Abstract
Background
The polycomb-group (PcG) proteins function as general regulators of stem cells. We previously reported that retrovirus-mediated overexpression of Bmi1, a gene encoding a core component of polycomb repressive complex (PRC) 1, maintained self-renewing hematopoietic stem cells (HSCs) during long-term culture. However, the effects of overexpression of Bmi1 on HSCs in vivo remained to be precisely addressed.
Methodology/Principal findings
In this study, we generated a mouse line where Bmi1 can be conditionally overexpressed under the control of the endogenous Rosa26 promoter in a hematopoietic cell-specific fashion (Tie2-Cre;R26StopFLBmi1). Although overexpression of Bmi1 did not significantly affect steady state hematopoiesis, it promoted expansion of functional HSCs during ex vivo culture and efficiently protected HSCs against loss of self-renewal capacity during serial transplantation. Overexpression of Bmi1 had no effect on DNA damage response triggered by ionizing radiation. In contrast, Tie2-Cre;R26StopFLBmi1 HSCs under oxidative stress maintained a multipotent state and generally tolerated oxidative stress better than the control. Unexpectedly, overexpression of Bmi1 had no impact on the level of intracellular reactive oxygen species (ROS).
Conclusions/Significance
Our findings demonstrate that overexpression of Bmi1 confers resistance to stresses, particularly oxidative stress, onto HSCs. This thereby enhances their regenerative capacity and suggests that Bmi1 is located downstream of ROS signaling and negatively regulated by it.
Introduction
Hematopoietic stem cells (HSCs) are defined as primitive cells that are capable of both self-renewal and differentiation into any of the hematopoietic cell lineages. Cell fate decisions of HSCs (self-renewal vs. differentiation) are precisely regulated to maintain their numbers and lifespan. Defects in these processes lead to hematopoietic insufficiencies and to the development of hematopoietic malignancies.
The polycomb-group (PcG) proteins play key roles in the initiation and maintenance of gene silencing through histone modifications. PcG proteins belong to two major complexes, Polycomb repressive complex 1 and 2 (PRC1 and PRC2). PRC1 monoubiquitylates histone H2A at lysine 119 and PRC2 trimethylates histone H3 at lysine 27 [1]. Of note, PcG proteins have been implicated in the maintenance of self-renewing stem cells [2]–[4]. Among PcG proteins, Bmi1, a core component of PRC1, plays an essential role in the maintenance of self-renewal ability of HSCs at least partially by silencing the Ink4a/Arf locus [5]–[8]. Bmi1 also maintains multipotency of HSCs by keeping developmental regulator gene promoters poised for activation [9]. Furthermore, Bmi1 has been implicated in the maintenance of the proliferative capacity of leukemic stem cells [5]. Consistent with these findings, levels of BMI1 expression in the human CD34+ cell fraction have been reported to correlate well with the progression and prognosis of myelodysplastic syndrome and chronic and acute myeloid leukemia [4], [10], suggesting a role of BMI1 in leukemic stem cells.
We previously reported that overexpression of Bmi1 using a retrovirus maintains self-renewal capacity of HSCs and markedly expands multipotent progenitors ex vivo, resulting in an enhancement of repopulating capacity of HSCs after culture. Likewise, forced expression of BMI1 was demonstrated to promote leukemic transformation of human CD34+ cells by BCR-ABL [11]. However, the effects of overexpression of Bmi1 on hematopoiesis remained to be precisely addressed.
In this study, we generated mice overexpressing Bmi1 in a hematopoietic cell-specific manner. We analyzed the effects of overexpression of Bmi1 on hematopoiesis under steady state conditions as well as under multiple stresses. Our findings revealed a protective function for Bmi1 in HSCs from stresses, such as ROS, that usually limit the lifespan of HSCs.
Results
Generation of Mice Overexpressing Bmi1 in Hematopoietic Cells
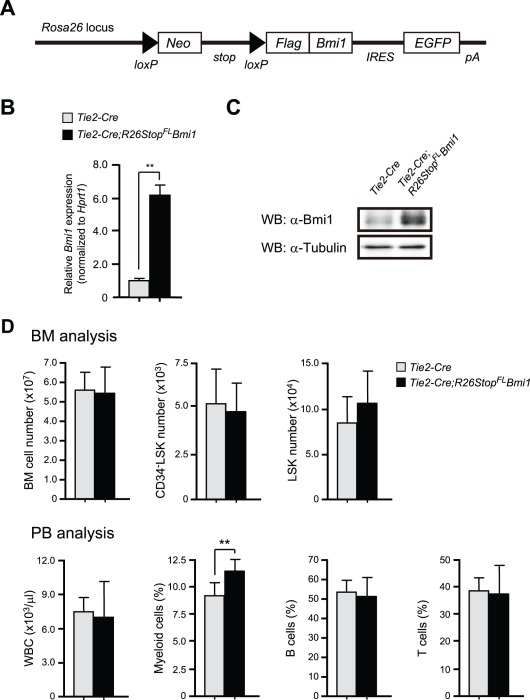
To generate tissue-specific Bmi1-transgenic mice, we knocked a loxP-flanked neor-stop cassette followed by Flag-tagged Bmi1, an frt-flanked IRES-eGFP cassette, and a bovine polyadenylation sequence into the Rosa26 locus ( Figure 1A ). The obtained mice (hereafter referred to as R26StopFLBmi1) were crossed with Tie2-Cre mice [12] to drive Bmi1 expression in a hematopoietic cell-specific manner. Quantitative RT-PCR analysis of bone marrow (BM) Lineage marker-Sca-1+c-Kit+ (LSK) cells confirmed 6-fold overexpression of Bmi1 in Tie2-Cre;R26StopFLBmi1 mice compared to the Tie2-Cre control mice ( Figure 1B ). Western blot analysis also verified overexpression of Bmi1 protein in BM c-Kit+ progenitor cells from Tie2-Cre;R26StopFLBmi1 mice ( Figure 1C ).
Figure 1. Generation of mice overexpressing Bmi1 in hematopoietic cells.
(A) Strategy for making a knock-in allele for Bmi1 downstream of the Rosa26 promoter. A loxP-flanked neor-stop cassette followed by Flag-tagged Bmi1, an frt-flanked IRES-eGFP cassette, and a bovine polyadenylation sequence was knocked-in the Rosa26 locus. (B) Quantitative RT-PCR analysis of Bmi1 in BM LSK cells from Tie2-Cre and Tie2-Cre;R26StopFLBmi1 mice. mRNA levels were normalized to Hprt1 expression. Expression levels relative to that in Tie2-Cre LSK cells are shown as the mean ± S.D. (n = 3). (C) Western blotting analysis of Bmi1 in c-Kit+ BM cells from Tie2-Cre and Tie2-Cre;R26StopFLBmi1 mice. α-tubulin was used as the loading control. (D) Hematopoietic analysis of 10-week-old Tie2-Cre and Tie2-Cre;R26StopFLBmi1 mice. Absolute numbers of BM cells, CD34-LSK cells, and LSK cells in bilateral femurs and tibiae are presented as the mean ± S.D. (upper panels, Tie2-Cre; n = 7, Tie2-Cre;R26StopFLBmi1; n = 8). PB analysis of 10-week-old Tie2-Cre and Tie2-Cre;R26StopFLBmi1 mice. White blood cell (WBC) counts and lineage contribution of myeloid, B, and T cells are shown as the mean ± S.D. (lower panels, Tie2-Cre; n = 7, Tie2-Cre;R26StopFLBmi1; n = 8). ** p<0.01.
