Abstract
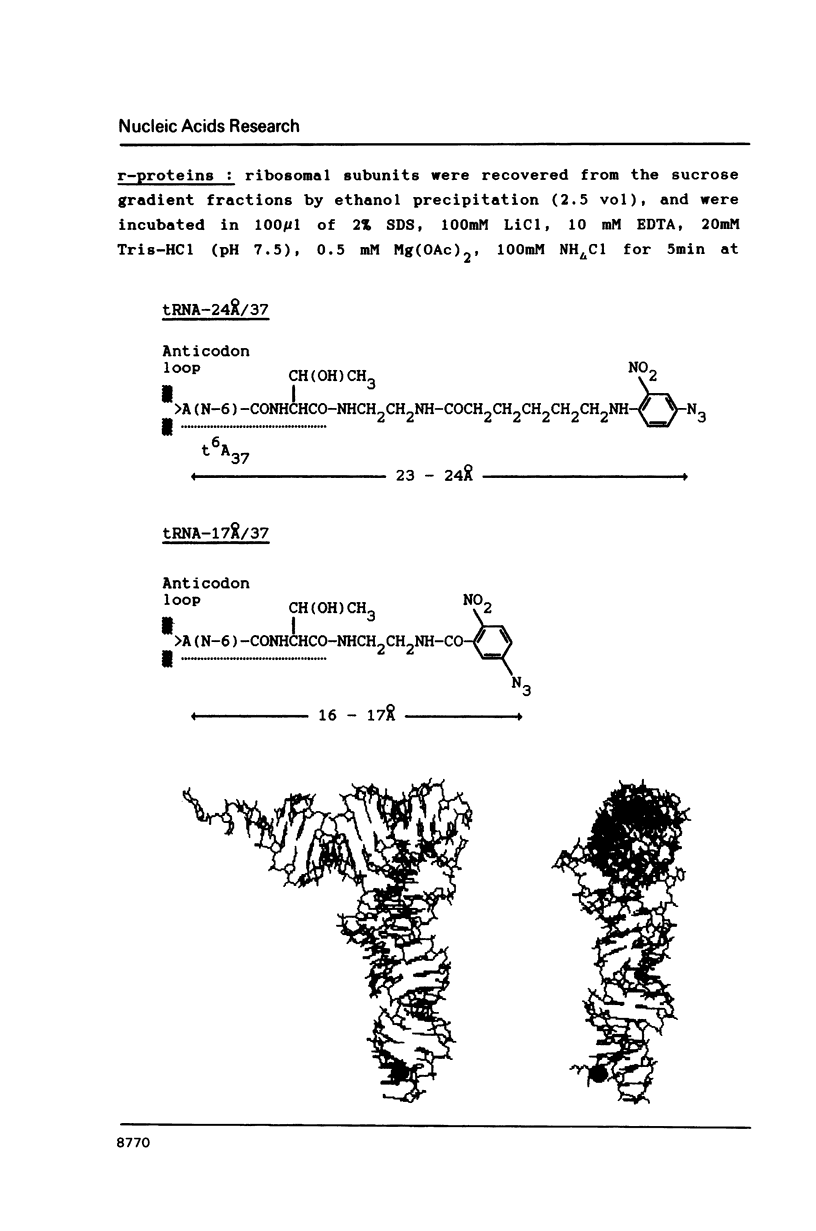
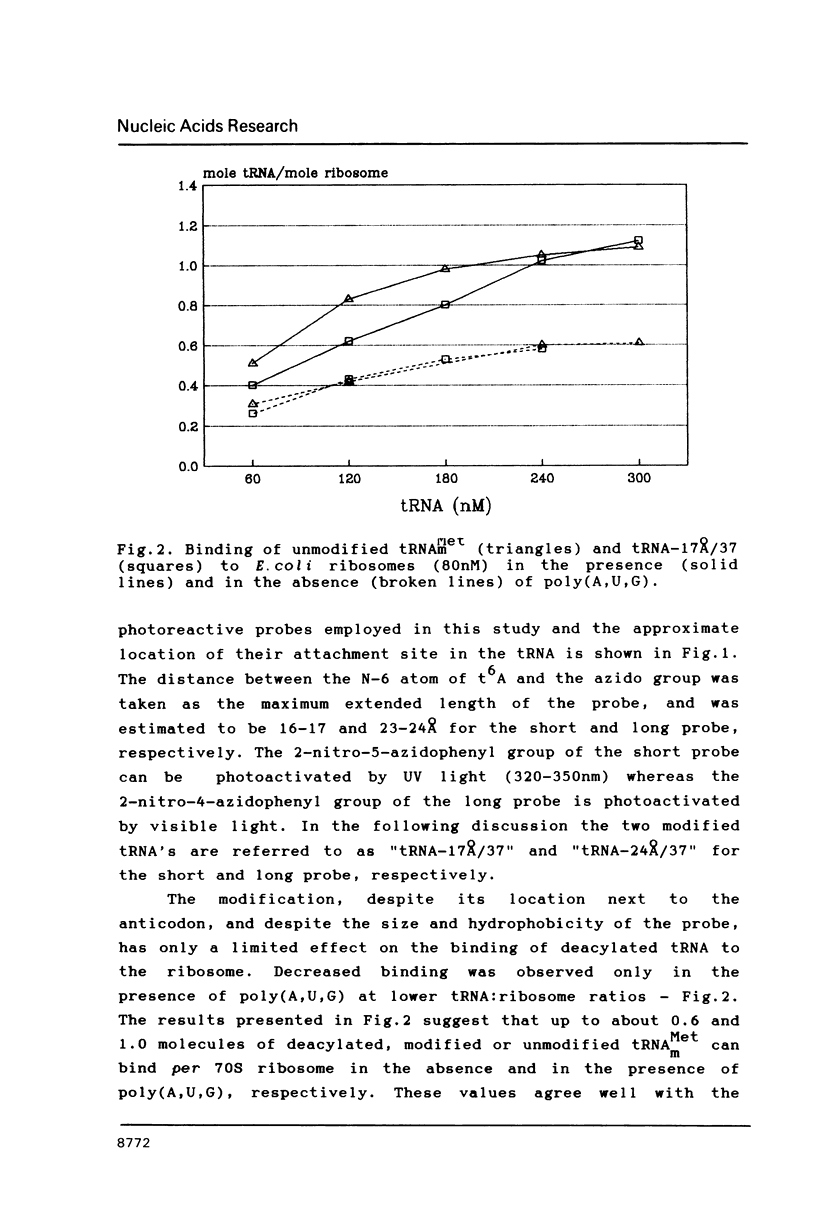
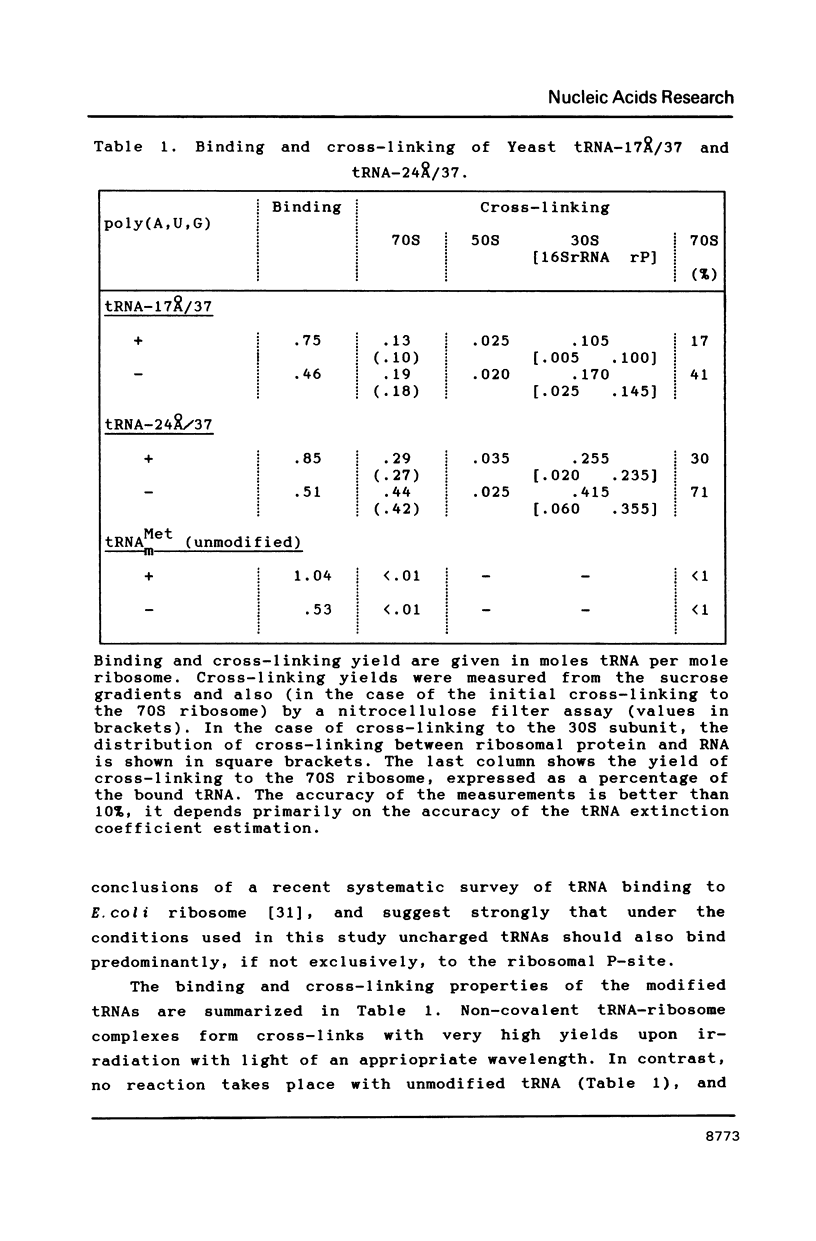
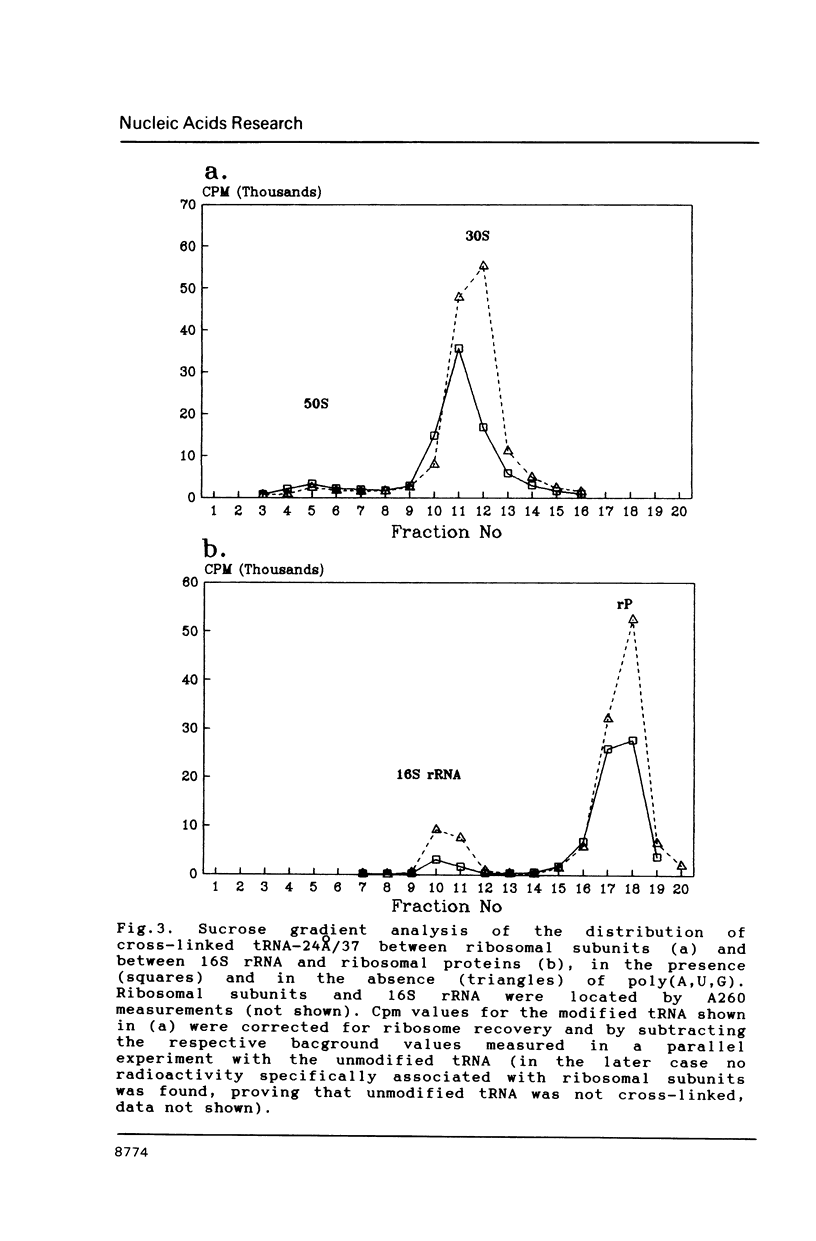
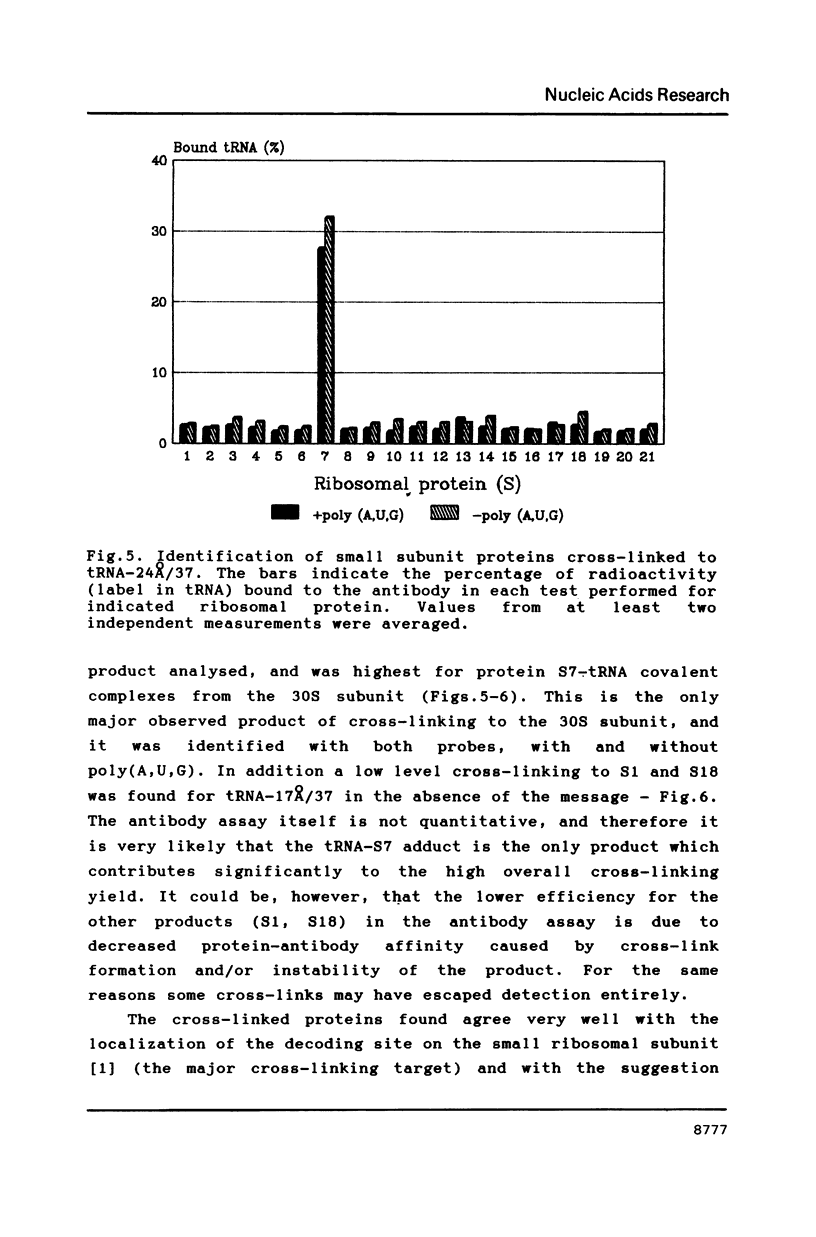
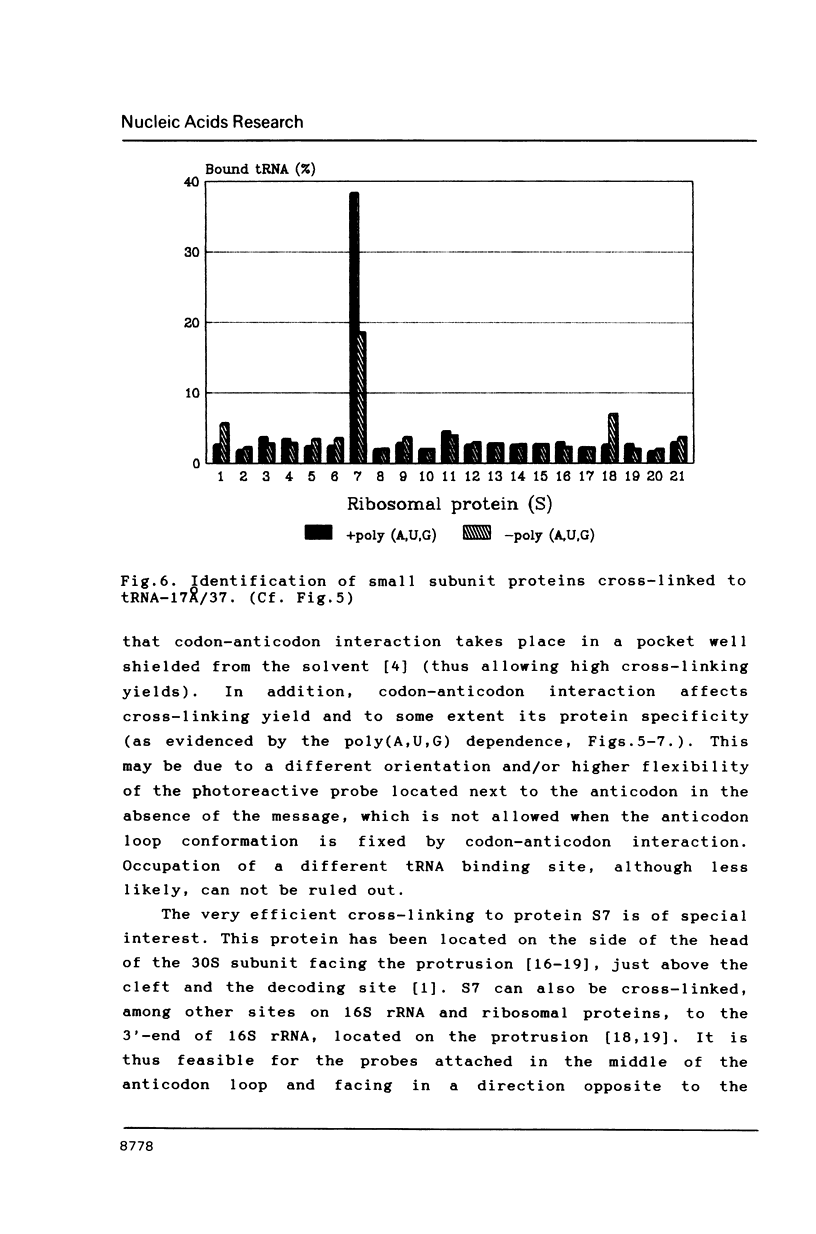
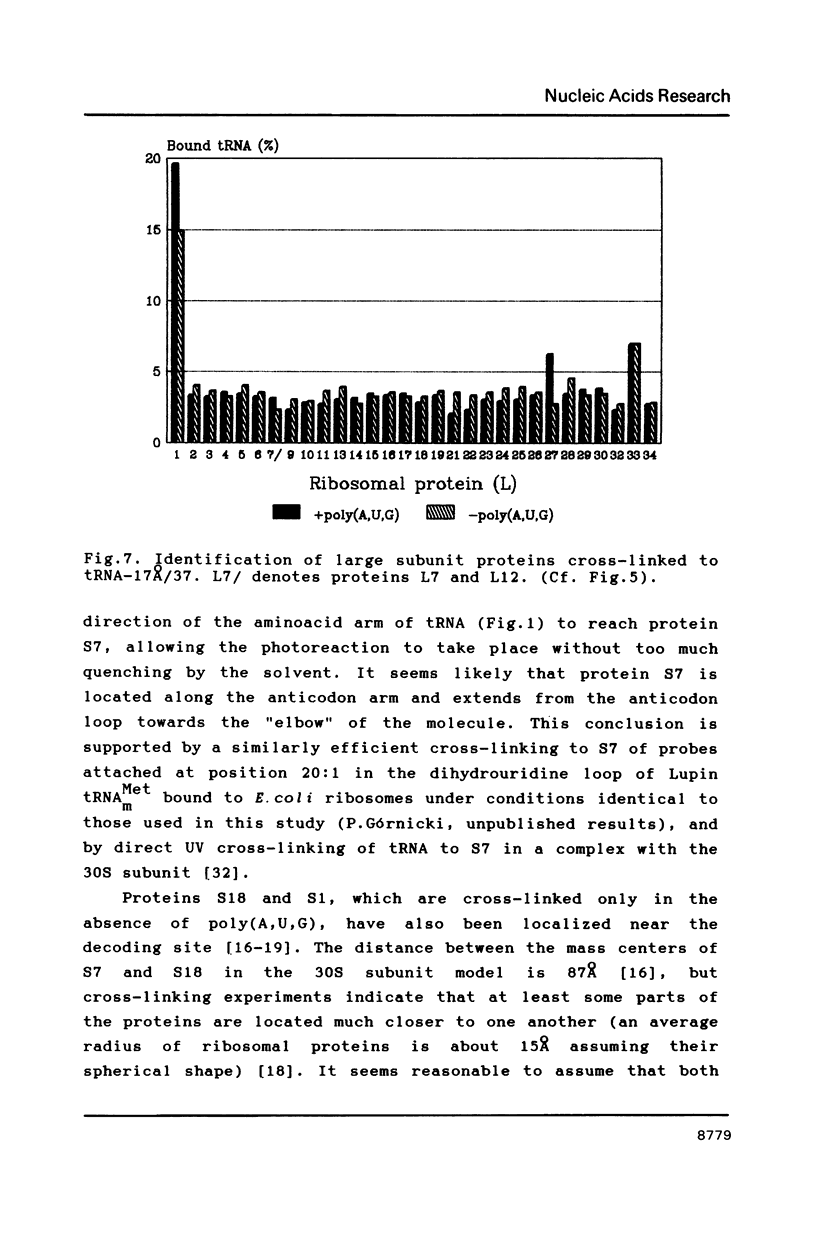
Two photoreactive azidonitrophenyl probes have been attached to Yeast methionine elongator tRNA by chemical modification of the N6-(threoninocarbonyl)adenosine located next to the 3'-end of the anticodon. The maximum distance between the purine ring and the azido group estimated for the two probes is 16-17 and 23-24A, respectively. Binding and cross-linking of the uncharged, modified tRNAs to E. coli ribosomes have been studied with and without poly(A,U,G) as a message, under conditions directing uncharged tRNAs preferentially to the P-site. The modified tRNAs retain their binding activity and upon irradiation bind covalently to the ribosome with very high yields. Protein S7 is the major cross-linking target for both modified tRNAs, in the presence or absence of poly(A,U,G). Protein L1 and to a lesser extent proteins L33 and L27 have been found to be cross-linked with the short probe. Cross-linking to 168 rRNA reaches significant levels only in the absence of the message.