Reif et al. (2009) | S | Genotype and proteomic data from participants in vaccination research | Adverse reaction to smallpox vaccination | Tree-based methods | Model with three cytokines and one SNP that associates with adverse event | [55] |
| Hsu et al. (2010) | MS | Genotype and gene-expression data (from human bone, lymphocyte and liver tissue + animal model tissue) | Osteoporosis-related traits | Pathway analysis | Three novel loci found that associate with traits in women and one confirmed locus from previous finding | [51] |
| Edwards et al. (2011) | MS | Genotype and gene-expression data from brain tissue from cases and controls | Parkinson’s disease | Pathway analysis | Axonal guidance, focal adhesion, and calcium signaling pathways associated with Parkinson’s disease | [52] |
CEU: European HapMap samples; LCL: Lymphoblastoid cell lines; MS: Multistage approach; S: Simultaneous analysis; SAHA: Suberoylanilide hydroxamic acid; YRI: Yoruba HapMap samples.
Multistage approach
Triangle model
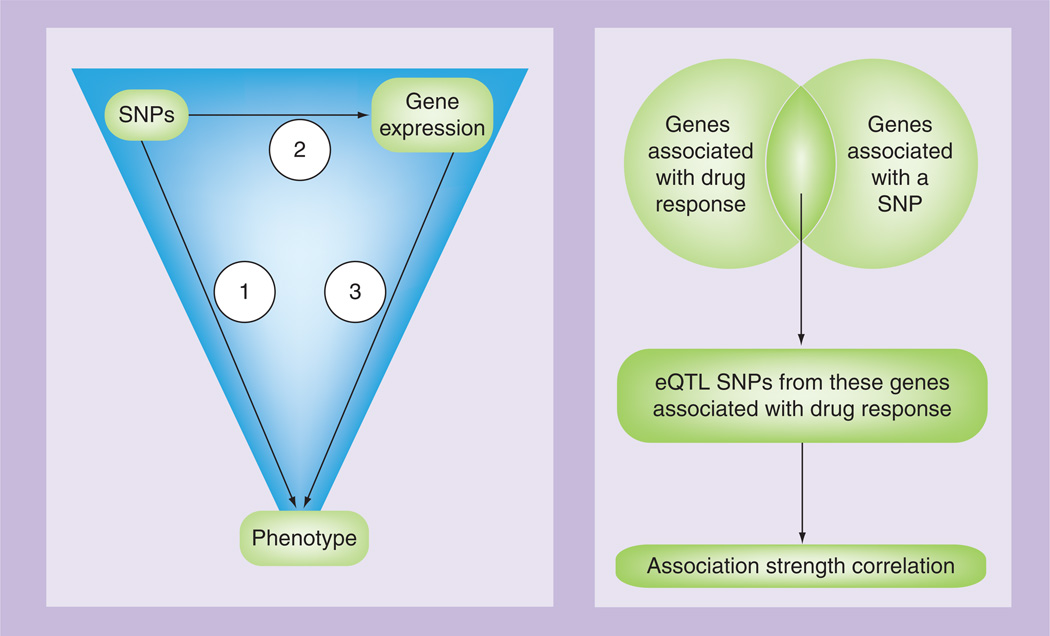
The main objective of the multistage approach is to divide the analysis into steps to find associations between the different datatypes and also with the datatypes and the trait. When the datatypes of interest are genotypes and gene-expression levels, this method is essentially mapping expression quantitative trait loci (eQTLs) to find ‘functional’ SNPs that are associated with the trait. The most commonly used integration technique thus far has been a three-stage or triangle method. Figure 1A illustrates the triangle method where SNPs are associated with the phenotype and filtered based on a genome-wide significance threshold; SNPs deemed significant from Step 1 are tested for association with gene-expression levels with a less stringent threshold owing to fewer statistical tests; and gene-expression levels detected in Step 2 are tested for correlation with the outcome of interest. Most of these analyses have been performed using genome-wide SNP genotypes and baseline gene-expression levels of HapMap LCLs to find associations with drug cytotoxicity measured as the IC50 [42–46].
Figure 1. Variations of the triangle method.
eQTL: Expression quantitative trait loci
In Choy et al., a variation of the triangle method is used in which each SNP was tested for association with baseline gene-expression levels and each gene-expression level was tested for association with drug response (Figure 1B) [40]. Next, eQTL SNPs from genes that were associated with both were tested for association with drug response. Finally, the correlation between the strength of each SNP association with drug response and gene expression was measured. The main difference between these two method variations is the level at which stringent filtering occurs. In the first method, SNPs that do not pass a genome-wide level of significance will not make it to Step 2. In the second method, the stringency lies in the fact that genes must associate with both a SNP and drug response. The impact of these two methods on power and Type I error rate is not immediately clear; however, more statistically significant results have been found with the first method (Figure 1A). For example, in Huang et al., the first method was used to find genetic variants that associate with the cytotoxicity of the chemotherapeutic agent daunorubicin. Six SNPs representing six different genes were detected using Utah residents with Northern and Western European ancestry HapMap samples (CEU), two of which were validated in an independent, non-HapMap LCL sample [44]. On the other hand, in the study that used the second method, the authors state that no eQTLs were significantly associated with any of the five drugs tested, but three were nominally associated [40].