Steady State Hematopoiesis in Tie2-Cre;R26StopFLBmi1 Mice
We first investigated the effect of overexpression of Bmi1 on hematopoiesis in a steady state. Unexpectedly, 10-week-old Tie2-Cre;R26StopFLBmi1 mice did not exhibit any significant differences in the numbers of total BM cells, CD34-LSK HSCs, LSK cells, multipotent progenitors (MPPs), common myeloid progenitors (CMPs), granulocyte/macrophage progenitors (GMPs), megakaryocyte/erythroid progenitors (MEPs), or common lymphoid progenitors (CLPs) compared to the Tie2-Cre control mice ( Figure 1D and Figure S1A). The number of white blood cells (WBC) in peripheral blood (PB) did not change upon forced expression of Bmi1. Only the proportion of PB Gr-1+/Mac-1+ myeloid cells in Tie2-Cre;R26StopFLBmi1 mice was significantly higher than in the control mice, although the difference was not drastic (a difference of only about 2%) ( Figure 1D ). Furthermore, Tie2-Cre;R26StopFLBmi1 mice did not show any significant differences in the numbers of total spleen cells, LSK cells in the spleen, total thymic cells, or CD4+CD8-, CD4-CD8+, or CD4+CD8+ cells in the thymus compared to the control mice (Figure S1A). These findings indicate that overexpression of Bmi1 does not largely compromise differentiation of HSCs. We further analyzed the cell cycle status of CD34-LSK HSCs by Pyronin Y staining, but again did not detect any changes (Figure S1B). These results indicate that overexpression of Bmi1 only slightly perturbs hematopoiesis under steady state conditions, suggesting that the level of endogenous Bmi1 is sufficient to repress the transcription of its target genes.
Colony-forming Capacity of Hematopoietic Stem and Progenitor Cells Overexpressing Bmi1
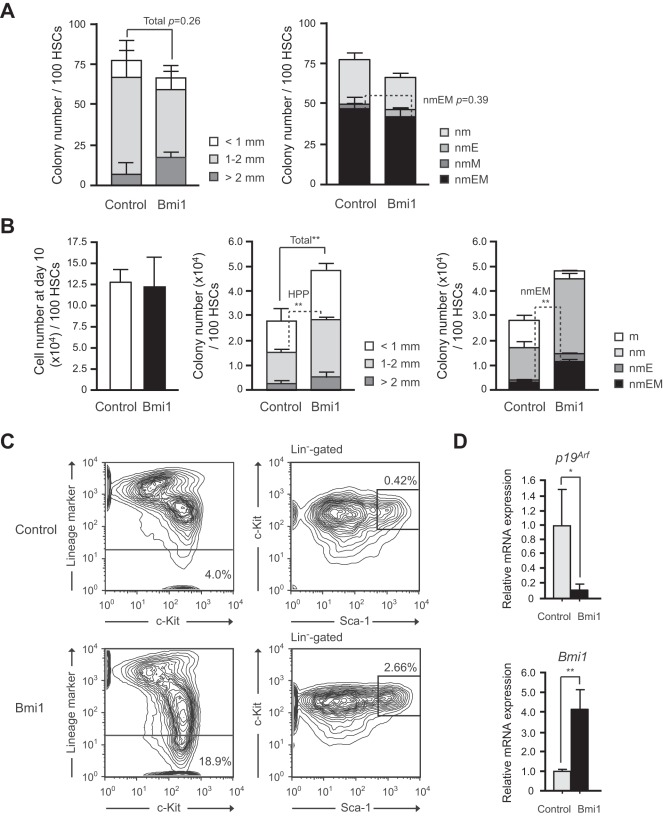
We next evaluated the proliferative and differentiation capacity of Tie2-Cre;R26StopFLBmi1 HSCs in vitro. Single CD34-LSK HSCs were clonally sorted into 96-microtiter plates with the medium supplemented with stem cell factor (SCF), thrombopoietin (TPO), interleukin-3 (IL-3), and erythropoietin (EPO) and allowed to form colonies. At day 14 of culture, the colonies were counted and individually collected for morphological examination. Both Tie2-Cre control and Tie2-Cre;R26StopFLBmi1 HSCs gave rise to comparable numbers of high proliferative potential (HPP) and low proliferative potential (LPP) colonies with a diameter greater than and less than 1 mm, respectively ( Figure 2A ). The morphological analysis of colonies revealed that the number of colony-forming unit (CFU)-neutrophil/macrophage/erythroblast/megakaryocyte (nmEM) was also comparable between the two groups ( Figure 2A ). CFU-nmEM is a major subpopulation among CD34−LSK HSCs and its frequency is well correlated with that of functional HSCs [13]. These findings indicate that overexpression of Bmi1 in freshly isolated CD34-LSK HSCs does not affect their colony-forming capacity or differentiation in vitro.
Figure 2. Effects of overexpression of Bmi1 on HSCs in vitro.
(A) Colony formation by HSCs isolated from Tie2-Cre (Control) and Tie2-Cre;R26StopFLBmi1 (Bmi1) mice. Single CD34-LSK cells were sorted into 96-well microtiter plates containing the SF-O3 medium supplemented with 10% FBS and multiple cytokines (10 ng/ml SCF, 10 ng/ml TPO, 10 ng/ml IL-3, and 3 u/ml EPO) and allowed to form colonies. At day 14 of culture, the colonies were counted and individually collected for morphological examination. Absolute numbers of LPP and HPP-CFCs which gave rise to colonies with a diameter less and greater than 1 mm, respectively are shown as the mean ± S.D. for triplicate cultures (left panel). Absolute numbers of each colony types were defined by the composition of colonies (right panel). Colonies were recovered and examined by microscopy to determine colony types. Composition of colonies is depicted as n, neutrophils; m, macrophages; E, erythroblasts; and M, megakaryocytes. (B) Colony formation by HSCs cultured for 10 days. CD34-LSK cells from Tie2-Cre (Control) and Tie2-Cre;R26StopFLBmi1 (Bmi1) mice were cultured in the SF-O3 serum-free medium supplemented with 50 ng/ml of SCF and TPO. At day 10 of culture, the cells were counted (left panel) and plated in methylcellulose medium to allow formation of colonies in the presence of 20 ng/ml SCF, 20 ng/ml TPO, 20 ng/ml IL-3, and 3 u/ml EPO. Absolute numbers of LPP and HPP-CFCs (middle panel) are shown as the mean ± S.D. for triplicate cultures. Absolute numbers of each colony type are shown in the right panel. (C) Flow cytometric analysis of CD34-LSK HSCs at day14 of culture. Representative flow cytometric profiles of LSK cells in cultures of CD34-LSK HSCs from Tie2-Cre (Control) and Tie2-Cre;R26StopFLBmi1 (Bmi1) mice are depicted. The proportion of Lin- and LSK cells in total cells are indicated. (D) Quantitative RT-PCR analysis of the expression of p19Arf, and Bmi1 in Tie2-Cre (Control) and Tie2-Cre;R26StopFLBmi1 (Bmi1) LSK cells. LSK cells were purified by cell sorting from CD34-LSK cultures in (C) at day 14 of culture. Each value was normalized to Hprt1 expression and the expression level of each gene in control cells was arbitrarily set to 1. Data are shown as the mean ± S.D. for triplicate analyses. * p<0.05, **p<0.01.
