ABSTRACT
Vibrio cholerae has two chromosomes (chrI and chrII) whose replication and segregation are under different genetic controls. The region covering the replication origin of chrI resembles that of the Escherichia coli chromosome, and both origins are under control of the highly conserved initiator, DnaA. The origin region of chrII resembles that of plasmids that have iterated initiator-binding sites (iterons) and is under control of the chrII-specific initiator, RctB. Both chrI and chrII encode chromosome-specific orthologs of plasmid partitioning proteins, ParA and ParB. Here, we have interfered with chrII replication, segregation, or both, using extra copies of sites that titrate RctB or ParB. Under these conditions, replication and segregation of chrI remain unaffected for at least 1 cell cycle. In this respect, chrI behaves similarly to the E. coli chromosome when plasmid maintenance is disturbed in the same cell. Apparently, no checkpoint exists to block cell division before the crippled chromosome is lost by a failure to replicate or to segregate. Whether blocking chrI replication can affect chrII replication remains to be tested.
IMPORTANCE
Chromosome replication, chromosome segregation, and cell division are the three main events of the cell cycle. They occur in an orderly fashion once per cell cycle. How the sequence of events is controlled is only beginning to be answered in bacteria. The finding of bacteria that possess more than one chromosome raises the important question: how are different chromosomes coordinated in their replication and segregation? It appears that in the evolution of the two-chromosome genome of V. cholerae, either the secondary chromosome adapted to the main chromosome to ensure its maintenance or it is maintained independently, as are bacterial plasmids. An understanding of chromosome coordination is expected to bear on the evolutionary process of chromosome acquisition and on the efficacy of possible strategies for selective elimination of a pathogen by targeting a specific chromosome.
Introduction
The presence of a multiplicity of chromosomes is a universal feature of eukaryotic cells. Each eukaryotic chromosome also has many origins. However, a subset of origins suffices to complete replication (1). Crippling an active origin is usually not a problem for replication, because forks emanating from adjacent active origins or activation of new dormant origins allow replication across the crippled origin (2, 3). Inactivation of a few origins may increase the time to complete duplication, but this does not affect replication initiation from other origins positioned on the same chromosome or on different chromosomes (4). Blocking of a large number of origins (~50) may, however, induce checkpoint factors that prolong the S phase and delay entry into mitosis. If a fork has to travel a long distance, it is more likely to encounter roadblocks such as abnormal DNA structures or opposing transcription, which might lead to significant fork stalling and the consequent checkpoint induction (5). The induction of a checkpoint response helps to stabilize the stalled fork and to suppress firing of dormant and late origins, apparently to avoid generation of further stalled forks till the damage is repaired (6, 7). The induction of checkpoint responses thus appears to be due to the stalling of replication forks, not due to blockage of origin firing.
Bacteria in most cases have only one chromosome, and the entire chromosome is duplicated starting from a single origin by two divergent replication forks (8). The forks travel thousands of kilobases without inducing a checkpoint response unless the DNA is significantly damaged by external agents, such as UV light or mitomycin (9). Although checkpoint response is not induced, forks stall nonetheless. Only when the degree of damage overwhelms the repair capacity of the cell is the checkpoint response induced, which blocks cell division to allow time for repairing the damage.
Bacterial plasmids replicate independently of the chromosome: blocking the replication of one seems not to affect the replication of the other. When replication initiation from the chromosomal origin is blocked, the chromosomes can be duplicated by replication initiated from an integrated plasmid copy (integrative suppression) (10). Plasmid-bearing cells can also be cured of plasmids without affecting chromosomal replication directly, although the plasmid loss can activate lethal toxins in some cases (11). Independence of plasmid and chromosomal replication is also evident from the fact that, whereas chromosomal replication initiates at a particular time of the cell cycle, plasmids normally initiate replication without regard to the cell cycle (12).
With the availability of many bacterial genome sequences, it has become clear that in about 10% of bacteria, the genome can be divided into separate chromosomes (13, 14). It is now possible to ask whether nonidentical bacterial chromosomes communicate with each other so as to ensure that their replication and segregation are completed in a timely fashion before the cell division, or whether they are maintained independently in the cell cycle, like the chromosome and plasmids of Escherichia coli. The two-chromosome bacterium, Vibrio cholerae, is ideally suited to address these questions, because much is known about how the individual chromosomes replicate (15–20) and segregate (21–24). Chromosome I (chrI; 3 Mbp) has the vast majority of genes essential for cell function. In contrast, chromosome II (chrII; 1 Mbp) has a few essential genes but mostly genes of unknown function and origin (25). The replication origin of chrI (oriI) is similar to the well-studied oriC of the E. coli chromosome, and oriI can replace oriC to maintain faithfully the E. coli chromosome (15, 26). Like oriC, oriI is controlled by the initiator DnaA (27). The origin of chrII (oriII) is different. It functions under control of a chrII-specific initiator, RctB. Moreover, oriII bears many similarities to plasmid origins that have iterated, plasmid-specific initiator binding sites (iterons). These sites are used both for replication initiation and its control (28). chrII iterons also play this dual role (16, 20).
A few additional features of the chrII replication control elements deserve mention. They are expected to endow chrII with the capacity to fire in a cell cycle-specific fashion, like other chromosomes, and unlike plasmids whose timing of replication initiation is generally random in the cell cycle (12, 18, 29). chrII iterons, unlike plasmid iterons, include GATC sequences, whose adenine is the substrate for methylation by the Dam methylase. RctB binds to iterons efficiently only when their GATC sequences are methylated on both the strands of duplex DNA (15). In addition to iterons, the control includes another kind of RctB-binding site, the 39-mers (30). One such site is present within a locus, called rctA, and serves as one of the key negative regulators of chrII replication.
chrII segregation has also been studied extensively (19, 21–24, 31). The chromosome has homologs of plasmid partitioning genes, called parABII, and several cis-acting sites, called parSII, in and around oriII. V. cholerae cells, deleted of parABII genes, grow poorly, as they lose chrII rapidly.