Pathway analysis
Another multistage approach that has been employed by several researchers is a pathway-based analysis. This method involves utilizing the pattern in which associated genotype and gene-expression variables fall into annotated genes within biological pathways to find modules that are over-represented for the phenotype of interest. This method was first utilized in gene-expression data analyses to identify genes that were coexpressed and may represent a functional unit associated with the outcome of interest [47]. Recently, this method has been applied to genome-wide SNP data by first mapping the SNPs to genes and then genes to pathways [48]. The primary benefits of a pathway analysis over the more traditional SNP association studies are improved power to find small effects, a more direct indication of the underlying biology, and the ease of replicating an entire pathway versus a single SNP [49]. Because the utility of this method was first shown in expression data and is now being applied to SNP data, using pathway analysis to integrate the two heterogeneous datatypes is a logical next step.
Notably, the three meta-dimensional examples shown in Table 1 that applied this approach used different human tissue to generate gene-expression data instead of LCLs [50–52]. The meta-dimensional pathway analysis by Emilsson et al. used correlation matrices of differential gene-expression levels in adipose tissue to detect transcriptional networks [50]. The network detected was found to be highly conserved in mouse and was enriched for genes in the inflammatory response and macrophage activation. Next, they integrated the pathway data with genotype data by selecting the strongest cis-eQTL for the genes in the network and tested them jointly for association to BMI and percentage body fat. They found modest levels of association with BMI. Edwards et al. integrated genotypes and gene-expression data from brain tissue to find over-represented pathways associated with Parkinson’s disease [52]. Their approach was simpler than the previously described study in that they searched for Kyoto Encyclopedia of Genes and Genomes pathways that were enriched for significant SNPs or gene-expression variables and then selected the pathways that were included in both sets for further testing. The top three pathways found were for axonal guidance, focal adhesion and calcium signaling. Finally, the study by Hsu et al. attempts to dissect the genetic architecture of osteoporosis-related traits by integrating expression data from both human and animal tissues with genome-wide genotype data to prioritize loci based on their potential functionality [51]. The prioritized loci were subsequently tested for enrichment of annotated biological pathways. Using this method, they were able to identify three novel regions and one previously identified locus that associated with these traits in women. They also found significant clustering of the prioritized loci in cell adhesion pathways.
Limitations of multistage approach
While these approaches are novel in their use of functional data to add information to the genotype data, there are still some limitations that should be considered. First, they are biased towards finding SNPs with large main effects on gene expression and phenotype variation. Models that include SNPs with small independent effects that interact with one another to affect the outcome would be missed [53]. Another limitation is that this approach would not detect models with SNPs and gene-expression levels acting independently to alter the phenotype. For example, models would be missed if they included a SNP that affected protein conformation but not expression levels, or if they included gene-expression levels that affected phenotype owing to epigenetic factors such as methylation or acetylation. Finally, a weakness specific to the pathway analysis is its reliance on previous biological knowledge from annotated databases. For example, in Hsu et al., two of the prioritized loci were not included in the pathway analysis owing to a deficit of biological annotation [51]. Annotated databases can be extremely useful by allowing researchers to overlay interpretable biological knowledge onto their often agnostic, genome-wide studies, but this may result in sacrificing information about previously undiscovered biology [54].
Simultaneous analysis
