Abstract
Genome-wide occupancy profiles of five components of the RNA Polymerase III (Pol III) machinery in human cells identified the expected tRNA and non-coding RNA targets and revealed many additional Pol III-associated loci, mostly near SINEs. Several genes are targets of an alternative TFIIIB containing Brf2 instead of Brf1 and have extremely low levels of TFIIIC. Strikingly, expressed Pol III genes, unlike non-expressed Pol III genes, are situated in regions with a pattern of histone modifications associated with functional Pol II promoters. TFIIIC alone associates with numerous ETC loci, via the B box or a novel motif. ETCs are often near CTCF binding sites, suggesting a potential role in chromosome organization. Our results suggest that human Pol III complexes associate preferentially with regions near functional Pol II promoters and that TFIIIC-mediated recruitment of TFIIIB is regulated in a locus-specific manner.
RNA Polymerase III (Pol III) is responsible for the synthesis of tRNAs and other non-protein-coding RNAs (ncRNAs) in eukaryotes1–3. Metazoan Pol III genes are classified into three types, each of which is transcribed by Pol III in concert with other multi-subunit general transcription factors. At tRNAs and other Pol III genes categorized as type 2, the recognition factor, TFIIIC, binds to DNA sequences termed A and B boxes, which are typically contained entirely within the body of the structural gene. TFIIIC recruits the initiation factor, TFIIIB, which consists of the TATA-binding protein (TBP), Bdp1, and Brf1 subunits. These proteins recruit the polymerase to begin transcription. At the type 1 gene encoding the 5S rRNA, transcription involves a dedicated transcription factor, TFIIIA, which recognizes special elements within the gene and recruits TFIIIC, TFIIIB, and Pol III.
Other Pol III-transcribed genes termed type 3 have upstream, gene-external promoters, much like Pol II-transcribed genes. At these loci, a proximal sequence element (PSE) is bound by the SNAPc complex, which is involved in transcription of small nuclear RNAs by either Pol II or Pol III. At Pol III promoters bound by SNAPc, TATA-bound TBP specifies Pol III transcription by recruiting the Brf2 subunit into an alternative TFIIIB comprising TBP, Bdp1, and Brf2, followed by Pol III recruitment1–3. In vitro, Brf2-dependent Pol III genes can be transcribed in the absence of Brf1 or TFIIIC3.
Whole-genome chromatin immunoprecipitation studies using microarrays have been performed on subunits of each of these general transcription factors in yeast. In S. cerevisiae, complete Pol III complexes are found at tRNA genes and at loci encoding a variety of ncRNAs4–6. At these regions, the general transcription factors TFIIIC, TFIIIB, and Pol III are present at a fairly constant ratio, suggesting that the amount of TFIIIC binding to promoters is predictive of the level of functional Pol III transcription complexes in vivo. However, while TFIIIC-mediated recruitment of TFIIIB shows little, if any, promoter specificity, TFIIIB recruitment is generally repressed by Maf1 in response to environmental signals7–9.
In contrast to conventional Pol III-transcribed genes, eight loci with incomplete transcription complexes consisting only of TFIIIC were identified in S. cerevisiae and termed ETC, for “Extra TFIIIC”5. ETC loci are characterized by a sequence motif that contains the B box and an additional three conserved nucleotides, suggesting that TFIIIC’s association with this special motif influences its potential to recruit the remainder of the Pol III machinery. The ETC loci are remarkably well-conserved both structurally and functionally across closely related yeast species5, indicating that these regions are biologically important. Similar loci were later identified in S. pombe, where they are more numerous and named COCs (“chromosome organizing clamps”) due to their apparent role in higher-order chromosome organization10. It is unknown whether ETC loci are present in metazoans.
Here, by analyzing subunits of the three major Pol III transcription factors, we examine the occupancy profile of the Pol III transcription machinery on a genome-wide scale. We identify new Pol III targets, distinct classes of ETC loci, and distinct binding profiles of Brf1- and Brf2-containing isoforms of TFIIIB. Further, we find binding of the Pol III machinery to target promoters to be strongly linked with (and perhaps dependent on) a nearby functional Pol II promoter and associated histone modifications.
RESULTS
Genome-wide occupancy of five Pol III transcription factors
We performed chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-seq) on three independent biological replicates of K562 cells, using antibodies against five proteins involved in RNA polymerase III transcription. These antibodies correspond to subunits of the promoter recognition factor TFIIIC (TFIIIC-110 subunit), the initiation factor, TFIIIB (Bdp1 and Brf1 subunits), and the polymerase itself (Rpc155 subunit). In addition, we used an antibody against Brf2, which is involved in the transcription of Pol III-transcribed genes with gene-external promoter architecture.
The overall quality and reproducibility of the data were high, with correlation coefficients between replicates typically ranging between 0.79 and 0.95. (Supplementary Fig. 1). The Brf2 data were lower in overall quality and reproducibility (correlation coefficients between 0.41 and 0.68), but previously known Brf2-bound loci were easily identifiable using high-stringency thresholds. Peak lists for the five proteins are shown in Supplementary Fig. 2.
Expected and novel targets of RNA polymerase III
The Rpc155 antibody yielded by far the most robust ChIP signals of the antibodies. Using a stringent threshold, we identified 1520 Pol III targets, which we then used as the reference list when comparing occupancies by the Pol III transcription factors. To determine the occupancy levels of all five proteins at each Rpc155-associated locus, we counted the sequence reads (“tags”) for every protein within a 300 bp window centered on the coordinate with maximal Rpc155 occupancy. By determining the relative occupancy of Pol III factors at each genomic region, we obtained information about the nature of the transcription complexes at the Pol III promoters in a manner analogous to that used for Pol II preinitiation complexes11.