Here, we have used rctA to interfere with chrII replication and segregation and monitored the fate of chrI replication and segregation and cell division. The rctA locus has one of the parSII sites of chrII for binding ParBII protein and one of the 39-mer sites for binding RctB. We first show that the two types of sites can be separated and used to interfere with chrII replication and segregation independently, when the sites were provided via a multicopy plasmid. The function of both types of sites could be restrained by transcription across them. Since chrII is essential for V. cholerae growth, transcription of the plasmid-borne rctA was required to maintain the clones without affecting cell viability, and by blocking transcription we could effectively block chrII replication and segregation. Under the latter conditions, chrI replication and segregation were found to remain unaffected, and cells could be born and divide without chrII. Apparently, there has been little adaptation in the maintenance of chrI to the presence of chrII. It remains possible that in the evolution of the two-chromosome genome of V. cholerae, either the secondary chromosome adapted to the main chromosome to ensure its maintenance or it is maintained independently, as are bacterial plasmids.
RESULTS
An inducible system to block chrII replication and segregation.
Our strategy to study communication between the two chromosomes of V. cholerae was to block replication and segregation of one chromosome at a time and determine the consequences for the replication and segregation of the other. It was possible to design a genetic system to interfere specifically with chrII maintenance. There is a locus, called rctA, which is a strong negative regulator of chrII replication (20). The locus also contains a site for chrII segregation, parSII (32). As shown below, rctA in a multicopy plasmid interferes with both replication and segregation of chrII, and both the activities could be restrained by transcription.
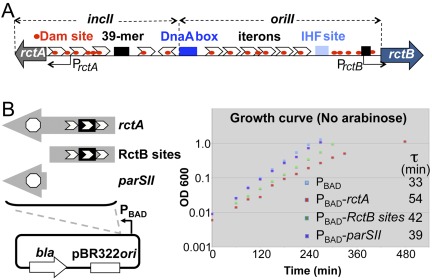
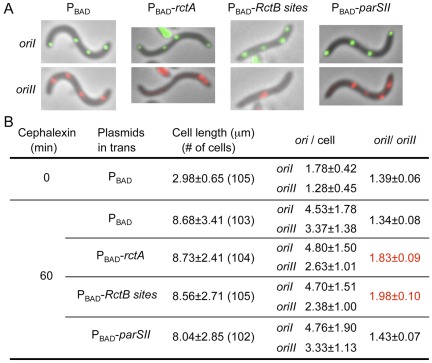
The RctB and ParBII sites in rctA do not overlap; they can easily be cloned separately (Fig. 1B). We separated the sites so that replication and segregation could be blocked individually. The sites were cloned under an arabinose-inducible promoter (PBAD), and the resultant plasmids were maintained in V. cholerae in the presence of a high concentration of arabinose (0.2%). Under these conditions, the growth rate of the cells was essentially identical (generation time of 28.5 ± 0.5 min), whether or not they carried the vector or any of the cloned rctA sites. When inhibitory activity of the sites was desired, a single colony grown in the presence of arabinose was inoculated into the growth medium without arabinose. The growth inhibition was found with cloned rctA sites, and it was greatest in cells carrying plasmids with intact rctA (Fig. 1B, τ). Cloned RctB sites caused less inhibition, but more than the parSII site. In any event, the continued growth of the cultures in the presence of rctA suggests that replication block may have been incomplete. Subsequent characterization of the cultures suggests that a subpopulation of the cells was affected only under the blocking conditions used.
FIG 1 .
Schematic map of the origin region of chromosome II (chrII) of V. cholerae and the effect of additional rctA on V. cholerae growth. (A) The region marked oriII is required in cis for initiation of replication, and the adjacent region marked incII is for controlling initiation. Initiation requires three trans-acting proteins, DnaA, IHF, and RctB. The binding sites for DnaA and IHF are shown by dark- and light-blue rectangles, respectively. The chrII-specific initiator, RctB, binds to two kinds of sites: the iterons (white arrowheads) and 39-mers (black rectangles). Iterons have built-in GATC sequences (red dots) that comprise the recognition site for the Dam methylase. Efficient binding of RctB to iterons requires the sites to be fully methylated. PrctB and PrctA are promoters responsible for transcribing rctB and rctA loci. The rctA locus is not translated. (B) A magnified view of the rctA locus, showing a parSII site for binding ParBII and three iteron-like sites within which also lies a 39-mer site. The sites were cloned under an inducible promoter, PBAD, to control transcription across them. The plots show the growth curves of wild-type cells (CVC209) with either the PBAD vector plasmid (CVC1111) or PBAD-rctA (CVC1142) or PBAD-RctB sites (CVC1143) or PBAD-parSII (CVC1144). The plot also includes generation times (τ) during the exponential growth phase of the cultures.
Additional rctA causes nucleoid condensation and lowers chrII replication.
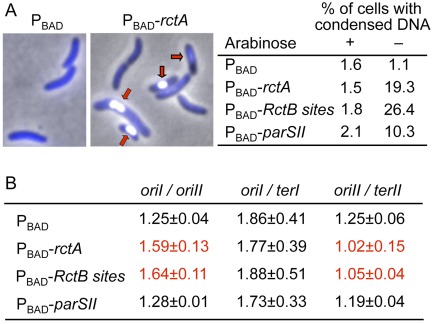
To determine the effect of additional rctA under inhibitory conditions, cells were grown to log phase (optical density at 600 nm [OD600] of ≤ 0.2) without arabinose, and their DNA was monitored by Hoechst staining and quantitative PCR (qPCR). Hoechst staining showed most cells to contain a single DNA mass (nucleoid): anucleate cells were <1% in the four cases examined (data not shown). However, there was a significant increase of cells with condensed DNA when any of the rctA sites were present (Fig. 2A). Cells with condensed DNA were seen in earlier studies when cells were born without chrII, and this was interpreted to be likely due to toxins that get activated upon the chrII loss (22). We also believe that the condensation was caused by the loss of chrII, as we show later (see Fig. 4). The percentage of condensed cells gives an approximate indication of the effectiveness of the replication and segregation blocks. The percentage of chrII-less cells seen by microscopy is also similar, as we show later (Fig. 3B).