We previously reported that overexpression of Bmi1 by retroviral transduction efficiently maintains hematopoietic stem and progenitor cells during long-term culture [7]. We re-evaluated the effect of forced expression of Bmi1 using Tie2-Cre;R26StopFLBmi1 HSCs. CD34-LSK cells were cultured for 10 days in a serum-free medium supplemented with SCF and TPO, a cytokine combination which supports the proliferation of HSCs and progenitors rather than their differentiation [14]. Although Tie2-Cre;R26StopFLBmi1 HSCs did not show any growth advantage over the control ( Figure 2B ), the Tie2-Cre;R26StopFLBmi1 HSC culture contained significantly more HPP-colony-forming cells (CFCs) and CFU-nmEM than the control ( Figure 2B ). Correspondingly, flow cytometric analysis revealed more LSK cells in the Tie2-Cre;R26StopFLBmi1 HSC culture than in the control culture at day 14 ( Figure 2C ). There was no significant difference in the frequency of apoptotic cells between the control and Tie2-Cre;R26StopFLBmi1 HSC cultures (Figure S2A). Of note, however, the Tie2-Cre;R26StopFLBmi1 HSC culture contained a significantly higher proportion of LSK cells in the G0/G1 stage of cell cycle than the control (Figure S2B). These findings suggest that overexpression of Bmi1 slows down cell cycle of immature hematopoietic cells in culture, leading to no growth advantages over the control cells in spite of an increase in immature progenitors in culture. As we reported previously, the Ink4a/Arf locus is a critical target of Bmi1 in HSCs [8]. Quantitative RT-PCR confirmed that p19Arf was closely repressed in transcription upon Bmi1 overexpression ( Figure 2D ). These results support our previous finding that HSCs overexpressing Bmi1 retain their self-renewal capacity better than the control HSCs under the culture stress.
Overexpression of Bmi1 Enhances Expansion of HSCs ex vivo and Protects HSCs During Serial Transplantation
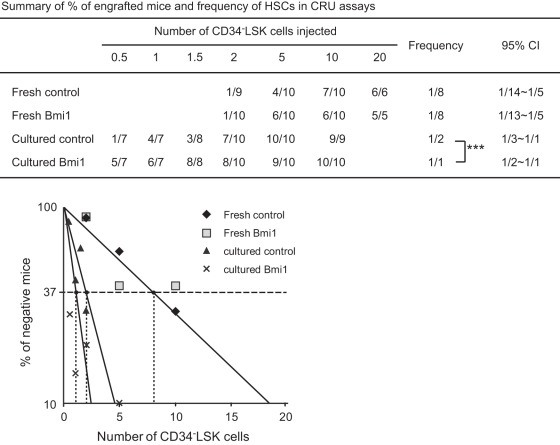
HSCs are exposed to oxidative stress during long-term culture in 20% O2 [15]. In order to precisely determine the effect of overexpression of Bmi1 on HSCs during culture, we next determined the frequency of functional HSCs contained in optimized serum-free culture by competitive repopulating unit (CRU) assay. We first transplanted limiting doses of fresh CD34-LSK cells from Tie2-Cre and Tie2-Cre;R26StopFLBmi1 mice along with 2×105 competitor BM cells. The frequency of long-term repopulating HSCs was 1 in 8 among fresh CD34-LSK cells from both Tie2-Cre and Tie2-Cre;R26StopFLBmi1 mice ( Figure 3 ). We then cultured CD34-LSK cells for 10 days in a serum-free medium supplemented with SCF and TPO in 20% O2. During the 10-day culture period, functional HSCs increased 4-fold (1 out of 2 CD34-LSK cells) in control CD34-LSK cells. Interestingly, overexpression of Bmi1 established 2-fold better expansion of HSCs than the control during culture ex vivo ( Figure 3 ). These findings are the first to show that Bmi1 has a role in the expansion of HSCs.
Figure 3. Overexpression of Bmi1 enhances expansion of HSCs ex vivo.
Competitive repopulating unit (CRU) assays using limiting numbers of CD34-LSK cells from Tie2-Cre (Control) mice and Tie2-Cre;R26StopFLBmi1 (Bmi1) mice. Freshly isolated CD34-LSK cells were immediately used for BM transplantation, or CD34-LSK cells were cultured in the SF-O3 serum-free medium supplemented with 50 ng/ml SCF and TPO for 10 days, and then a fraction of the culture cells corresponding to the indicated number (0.5∼10) of initial CD34-LSK cells was subjected to BM transplantation. The test cells (CD45.2) were transplanted along with 2×105 competitor BM cells (CD45.1) into CD45.1 recipient mice lethally irradiated at a dose of 9.5 Gy. Percent chimerism of donor cells in the recipient PB was determined at 16 weeks after transplantation. The mice with chimerism more than 1% in all three lineages (myeloid, B, and T cells) were considered successfully engrafted and the others were defined as negative mice. The frequency of HSCs was calculated using L-Calc software. The proportion of engrafted mice, frequency of functional HSCs, and the 95% confidence interval (CI) are summarized in the table and each data is plotted in the bottom panel. *** p<0.001.
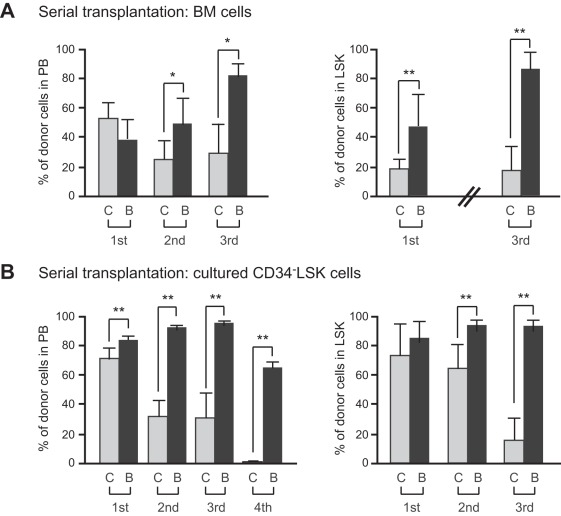
HSCs are exposed to various stresses including replicative and oxidative stresses during serial transplantation and eventually lose self-renewal capacity [16], [17]. We hypothesized that the effects of overexpression of Bmi1 on HSCs would manifest under stressful conditions such as serial transplantations. Therefore, we performed competitive repopulation assays using 5×105 fresh BM cells along with 5×105 competitor BM cells ( Figure 4A ) or the total cells produced from 20 CD34-LSK cells after a 10-day culture period along with 2×105 competitor BM cells ( Figure 4B ). The flow cytometric analysis of PB revealed little or no difference in the chimerism of donor cells between Tie2-Cre and Tie2-Cre;R26StopFLBmi1 cells at 12 weeks after the primary transplantations. However, in the secondary and tertiary transplantations, the chimerism of Tie2-Cre cells significantly declined while that of Tie2-Cre;R26StopFLBmi1 cells drasticaly increased. Tie2-Cre cells after 10-day culture failed to reconstitute hematopoiesis in the quaternary transplantation, while Tie2-Cre;R26StopFLBmi1 cells still established robust repopulation ( Figure 4B ). The chimerism of donor cells in BM LSK cells mirrored the changes in the PB. These results clearly indicate that overexpression of Bmi1 protects HSCs against the loss of self-renewal capacity during serial transplantation. The findings thus far suggest that overexpression of Bmi1 confers stress resistance onto HSCs.
Figure 4. Overexpression of Bmi1 protects HSCs during serial transplantation.