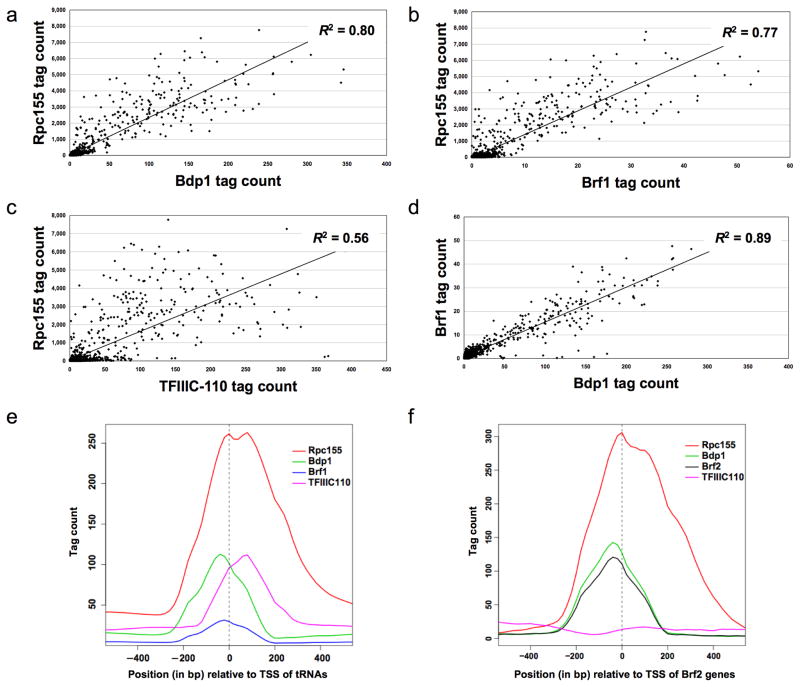
The general transcription factors show a high degree of co-occupancy at Pol III loci (Fig. 1A–D). The TFIIIB components Brf1 and Bdp1 were each highly correlated (R2 = 0.77 and 0.80) with Rpc155 at Pol III loci, validating our Rpc155-associated loci as meaningful targets of the complete Pol III machinery. Fittingly for two subunits of the same factor, Bdp1 and Brf1 showed a correlation (R2 = 0.89) comparable to that between replicate IPs of the same protein. At Rpc155 targets, TFIIIC was markedly less well correlated (R2 = 0.56) with polymerase than was TFIIIB. As will be discussed further below, this suggests that the degree of TFIIIC occupancy is not strictly related to the level of Pol III transcription.
Figure 1.
Pairwise comparison of occupancy by proteins at Pol III loci. Each plot compares the average number of sequence reads (“tag count”) for the indicated proteins within the 300bp window centered at each Rpc155 summit. (a) Rpc155 vs Bdp1. (b) Rpc155 vs Brf1. (c) Rpc155 vs TFIIIC-110. (d) Brf1 vs Bdp1. (e) Distribution of Bdp1, Brf1, TFIIIC-110, and Rpc155 association with respect to transcription start sites (TSS) of tRNAs. (f) Distribution of Bdp1, Brf2, TFIIIC-110, and Rpc155 association with respect to TSS of Brf2-associated genes.
We plotted the relative position of each protein with respect to transcription start sites (TSS) of Pol III genes (Fig. 1E and F). The peak of Rpc155 covers a region spanning from 5’ of the TSS into the structural gene. TFIIIC occupancy peaks downstream of the TSS and within the gene, reflecting the gene-internal position of the DNA sequences recognized by this factor. The TFIIIB components Brf1, Brf2, and Bdp1 have occupancy peaks upstream of the TSS, in accord with their role in initiation. Thus, the mapping afforded by the sequencing data is of sufficient resolution to distinguish closely spaced binding sites even within small Pol III genes.