FIG 2 .
Effect of additional rctA on V. cholerae nucleoid volume and replication initiation efficiency. The strains are the same as those shown in Fig. 1B. (A) DAPI (4′,6-diamidino-2-phenylindole)-stained image of V. cholerae cells carrying either a PBAD vector plasmid or the same vector with a cloned copy of rctA under the PBAD control (PBAD-rctA). The red arrows indicate cells with condensed nucleoids. The table shows the percentage of cells with condensed nucleoids. (B) Marker frequency analysis by qPCR. The frequencies of the four markers, oriI, oriII, terI, and terII, were determined from three cultures grown from independent colonies in each case, and the means of the three ratios, with one standard deviation, are shown. The ratios considered significantly different from the vector-only control (top line) are shown in red.
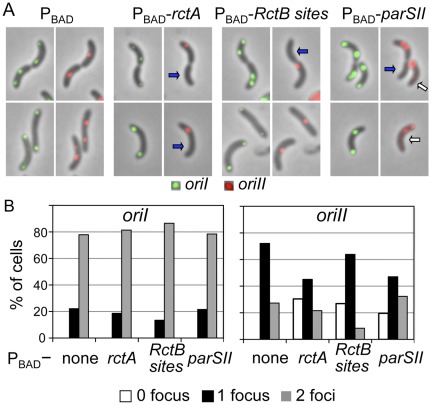
FIG 3 .
Effect of additional rctA on V. cholerae origin numbers in individual cells. The plasmids supplying additional rctA sites are the same as in Fig. 1B. (A) Origins were visualized by inserting a P1parS site 135 kb away (clockwise) from oriI and a pMTparS site about 40 kb away from oriII (clockwise) and supplying from plasmid pRN011 the corresponding fluorescent-tagged proteins GFP-P1ParB and mCherry-pMTParB that bind to those sites. The oriI foci are shown in green and oriII foci in red. The arrows indicate cells either without oriII (blue) or mislocalized oriII (white). (B) Distribution of cell numbers with different numbers of origin foci.
qPCR was used to determine the frequency of four markers: oriI, oriII, terI, and terII (Fig. 2B). The ratio of oriI to oriII increased and that of oriII to terII decreased in the presence of RctB sites but not in the presence of the parSII site, consistent with a specific block of oriII firing. A segregation defect alone is not expected to change the oriI and oriII values. The results of Fig. 2 provided the initial indication that blocking chrII replication is without effect on chrI replication.
Additional rctA does not block oriI firing.
Replication initiation of the two chromosomes was monitored by counting the number of origins in individual cells. In these experiments, oriI and oriII were labeled with two different fluorescent markers so that they could be visualized in the same cell simultaneously (33). Under our culturing conditions, cells showed one or two foci for both oriI and oriII (Fig. 3A). The distributions of cells with one and two oriI foci were similar whether or not cloned rctA sites were present (Fig. 3B). Most cells had two oriI foci, indicating that initiation of chrI occurred early in the cell cycle. In comparison, cells with two oriII foci were less frequent, consistent with an earlier finding that oriII fires later in the cell cycle than oriI (18). When any of the cloned rctA sites was present, up to about 30% of cells with oriI failed to show any oriII focus. The appearance of oriII-less cells in the presence of cloned RctB sites indicates that cell division can proceed without chrII replication. oriII-less cells also appeared in the presence of the cloned parSII site, indicating that a single centromeric site in a multicopy vector can cause chrII missegregation. It is also possible that the parSII effect is mediated through chrII replication since the presence of the site has been shown recently to alleviate some of the negative regulatory activity on replication by rctA (34). Our results appear to be more consistent with missegregation, because the fraction of cells with two foci over the total number of cells was similar whether the cells had the vector or the parSII site (Fig. 3B, none versus parSII values in the oriII panel). In the case of a replication block, the frequency of cells with two foci should have decreased, as was the case in cells with cloned RctB sites. Examples of cells with only oriII and no oriI foci were rare. In summary, the results support the view that interfering with chrII replication and segregation does not influence similar processes in chrI nor block cell division.
Loss of chrII disorganizes chrI.
Cytological studies in Caulobacter crescentus, E. coli, and Bacillus subtilis have shown the chromosome to be highly organized (35–38). As in C. crescentus and sporulating B. subtilis, oriI of V. cholerae is found near a cell pole where it also duplicates (19, 21, 24, 31, 39). One daughter oriI stays at the place of birth, and the other moves to the opposite pole. The segregation pattern of chrII is like that of the E. coli chromosome: oriII is found near the cell center, and after duplication, the two daughter origins move to the cell quarter positions. Since chrI spans the entire length of the bacterium, it overlaps chrII over nearly half of the cell length around the cell center. Although interfering with replication and segregation of oriII did not disturb the overall distribution or the number of oriI foci (Fig. 3B), the possibility remains that the location of oriI is disturbed in the absence of chrII. The location of oriI was mapped in cells with and without oriII (Fig. 4).
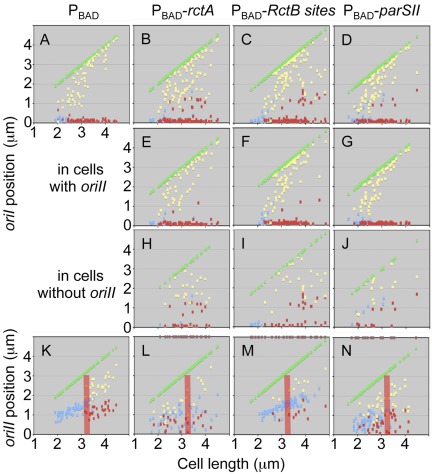
FIG 4 .