Owing to the relatively nascent stages of using novel data mining techniques to find the etiology of complex human traits in high-throughput genetic and genomic data, simultaneous analyses are far less common than the multistage approach, which typically uses more traditional statistics. An effective simultaneous approach must be able to efficiently move through the search space to select the important quantitative or discrete variables and put them into a predictive model without bias towards either data type. For this section, we will go over one study that has applied a machine learning technique to meta-dimensional data and then discuss other methods that show promise for a simultaneous meta-dimensional analysis.
Tree-based methods
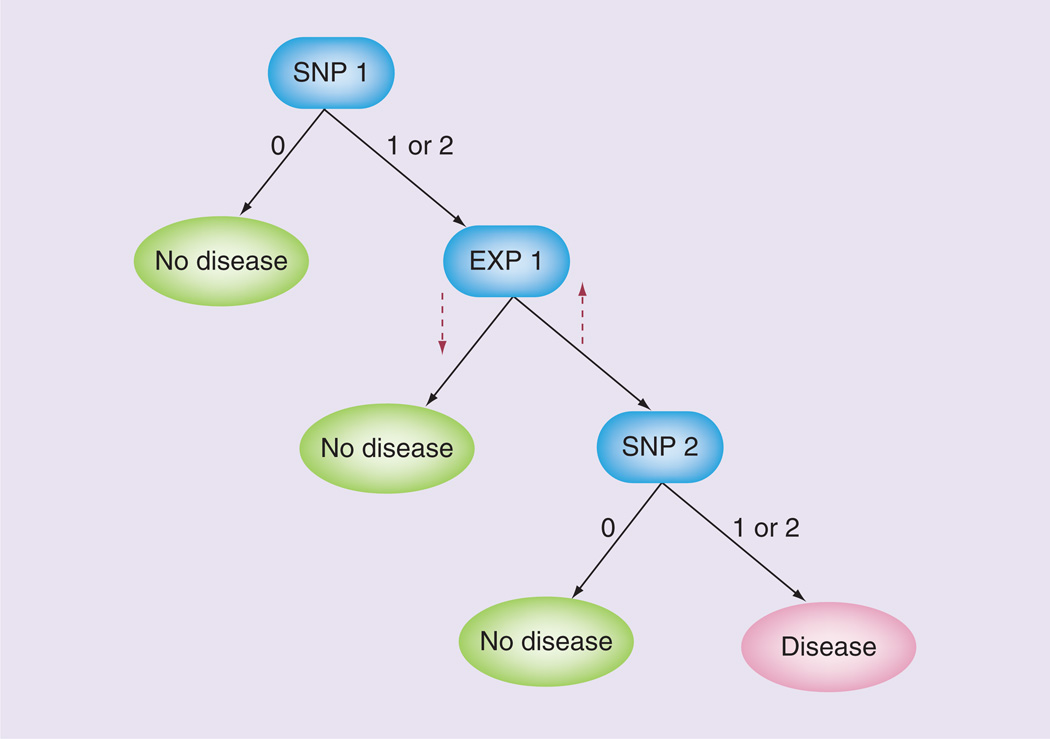
Owing to the extremely large number of variables associated with high-throughput data and the expansion of the search space as higher order models are considered, nonexhaustive data mining techniques, or methods that search for important patterns in the data without testing every possible combination of variables, are an attractive approach. Reif et al. perform an analysis to find meta-dimensional models that include both SNP genotypes and proteomic data in the form of serum cytokine levels to predict adverse reaction to smallpox vaccination [55]. First, they use Random Forests™ (RFs) to filter their data [56]. Briefly, RFs are a collection of classification or regression trees (Figure 2). Each tree is trained using a bootstrap sample of individuals from the dataset. For each tree node, the attribute, or independent variable, is selected from a subset of all attributes based on how well it reduces an impurity measure. Individuals not used for tree generation (‘out-of-bag’ individuals) are used to calculate tree prediction error and assign an ‘importance’ constant to each variable based on the effect of permuting the values [57]. This internal validation method helps to prevent overfitting. As the authors state, RFs are an appealing method because they can handle quantitative proteomic data and discrete genotype data. Notably, the fact that RFs rank the importance of each variable allows this method to be used for efficient variable selection. After RF filtering, the authors build decision trees from the most important variables to generate a more interpretable model. The final best tree from their analysis contained three proteomic variables and one SNP variable and had 75% prediction accuracy based on tenfold crossvalidation. Although the proteomic variables overall had higher importance values and dominated the best model, it is unclear whether RFs are biased towards one data type or if this is owing to the large role cytokines play in immune response.
Figure 2. Decision tree example.
For the SNP variables, the genotypes are represented as: 0: no minor alleles; 1: one minor allele; and 2: two minor alleles. The up and down dashed arrows indicate increased and decreased gene expression, respectively.
EXP: Gene expression
One limitation to decision trees and RFs is that they do not scale up to high-throughput, genome-wide data. To address this issue, Random Jungle (a faster version of RF) was developed [58]. Another limitation is that, although RFs are more robust to models that include interactions than single trees, for the initial split to be informative, at least some marginal effects are necessary.
Bayesian networks
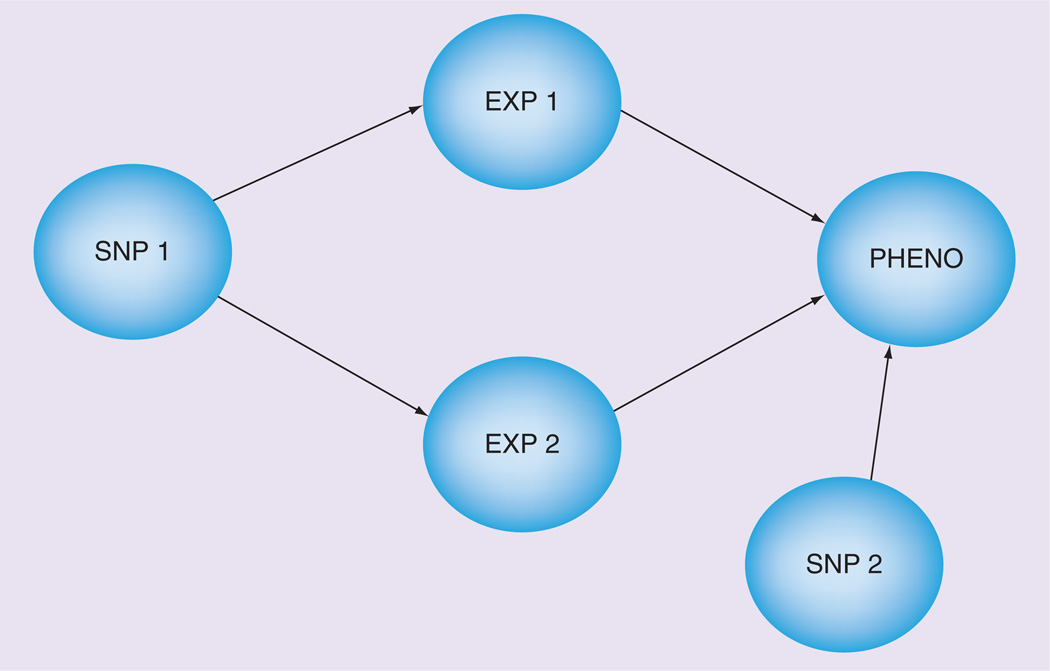
A Bayesian network (BN) is a directed acyclic graph that represents the joint probability distribution of random variables. BNs are appealing for meta-dimensional analyses for several reasons. First, BNs allow for the representation of conditional relationships and can be used to distinguish between indirect and direct associations, as shown in Figure 3. This is useful for the integration of genotype and gene-expression data where the genotypes may be operating directly on the phenotype or indirectly through gene expression. This aspect of the BN may also help in inferring causality [8]. Also, BNs assign probabilities to the variables, which provide a level of confidence or belief about the network. In addition, Bayesian methods are flexible in that they can be relatively agnostic by using noninformative priors or they can incorporate information from previous biological knowledge into the prior distribution to assist the network search [59]. Finally, similar to RFs, they are able to handle both quantitative and discrete input variables. Bayesian methods have been used to analyze genetic data to detect interacting networks of genes that associate with human traits [60].