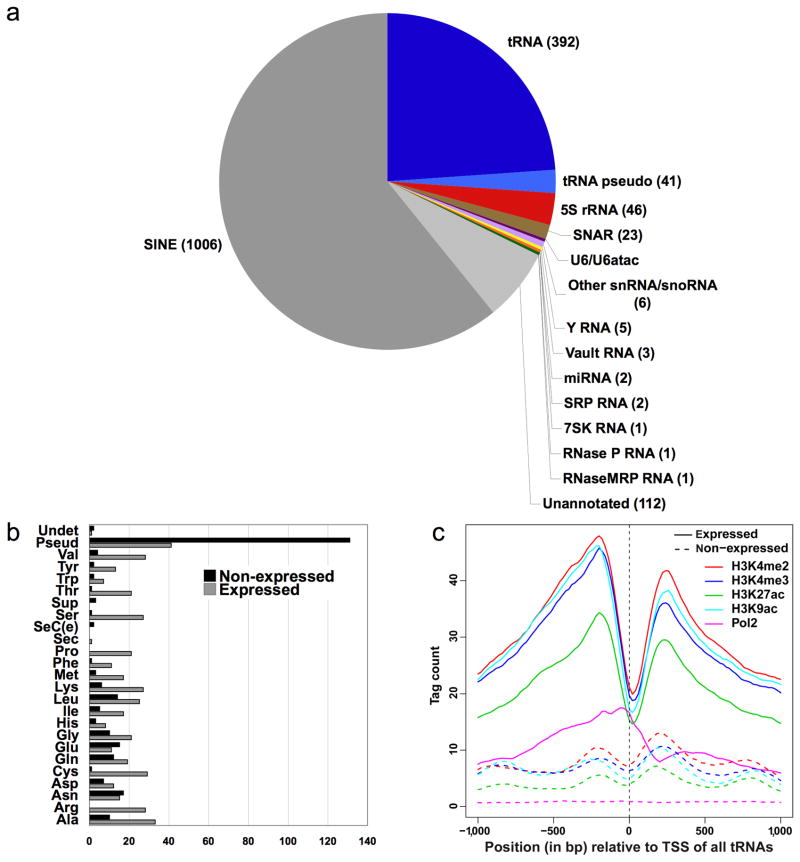
As expected1, the complete Pol III machinery is found at genes encoding tRNAs, 5S rRNA, U6, hY, 7SK RNA, and RNase P RNA (Fig. 2A), representing all three types of Pol III promoters. In general, TFIIIC, TFIIIB, and Pol III association with these loci is robust (> 100-fold enrichment of Pol III in many cases). Of the 513 tRNA genes, 392 are targets of Rpc155 by our stringent peak-calling criteria. In addition, we detected Pol III occupancy at 41 out of the 172 tRNA pseudogenes in the human genome. Because of the clustering of many tRNA genes, some Rpc155 peaks contain more than one tRNA target. The percentage of expressed genes for each type of tRNA is variable, with, for example, almost all cysteine tRNA genes, but fewer than half of glutamic acid tRNA genes, being occupied by the polymerase (Fig. 2B).
Figure 2.
Properties of Pol III targets. (a) Pie chart depicting the categories of Pol III-occupied loci. (b) Occupancy of tRNA genes by isotype. (c) Positions of histone modifications and Pol II occupancy relative to expressed (i.e., Pol III-occupied, solid lines) or non-expressed (dashed lines) tRNA genes.
In addition to the expected targets, many additional loci are associated with Pol III (Supplementary Fig. 3). These include NF90-associated RNAs encoded by snaR loci, which possess B boxes and Pol III termination sequences and are transcribed by Pol III in vitro12. We do not observe Pol III at any annotated miRNA genes (except for miR-1975 and miR-886, which are unlikely to be relevant, as they overlap completely with hY5 and vault RNAs that are known Pol III targets); this includes miRNA genes clustered on chromosome 19 that have been the subject of conflicting reports13,14. Small but reproducible amounts of Pol III are associated with U91, which overlaps the ncRNA SCARNA18, and with U13 snoRNA. A striking majority (90%) of the otherwise non-annotated Pol III-associated loci are near SINE elements. In general, these SINE-linked loci have considerably lower levels of Pol III association than observed at tRNA genes.
Active Pol III genes are near Pol II-dependent histone marks
In this paper, we define expressed and non-expressed tRNAs by virtue of Pol III association. While it is formally possible that Pol III association might not result in RNA synthesis, this is unlikely because tRNA levels are very high. We analyzed the proximity of expressed (i.e., Pol III-occupied) and non-expressed tRNAs with regard to nearby histone modifications. There is a marked dip in all histone marks corresponding exactly to the TSS of expressed tRNAs, suggesting a strong and localized nucleosome depletion over the TSS (Fig. 2C, solid lines). This apparent nucleosome depletion is similar to that observed at tRNA genes in S. cerevisiae15,16, and it is likely due to nucleosome exclusion by the extremely high levels of transcription at Pol III genes.
Although Pol III peaks are often in proximity to Pol II peaks17, the functional relationship of this observation is unclear. Strikingly, for the 392 expressed Pol III genes, there is a high correlation with a pattern of histone marks typical of Pol II TSS regions and with occupancy by Pol II itself. This proximity is directional; Pol II occupancy peaks 5’ of expressed Pol III genes. In contrast, no such pattern is observed for the non-expressed subset of tRNA genes (Fig. 2C, dashed lines). Together, these results suggest that the histone modification pattern at Pol II TSS regions is an important determinant of Pol III expression.