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abdurashidova G. G., Tsvetkova E. A., Budowsky E. I. Nucleotide residues of tRNA, directly interacting with proteins within the complex of the 30 S subunit of E. coli ribosome with poly(U) and NAcPhe-tRNA(Phe). FEBS Lett. 1989 Jan 30;243(2):299–302. doi: 10.1016/0014-5793(89)80149-8. [DOI] [PubMed] [Google Scholar]
- Beauclerk A. A., Cundliffe E. Sites of action of two ribosomal RNA methylases responsible for resistance to aminoglycosides. J Mol Biol. 1987 Feb 20;193(4):661–671. doi: 10.1016/0022-2836(87)90349-4. [DOI] [PubMed] [Google Scholar]
- Brimacombe R., Atmadja J., Stiege W., Schüler D. A detailed model of the three-dimensional structure of Escherichia coli 16 S ribosomal RNA in situ in the 30 S subunit. J Mol Biol. 1988 Jan 5;199(1):115–136. doi: 10.1016/0022-2836(88)90383-x. [DOI] [PubMed] [Google Scholar]
- Capel M. S., Engelman D. M., Freeborn B. R., Kjeldgaard M., Langer J. A., Ramakrishnan V., Schindler D. G., Schneider D. K., Schoenborn B. P., Sillers I. Y. A complete mapping of the proteins in the small ribosomal subunit of Escherichia coli. Science. 1987 Dec 4;238(4832):1403–1406. doi: 10.1126/science.3317832. [DOI] [PubMed] [Google Scholar]
- Ciesiolka J., Gornicki P., Ofengand J. Identification of the site of cross-linking in 16S rRNA of an aromatic azide photoaffinity probe attached to the 5'-anticodon base of A site bound tRNA. Biochemistry. 1985 Aug 27;24(18):4931–4938. doi: 10.1021/bi00339a031. [DOI] [PubMed] [Google Scholar]
- Denman R., Nègre D., Cunningham P. R., Nurse K., Colgan J., Weitzmann C., Ofengand J. Effect of point mutations in the decoding site (C1400) region of 16S ribosomal RNA on the ability of ribosomes to carry out individual steps of protein synthesis. Biochemistry. 1989 Feb 7;28(3):1012–1019. doi: 10.1021/bi00429a014. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Moine H., Mougel M., Dondon J., Grunberg-Manago M., Ebel J. P., Ehresmann B. Cross-linking of initiation factor IF3 to Escherichia coli 30S ribosomal subunit by