Effect of additional rctA on V. cholerae origin position. The plots show the positions of oriI and oriII foci in different cells. The cells were arranged according to increasing lengths along the abscissa. One pole is placed on the abscissa and the other is shown by green dots. Focus positions were measured from the pole placed on the abscissa. This pole was chosen because the nearest focus from this pole was closer than the nearest focus from the opposite pole. Small cells usually showed one focus (blue dot) and larger cells two foci (red and yellow dots). (A to J) Plots of oriI positions. Cells with oriI did not always show oriII foci. Cells with oriI irrespective of the presence of oriII foci are in panels A to D. These cells were separated into two groups: ones with oriII foci (E to G) and ones without oriII foci (H to J). (K to N) The plot of oriII positions. The pink bars mark the cell size at which two-focus cells appear in the wild type (K). The bars serve as a reference in other panels (L to N). The cell size at which the two foci appeared varied in the mutants. The brown dots on the top axis in panels L to N show the sizes of cells without oriII (these are the same cells as shown in panels H to J). The dots show that oriII-less cells can be of all possible sizes.
In cells with vector only, oriI was found primarily at or near the cell poles, as expected from earlier studies (Fig. 4A). When extra rctA sites were present, there was a significant increase in the fraction of oriI foci more centrally located, and this was particularly pronounced in cells devoid of oriII foci (compare Fig. 4E and H). These results are consistent with nucleoid condensation in the absence of chrII and the consequent mislocalization of oriI. How the loss of chrII causes condensation of chrI is not known.
In vector-only cells, a centrally located single focus of oriII was found in small cells and, when the cells exceeded a certain size, two oriII foci were found mostly near cell quarter positions (Fig. 4K). The cell size at which one focus becomes two was no longer distinct when cells carried the parSII clone (Fig. 4K versus L and N). The mislocalization of oriII apparently did not disturb oriI localization (Fig. 4E and G). Excess RctB sites seem to only delay oriII firing, as single oriII foci were found in relatively large cells (Fig. 4M).
We confirmed, by time-lapse photography, that the failure of chrII to replicate perturbed neither the replication of chrI nor cell division (Fig. 5). In summary, the results show that RctB sites and the parSII site perturb chrII replication initiation and segregation, respectively. These perturbations do not affect oriI firing or localization unless chrII is lost from the cell. The loss appears to cause condensation of chrI, leading to nonpolar localization of oriI.
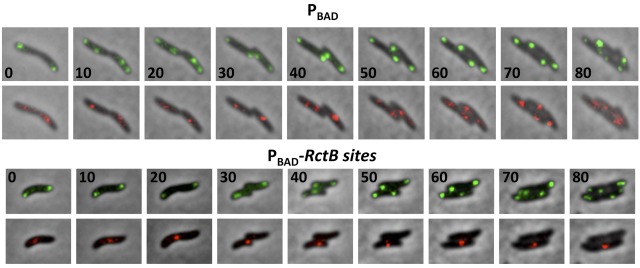
FIG 5 .
Effect of additional RctB sites on origin dynamics in V. cholerae cells by time-lapse microscopy. The figure shows cell growth and segregation of oriI and oriII foci in cells with either the PBAD plasmid (CVC1111) or the PBAD-RctB sites plasmid (CVC1143). The oriI foci are shown in green and oriII foci in red. The cells were grown on an agarose pad by continually feeding them from underneath with fresh medium (MM) using a homemade flow cell. The numbers indicate the time in minutes at which the cells were imaged. In the time interval tested, oriI foci usually duplicate two times and oriII foci once (oriII foci duplicate later in the cell cycle, thwarting capturing of two full replication cycles for chrII because of mCherry bleaching).
Nucleoid condensation follows upon loss of chrII.
To further demonstrate that the replication and segregation cycles of chrI can be independent of chrII, and to avoid the complexity of nucleoid condensation upon chrII loss, the effect of rctA sites were tested in cephalexin-treated cells. The drug blocks cell division and thus can prevent the loss of chrII when its replication is blocked. The cell division block was obvious, as cells elongated as expected (about 3-fold in an hour after drug treatment, covering about two generations of growth without the drug) (Fig. 6). In cephalexin-treated control cells (with PBAD), up to four foci of oriI and of oriII could be seen distributed over their entire lengths. In the presence of any or all of the rctA sites, the number and distribution of oriI foci were not significantly affected. The number of oriII foci was reduced, particularly when the RctB sites were present. These results confirm that ongoing replication and segregation of chrII are not required for the replication and segregation of chrI to proceed in the same cell, at least for about two generations.
FIG 6 .
Effect of additional rctA on origin numbers in cephalexin-treated V. cholerae cells. Cells were treated with cephalexin to prevent chrII loss upon replication/segregation block. Cells are the same as those used in Fig. 3 and 4. (A) The oriI foci are shown in green and oriII foci in red. Note that the oriII foci number reduced when additional RctB sites but not parSII sites were provided. (B) The table shows increase of cell length upon cephalexin treatment and decrease in oriII focus number in the presence of RctB sites. The errors in oriI-to-oriII ratios (σf) represent standard errors of the mean (SE) using the relationship σ2f= (1/b2) σ2a + (a2/b4) σ2b, where σa and σb are the SE of mean copy numbers of oriI (a) and oriII (b).
DISCUSSION
In eukaryotes, the mechanisms that maintain chromosomes are common to all the chromosomes. In V. cholerae, the genetic programs that maintain its two chromosomes are separate. Although there are genes, such as dnaA, ihf, and dam, that are used by both the chromosomes for replication initiation, the factors responsible for controlling replication initiation frequency are different for the two chromosomes (27). Similarly, the genetic programs for segregation of the two chromosomes are separate (22, 40). The possibility still remained that the two chromosomes coordinate their replication and segregation (15, 16). Here we show that the cell cycle progresses even when chrII replication or segregation is blocked. This makes it unlikely that there could be any checkpoint mechanism to ensure completion of replication and segregation of both the chromosomes before cell division. The system thus could be prone to chromosome loss. It is all the more likely since vibrios are among the most rapidly growing bacteria, having a generation time that can be as short as 9 min (41). Chromosome loss, however, might be justified if it served some altruistic purpose, such as feeding neighboring cells in a community of cells, as in a biofilm (42). This is unlikely to happen for long, because chrI loss would not generate new transcription and translation machineries, and chrII loss would trigger degradation of chrI DNA (22). On the other hand, depending on the cell density, DNA from lysing cells could be nutrients for cells in the same habitat or help in adaptation since V. cholerae can develop natural competence for genetic transformation (43, 44).