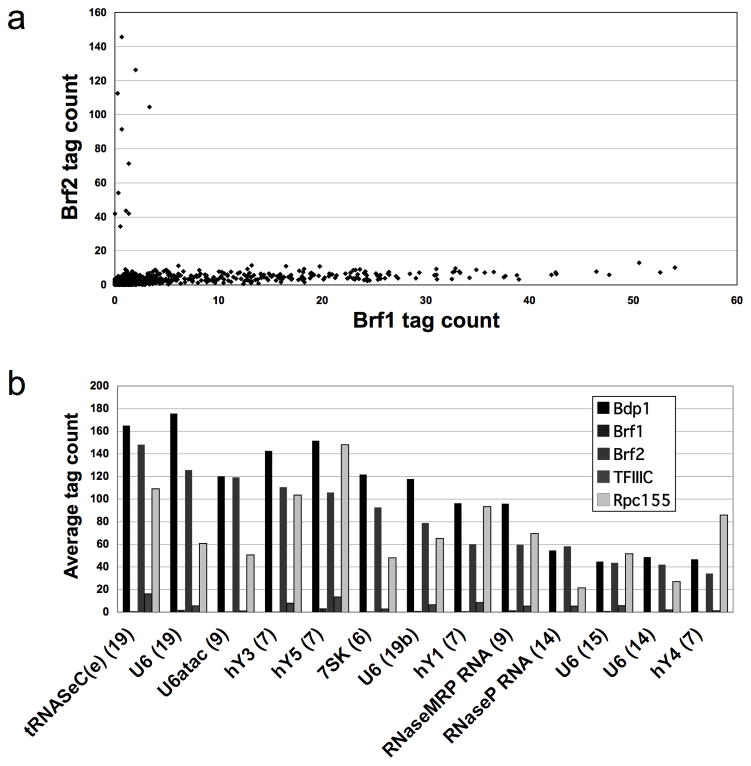
Brf2 sites lack Brf1 and are very low in TFIIIC
Brf2 occupies U6 and several other loci with TATA-containing promoters located upstream of the sequences encoding mature RNA. In vitro, Brf2 is recruited by TBP in a TFIIIC-independent manner to these promoters, forming a complex containing Bdp1 but lacking Brf13. In accord with this biochemical observation, Brf1 and Brf2 occupancies are uncorrelated at Pol III-occupied loci (Fig. 3A). Furthermore, Brf1 occupancy at Brf2 sites is very low or absent (Fig. 3B), indicating that Brf1 is not required for transcription of Brf2-dependent genes in vivo. TFIIIC association is also very low at Brf2 targets, although not entirely absent (~2% the level observed at Brf1-associated loci), suggesting that TFIIIC is unnecessary for Pol III recruitment to Brf2 loci in vivo (Fig. 3B). Notably, the Brf2-occupied locus with the highest TFIIIC binding (though still only a small fraction of the TFIIIC level at Brf1 targets), is tRNASec, which in X. laevis uses both a gene-internal B box and the gene-external promoter structure typical of type 3 genes18.
Figure 3.
Distinctions between TFIIIB isoforms. (a) Pairwise comparison of Brf1 and Brf2 occupancy at Rpc155 peaks. The average number of sequence reads for Brf1 and Brf2 are compared for the 300bp window centered at each Rpc155 summit. (b) Tag counts for all five proteins at Brf2 targets. Tag counts are shown for each protein within a 300bp window centered at the summit of every indicated Brf2 peak (the chromosome number of each locus is indicated in parentheses). We show only those peaks with above-background levels of Rpc155. Rpc155 tag counts were divided by 25 to allow use of a single scale for viewing all proteins.
Numerous ETC loci in human cells
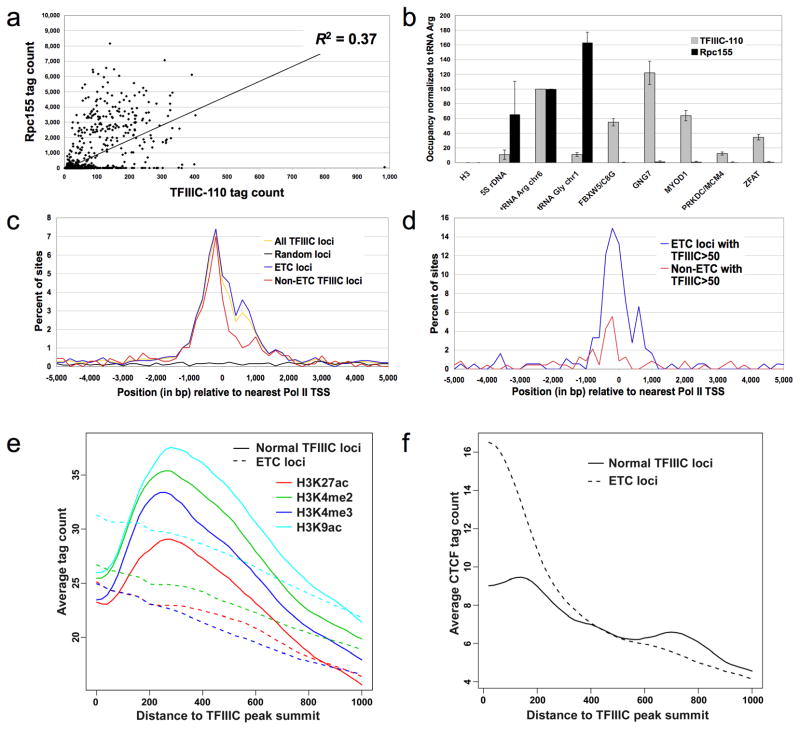
TFIIIC peaks vastly outnumber the peaks for the other transcription factors; our TFIIIC target list consists of 5474 loci, in contrast with 1520 at Rpc155, even though fold-enrichments and hence assay sensitivity are higher for Rpc155. Unlike TFIIIB loci, TFIIIC targets are highly variable in the extent to which they are occupied by the other components of the Pol III machinery (Fig. 4A), with only a limited correlation (R2 = 0.37) between Rpc155 and TFIIIC at TFIIIC targets. To identify human ETC loci, we first restricted our TFIIIC peak list to an even higher level of stringency (see methods). As the median ratio of TFIIIC to Rpc155 at tRNA genes is 0.06, we defined an ETC locus as having a TFIIIC/Rpc155 ratio higher than 2.04, three standard deviations above this median ratio. ETCs by this definition do not need to be completely lacking in Pol III occupancy. We validated binding of Rpc155 and TFIIIC at several normal and ETC targets using quantitative PCR analysis in real time (Fig. 4B). Under these strict criteria (only 3 tRNAs pass), we identify 1865 ETCs (listed in Supplementary Fig. 4) and suspect there are several thousand in the genome.
Figure 4.