Figure 3. Bayesian network example with direct and indirect effects.
EXP: Gene expression; PHENO: Phenotype.
One of the main drawbacks of BNs is the computational burden of evaluating networks across the search space [61]. To address this, Bayesian techniques often apply simulation techniques, such as the Gibbs sampler employed by the WinBUGS program [62], to allow for faster integration. Even with these faster methods, high-throughput data analysis may require a filtering technique to be computationally feasible.
Evolutionary computation methods
The final technique we will discuss that shows potential for meta-dimensional analysis is the use of evolutionary computation, either genetic programming (GP) or grammatical evolution (GE), which takes advantage of characteristics of biological evolution in order to optimize specific types of computer programs [63]. For meta-dimensional analyses, these computer programs will be in the form of solutions that contain quantitative or discrete variables that are modeled to predict a phenotype. The two types of solutions that will be discussed here are symbolic regression formulas and artificial neural networks (ANNs). The basic GP algorithm is as follows:
-
▪
A random population of solutions is generated and tested using the data to assign a ‘fitness’ to each network;
-
▪
The fittest solutions undergo evolutionary operations such as mutation, crossover and reproduction so that their ‘genes’ carry on to the next generation;
-
▪
This process is repeated for a prespecified number of generations so that, optimally, the final generation contains very fit (or highly predictive) solutions. Often this process is done using n-fold crossvalidation to prevent overfitting.
Symbolic regression solutions are mathematical formulas that map patterns in the input variables (SNPs and expression variables) back to some output (phenotype) [64–66]. These formulas are traditionally optimized using GP to find variables and mathematical operators that come together to form predictive models. These models can be very simple or complex depending on the operators used to initialize the process. Symbolic discriminant analysis is a method that uses evolutionary computation to optimize symbolic regression formulas to discriminate between values of a dichotomous outcome [64]. Symbolic discriminant analysis has been applied to the analysis of high-throughput gene-expression data [67]. In the review by Reif et al., symbolic discriminant analysis was used to analyze in silico data that included genotypic and proteomic variables with varying levels of interactions between the datatypes [6]. The classification errors for all models were lowest when the model included some main effects. Notably, the data was simulated with very few variables and does not represent a high-throughput scale.
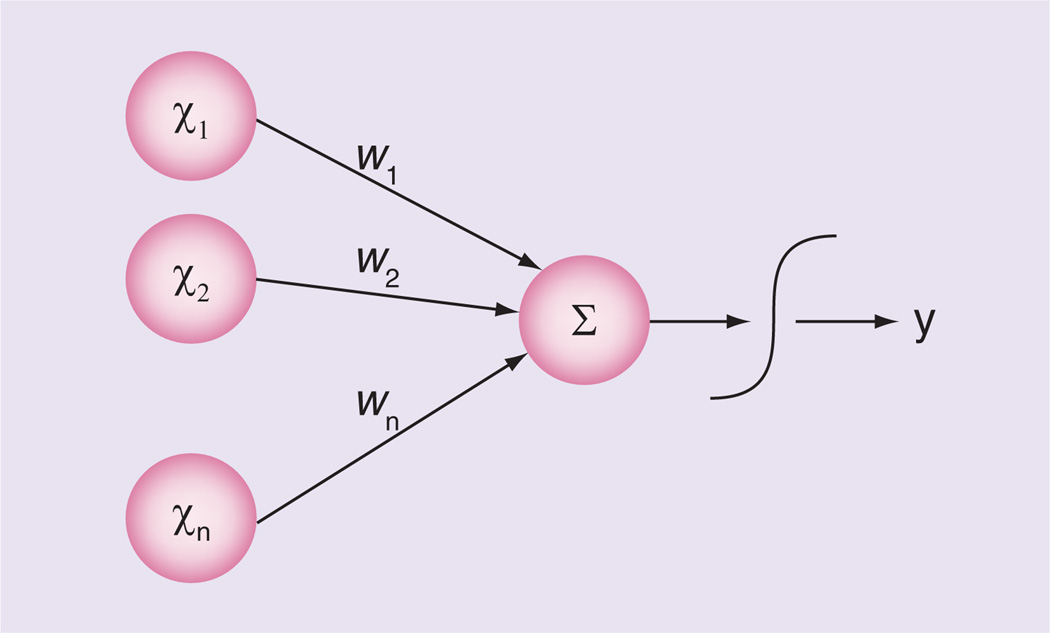
The ANN is a pattern recognition method that was originally designed to model learning processes in the brain. In short, ANNs are directed graphs that consist of an input layer, hidden layers and an output layer. The input layer nodes are independent variables; the hidden layer nodes are processing elements that send their signal to other hidden layer nodes or the output node (Figure 4). The arcs connecting the nodes are all associated with a weight constant [68]. Traditionally, ANNs are optimized using a gradient descent algorithm such as back-propagation that iteratively alters the weights until fitness is no longer improved. Back-propagation requires that the user prespecifies the input variables and the structure of the network. Using GP to optimize ANNs allows for simultaneous variable selection, architecture, and weight optimization [69].