(A) Serial transplantation of BM cells. BM cells (5×105) from Tie2-Cre (denoted as “C”) and Tie2-Cre;R26StopFLBmi1 (denoted as “B”) mice (CD45.2) along with 5×105 competitor BM cells (CD45.2) were transplanted into CD45.1 recipient mice lethally irradiated at a dose of 9.5 Gy. For serial transplantation, BM cells were collected from all recipient mice at 12–20 weeks after transplantation and pooled together. Then, 5×106 BM cells were transplanted into lethally irradiated recipient mice without competitor cells. Third and fourth transplantation were similarly performed using 5×106 pooled BM cells. Percent chimerism of donor cells in the recipient PB and BM LSK cells was determined at 16 weeks post-transplantation. Results are shown as the mean ± S.D. (n = 6, 3rd transplantation; n = 4). (B) Serial transplantation of cultured CD34-LSK cells. CD34-LSK cells were cultured in the SF-O3 serum-free medium supplemented with 50 ng/ml of SCF and TPO for 10 days. Then, the cells in culture corresponding to the 20 initial CD34-LSK cells were injected into a recipient mouse along with 2×105 competitor BM cells (CD45.2) as described in (A) (n = 6, 4th transplantation; n = 5). * P<0.05, ** P<0.01.
Overexpression of Bmi1 has no Impact on Radioprotection
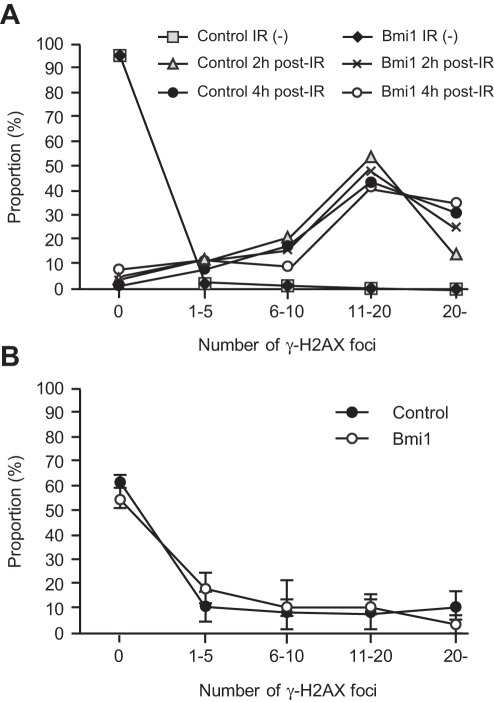
DNA damage is intimately linked to stem cell aging. Heritable DNA damage accrued in stem cells leads to stem cell senescence or apoptosis, which over time can lead to the depletion of the stem cell pool and reduced regenerative capacity of stem cells [18]. Bmi1 is rapidly recruited to sites of DNA damage and is required for DNA damage-induced ubiquitination of histone H2A at lysine 119. Loss of Bmi1 leads to impaired repair of DNA double-strand breaks (DSBs) by homologous recombination [19], [20]. In glioblastoma multiforme (GBM) cells, Bmi1 was co-purified with DSB response proteins, such as ATM and the histone γH2AX, and non-homologous end joining (NHEJ) repair proteins. Of interest, BMI1 overexpression in normal neural stem cells enhanced ATM recruitment to the chromatin, the rate of γH2AX foci resolution, and resistance to radiation [21]. In order to understand the role of overexpressed Bmi1 in HSCs, we examined the radioresistance of HSCs by quantifying the number of γH2AX foci following genotoxic stress, a metric which reflects DNA DSBs.
We purified CD34-LSK cells from Tie2-Cre and Tie2-Cre;R26StopFLBmi1 mice and irradiated them at a dose of 2 Gy. At 2 and 4 hours after irradiation, cells were stained with anti-γH2AX. We expected rapid resolution of γH2AX by overexpression of Bmi1, but no significant difference was observed in the number of γH2AX foci between Tie2-Cre and Tie2-Cre;R26StopFLBmi1 HSCs ( Figure 5A ). HSCs recovered from the recipients of tertiary transplantation did not show any difference in the number of γH2AX foci, either ( Figure 5B ). We then tested hematopoietic recovery after irradiation in mice. We irradiated recipient mice reconstituted with Tie2-Cre and Tie2-Cre;R26StopFLBmi1 BM cells at a dose of 5 Gy, and monitored hematopoietic recovery for 4 weeks. The recovery of hematopoietic components in PB as well as BM LSK cells was comparable between the two groups (Figure S3). These findings suggest that overexpression of Bmi1 does not afford an advantage to HSCs in their ability to resist genotoxic stress.
Figure 5. DNA damage response of Tie2-Cre;R26StopFLBmi1 HSCs.
(A) DNA damage response of CD34-LSK cells from Tie2-Cre (Control) and Tie2-Cre;R26StopFLBmi1 (Bmi1) mice in vitro. Purified CD34-LSK cells were irradiated (IR) at a dose of 2 Gy. At 2 and 4 hours after irradiation, cells were stained with anti-γH2AX. Numbers of γH2AX foci expressed per cell are depicted. (B) DNA damage response of CD34-LSK cells from Tie2-Cre (Control) and Tie2-Cre;R26StopFLBmi1 (Bmi1) mice in vivo. LSK cells were purified from the recipients of tertiary transplantation and stained with anti-γH2AX. Numbers of γH2AX foci expressed per cell are depicted as the mean ± S.D. (n = 3).
Overexpression of Bmi1 Confers Resistance to Oxidative Stress on HSCs
HSCs contain lower levels of reactive oxygen species (ROS) than their mature progeny in order to maintain their quiescent state. ROS reportedly act through p38 mitogen-activated protein kinase (MAPK) to limit the lifespan of HSCs [16], [22]. It has been demonstrated that prolonged treatment with the antioxidant N-acetyl-L-cysteine (NAC) or an inhibitor of p38 MAPK extends the lifespan of HSCs in serial transplantation assays, suggesting that oxidative stress is one of the major factors that affects HSC function during these assays [16], [17], [23]. Given that Tie2-Cre;R26StopFLBmi1 HSCs retain self-renewal capacity during serial transplantation, overexpression of Bmi1 may bestow a protective effect onto HSCs against oxidative stress.