Relationship of TFIIIC occupancy to Pol III occupancy and identification of ETC loci. (a) Correlation of Rpc155 and TFIIIC-110 occupancy at TFIIIC-110 peaks. Comparison of the number of Rpc155 and TFIIIC-110 sequence reads within the 300 bp window centered on each TFIIIC-110 peak summit. (b) Pol III and TFIIIC-100 association (± s.d.) at typical (left) and ETC (right) loci by quantitative PCR in real-time. (c) ETC loci are near the TSS of Pol II genes. Frequency distribution of TFIIIC-110 peak summits with respect to the nearest Pol II TSS. (d) Frequency distribution of ETC and non-ETC TFIIIC-110 peaks with the highest TFIIIC levels (average tag counts >50 in the 300bp-window around the peak summit) with respect to Pol II TSS. (e) Positions of histone modifications relative to TFIIIC-110 peak summits at non-ETC (solid lines) or ETC (dashed lines). (f) CTCF tag count as a function of distance from TFIIIC-110 peak summits for non-ETC (solid lines) or ETC (dashed lines) loci.
ETCs are often near Pol II genes and CTCF-bound regions
The distribution of TFIIIC-occupied loci of both the ETC and the non-ETC types reveals a positional bias toward the transcription start sites of Pol II genes (Fig. 4C). ETC loci show a wide dynamic range in the amount of TFIIIC binding, with some sites emerging as especially pronounced ETCs. The 181 ETC loci with the highest levels of TFIIIC occupancy (>50 sequence reads per site) are strikingly well-correlated with the TSS of Pol II genes, with 68% being located within 1kb of a Pol II TSS (Fig. 4D). This is reminiscent of the S. cerevisiae ETCs, which are ~200–300 bp upstream of a neighboring Pol II transcribed gene. GO categories of Pol II genes adjacent to ETC loci show significant overrepresentation of nuclear-localized proteins. In addition, ETCs are significantly overrepresented in regions upstream of closely spaced, divergently transcribed pairs of Pol II genes. Of the 1431 shared upstream sequences for Pol II genes (defined as having TSS less than 1kb apart), 86 harbor ETC loci (P < 10−300).
As expected from the proximity of strong ETCs to mRNA initiation sites, the ETC population shows histone modifications associated with functional Pol II promoters, with highest levels of these modifications near the TFIIIC peaks (Fig. 4E). This pattern of histone modifications at ETC loci (dashed lines) differs from the pattern at expressed Pol III genes (solid lines), in which histone modifications appear very low near the promoter due to nucleosome loss accompanying high levels of Pol III transcription. Lastly, in accord with the observation that TFIIIC targets in yeast play a role in genome organization10,19,20 and act as transcriptional insulator elements21–23, TFIIIC association at ETC loci is highly correlated with association of CTCF (Fig. 4F), a protein that interacts with cohesins and is involved in insulation, looping, and chromosome conformation24,25. This observation suggests that TFIIIC may play a role in chromosome organization in humans.
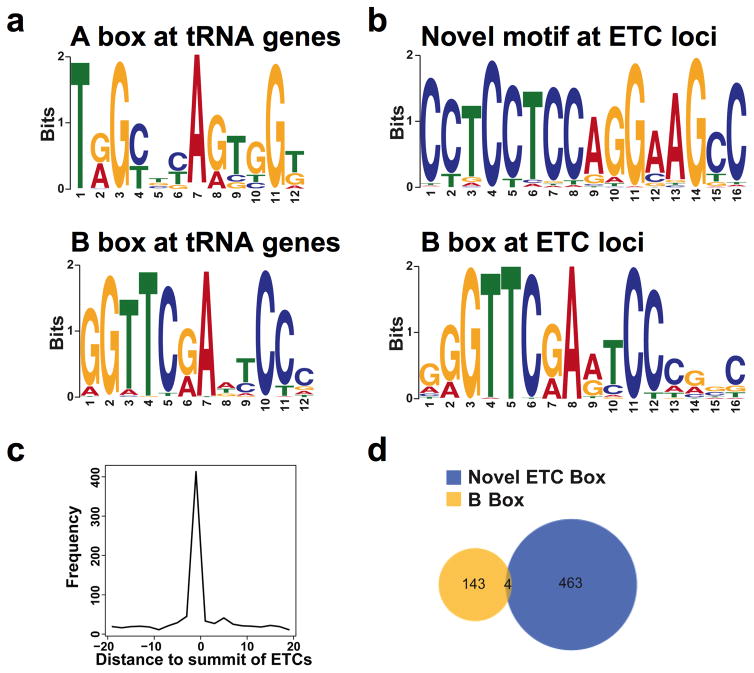
A novel sequence motif at a subset of ETC loci
De novo motif searching revealed that in contrast with tRNAs, which have A and B boxes, the ETC loci are characterized by one of two significantly overrepresented sequence motifs: the standard B box or a novel motif that may be loosely related to a motif for Ets transcription factors (Fig. 5A and B). The ETC motif coincides with the summit of TFIIIC peaks at many ETCs (Fig. 5C). ETC and B box motifs are mostly mutually exclusive, with only 4 ETC loci having both motifs (Fig. 5D). The existence of an ETC-specific motif is analogous to the situation in S. cerevisiae, where ETC loci possess a novel motif extending the canonical B box. In both organisms, TFIIIC could associate with these ETC-specific motifs in a manner structurally unconducive to the assembly of complete Pol III complexes.
Figure 5.