Figure 4. Single-layer artificial neural network.
The Analysis Tool for Heritable and Environmental Network Associations (ATHENA) is a software package that uses GE to optimize symbolic regression formulas and ANN (i.e., GE + ANN: GENN). Both methods have been able to detect various types of genetic models, including highly epistatic interactions, using in silico genotype data [70–75]. GENN has been used in biological data to find interactions between eQTLs [76] and to model environmental and SNP genotypes to predict age-related macular degeneration [77]. Both GE symbolic regression formulas and GENN can accept quantitative and categorical variables to predict binary or continuous outcomes.
Using GP or GE to optimize either of these methods (symbolic regression or ANN) would be useful for meta-dimensional analysis because there is no need for a priori model specification, it performs a nonexhaustive search of the solution space, it can be easily parallelized for faster computation, and it allows for the discovery of any genetic model, including those with no main effects. One of the main weaknesses of GP is that, unlike BNs, it is not clear how to distinguish between indirect or direct effects in the final model. Also, there are no values generated that can be interpreted as probabilities or importance measures for the variables. Together, these limitations make the GP models less interpretable and harder to map back to specific biological functions. Also, as with the other two methods, the search space becomes infinitely large with high-throughput data and a filtering method should be used to increase the likelihood of model detection.
Future perspective
In the realm of biological research, the technology is advancing faster than the methods we use to analyze the data it generates. This requires that any new computational technique be flexible enough to adapt to different types of high-throughput data. Many of the methods discussed here use microarray data for gene expression and SNP genotypes for DNA variation; however, other measures of biological variability, such as copy number variation, RNA-sequence data and whole-genome sequence data, should be taken into consideration during the method development process.
Of note, there are computational methods not mentioned in this review that should also be considered as potential candidates for meta-dimensional disease analysis. For example, interactome network topology methods, which examine specific features of networks generated from meta-dimensional data that are significantly different from a null network, could be used to detect biological modules that correlate with a particular human trait [78]. Other types of methods that could be used are clustering or coclustering techniques that attempt to reduce the complexity of the data by generating new subgroups, or clusters, with similar attributes both within and between high-throughput datatypes [9]. Association between the subgroups and the complex human trait of interest could then be analyzed.
Models found by any of the methods reviewed in this article need to be validated statistically using independent datasets and functionally at the bench using in vitro and in vivo techniques. Exact statistical replication of the complex models resulting from some of the data mining techniques is extremely unlikely owing to environmental effects and genetic heterogeneity. Therefore, validation will require a more liberal interpretation of finding models with a high level of similarity across studies. Functional validation will be crucial to provide additional evidence that these are ‘true’ findings. Ultimately, comprehensive knowledge about the etiology that underlies complex human traits will allow for better treatment and prevention strategies in the future.
Executive summary.
Complex human trait etiology
-
▪
Genome-wide association studies have found many loci that associate with complex human traits, but most of the estimated heritability remains unexplained.
Meta-dimensional data analysis
-
▪
A study that combines different types of high-throughput data into one analysis may be able to find disease models that would not have been discovered with single data-type analysis.
Multistage approaches
-
▪
Triangle model: find expression quantitative trait loci that associate with complex traits.
-
▪
Pathway approach: use genotype and gene-expression data discovering more about the biology by finding annotated pathways that are over-represented.
Simultaneous analysis approaches
-
▪
Tree-based approaches: Random Forests™ and decision trees.
-
▪
Bayesian networks
-
▪
Evolutionary computing methods.
Acknowledgments
ER Holzinger was supported by NIH/National Institute of General Medical Sciences training grant T32 GM080178. MD Ritchie was supported by NIH grants LM010040 and Pharmacogenomics Research Network Statistical Analysis Resource (P-STAR). P-STAR is supported by funding from National Institute of General Medical Sciences and is part of the Pharmacogenomics Research Network. P-STAR is a component of HL065962.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
References
Papers of special note have been highlighted as:
▪ of interest
- 1.Maher B. Personal genomes: the case of the missing heritability. Nature. 2008;456(7218):18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 2.Grant GR, Manduchi E, Stoeckert CJ., Jr Analysis and management of microarray gene expression data. Curr. Protoc. Mol. Biol. 2007;Chapter 19(Unit 19.6) doi: 10.1002/0471142727.mb1906s77. [DOI] [PubMed] [Google Scholar]
- 3.Ozsolak F, Platt AR, Jones DR, et al. Direct RNA sequencing. Nature. 2009;461(7265):814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- 4.Nilsson T, Mann M, Aebersold R, et al. Mass spectrometry in high-throughput proteomics: ready for the big time. Nat. Methods. 2010;7(9):681–685. doi: 10.1038/nmeth0910-681. [DOI] [PubMed] [Google Scholar]
- 5.Carless M. Investigation of genomic methylation status using methylation-specific and bisulfite sequencing polymerase chain reaction. Methods Mol. Biol. 2009;523:217–234. doi: 10.1007/978-1-59745-190-1_15. [DOI] [PubMed] [Google Scholar]
- 6. Reif DM, White BC, Moore JH. Integrated analysis of genetic, genomic and proteomic data. Expert Rev. Proteomics. 2004;1(1):67–75. doi: 10.1586/14789450.1.1.67. ▪ Reviews in detail the benefits of integrating different types of data for genetic studies.