In the family of Vibrionaceae, whose members each have two chromosomes, the secondary chromosome may have evolved from a plasmid ancestor that incorporated a few essential genes and changed its replication program to initiate at a specific time in the cell cycle, as chromosomes generally do (45). Since the dynamics of chrI are not altered when the dynamics of chrII are perturbed (Fig. 7), the two chromosomes could be maintained by independent mechanisms without rapid communication between them, as is the case between the chromosome and several well-studied plasmids in E. coli and in eukaryotic cells, in which the blockage of initiation in one chromosome appears not to be sensed by other chromosomes in the same cell. Our results are consistent with an earlier finding that in ΔparAB2 cells, oriI localization is not perturbed (22) and a more recent finding that a viable V. cholerae can be made by fusing its two chromosomes, which also deletes the chrII-specific genes for replication and segregation (46). The latter study is a clear indication for nonessentiality of chrII replication and segregation for similar functions of chrI.
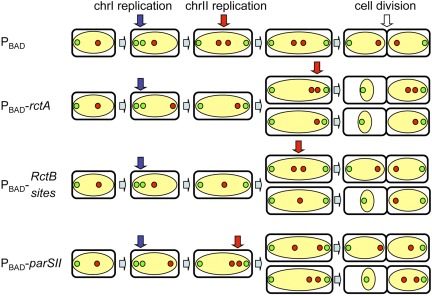
FIG 7 .
A cartoon summarizing the effect of additional rctA sites on chromosome dynamics of V. cholerae. The nucleoid is shown in yellow, oriI in green dots, and oriII in red dots. In wild-type cells (top), oriI replicates near a cell pole and oriII near the cell center but at a time later than oriI. Replication and segregation of oriI are not disturbed by any of the rctA sites unless chrII is lost from cells (see Fig. 4E to G versus H to J). The mislocalization of oriI apparently results from chromosome condensation that follows upon the loss of chrII due to unknown reasons. The presence of RctB sites delays oriII firing, since a single oriII focus was seen in longer cells (Fig. 4M). The presence of parSII sites does not delay oriII firing but mislocalizes oriII, as two oriII foci were seen even in small cells (Fig. 4N). The results in the presence of intact rctA are a composite of the effects seen with RctB and parSII sites separately (Fig. 4L).
Chromosomes I and II initiate replication at different times so that their replications terminate more or less at the same time (18). Even if the two chromosomes are independently maintained, there must have been a one-way adaptation of chrII to suit the dynamics of chrI and/or to align its replication initiation to the cell cycle. There is also some understanding on how the replication initiation of chrII occurs at a specific time of the cell cycle, whereas the timing of plasmid replication is random in the cell cycle (30).
The plasmid-like maintenance strategy of chrII is suggested not only by the similarity in the organization of their replication origins and the nature of partitioning genes but also by the presence of toxin-antitoxin (TA) modules in both. The TA systems can severely compromise growth of plasmid-free cells, should they arise due to replication or segregation error. V. cholerae cells die rapidly if they are born without chrII, because of the loss of essential genes present in chrII and possibly due to the activation of TA systems (22, 47, 48). The intrinsic maintenance mechanisms must be efficient in the first place to avoid activation of TA systems. Although the TA systems can increase the apparent stability of the genome, its frequent activation is likely to slow down the clonal growth of the culture and may put the species at a disadvantage in a competitive environment.
The only phenotype we see upon loss of chrII is the condensation of the chrI nucleoid. The phenotype is the same as was reported earlier when chrII maintenance was perturbed by deleting its segregation genes (22, 48). The condensation could be brought about by activation of TA systems or by the vacuum created by the loss of chrII (49). It also remains possible that chrII encodes some gene product that directly controls chromosome architecture. The mechanism of condensation remains to be elucidated.
Although rctA was used as a tool here to block chrII dynamics, the results also shed light on its function. They show that a single centromeric site in trans can cause chrII missegregation, a phenomenon attributed to partition-mediated incompatibility (50). This is remarkable, considering that chrII has at least nine such sites (32). A single locus capable of controlling both replication and segregation of a chromosome is not that common, although examples of partitioning genes controlling replication by a different mechanism have been found (40, 51). The phenotypes of intact rctA could be additive of those conferred by RctB sites and parSII sites separately: the generation time increased most when rctA was intact (Fig. 1B). However, the additive effect was not obvious in other experiments (Fig. 2 to 6). It has been shown recently that the parSII site in rctA can neutralize some of the negative regulatory activities that regulate chrII replication (34). This can explain how the phenotype of RctB sites can be stronger in the absence of the parSII site (Fig. 3 and 6). However, it is clear from the present study that in the intact rctA locus, the RctB sites remain quite active in trans in reducing chrII replication initiation. More studies will be required to understand the interplay between the two types of sites of the locus more fully. It remains to be determined whether blocking chrI blocks chrII or whether chrII is maintained independently following the plasmid paradigm.
MATERIALS AND METHODS
Strains and growth conditions.
Strains are listed in Table 1, and the primers used are listed in Table S1 in the supplemental material. E. coli DH5Δlac (BR2846) and C600 (CVC1221) were used for standard plasmid manipulations and for plasmid propagation. V. cholerae strains are derivatives of N16961 (CVC209) or N16961 hapR+ (CVC1118). Cells were grown in minimal medium (MM; M63 buffer, fructose [0.2%], and Casamino Acids [0.1%]). Antibiotics were used at the following concentrations: ampicillin, 100 µg/ml; chloramphenicol, 25 µg/ml for E. coli, 6.5 µg/ml for V. cholerae; kanamycin, 25 µg/ml; and spectinomycin, 40 µg/ml.