trans-diamminedichloroplatinum(II): characterization of two cross-linking sites in 16S rRNA; a possible way of functioning for IF3. Nucleic Acids Res. 1986 Jun 25;14(12):4803–4821. doi: 10.1093/nar/14.12.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller W., Hodgson A. Conformation of the anticodon loop intRNA. Nature. 1967 Aug 19;215(5103):817–821. doi: 10.1038/215817a0. [DOI] [PubMed] [Google Scholar]
- Gnirke A., Geigenmüller U., Rheinberger H. J., Nierhaus L. H. The allosteric three-site model for the ribosomal elongation cycle. Analysis with a heteropolymeric mRNA. J Biol Chem. 1989 May 5;264(13):7291–7301. [PubMed] [Google Scholar]
- Gornicki P., Baudin F., Romby P., Wiewiorowski M., Kryzosiak W., Ebel J. P., Ehresmann C., Ehresmann B. Use of lead(II) to probe the structure of large RNA's. Conformation of the 3' terminal domain of E. coli 16S rRNA and its involvement in building the tRNA binding sites. J Biomol Struct Dyn. 1989 Apr;6(5):971–984. doi: 10.1080/07391102.1989.10506525. [DOI] [PubMed] [Google Scholar]
- Gornicki P., Ciesiolka J., Ofengand J. Cross-linking of the anticodon of P and A site bound tRNAs to the ribosome via aromatic azides of variable length: involvement of 16S rRNA at the A site. Biochemistry. 1985 Aug 27;24(18):4924–4930. doi: 10.1021/bi00339a030. [DOI] [PubMed] [Google Scholar]
- Gornicki P., Nurse K., Hellmann W., Boublik M., Ofengand J. High resolution localization of the tRNA anticodon interaction site on the Escherichia coli 30 S ribosomal subunit. J Biol Chem. 1984 Aug 25;259(16):10493–10498. [PubMed] [Google Scholar]
- Gravel M., Melançon P., Brakier-Gingras L. Cross-linking of streptomycin to the 16S ribosomal RNA of Escherichia coli. Biochemistry. 1987 Sep 22;26(19):6227–6232. doi: 10.1021/bi00393a041. [DOI] [PubMed] [Google Scholar]
- Gulle H., Hoppe E., Osswald M., Greuer B., Brimacombe R., Stöffler G. RNA-protein cross-linking in Escherichia coli 50S ribosomal subunits; determination of sites on 23S RNA that are cross-linked to proteins L2, L4, L24 and L27 by treatment with 2-iminothiolane. Nucleic Acids Res. 1988 Feb 11;16(3):815–832. doi: 10.1093/nar/16.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingerty B., Brown R. S., Jack A. Further refinement of the structure of yeast tRNAPhe. J Mol Biol. 1978 Sep 25;124(3):523–534. doi: 10.1016/0022-2836(78)90185-7. [DOI] [PubMed] [Google Scholar]