- 7.Hamid JS, Hu P, Roslin NM, et al. Data integration in genetics and genomics: methods and challenges. Hum. Genomics Proteomics. 2009 doi: 10.4061/2009/869093. pii: 869093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sieberts SK, Schadt EE. Moving toward a system genetics view of disease. Mamm. Genome. 2007;18(6–7):389–401. doi: 10.1007/s00335-007-9040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat. Rev. Genet. 2010;11(7):476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bahlo M, Stankovich J, Danoy P, et al. Saliva-derived DNA performs well in large-scale, high-density single-nucleotide polymorphism microarray studies. Cancer Epidemiol. Biomarkers Prev. 2010;19(3):794–798. doi: 10.1158/1055-9965.EPI-09-0812. [DOI] [PubMed] [Google Scholar]
- 11.Silander K, Saarela J. Whole genome amplification with Phi29 DNA polymerase to enable genetic or genomic analysis of samples of low DNA yield. Methods Mol. Biol. 2008;439:1–18. doi: 10.1007/978-1-59745-188-8_1. [DOI] [PubMed] [Google Scholar]
- 12.Ivarsson M, Carlson J. Extraction, quantitation, and evaluation of function DNA from various sample types. Methods Mol. Biol. 2011;675:261–277. doi: 10.1007/978-1-59745-423-0_14. [DOI] [PubMed] [Google Scholar]
- 13.Ponten F, Gry M, Fagerberg L, et al. A global view of protein expression in human cells, tissues, and organs. Mol. Syst. Biol. 2009;5:337. doi: 10.1038/msb.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lundberg E, Fagerberg L, Klevebring D, et al. Defining the transcriptome and proteome in three functionally different human cell lines. Mol. Syst. Biol. 2010;6:450. doi: 10.1038/msb.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilke RA, Xu H, Denny JC, et al. The emerging role of electronic medical records in pharmacogenomics. Clin. Pharmacol. Ther. 2011;89(3):379–386. doi: 10.1038/clpt.2010.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zatloukal K, Hainaut P. Human tissue biobanks as instruments for drug discovery and development: impact on personalized medicine. Biomark. Med. 2010;4(6):895–903. doi: 10.2217/bmm.10.104. [DOI] [PubMed] [Google Scholar]
- 17.Swede H, Stone CL, Norwood AR. National population-based biobanks for genetic research. Genet. Med. 2007;9(3):141–149. doi: 10.1097/gim.0b013e3180330039. [DOI] [PubMed] [Google Scholar]
- 18.Kaiser J. Biobanks. Population databases boom, from Iceland to the U.S. Science. 2002;298(5596):1158–1161. doi: 10.1126/science.298.5596.1158. [DOI] [PubMed] [Google Scholar]
- 19.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin. Pharmacol. Ther. 2008;84(3):362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Botling J, Micke P. Fresh frozen tissue: RNA extraction and quality control. Methods Mol. Biol. 2011;675:405–413. doi: 10.1007/978-1-59745-423-0_25. [DOI] [PubMed] [Google Scholar]
- 21.Ericsson C, Nister M. Blood plasma handling for protein analysis. Methods Mol. Biol. 2011;675:333–341. doi: 10.1007/978-1-59745-423-0_20. [DOI] [PubMed] [Google Scholar]
- 22.Hallmans G, Vaught JB. Best practices for establishing a biobank. Methods Mol. Biol. 2011;675:241–260. doi: 10.1007/978-1-59745-423-0_13. [DOI] [PubMed] [Google Scholar]
- 23.Louie LG, King MC. A novel approach to establishing permanent lymphoblastoid cell lines: Epstein–Barr virus transformation of cryopreserved lymphocytes. Am. J. Hum. Genet. 1991;48(3):637–638. [PMC free article] [PubMed] [Google Scholar]
- 24.Gibbs RA, Belmont JW, Hardenbol P, et al. The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 25.International HapMap Consortium. Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein TE, Chang JT, Cho MK, et al. Integrating genotype and phenotype information: an overview of the PharmGKB project. Pharmacogenetics Research Network and Knowledge Base. Pharmacogenomics J. 2001;1(3):167–170. doi: 10.1038/sj.tpj.6500035. [DOI] [PubMed] [Google Scholar]
- 28.Duan S, Huang RS, Zhang W, et al. Genetic architecture of transcript-level variation in humans. Am. J. Hum. Genet. 2008;82(5):1101–1113. doi: 10.1016/j.ajhg.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang W, Duan S, Kistner EO, et al. Evaluation of genetic variation contributing to differences in gene expression between populations. Am. J. Hum. Genet. 2008;82(3):631–640. doi: 10.1016/j.ajhg.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang W, Duan S, Bleibel WK, et al. Identification of common genetic variants that account for transcript isoform variation between human populations. Hum. Genet. 2009;125(1):81–93. doi: 10.1007/s00439-008-0601-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheung VG, Spielman RS. The genetics of variation in gene expression. Nat. Genet. 2002;32(Suppl.):522–525. doi: 10.1038/ng1036. [DOI] [PubMed] [Google Scholar]
- 32.Cheung VG, Conlin LK, Weber TM, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat. Genet. 2003;33(3):422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 33.Cheung VG, Spielman RS, Ewens KG, et al. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437(7063):1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spielman RS, Bastone LA, Burdick JT, et al. Common genetic variants account for differences in gene expression among ethnic groups. Nat. Genet. 2007;39(2):226–231. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheung VG, Spielman RS. Genetics of human gene expression: mapping DNA variants that influence gene expression. Nat. Rev. Genet. 2009;10(9):595–604. doi: 10.1038/nrg2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shukla SJ, Dolan ME. Use of CEPH and non-CEPH lymphoblast cell lines in pharmacogenetic studies. Pharmacogenomics. 2005;6(3):303–310. doi: 10.1517/14622416.6.3.303. ▪ Reviews in detail studies that have used cell line models to find pharmacogenetic loci and the benefits and limitations of this approach.