TABLE 1 .
Strains and plasmids used in this study
| Bacteria or plasmid | Relevant description | Source or reference |
|---|---|---|
| E. coli | ||
| BR2846 | DH5∆lac = K-12 recA Δ(argF-lac)U169 | 53 |
| CVC1221 | C600 = F− thi∆1 thr-1 supE44 leuB6 lacY1 tonA21 mcrA− mcrB− λ− | NAIST lab stock |
| V. cholerae | ||
| CVC209 | N16961 El tor; Strr | M. Waldor |
| CVC1111 | CVC209/pRN012 (PBAD) | This study |
| CVC1118 | N16961 hapR+ | 40 |
| CVC1140 | CVC1118 + P1parS-Kn (at +135 kb on chrI) | This study |
| CVC1142 | CVC209/pTVC58 (PBAD-rctA) | This study |
| CVC1143 | CVC209/pRN007 (PBAD-RctB sites) | This study |
| CVC1144 | CVC209; Strr/pRN008 (PBAD-parSII) | This study |
| CVC1146 | CVC1140 + pMTparS-Sp (at +40 kb on chrII) | This study |
| CVC1147 | CVC1146/pRN012 (PBAD) | This study |
| CVC1148 | CVC1146/pTVC58 (PBAD-rctA) | This study |
| CVC1149 | CVC1146/pRN007 (PBAD-RctB sites) | This study |
| CVC1150 | CVC1146/pRN008 (PBAD-parSII) | This study |
| Plasmids | ||
| pALA1840 | Source of pMTparS-Sp cassette; Apr Spr | 54 |
| pPS2 | Coordinates 39216 to 41197 of chrII in pDS132; Cmr | 29 |
| pPS4 | Coordinates 133582 to 135584 of chrI cloned at PstI site of pDS132; Cmr | 40 |
| pPS45 | Source of P1parS-Kn cassette; Apr Knr | 40 |
| pPS47 | Source of P1parS-Kn cassette; Cmr Knr | 40 |
| pPS69 | pALA2705 modified to incorporate XhoI and SacII sites + mCherry-mreB PCR XhoI-SacII fragment; Apr | 29 |
| pRN007 | pTVC58 except that rctA is replaced with RctB sites (PBAD-RctB sites) | This study |
| pRN008 | pTVC58 except that rctA is replaced with a parSII site (PBAD-parSII) | This study |
| pRN009 | pPS2 + NsiI fragment from pPS45 containing pMTparS-Sp cassette cloned at the NsiI site; Cmr Spr | This study |
| pRN010 | pPS69 mreB gene between AatII-SacII replaced with pMTparB AatII-SacII PCR fragment | This study |
| pRN011 | pST52 + lacIq-mCherry-pMTparB-gfp-P1parB fragment (NsiI-HindIII) from pRN010 | This study |
| pTVC58 | pMLB1109 + araC-PBAD-rctA from pTVC38 as EcoRI-SmaI PCR fragment (PBAD-rctA); Apr | 20 |
Plasmids with rctA, RctB sites, and parSII under PBAD control.
pTVC58 was used as the source of PBAD-rctA. The rctA open reading frame (ORF) was removed by digesting pTVC58 with NheI and SmaI, and after blunting with Klenow, the plasmid backbone was self-ligated. This generated pRN012, which was used here as the PBAD vector control. To supply the RctB sites within the rctA ORF, the sites were amplified by PCR using primer pairs RP004 and RP097 and N16961 DNA as the template. RP004 has SmaI and NheI sites, and RP097 has only SmaI sites at their 5′ ends. The PCR product was digested with NheI and SmaI and ligated to similarly digested pTVC58. This generated pRN007 (PBAD-RctB sites). To clone the parSII within rctA, RP007 and RP008 oligonucleotides were annealed and ligated to NheI- and SmaI-digested pTVC58. This generated pRN008 (PBAD-parSII). These same four plasmids (PBAD, PBAD-rctA, PBAD-RctB sites, and PBAD-parSII) have been used in all the experiments reported in this study.
Chromosomal integration of P1parS and pMTparS.
To localize oriI, P1parS as a P1parS-Kn cassette was inserted into the hapR+ strain CVC1118 at approximately +135 kb in the intergenic region between VC0142 and VC0143. The resultant strain (CVC1140) was constructed by allelic exchange after transferring by conjugation pPS47 carrying the cassette into CVC1118. To localize oriII, the pMTparS-Sp cassette was integrated in the intergenic region between VCA032 and VCA033 of CVC1140, both encoding hypothetical proteins. To clone the pMTparS-Sp cassette, pPS2 carrying the intergenic region (chrII DNA coordinates 39216 to 41197) was digested with NsiI and ligated to the pMTparS-Sp cassette obtained from pALA1840 by PCR using RP012 and RP013 primers containing the NsiI site. From the resultant plasmid (pRN009), pMTparS-Sp with flanking chrII DNA was amplified by PCR using primers RN033 and RN038. The PCR product was introduced into chrII by natural transformation (40). The strain thus made (CVC1146) has two different parS sites, one in chrI (P1parS) and another in chrII (pMTparS).
Construction of plasmid carrying genes for two different fluorescent ParB protein fusions.
The pMTparB gene was amplified from the pFHC2973 plasmid using primers RN005 and RN006 (33). The PCR product was digested with AatII and SacII and subsequently cloned under an isopropyl-β-thiogalactopyranoside (IPTG)-inducible promoter present in pPS69. The pPS69 has an operon with two fluorescent fusion protein genes, mCherry-mreB and gfp-P1parB, under pTrc promoter (29). The pMTparB fragment was cloned in the orientation AatII to SacII between the mCherry and gfp genes, replacing mreB. In the resultant plasmid (pRN010), the order of elements downstream of pTrc is mCherry-pMTparB-gfp-P1parB. To clone the pTrc-mCherry-pMTparB-gfp-P1parB cassette, pRN010 was digested with NsiI and HindIII, and the cassette was ligated to BglII-digested pPT52, a pBR322-compatible vector (52). The resulting plasmid was called pRN011.