ETC motifs. (a) Known A and B box motifs found by de novo motif searching of tRNA genes. (b) Motifs found by de novo searching of ETC loci are a novel motif (top) and the B box motif (bottom). (c) Frequency plot of the novel ETC motif position as a function of distance from the TFIIIC-110 peak summit. (d) Venn diagram illustrating very little overlap between TFIIIC-110 peaks with the B box motif and those sites with the ETC motif.
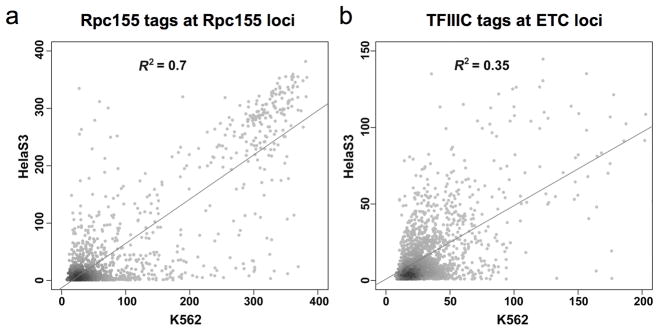
ETC loci show cell-type specificity
To determine whether Pol III occupancy varies with cell type, we examined the genome-wide association of Pol III factors in a second cell line, HeLa S3. Interestingly, while binding of Pol III factors at tRNA loci is highly correlated between cell lines (Fig. 6A), there are differences in TFIIIC occupancy at ETC loci between HeLa S3 and K562 cells (Fig. 6B). This observation suggests that TFIIIC binding and/or TFIIIB recruitment at ETC loci, but not tRNA genes, is influenced by cell-type-specific factors.
Figure 6.
Cell-specificity of ETC loci. (a) Correlation of Rpc155 tag counts in K562 and HeLa cells at Rpc155-occupied loci. (b) Correlation of TFIIIC-110 tag counts in K562 and HeLa cells at ETC loci.
DISCUSSION
Genome-wide profiling reveals ~1500 Pol III-associated loci in K562 cells. In general, the expected 400–500 Pol III-transcribed genes encoding tRNAs and ncRNAs show very high levels of all three Pol III factors. In contrast, ~1000 Pol III-associated loci have not been previously described, and the vast majority of these (90%) are located near SINEs. The SINE-associated loci show much lower levels of Pol III factors as compared to tRNA genes, although these levels are clearly above the background. The transcriptional products of and biological functions of these Pol III-associated loci near SINEs are unknown. The prevalence of SINEs near these Pol III-associated loci might reflect DNA sequences resembling B or A blocks in a subset of SINEs.
Most, but not all, tRNA genes are occupied by the complete Pol III transcription machinery, and Pol III association at expressed vs. non-expressed genes differs by a factor of 100. Strikingly, the genomic regions in the vicinity of expressed and non-expressed Pol III genes are different. Expressed Pol III genes are located close to regions that have histone modifications characteristic of functional Pol II promoters and Pol II itself. In contrast, this distinctive chromatin signature is absent from non-expressed Pol III genes. These observations suggest that Pol III factors bind preferentially to genomic regions with a histone modification pattern generated by a functional Pol II promoter. We think it unlikely that non-expressed Pol III genes have defective TFIIIC recognition sites, because they generally possess high-quality B box sequences. Conversely, TFIIIC associates with fewer than 2% of the B box motifs in the human genome, indicating that the presence of a B box alone is insufficient for binding in vivo. In addition, the fact that ETC loci are located near Pol II promoters and associated histone modification strongly argues that TFIIIC association depends on Pol II-generated chromatin regions.
The characteristic pattern of histone modifications at Pol II promoters depends on Pol II preinitiation complexes but not on extensive elongation, because Pol II is often paused just downstream of many promoters in a manner that precludes any appreciable transcription26,27. This suggests that TFIIIC binding to regions near functional Pol II promoters is largely, and perhaps completely, independent of Pol II transcriptional activity (i.e. mRNA synthesis). In accord with this suggestion, treatment of human cells with alpha-amanitin, an inhibitor that blocks Pol II transcription after preinitiation complex formation, has limited effects on Pol III transcription17. For these reasons, we speculate that TFIIIC binding is strongly enhanced by the chromatin structure generated by nearby functional Pol II promoters. In principle, TFIIIC binding might be increased by promoter accessibility due to histone acetylation reducing histone-DNA contacts, direct interactions with modified histones via an effector domain(s) in a TFIIIC subunit or associated protein, or a histone variant (e.g. H2AZ) near Pol II promoter regions.
Accessibility to the recognition factor TFIIIC is not, however, the only determinant of Pol III association in human cells. In S. cerevisiae, TFIIIC levels are highly correlated with Pol III occupancy, and recruitment of the complete Pol III machinery exhibits a one-to-one correspondence with TFIIIC binding. In contrast, the human TFIIIC/Pol III ratio is considerably more variable than it is in yeast, and a wide range of TFIIIC levels may recruit a given amount of polymerase. The presence of a Brf2 mechanism for Pol III recruitment in human cells is one obvious alternative pathway, allowing transcription with little or no TFIIIC. At the opposite extreme are the ETC loci, which recruit little or no polymerase despite having high occupancy by TFIIIC. Even at standard type 2 Pol III genes, the ratio of TFIIIC to polymerase varies over a wide range, in contrast to the TFIIIB/Pol III ratio, which is relatively consistent.