- Hui A., de Boer H. A. Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4762–4766. doi: 10.1073/pnas.84.14.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob W. F., Santer M., Dahlberg A. E. A single base change in the Shine-Dalgarno region of 16S rRNA of Escherichia coli affects translation of many proteins. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4757–4761. doi: 10.1073/pnas.84.14.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriatsoulis A., Maly P., Greuer B., Brimacombe R., Stöffler G., Frank R., Blöcker H. RNA-protein cross-linking in Escherichia coli ribosomal subunits: localization of sites on 16S RNA which are cross-linked to proteins S17 and S21 by treatment with 2-iminothiolane. Nucleic Acids Res. 1986 Feb 11;14(3):1171–1186. doi: 10.1093/nar/14.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987 Jun 4;327(6121):389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986 Dec 26;47(6):985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- Noll M., Hapke B., Schreier M. H., Noll H. Structural dynamics of bacterial ribosomes. I. Characterization of vacant couples and their relation to complexed ribosomes. J Mol Biol. 1973 Apr 5;75(2):281–294. doi: 10.1016/0022-2836(73)90021-1. [DOI] [PubMed] [Google Scholar]
- Parthasarathy R., Ohrt J. M., Chheda G. B. Modified nucleosides and conformation of anticodon loops: crystal structure of t6A and g6A. Biochemistry. 1977 Nov 15;16(23):4999–5008. doi: 10.1021/bi00642a010. [DOI] [PubMed] [Google Scholar]
- Schüler D., Brimacombe R. The Escherichia coli 30S ribosomal subunit; an optimized three-dimensional fit between the ribosomal proteins and the 16S RNA. EMBO J. 1988 May;7(5):1509–1513. doi: 10.1002/j.1460-2075.1988.tb02970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S., Weiser B., Noller H. F. Model for the three-dimensional folding of 16 S ribosomal RNA. J Mol Biol. 1988 Nov 20;204(2):447–481. doi: 10.1016/0022-2836(88)90588-8. [DOI] [PubMed] [Google Scholar]