Fluorescence microscopy.
Cells were grown in MM at 37°C to log phase (OD600 between 0.1 and 0.2) and concentrated 100-fold by centrifugation and resuspension in the same medium. They were stained for DNA with Hoechst-33342 at a final concentration of 0.5 µg/ml at room temperature and observed under a fluorescence microscope within 10 min, as described (40).
Time-lapse microscopy.
Time-lapse microscopy was done using a grooved microscope slide (homemade) that was overlaid with an agarose pad. The slide with the pad was placed on the microscope stage and heated at 37°C. The pad was equilibrated for about an hour with MM containing appropriate antibiotics and inducer by continually running the medium through the grooves by gravity flow. About 5 µl of log-phase cells was placed on the pad and overlaid with an untreated coverslip. Imaging was done every 10 min, where the intensity of the illuminating light was increased in steps, as required.
Flow cytometry.
Log-phase cells were processed for flow cytometric analysis as previously described (31).
Marker frequency analysis.
Frequencies of four markers, oriI, oriII, terI, and terII, from exponentially growing cells without arabinose were determined as previously described (15).
SUPPLEMENTAL MATERIAL
Primers used in this study.
ACKNOWLEDGMENTS
We thank Stuart Austin for generously providing many of the reagents of fluorescence microscopy, Marco Fioani for advice on eukaryotic DNA replication control, and Michael Yarmolinsky for critical reading of the manuscript.
This work was supported by the Intramural Research Program, Center for Cancer Research of the National Cancer Institute.
Footnotes
Citation Kadoya R, Chattoraj DK. 2012. Insensitivity of chromosome I and the cell cycle to blockage of replication and segregation of Vibrio cholerae chromosome II. mBio 3(3):e00067-12. doi:10.1128/mBio.00067-12.
REFERENCES
- 1. Dershowitz A, Newlon CS. 1993. The effect on chromosome stability of deleting replication origins. Mol. Cell. Biol. 13:391–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blow JJ, Ge XQ. 2009. A model for DNA replication showing how dormant origins safeguard against replication fork failure. EMBO Rep. 10:406–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Karnani N, Dutta A. 2011. The effect of the intra-S-phase checkpoint on origins of replication in human cells. Genes Dev. 25:621–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Borde V, Goldman AS, Lichten M. 2000. Direct coupling between meiotic DNA replication and recombination initiation. Science 290:806–809 [DOI] [PubMed] [Google Scholar]
- 5. Branzei D, Foiani M. 2010. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11:208–219 [DOI] [PubMed] [Google Scholar]
- 6. Lopes M, et al. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412:557–561 [DOI] [PubMed] [Google Scholar]
- 7. Szyjka SJ, et al. 2008. Rad53 regulates replication fork restart after DNA damage in Saccharomyces cerevisiae. Genes Dev. 22:1906–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Breier AM, Weier HU, Cozzarelli NR. 2005. Independence of replisomes in Escherichia coli chromosomal replication. Proc. Natl. Acad. Sci. U. S. A. 102:3942–3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cox MM, et al. 2000. The importance of repairing stalled replication forks. Nature 404:37–41 [DOI] [PubMed] [Google Scholar]
- 10. Nishimura Y, Caro L, Berg CM, Hirota Y. 1971. Chromosome replication in Escherichia coli. IV. Control of chromosome replication and cell division by an integrated episome. J. Mol. Biol. 55:441–456 [DOI] [PubMed] [Google Scholar]
- 11. Yarmolinsky MB. 1995. Programmed cell death in bacterial populations (comment). Science 267:836–837 [DOI] [PubMed] [Google Scholar]
- 12. Leonard AC, Helmstetter CE. 1988. Replication patterns of multiple plasmids coexisting in Escherichia coli. J. Bacteriol. 170:1380–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Egan ES, Fogel MA, Waldor MK. 2005. Divided genomes: negotiating the cell cycle in prokaryotes with multiple chromosomes. Mol. Microbiol. 56:1129–1138 [DOI] [PubMed] [Google Scholar]
- 14. Jha JK, Baek JH, Venkova-Canova T, Chattoraj DK. 28 January 2012. Chromosome dynamics in multichromosome bacteria. Biochim. Biophys. Acta. [Epub ahead of print.]. [Epub ahead of print.]10-1016/j.bbagrm.2012.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Demarre G, Chattoraj DK. 2010. DNA adenine methylation is required to replicate both Vibrio cholerae chromosomes once per cell cycle. PLoS Genet. 6:e1000939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Egan ES, Waldor MK. 2003. Distinct replication requirements for the two Vibrio cholerae chromosomes. Cell 114:521–530 [DOI] [PubMed] [Google Scholar]
- 17. Pal D, Venkova-Canova T, Srivastava P, Chattoraj DK. 2005. Multipartite regulation of rctB, the replication initiator gene of Vibrio cholerae chromosome II. J. Bacteriol. 187:7167–7175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rasmussen T, Jensen RB, Skovgaard O. 2007. The two chromosomes of Vibrio cholerae are initiated at different time points in the cell cycle. EMBO J. 26:3124–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Srivastava P, Chattoraj DK. 2007. Selective chromosome amplification in Vibrio cholerae. Mol. Microbiol. 66:1016–1028 [DOI] [PubMed] [Google Scholar]
- 20. Venkova-Canova T, Srivastava P, Chattoraj DK. 2006. Transcriptional inactivation of a regulatory site for replication of Vibrio cholerae chromosome II. Proc. Natl. Acad. Sci. U. S. A. 103:12051–12056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fogel MA, Waldor MK. 2006. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 20:3269–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yamaichi Y, Fogel MA, Waldor MK. 2007. par genes and the pathology of chromosome loss in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 104:630–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fiebig A, Keren K, Theriot JA. 2006. Fine-scale time-lapse analysis of the biphasic, dynamic behaviour of the two Vibrio cholerae chromosomes. Mol. Microbiol. 60:1164–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Saint-Dic D, Frushour BP, Kehrl JH, Kahng LS. 2006. A parA homolog selectively influences positioning of the large chromosome origin in Vibrio cholerae. J. Bacteriol. 188:5626–5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harrison PW, Lower RP, Kim NK, Young JP. 2010. Introducing the bacterial “chromid”: not a chromosome, not a plasmid. Trends Microbiol. 18:141–148 [DOI] [PubMed] [Google Scholar]