The different patterns of histone marks near ETC-type and transcriptionally active TFIIIC sites suggest that TFIIIC’s ability to recruit TFIIIB to type 2 genes might depend on a particular histone modification pattern. Indeed, Myc-dependent activation of Pol III transcription is associated with targeted histone acetylation and increased association of TFIIIB28. In a similar vein, transcription of type 3 genes is influenced by the CHD8 chromatin modifying protein29. Interestingly, TFIIIC either possesses histone acetylase activity30,31 and/or recruits the p300 histone acetylase32, such that it could actively participate in generating a chromatin state appropriate for TFIIIB recruitment. Association of the initiator TFIIIB is an important rate-limiting step in Pol III gene expression in humans, because the excellent correlation of TFIIIB and polymerase levels suggests that Pol III occupancy follows linearly from TFIIIB recruitment. In this regard, TFIIIB is a target of regulation Maf133, Rb34, p5335, Erk36, and Myc28.
The presence of TFIIIC at many loci without TFIIIB or Pol III, much like similar loci in yeast, suggests a role for human TFIIIC beyond its function in Pol III transcription. TFIIIC bound to the intergenic region of closely spaced, divergently transcribed Pol II genes might be important for the regulation and separate expression of the two genes. In yeast, TFIIIC-bound loci function as heterochromatin barriers and insulators10,21,22 and also participate in higher-order chromosome organization10,37. The correlation of TFIIIC peaks with CTCF is noteworthy and suggests the possible involvement of human TFIIIC in chromosome organization.
METHODS
Cell culture and formaldehyde crosslinking
We cultured three separate preparations of K562 cells in RPMI supplemented with 10% (v/v) FBS to a density of 2×107 cells/ml and crosslinked the cells with 1% (v/v) formaldehyde for 10 minutes before harvesting and freezing them.
Chromatin immunoprecipitation and quantitative PCR
We performed chromatin preparation and chromatin immunoprecipitations as previously described38 on material derived from three independent biological replicates of 2×107 cells each. The antibodies used were 2663 for Bdp139, 128 for Brf140, 4286 for TFIIIC11034, and 1900 for Rpc15539. Antibody 4295 against Brf2 was raised by immunizing rabbits with keyhole limpet hemocyanin coupled to synthetic peptides VSRSQQRGLRRVRDLC and SDSEIEQYLRTPQEVR, corresponding to human Brf2 residues 66–80 and 385–400, respectively. 2/3 of each IP was used for Illumina sequencing. We performed quantitative PCR in real time as previously described5. We obtained occupancy values for genomic loci by subtracting the value obtained for the control H3 locus and then expressing the results relative to the level at the arginine tRNA locus on chromosome VI that was arbitrarily defined as 100.
Illumina sequencing
We subjected input control and IP DNA to amplification and sequencing on an Illumina Genome Analyzer as previously described41. The IP DNA from each replicate was amplified separately and sequenced in different lanes, yielding an average read count of more than 15–19 million.
Data analysis and peak identification
We analyzed only those matches aligning to unique positions in the genome. For most factors, we obtained 9–10 million uniquely aligned reads per K562 replicate and 14–15 million per HeLa replicate. TFIIIC-110 gave a lower percentage of uniquely mappable reads (approximately 4 million per replicate), most likely because TFIIIC recognizes a DNA region with a high degree of sequence identity across tRNAs. We used the MACS peak-finding software42 to analyze the sequencing data and identify targets of each proteins. For each factor, sequence data from the three replicates were analyzed both individually and also as a single merged data set. We considered a peak to be a target if it had a pValue in the combined data set of 1×10−7 or better, with the additional requirement that two or more replicates individually yield pValues of 1×10−5 or lower. When looking for ETCs, we increased the stringency of the cutoff used to define TFIIIC sites by requiring that a TFIIIC peak be defined by an overall pValue of 1×10−7 or better and a pValue of 1×10−5 or better in all three replicates. We visualized peaks using the Affymetrix Integrated Genome Browser. We obtained genome information from the UCSC Genome Browser and the Eddy lab tRNA database. Data for histone modifications and CTCF came from the Broad Institute database, and Pol II ChIP-sequencing data was from the Yale ENCODE sequencing project data on the UCSC genome database. We identified sequence motifs using the MEME suite43.
Supplementary Material
Acknowledgments
We thank Tao Liu and Xiaole Liu for initial comments about the sequencing data, Joe Mellor for helpful advice on programming in Python, and Nathan Lamarre-Vincent, Xiaochun Fan, Heather Hirsch, and Marianne Lindahl Allen for discussion. We acknowledge Ghia Euskirchen, Hannah Monahan, Minyi Shi, and Phil Lacroute for help with DNA sequencing and Mike Wilson for help with database submission. Funding for this work was provided by NIH grants GM 30186 (K.S.), HG 4558 (M.S. and K.S), and HG 4695 (Z.W.).
Footnotes
Accession codes
The DNA sequencing datasets described in this work are available at http://genome.ucsc.edu/ENCODE/.