- 37. Zhang W, Dolan ME. Use of cell lines in the investigation of pharmacogenetic loci. Curr. Pharm. Des. 2009;15(32):3782–3795. doi: 10.2174/138161209789649475. ▪ Reviews in detail studies that have used cell line models to find pharmacogenetic loci and the benefits and limitations of this approach.
- 38.Welsh M, Mangravite L, Medina MW, et al. Pharmacogenomic discovery using cell-based models. Pharmacol. Rev. 2009;61(4):413–429. doi: 10.1124/pr.109.001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sie L, Loong S, Tan EK. Utility of lymphoblastoid cell lines. J. Neurosci. Res. 2009;87(9):1953–1959. doi: 10.1002/jnr.22000. [DOI] [PubMed] [Google Scholar]
- 40.Choy E, Yelensky R, Bonakdar S, et al. Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genet. 2008;4(11):e1000287. doi: 10.1371/journal.pgen.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stark AL, Zhang W, Mi S, et al. Heritable and non-genetic factors as variables of pharmacologic phenotypes in lymphoblastoid cell lines. Pharmacogenomics J. 2010;10(6):505–512. doi: 10.1038/tpj.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang RS, Duan S, Bleibel WK, et al. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proc. Natl Acad. Sci. USA. 2007;104(23):9758–9763. doi: 10.1073/pnas.0703736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang RS, Duan S, Shukla SJ, et al. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach. Am. J Hum. Genet. 2007;81(3):427–437. doi: 10.1086/519850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang RS, Duan S, Kistner EO, et al. Genetic variants contributing to daunorubicin-induced cytotoxicity. Cancer Res. 2008;68(9):3161–3168. doi: 10.1158/0008-5472.CAN-07-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang RS, Duan S, Kistner EO, Hartford CM, Dolan ME. Genetic variants associated with carboplatin-induced cytotoxicity in cell lines derived from Africans. Mol. Cancer Ther. 2008;7(9):3038–3046. doi: 10.1158/1535-7163.MCT-08-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hartford CM, Duan S, Delaney SM, et al. Population-specific genetic variants important in susceptibility to cytarabine arabinoside cytotoxicity. Blood. 2009;113(10):2145–2153. doi: 10.1182/blood-2008-05-154302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pedroso I. Gaining a pathway insight into genetic association data. Methods Mol. Biol. 2010;628:373–382. doi: 10.1007/978-1-60327-367-1_20. [DOI] [PubMed] [Google Scholar]
- 48. Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. Nat. Rev. Genet. 2010;11(12):843–854. doi: 10.1038/nrg2884. ▪ Describes many of the novel approaches for finding complex interaction models in human genetics studies.
- 49.Luo L, Peng G, Zhu Y, et al. Genome-wide gene and pathway analysis. Eur. J. Hum. Genet. 2010;18(9):1045–1053. doi: 10.1038/ejhg.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emilsson V, Thorleifsson G, Zhang B, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452(7186):423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 51.Hsu YH, Zillikens MC, Wilson SG, et al. An integration of genome-wide association study and gene expression profiling to prioritize the discovery of novel susceptibility loci for osteoporosis-related traits. PLoS Genet. 2010;6(6):e1000977. doi: 10.1371/journal.pgen.1000977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edwards YJ, Beecham GW, Scott WK, et al. Identifying consensus disease pathways in Parkinson’s disease using an integrative systems biology approach. PLoS ONE. 2011;6(2):e16917. doi: 10.1371/journal.pone.0016917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Culverhouse R, Suarez BK, Lin J, Reich T. A perspective on epistasis: limits of models displaying no main effect. Am. J. Hum. Genet. 2002;70(2):461–471. doi: 10.1086/338759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritchie MD. Using biological knowledge to uncover the mystery in the search for epistasis in genome-wide association studies. Ann. Hum. Genet. 2011;75(1):172–182. doi: 10.1111/j.1469-1809.2010.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reif DM, Motsinger-Reif AA, McKinney BA, et al. Integrated analysis of genetic and proteomic data identifies biomarkers associated with adverse events following smallpox vaccination. Genes Immun. 2009;10(2):112–119. doi: 10.1038/gene.2008.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baurley JW, Conti DV, Gauderman WJ, Thomas DC. Discovery of complex pathways from observational data. Stat. Med. 2010;29(19):1998–2011. doi: 10.1002/sim.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Breiman L. Random Forests. Machine Learning. 2001;45(1):5–32. [Google Scholar]