- 26. Koch B, Ma X, Løbner-Olesen A. 2010. Replication of Vibrio cholerae chromosome I in Escherichia coli: dependence on dam methylation. J. Bacteriol. 192:3903–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Duigou S, et al. 2006. Independent control of replication initiation of the two Vibrio cholerae chromosomes by DnaA and RctB. J. Bacteriol. 188:6419–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paulsson J, Chattoraj DK. 2006. Origin inactivation in bacterial DNA replication control. Mol. Microbiol. 61:9–15 [DOI] [PubMed] [Google Scholar]
- 29. Srivastava P, Demarre G, Karpova TS, McNally J, Chattoraj DK. 2007. Changes in nucleoid morphology and origin localization upon inhibition or alteration of the actin homolog, MreB, of Vibrio cholerae. J. Bacteriol. 189:7450–7463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Venkova-Canova T, Chattoraj DK. 2011. Transition from a plasmid to a chromosomal mode of replication entails additional regulators. Proc. Natl. Acad. Sci. U. S. A. 108:6199–6204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Srivastava P, Fekete RA, Chattoraj DK. 2006. Segregation of the replication terminus of the two Vibrio cholerae chromosomes. J. Bacteriol. 188:1060–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yamaichi Y, Fogel MA, McLeod SM, Hui MP, Waldor MK. 2007. Distinct centromere-like parS sites on the two chromosomes of Vibrio spp. J. Bacteriol. 189:5314–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nielsen HJ, Ottesen JR, Youngren B, Austin SJ, Hansen FG. 2006. The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol. Microbiol. 62:331–338 [DOI] [PubMed] [Google Scholar]
- 34. Yamaichi Y, Gerding MA, Davis BM, Waldor MK. 2011. Regulatory cross-talk links Vibrio cholerae chromosome II replication and segregation. PLoS Genet. 7:e1002189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ben-Yehuda S, Rudner DZ, Losick R. 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299:532–536 [DOI] [PubMed] [Google Scholar]
- 36. Nielsen HJ, Li Y, Youngren B, Hansen FG, Austin S. 2006. Progressive segregation of the Escherichia coli chromosome. Mol. Microbiol. 61:383–393 [DOI] [PubMed] [Google Scholar]
- 37. Viollier PH, et al. 2004. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc. Natl. Acad. Sci. U. S. A. 101:9257–9262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang X, Liu X, Possoz C, Sherratt DJ. 2006. The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev. 20:1727–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shebelut CW, Guberman JM, van Teeffelen S, Yakhnina AA, Gitai Z. 2010. Caulobacter chromosome segregation is an ordered multistep process. Proc. Natl. Acad. Sci. U. S. A. 107:14194–14198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kadoya R, Baek JH, Sarker A, Chattoraj DK. 2011. Participation of chromosome segregation protein ParAI of Vibrio cholerae in chromosome replication. J. Bacteriol. 193:1504–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Aiyar SE, Gaal T, Gourse RL. 2002. rRNA promoter activity in the fast-growing bacterium Vibrio natriegens. J. Bacteriol. 184:1349–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Heidelberg JF, et al. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Blokesch M, Schoolnik GK. 2008. The extracellular nuclease Dns and its role in natural transformation of Vibrio cholerae. J. Bacteriol. 190:7232–7240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Blokesch M, Schoolnik GK. 2007. Serogroup conversion of Vibrio cholerae in aquatic reservoirs. PLoS Pathog. 3:e81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Waldor MK, RayChaudhuri D. 2000. Treasure trove for cholera research. Nature 406:469–470 [DOI] [PubMed] [Google Scholar]
- 46. Val ME, Skovgaard O, Ducos-Galand M, Bland MJ, Mazel D. 2012. Genome engineering in Vibrio cholerae: a feasible approach to address biological issues. PLoS Genet. 8:e1002472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Christensen-Dalsgaard M, Gerdes K. 2006. Two higBA loci in the Vibrio cholerae superintegron encode mRNA cleaving enzymes and can stabilize plasmids. Mol. Microbiol. 62:397–411 [DOI] [PubMed] [Google Scholar]
- 48. Yuan J, Yamaichi Y, Waldor MK. 2011. The three Vibrio cholerae chromosome II-encoded ParE toxins degrade chromosome I following loss of chromosome II. J. Bacteriol 193:611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jun S, Mulder B. 2006. Entropy-driven spatial organization of highly confined polymers: lessons for the bacterial chromosome. Proc. Natl. Acad. Sci. U. S. A. 103:12388–12393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bouet JY, Rech J, Egloff S, Biek DP, Lane D. 2005. Probing plasmid partition with centromere-based incompatibility. Mol. Microbiol. 55:511–525 [DOI] [PubMed] [Google Scholar]
- 51. Murray H, Errington J. 2008. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell 135:74–84 [DOI] [PubMed] [Google Scholar]
- 52. Som T, Tomizawa J. 1982. Origin of replication of Escherichia coli plasmid RSF 1030. Mol. Gen. Genet. 187:375–383 [DOI] [PubMed] [Google Scholar]
- 53. Sozhamannan S, Chattoraj DK. 1993. Heat shock proteins DnaJ, DnaK, and GrpE stimulate P1 plasmid replication by promoting initiator binding to the origin. J. Bacteriol. 175:3546–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Youngren B, Radnedge L, Hu P, Garcia E, Austin S. 2000. A plasmid partition system of the P1-P7par family from the pMT1 virulence plasmid of Yersinia pestis. J. Bacteriol. 182:3924–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in this study.