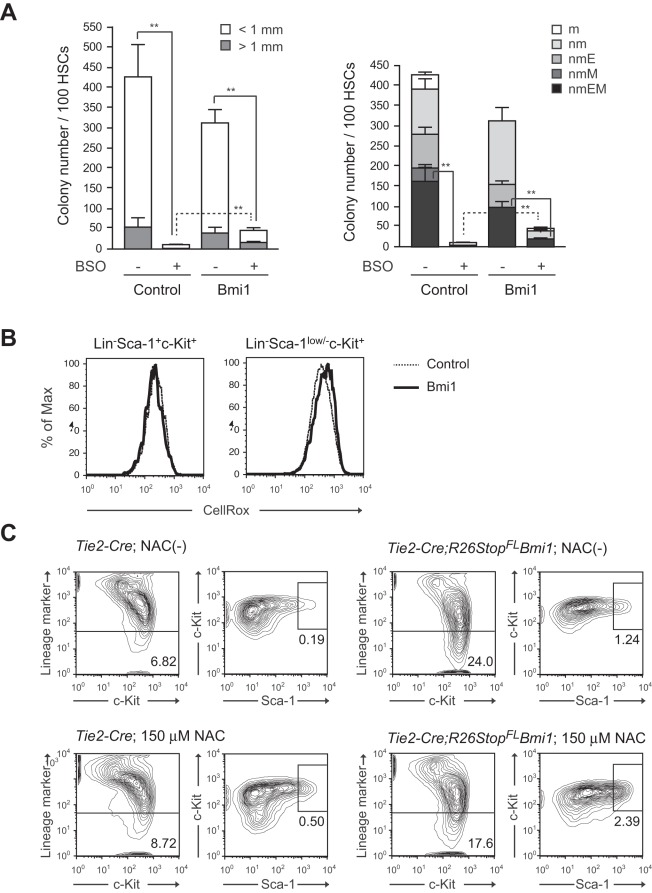
To address this question, we cultured HSCs in the presence of buthionine sulfoximine (BSO), which depletes intracellular glutathione and thereby increases intracellular ROS levels. We found that highly purified CD34-LSK HSCs were susceptible to an increase in ROS levels because treatment with BSO significantly suppressed their growth and induced cell death (data not shown). After 3 days of BSO treatment, surviving cells were subjected to colony-forming assays. Both Tie2-Cre control and Tie2-Cre;R26StopFLBmi1 HSCs cultured with BSO gave rise to significantly fewer colonies than HSCs cultured without BSO. Interestingly, Tie2-Cre;R26StopFLBmi1 HSCs gave rise to a significantly more colonies than the control HSCs ( Figure 6A ). Notably, the number of HPP colonies was reduced 48-fold after treatment of control HSCs with BSO, but only 3-fold upon overexpression of Bmi1. The frequency of CFU-nmEM was also less perturbed following treatment with BSO in HSCs overexpressing Bmi1. These results indicate a role for Bmi1 in the resistance to oxidative stress.
Figure 6. Overexpression of Bmi1 confers oxidative stress on HSCs.
(A) Colony formation by HSCs cultured for 3 days. CD34-LSK cells from Tie2-Cre (Control) and Tie2-Cre;R26StopFLBmi1 (Bmi1) mice were cultured in the SF-O3 serum-free medium supplemented with 50 ng/ml SCF, TPO and 0.05 mM of BSO. At day 3 of culture, the cells were plated in methylcellulose medium to allow formation of colonies in the presence of 20 ng/ml SCF, 20 ng/ml TPO, 20 ng/ml IL-3, and 3 u/ml EPO. Absolute numbers of LPP and HPP-CFCs (left panel) are shown as the mean ± S.D. for triplicate cultures. Absolute numbers of each colony types are shown in the right panel. Data are shown as the mean ± S.D. for triplicate analyses. Statistical analyses were performed on the total colony numbers (left panel) and nmEM colony numbers (right panel), respectively. **p<0.01. (B) Levels of ROS in cells overexpressing Bmi1. CD34-LSK cells from Tie2-Cre (Control) and Tie2-Cre;R26StopFLBmi1 (Bmi1) mice were cultured in the SF-O3 serum-free medium supplemented with 50 ng/ml SCF and TPO. Representative flow cytometric profiles of LSK and Lineage marker-Sca-1low/−c-Kit+ cells in cultures at day 14 are depicted. (C) Effects of NAC on Bmi1 culture. CD34-LSK cells from Tie2-Cre and Tie2-Cre;R26StopFLBmi1 mice were cultured in the SF-O3 serum-free medium supplemented with 50 ng/ml SCF and TPO in the presence and absence of 150 µM NAC. Representative flow cytometric profiles of LSK cells in cultures at day 14 are depicted. The proportion of Lin- and LSK cells in total cells are indicated.
Bmi1 regulates mitochondrial function by regulating the expression of a cohort of genes related to mitochondrial function and ROS generation. Bmi1-deficient cells have impaired mitochondrial function, which causes a marked increase in the intracellular levels of ROS [24]. Based on these observations, we then measured the intracellular ROS levels in CD34-LSK cells at day 14 of culture. Unexpectedly, overexpression of Bmi1 did not affect the levels of ROS in either LSK HSCs/MPPs or Lin-Sca-1low/−c-Kit+ downstream progenitors ( Figure 6B ). Overexpression of Bmi1 had no significant effect on the ROS levels even in the presence of BSO (Figure S4). Likewise, treatment of cells with the antioxidant NAC promoted cell growth and increased the proportion of LSK cells in both control and Tie2-Cre;R26StopFLBmi1 culture similarly ( Figure 6C and data not shown). These results indicate that an excess of Bmi1 does not regulate the generation or scavenging of ROS, but confers resistance to higher levels of ROS on HSCs through unknown mechanisms.
Discussion
In this study, we generated a new mouse line where Bmi1 can be conditionally overexpressed in a hematopoietic cell-specific fashion and analyzed the effect of overexpression of Bmi1 in detail. Overexpression of Bmi1 did not significantly affect steady state hematopoiesis, but it efficiently protected HSCs from stresses. Our findings suggest that overexpression of Bmi1 confers resistance to stresses on HSCs, thereby augmenting their regenerative capacity.
Recent findings have established that the regulation of oxidative stress in HSCs is critical for the maintenance of HSCs. In this study, we demonstrated that overexpression of Bmi1 protects HSCs from loss of self-renewal capacity at least in part by increasing the capacity of HSCs to resist oxidative stress. It has been reported that Bmi1-deficient mice have an increased level of intracellular ROS due to de-regulated expression of genes related to mitochondrial function and ROS generation [24], [25]. However, an excess of Bmi1 in this study had no effect on the levels of intracellular ROS. Thus, it is hypothesized that Bmi1 is negatively regulated downstream of the ROS signal and an excess of Bmi1 overcomes this negative regulation. Indeed, ROS reportedly primes Drosophila hematopoietic progenitors for differentiation and this process involves downregulation of PcG activity [26]. ROS signaling activates p38 and eventually releases the transcriptional repression of p16Ink4a and p19Arf, critical targets of Bmi1 [16]. Furthermore, recent studies including ours have revealed that PcG proteins are downregulated and dissociate from the Ink4a/Arf locus when cells are exposed to intra- or extracellular stress, including tissue culture- and oncogene-induced stress [27], [28]. Together, this accumulating evidence suggests that Bmi1 is dynamically regulated in response to oxidative stress, probably downstream of p38. Our preliminary data demonstrated that activated p38 directly phosphorylates Bmi1 in vitro (Oshima and Iwama., unpublished data). Thus, it is possible that p38, which is activated by oxidative stress, attenuates Bmi1 function via direct phosphorylation of Bmi1. How oxidative stress restricts the expression and function of Bmi1 is an important issue to be addressed.
Of note, the effect of Bmi1 overexpression in serial transplantation resembles that of overexpression of Ezh2, a gene encoding a core component of PRC2 [29]. Overexpression of PcG genes, Bmi1 and Scmh1, also induces tolerance of cortical neurons to ischemia [30]. Thus, various cellular stresses may target PcG complexes to release transcriptional repression of PcG-regulated genes, such as tumor suppressor and developmental regulator genes, thereby affecting stemness. All these findings support the notion that enforcement of PcG function is a key for successful regenerative therapies.
Meanwhile, the role of PcG proteins in resistance to oxidative stress is also implicated in cancer. Expression of PcG proteins including BMI1 and EZH2 are often up-regulated in various cancers, particularly in their cancer stem cell fractions [31]. Interestingly, cancer stem cells in some tumors appear to be susceptible to ROS, similar to normal stem cells, and thus develop mechanisms to keep the levels of ROS low [32]. Interference of EZH2 function by the small-molecule histone methyltransferases inhibitor, DZNep, is reported to increase ROS levels in acute myeloid leukemia cells like in Bmi1-deficient mice [33]. Conversely, our findings in this study suggest that an excess of PcG proteins often observed in aggressive cancer could help cancer stem cells tolerate oxidative stress. In this regard, overexpression of PcG proteins could also be therapeutic targets in cancers including leukemia. Finally, no Tie2-Cre;R26StopFLBmi1 mice developed hematological malignancies during the observation period, up to 18 months after birth. Only one recipient mice with Tie2-Cre;R26StopFLBmi1 BM cells developed acute lymphocytic leukemia in the tertiary transplantation. These findings suggest that Bmi1 by itself is not sufficient to induce hematological malignancies.