- 58.Motsinger AA, Ritchie MD, Reif DM. Novel methods for detecting epistasis in pharmacogenomics studies. Pharmacogenomics. 2007;8(9):1229–1241. doi: 10.2217/14622416.8.9.1229. [DOI] [PubMed] [Google Scholar]
- 59.Schwarz DF, Konig IR, Ziegler A. On safari to Random Jungle: a fast implementation of Random Forests for high-dimensional data. Bioinformatics. 2010;26(14):1752–1758. doi: 10.1093/bioinformatics/btq257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang X, Barmada MM, Visweswaran S. Identifying genetic interactions in genome-wide data using Bayesian networks. Genet. Epidemiol. 2010;34(6):575–581. doi: 10.1002/gepi.20514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carniak E. Bayesian networks without tears. AI Magazine. 1991 Winter;:50–63. [Google Scholar]
- 62.Lunn D, Andrew T, Best N, Spiegelhalter D. WinBUGS – a Bayesian modelling framework: concepts, structure, and extensibility. Stat. Comput. 2000;10:325–337. [Google Scholar]
- 63.Koza J. Genetic Programmming: On the Programming of Computers by Means of Natural Selection. Cambridge, MA, USA: MIT Press; 1992. [Google Scholar]
- 64.Moore JH, Barney N, Tsai CT, et al. Symbolic modeling of epistasis. Hum. Hered. 2007;63(2):120–133. doi: 10.1159/000099184. [DOI] [PubMed] [Google Scholar]
- 65.Bautu E, Bautu A, Luchian H. Symbolic regression on noisy data with genetic and gene expression programming. IEEE Computer Society. 2005;321 [Google Scholar]
- 66.Schmidt M, Lipson H. Distilling free-form natural laws from experimental data. Science. 2009;324(5923):81–85. doi: 10.1126/science.1165893. [DOI] [PubMed] [Google Scholar]
- 67.Moore JH, Parker JS, Olsen NJ, Aune TM. Symbolic discriminant analysis of microarray data in autoimmune disease. Genet. Epidemiol. 2002;23(1):57–69. doi: 10.1002/gepi.1117. [DOI] [PubMed] [Google Scholar]
- 68.Anderson JA. An Introduction to Neural Networks. Cambridge, MA, USA: MIT Press; 1995. [Google Scholar]
- 69.Koza JR, Rice JP. Genetic generation of both the weights and architecture for a neural network. IEEE Trans. 1991;2:397–404. [Google Scholar]
- 70.Holzinger ER, Dudek SM, Torstenson ES, Ritchie MD. ATHENA optimization: the effect of initial parameter settings across different genetic models. Lect. Notes Comput. Sci. 2011;6623:48–58. [Google Scholar]
- 71.Turner SD, Ritchie MD, Bush WS. Conquering the needle-in-a-haystack: how correlated input variables beneficially alter the fitness landscape for neural networks. Lect. Notes Comput. Sci. 2009;5483:80–91. [Google Scholar]
- 72.Motsinger-Reif AA, Dudek SM, Hahn LW, Ritchie MD. Comparison of approaches for machine-learning optimization of neural networks for detecting gene–gene interactions in genetic epidemiology. Genet. Epidemiol. 2008;32(4):325–340. doi: 10.1002/gepi.20307. [DOI] [PubMed] [Google Scholar]
- 73.Motsinger-Reif AA, Fanelli TJ, Davis AC, Ritchie MD. Power of grammatical evolution neural networks to detect gene–gene interactions in the presence of error. BMC Res. Notes. 2008;1:65. doi: 10.1186/1756-0500-1-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Holzinger ER, Buchanan CC, Dudek SM, et al. Initialization parameter sweep in ATHENA: optimizing neural networks for detecting gene–gene interactions in the presence of small main effects. Genet. Evol. Comput. Conf. 2010;12:203–210. doi: 10.1145/1830483.1830519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Turner SD, Dudek SM, Ritchie MD. ATHENA: a knowledge-based hybrid backpropagation–grammatical evolution neural network algorithm for discovering epistasis among quantitative trait loci. BioData Min. 2010;3(1):5. doi: 10.1186/1756-0381-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Turner SD, Dudek SM, Ritchie MD. Grammatical evolution of neural networks for discovering epistasis among quantitative trait loci. Lect. Notes Comput. Sci. 2010;6023:86–97. doi: 10.1186/1756-0381-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Spencer KL, Olson LM, Schnetz-Boutaud N, et al. Using genetic variation and environmental risk factor data to identify individuals at high risk for age-related macular degeneration. PLoS ONE. 2011;6(3):e17784. doi: 10.1371/journal.pone.0017784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tieri P, de la Fuente A, Termanini A, Franceschi C. Integrating omics data for signaling pathways, interactome reconstruction, and functional analysis. Methods Mol. Biol. 2011;719:415–433. doi: 10.1007/978-1-61779-027-0_19. [DOI] [PubMed] [Google Scholar]
Websites
- 101.Hindorff LA, Junkins HA, Hall PN, Mehta JP, Manolio TA. A catalog of published genome-wide association studies. www.genome.gov/gwastudies.
- 102.Coriell Institute for Medical Research. Camden, NJ, USA: http://ccr.coriell.org. [Google Scholar]