- Stiege W., Stade K., Schüler D., Brimacombe R. Covalent cross-linking of poly(A) to Escherichia coli ribosomes, and localization of the cross-link site within the 16S RNA. Nucleic Acids Res. 1988 Mar 25;16(6):2369–2388. doi: 10.1093/nar/16.6.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöffler-Meilicke M., Stöffler G. The topography of ribosomal proteins on the surface of the 30S subunit of Escherichia coli. Biochimie. 1987 Oct;69(10):1049–1064. doi: 10.1016/0300-9084(87)90005-8. [DOI] [PubMed] [Google Scholar]
- Walleczek J., Schüler D., Stöffler-Meilicke M., Brimacombe R., Stöffler G. A model for the spatial arrangement of the proteins in the large subunit of the Escherichia coli ribosome. EMBO J. 1988 Nov;7(11):3571–3576. doi: 10.1002/j.1460-2075.1988.tb03234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Dahlberg A. E., Atkins J. F., Gesteland R. F. Reading frame switch caused by base-pair formation between the 3' end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988 May;7(5):1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann H. G. Architecture of prokaryotic ribosomes. Annu Rev Biochem. 1983;52:35–65. doi: 10.1146/annurev.bi.52.070183.000343. [DOI] [PubMed] [Google Scholar]