Methods
Ethics Statement
All experiments using the mice were performed in accordance with our institutional guidelines for the use of laboratory animals and approved by the review board for animal experiments of Chiba University (approval ID: 21–150).
Generation of Mice
To generate tissue-specific Bmi1-transgenic mice, we used the plasmid R26StopFL, a modified version of pROSA26-1 with a loxP-flanked neor-stop cassette, an frt-flanked IRES-eGFP cassette, and a bovine polyadenylation sequence [34]. We cloned a cDNA encoding a flag-tagged Bmi1 upstream of the IRES sequence (R26StopFLBmi1). R1 ES cells were transfected, cultured, and selected as previously described [35]. For conditional expression of Bmi1, the RosaStopFLBmi1 mice were crossed with Tie2-Cre mice. C57BL/6 (CD45.2) mice were purchased from Japan SLC (Shizuoka, Japan). C57BL/6 mice congenic for the Ly5 locus (CD45.1) were purchased from Sankyo-Lab Service (Tsukuba, Japan). Mice were bred and maintained in the Animal Research Facility of the Graduate School of Medicine, Chiba University in accordance with institutional guidelines. This study was approved by the institutional review committees of Chiba University (approval numbers 21–65 and 21–150).
Flow Cytometric Analysis and Cell Sorting
Mouse CD34–LSK HSCs were purified from BM of 8–12-week-old mice. Mononuclear cells were isolated on Ficoll-Paque PLUS (GE Healthcare). Cells were stained with an antibody cocktail consisting of biotinylated anti-Gr-1, Mac-1, interleukin (IL)-7Rα, B220, CD4, CD8α, and Ter119 monoclonal antibodies. The monoclonal antibodies were purchased from eBioScience or BioLegend. Lineage-positive cells were depleted with goat anti-rat IgG microbeads (Miltenyi Biotec) through an LS column (Miltenyi Biotec). Cells were further stained with Alexa Fluor® 647 or eFluor® 660-conjugated anti-CD34, phycoerythrin (PE)-conjugated anti-Sca-1, and phycoerythrin/Cy7 (PE/Cy7)-conjugated anti-c-Kit antibodies. Biotinylated antibodies were detected with allophycocyanin/Cy7 (APC/Cy7)-conjugated streptavidin. Dead cells were eliminated by staining with Propidium iodide (1 µg/ml, Sigma). Analysis and sorting were performed on a FACS Aria II (BD Bioscience).
Cell Cycle Analysis
Fresh BM cells (1×107, CD45.2) were transplanted into 8-week-old CD45.1 mice irradiated at a dose of 9.5 Gy without competitor cells. Four months later, BM mononuclear cells were isolated on Ficoll-Paque PLUS. Cells were stained with an antibody cocktail consisting of biotinylated anti-Gr-1, Mac-1, IL-7Rα, B220, CD4, CD8α, Ter119, and CD45.1 monoclonal antibodies. Cells were further stained with Alexa Fluor® 700-conjugated anti-CD34, pacific blue-conjugated anti-Sca-1, and APC-conjugated anti-c-Kit antibodies. Biotinylated antibodies were detected with APC/Cy7-conjugated streptavidin. Analysis was performed on a FACS Aria II. To analyze the cell-cycle status, cells were incubated with 1 µg/ml Pyronin Y (Sigma) at 37°C for 45 min with protection from light. Bulk sorted CD34-LSK cells were incubated in SF-O3 supplemented with 50 µM?2-β-mercaptoethanol, 0.2% BSA, 1% GPS, 50 ng/ml SCF, 50 ng/ml TPO for 10 days at 37°C in a 5% CO2 atmosphere. At day 10 of culture, the cell cycle profiles of culture cells were analyzed using an APC BrdU Flow Kit (BD Pharmingen). The cells were incubated with 10 µM BrdU at 37°C for 30 min and then stained with an antibody cocktail consisting of biotinylated anti-Gr-1, Mac-1, IL-7Rα, B220, CD4, CD8α, and Ter119 monoclonal antibodies. Cells were further stained with PE-conjugated anti-Sca-1, and PE/Cy7-conjugated anti-c-Kit antibodies. Biotinylated antibodies were detected with APC/Cy7-conjugated streptavidin. Analysis was performed on a FACS Canto II (BD Bioscience).
Colony Assay
Colony assays were performed in methylcellulose-containing Iscove’s modified Dulbecco’s medium (Methocult M3234; Stemcell Technologies) supplemented with 20 ng/ml mouse SCF, 20 ng/ml mouse IL-3, 20 ng/ml human TPO, and 3 U/ml human EPO (Peprotech), and incubated at 37°C in a 5% CO2 atmosphere. The number of HPP- and LPP-colony-forming cells (CFCs), which generate a colony with a diameter ≥1 mm and <1 mm, respectively, were evaluated by counting colonies at day 10–14 of culture. Colonies were individually collected, cytospun onto glass slides, and subjected to Hemacolor (MERCK) staining for morphological examination. To evaluate the proliferative and differentiation capacity of Tie2-Cre;R26StopFLBmi1 HSCs in vitro, single CD34-LSK HSCs were clonally sorted into 96-microtiter plates containing 100 µl SF-O3 (Sanko Junyaku) supplemented with 50 µM 2-β-mercaptoethanol, 10% FBS, 1% L-glutamine, penicillin, streptomycin solution (GPS; Sigma), 10 ng/ml mouse SCF, 10 ng/ml human TPO, 10 ng/ml mouse IL-3, and 3 unit/ml human EPO (PeproTech). At day 14 of culture, the colonies were counted and individually collected for morphological examination. To evaluate the tolerance of test cells against oxidative stress, CD34-LSK cells were cultured in the presence of DL-Buthionin-(S,R)-sulfoximine (BSO, Sigma) or N-Acetyl-L-cysteine (NAC, Sigma) for the indicated time periods, then subjected to colony assays or flow cytometric analyses.
Serial Transplantation and CRU Assays
Fresh BM cells (5×105, CD45.2) or 10-day cultured CD34-LSK cells (CD45.2) corresponding to 20 initial CD34-LSK cells were transplanted into 8-week-old recipient mice (CD45.1) irradiated at a dose of 9.5 Gy together with 5×105 and 2×105 BM competitor cells from 8-week-old CD45.1 mice, respectively. For serial transplantation, BM cells were collected from all recipient mice at 12–20 weeks after transplantation and pooled together. Then, 5×106 BM cells were transplanted into 8-week-old B6-CD45.1 mice irradiated at a dose of 9.5 Gy without competitor cells. Third and fourth transplantation were similarly performed using 5×106 pooled BM cells. Peripheral blood (PB) cells of the recipient mice were analyzed with a mixture of antibodies that included PE/Cy7-conjugated anti-CD45.1, pacific blue-conjugated anti-CD45.2, PE-conjugated anti-Mac-1 and anti-Gr-1, APC-conjugated anti-B220, and APC/Cy7-conjugated anti-CD4 and anti-CD8α antibodies. Cells were analyzed on a FACS Canto II. Percent donor chimerism was calculated as (% donor cells) ×100/(% donor cells + % recipient cells). To obtain the competitive repopulating units (CRUs), CRU assays were performed with a limiting number of test cells and the data were analyzed using L-Calc software (StemCell Technologies). Peripheral blood cell counts were made using an automated cell counter, Celltec α (Nihon Kohden).