References
- 1.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Geiduschek EP, Kassavetis GA. The RNA polymerase III transcription apparatus. J Mol Biol. 2001;310:1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- 3.Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 4.Harismendy O, et al. Genome-wide location of yeast RNA polymerase III transcription machinery. EMBO J. 2003;22:4738–4747. doi: 10.1093/emboj/cdg466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moqtaderi Z, Struhl K. Genome-wide occupancy of the RNA polymerase III machinery in Saccharomyces cerevisiae reveals loci with incomplete transcription complexes. Mol Cell Biol. 2004;24:4118–4127. doi: 10.1128/MCB.24.10.4118-4127.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts DN, Stewart AJ, Huff JT, Cairns BR. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc Natl Acad Sci USA. 2003;100:14695–14700. doi: 10.1073/pnas.2435566100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oficjalska-Pham D, et al. General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1. Mol Cell. 2006;22:623–32. doi: 10.1016/j.molcel.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Roberts DN, Wilson B, Huff JT, Stewart AJ, Cairns BR. Dephosphorylation and genome-wide association of Maf1 with Pol III-transcribed genes during repression. Mol Cell. 2006;22:633–44. doi: 10.1016/j.molcel.2006.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Upadhya R, Lee J, Willis IM. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol Cell. 2002;10:1489–94. doi: 10.1016/s1097-2765(02)00787-6. [DOI] [PubMed] [Google Scholar]
- 10.Noma KI, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 11.Kuras L, Kosa P, Mencia M, Struhl K. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science. 2000;288:1244–1248. doi: 10.1126/science.288.5469.1244. [DOI] [PubMed] [Google Scholar]
- 12.Parrott AM, Mathews MB. Novel rapidly evolving hominid RNAs bind nuclear factor 90 and display tissue-restricted distribution. Nucl Acids Res. 2007;35:6249–6258. doi: 10.1093/nar/gkm668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 14.Bortolin-Cavaille ML, Dance M, Weber M, Cavaille J. C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucl Acids Res. 2009;37:3464–3473. doi: 10.1093/nar/gkp205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee W, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 16.Mavrich TN, et al. A barrier nucleosome model for statistical positioning of nucleosome throughout the yeast genome. Genome Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raha D, et al. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc Natl Acad Sci USA. 2010;107 doi: 10.1073/pnas.0911315106. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carbon P, Krol A. Transcription of the Xenopus laevis selenocysteine tRNA(Ser)Sec gene: a system that combines an internal B box and upstream elements also found in U6 snRNA genes. EMBO J. 1991;10:599–606. doi: 10.1002/j.1460-2075.1991.tb07987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Ambrosio C, et al. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 2008;22:2204–2214. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simms TA, et al. TFIIIC Binding Sites Function as both Heterochromatin Barriers and Chromatin Insulators in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7:2078–2086. doi: 10.1128/EC.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valenzuela L, Dhillon N, Kamakaka RT. Transcription independent insulation at TFIIIC-dependent insulators. Genetics. 2009;183:131–148. doi: 10.1534/genetics.109.106203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes & Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parelho V, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 26.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kenneth NS, et al. TRRAP and GCN5 are used by c-Myc to activate RNA polymerase III transcription. Proc Natl Acad Sci U S A. 2007;104:14917–22. doi: 10.1073/pnas.0702909104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan CC, et al. CHD8 Associates with Human Staf and contributes to efficient U6 RNA polymerase III transcription. Mol Cell Biol. 2007;27:8729–8738. doi: 10.1128/MCB.00846-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsieh YJ, Kundu TK, Wang Z, Kovelman R, Roeder RG. The TFIIIC90 subunit of TFIIIC interacts with multiple components of the RNA polymerase III machinery and contains a histone-specific acetyltransferase activity. Mol Cell Biol. 1999;19:7697–704. doi: 10.1128/mcb.19.11.7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kundu TK, Wang Z, Roeder RG. Human TFIIIC relieves chromatin-mediated repression of RNA polymerase III transcription and contains an intrinsic histone acetyltransferase activity. Mol Cell Biol. 1999;19:1605–1615. doi: 10.1128/mcb.19.2.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mertens C, Roeder RG. Different functional modes of p300 in activation of RNA polymerase transcription from chromatin templates. Mol Cell Biol. 2008;28:5764–5776. doi: 10.1128/MCB.01262-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rollins J, Veras I, Cabarcas S, Willis I, Schramm L. Human Maf1 negatively regulates RNA polymerase III transcription via the TFIIB family members Brf1 and Brf2. Int J Biol Sci. 2007;3:292–302. doi: 10.7150/ijbs.3.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sutcliffe JE, Brown TR, Allison SJ, Scott PH, White RJ. Retinoblastoma protein disrupts interactions required for RNA polymerase III transcription. Mol Cell Biol. 2000;20:9192–9202. doi: 10.1128/mcb.20.24.9192-9202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crighton D, et al. p53 represses RNA polymerase III transcription by targeting TBP and inhibiting promoter occupancy by TFIIIB. EMBO J. 2003;22:2810–2820. doi: 10.1093/emboj/cdg265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Felton-Edkins ZA, et al. The mitogen-activated protein (MAP) kinase ERT induces tRNA synthesis by phosphorylating TFIIIB. EMBO J. 2003;22:2422–2432. doi: 10.1093/emboj/cdg240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hiraga SI, Botsios S, Donaldson AD. Histone H3 lysine 56 acetylation by Rtt1090 is crucial for nucleosome positioning. J Cell Biol. 2008;183:641–651. doi: 10.1083/jcb.200806065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miotto B, Struhl K. HBO1 histone acetylase is a co-activator of the replication licensing factor Cdt1. Genes & Dev. 2008;22:2633–2638. doi: 10.1101/gad.1674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fairley JA, Scott PH, White RJ. TFIIIB is phosphorylated, disrupted and selectively released from tRNA promoters during mitosis in vivo. EMBO J. 2003;22:5841–5850. doi: 10.1093/emboj/cdg544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cairns CA, White RJ. p53 is a general repressor of RNA polymerase III transcription. EMBO J. 1998;17:3112–3123. doi: 10.1093/emboj/17.11.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, et al. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol. 2009;16:847–852. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.