Apoptosis Analysis
Bulk sorted CD34-LSK cells were incubated in SF-O3 supplemented with 50 µM?2-β-mercaptoethanol, 0.2% BSA, 1% GPS, 50 ng/ml SCF, 50 ng/ml TPO for 10 days at 37°C in a 5% CO2 atmosphere. At day 10 of culture, the cultured cells were incubated with APC-conjugated anti-Annexin V (BD Pharmingen) and propidium iodide at room temperature for 15 min with protection from light. Analysis was performed on FACS Canto II.
Immunostaining of γH2A.X
Cells were incubated in a culture medium drop on slide glasses pre-treated with poly-L-lysine (Sigma) for 2 hours. After fixation with 2% paraformaldehyde and blocking in 4% sheep serum for 30 min at room temperature, cells were incubated with purified anti-phospho-Histone H2A.X (Ser139) antibody (Cell Signaling Technology) for 12 hours at 4°C. The cells were then washed and incubated with Alexa Flour 555-conjugated anti-rabbit IgG goat polyclonal antibody (Invitrogen) for 60 min at room temperature. DNA was counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Images were taken with a Keyence BZ-9000 fluorescence microscope.
RT-PCR
Total RNA was isolated using TRIZOL LS solution or TRIZOL solution (Invitrogen) and reverse transcribed by the ThermoScript RT-PCR system (Invitrogen) with an oligo-dT primer. Real-time quantitative polymerase chain reaction (PCR) was performed with an ABI prism 7300 Thermal Cycler (Applied Biosystems) using FastStart Universal Probe Master (Roche). The combination of primer sequences and probe numbers are as follows: for p16Ink4a, probe #91, 5′-AATCTCCGCGAGGAAAGC-3′, and 5′-GTCTGTCTGCAGCGGACTC-3′; for p19Arf, probe #106, 5′-GGGTTTTCTTGGTGAAGTTCG-3′, 5′- TTGCCCATCATCATCACCT-3′, and for Bmi1, probe #95, 5′-AAACCAGACCACTCCTGAACA-3′ and 5′-TCTTCTTCTCTTCATCTCATTTTTGA-3′.
Western Blotting
Total cell lysate was resolved by SDS-PAGE and transferred to a PVDF membrane. The blots were probed with a mouse anti-Bmi1 (clone 8A9, kindly provided by Dr. N. Nozaki, MAB Institute, Co. Ltd., Japan), and a horseradish peroxidase-conjugated secondary antibody. The protein bands were detected with an enhanced chemiluminescence reagent (SuperSignal, Pierce Biotechnology).
Detection of ROS
Cells were stained with an antibody cocktail consisting of biotinylated anti-Gr-1, Mac-1, IL-7Rα, B220, CD4, CD8α, and Ter119 monoclonal antibodies. Cells were further stained with PE-conjugated anti-Sca-1, and PE/Cy7-conjugated anti-c-Kit antibodies. Biotinylated antibodies were detected with APC/Cy7-conjugated streptavidin. After staining with antibodies, cells were incubated with CellROX™ Deep Red Reagent (5 µM, Invitrogen) at 37°C for 30 min with protection from light. Dead cells were eliminated by staining with propidium iodide (1 µg/ml, Sigma). Analysis was performed on a FACS Aria II.
Supporting Information
Steady state hematopoiesis in Tie2-Cre;R26StopFLBmi1 mice. (A) Hematopoietic analysis of 10-week-old Tie2-Cre and Tie2-Cre;R26StopFLBmi1 mice. Absolute numbers of CMPs, GMPs, MEPs, and CLPs in bilateral femurs and tibiae (upper panels), total spleen cells and LSK cells in the spleen (middle panel), and total thymic cells, CD4+CD8- cells, CD4-CD8+ cells, and CD4+CD8+ cells in the thymus (lower panels) are shown as the mean ± S.D. (Tie2-Cre; n = 8, Tie2-Cre;R26StopFLBmi1; n = 7). (B) Cell cycle status of CD34-LSK cells examined by Pyronin Y incorporation. Proportion of CD34-LSK cells in the G0 phase of the cell cycle (Pyronin Y-) was shown as the mean ± S.D. (n = 4) (left panel). Representative flow cytometric profiles are also depicted (right panel).
(EPS)
Apoptosis and cell cycle status of Tie2-Cre ; R26StopFLBmi1 LSK cells in culture. (A) The proportion of apoptotic cells in the LSK fraction in culture. CD34-LSK cells from Tie2-Cre (Control) and Tie2-Cre;R26StopFLBmi1 (Bmi1) mice were cultured in the SF-O3 serum-free medium supplemented with 50 ng/ml SCF and TPO. At day 10 of culture, apoptotic cells were detected by staining culture cells with anti-Annexin V and propidium iodide (PI). The percentage of Annexin V+PI- apoptotic cells in the LSK fraction is shown as the mean ± S.D. (n = 5). (B) The cell cycle status of LSK cells overexpressing Bmi1. CD34-LSK cells from Tie2-Cre (Control) and Tie2-Cre;R26StopFLBmi1 (Bmi1) mice were cultured in the SF-O3 serum-free medium supplemented with 50 ng/ml SCF and TPO. At day 10 of culture, the cells were incubated with 10 µM BrdU at 37°C for 30 min and then analyzed using a BrdU Flow Kit. Data are shown as the mean ± SD (n = 4).
(EPS)
Hematopoietic recovery in recipients of Tie2-Cre;R26StopFLBmi1 HSCs after irradiation. Fresh BM cells from Tie2-Cre and Tie2-Cre;R26StopFLBmi1 mice (1×107, CD45.2) were transplanted into 8-week-old CD45.1 mice irradiated at a dose of 9.5 Gy without competitor cells. Four months later, the recipient mice were irradiated at a dose of 5 Gy. Changes in the PB cell count were monitored for 4 weeks (A) and the absolute number of BM LSK cells in bilateral femurs and tibiae was examined at 4 weeks post-irradiation (B). Data are shown as the mean ± SD (n = 5).
(EPS)
ROS levels in Tie2-Cre;R26StopFLBmi1 cells in culture. Levels of ROS in cells overexpressing Bmi1 in culture. CD34-LSK cells from Tie2-Cre (Control) and Tie2-Cre;R26StopFLBmi1 (Bmi1) mice were cultured in the SF-O3 serum-free medium supplemented with 50 ng/ml SCF and TPO. Cells from day 11 or 12 of culture were further cultured for 2 days in the presence of 0.2 mM BSO, then levels of ROS in Lin-Sca-1+c-Kit+ cells and Lin-Sca-1low/−c-Kit+ cells were analyzed using CellROXTM Deep Red Reagent. Data are shown as dots and the mean values are indicated by bars (n = 4).
(EPS)
Acknowledgments
We thank Naohito Nozaki for the anti-Bmi1 antibody, George Wendt for critical reading of the manuscript, and Mieko Tanemura for laboratory assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported in part by MEXT KAKENHI and the Global COE Program (Global Center for Education and Research in Immune System Regulation and Treatment), MEXT, Japan, a grant for Core Research for Evolutional Science and Technology (CREST) from the Japan Science and Technology Corporation (JST), a grant from the Tokyo Biochemical Research Foundation, and a grant from Astellas Foundation for Research on Metabolic Disorders. No additional external funding was received for this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 2.Iwama A, Oguro H, Negishi M, Kato Y, Nakauchi H. Epigenetic regulation of hematopoietic stem cell self-renewal by polycomb group genes. Int J Hematol. 2005;81:294–300. doi: 10.1532/IJH97.05011. [DOI] [PubMed] [Google Scholar]
- 3.Konuma T, Oguro H, Iwama A. Role of the polycomb group proteins in hematopoietic stem cells. Dev Growth Differ. 2010;52:505–516. doi: 10.1111/j.1440-169X.2010.01191.x. [DOI] [PubMed] [Google Scholar]
- 4.Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 6.Park IK, Qian D, Kiel M, Becker MW, Pihalja M, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 7.Iwama A, Oguro H, Negishi M, Kato Y, Morita Y, et al. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity. 2004;21:843–851. doi: 10.1016/j.immuni.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Oguro H, Iwama A, Morita Y, Kamijo T, van Lohuizen M, et al. Differential impact of Ink4a and Arf on hematopoietic stem cells and their bone marrow microenvironment in Bmi1-deficient mice. J Exp Med. 2006;203:2247–2253. doi: 10.1084/jem.20052477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oguro H, Yuan J, Ichikawa H, Ikawa T, Yamazaki S, et al. Poised lineage specification in multipotent hematopoietic stem and progenitor cells by the polycomb protein Bmi1. Cell Stem Cell. 2010;6:279–286. doi: 10.1016/j.stem.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Mihara K, Chowdhury M, Nakaju N, Hidani S, Ihara A, et al. Bmi-1 is useful as a novel molecular marker for predicting progression of myelodysplastic syndrome and patient prognosis. Blood. 2006;107:305–308. doi: 10.1182/blood-2005-06-2393. [DOI] [PubMed] [Google Scholar]
- 11.Rizo A, Horton SJ, Olthof S, Dontje B, Ausema A, et al. BMI1 collaborates with BCR-ABL in leukemic transformation of human CD34+ cells. Blood. 2010;116:4621–4630. doi: 10.1182/blood-2010-02-270660. [DOI] [PubMed] [Google Scholar]
- 12.Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, et al. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 13.Takano H, Ema H, Sudo K, Nakauchi H. Asymmetric division and lineage commitment at the level of hematopoietic stem cells: inference from differentiation in daughter cell and granddaughter cell pairs. J Exp Med. 2004;199:295–302. doi: 10.1084/jem.20030929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ema H, Takano H, Sudo K, Nakauchi H. In vitro self-renewal division of hematopoietic stem cells. J Exp Med. 2000;192:1281–1288. doi: 10.1084/jem.192.9.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shima H, Takubo K, Iwasaki H, Yoshihara H, Gomei Y, et al. Reconstitution activity of hypoxic cultured human cord blood CD34-positive cells in NOG mice. Biochem Biophys Res Commun. 2009;378:467–472. doi: 10.1016/j.bbrc.2008.11.056. [DOI] [PubMed] [Google Scholar]
- 16.Ito K, Hirao A, Arai F, Takubo K, Matsuoka S, et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med. 2006;12:446–451. doi: 10.1038/nm1388. [DOI] [PubMed] [Google Scholar]
- 17.Yahata T, Takanashi T, Muguruma Y, Ibrahim AA, Matsuzawa H, et al. Accumulation of oxidative DNA damage restricts the self-renewal capacity of human hematopoietic stem cells. Blood. 2011;118:2941–2950. doi: 10.1182/blood-2011-01-330050. [DOI] [PubMed] [Google Scholar]
- 18.Rossi DJ, Jamieson CH, Weissman IL. Stems cells and the pathways to aging and cancer. Cell. 2008;132:681–696. doi: 10.1016/j.cell.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 19.Chagraoui J, Hébert J, Girard S, Sauvageau G. An anticlastogenic function for the Polycomb Group gene Bmi1. Proc Natl Acad Sci USA. 2011;108:5284–5289. doi: 10.1073/pnas.1014263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ginjala V, Nacerddine K, Kulkarni A, Oza J, Hill SJ, et al. BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol Cell Biol. 2011;31:1972–1982. doi: 10.1128/MCB.00981-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Facchino S, Abdouh M, Chatoo W, Bernier G. BMI1 confers radioresistance to normal and cancerous neural stem cells through recruitment of the DNA damage response machinery. J Neurosci. 2010;30:10096–10111. doi: 10.1523/JNEUROSCI.1634-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shao L, Li H, Pazhanisamy SK, Meng A, Wang Y, et al. Reactive oxygen species and hematopoietic stem cell senescence. Int J Hematol. 2010;94:24–32. doi: 10.1007/s12185-011-0872-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jang YY, Sharkis SJ. A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood. 2007;110:3056–3063. doi: 10.1182/blood-2007-05-087759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, Liu C, Chen J, Song S, Lee IH, et al. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature. 2009;459:387–392. doi: 10.1038/nature08040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rizo A, Olthof S, Han L, Vellenga E, de Haan G, et al. Repression of BMI1 in normal and leukemic human CD34+ cells impairs self-renewal and induces apoptosis. Blood. 2009;114:1498–1505. doi: 10.1182/blood-2009-03-209734. [DOI] [PubMed] [Google Scholar]
- 26.Owusu-Ansah E, Banerjee U. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature. 2009;461:537–541. doi: 10.1038/nature08313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, et al. The polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Negishi M, Saraya A, Mochizuki S, Helin K, Koseki H, et al. A novel zinc finger protein Zfp277 mediates transcriptional repression of the Ink4a/Arf locus through polycomb repressive complex 1. PLoS One. 2010;5:e12373. doi: 10.1371/journal.pone.0012373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamminga LM, Bystrykh LV, de Boer A, Houwer S, Douma J, et al. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood. 2006;107:2170–2179. doi: 10.1182/blood-2005-09-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stapels M, Piper C, Yang T, Li M, Stowell C, et al. Polycomb group proteins as epigenetic mediators of neuroprotection in ischemic tolerance. Sci Signal. 2010;3:ra15. doi: 10.1126/scisignal.2000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 32.Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J, Bi C, Cheong LL, Mahara S, Liu SC, et al. The histone methyltransferase inhibitor, DZNep, up-regulates TXNIP, increases ROS production, and targets leukemia cells in AML. Blood. 2011;118:2830–2839. doi: 10.1182/blood-2010-07-294827. [DOI] [PubMed] [Google Scholar]
- 34.Sasaki Y, Derudder E, Hobeika E, Pelanda R, Reth M, et al. Canonical NF-kappaB activity, dispensable for B cell development, replaces BAFF-receptor signals and promotes B cell proliferation upon activation. Immunity. 2006;24:729–739. doi: 10.1016/j.immuni.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 35.Fukamachi H, Fukuda K, Suzuki M, Furumoto T, Ichinose M, et al. Mesenchymal transcription factor Fkh6 is essential for the development and differentiation of parietal cells. Biochem Biophys Res Commun. 2001;280:1069–1076. doi: 10.1006/bbrc.2001.4247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Steady state hematopoiesis in Tie2-Cre;R26StopFLBmi1 mice. (A) Hematopoietic analysis of 10-week-old Tie2-Cre and Tie2-Cre;R26StopFLBmi1 mice. Absolute numbers of CMPs, GMPs, MEPs, and CLPs in bilateral femurs and tibiae (upper panels), total spleen cells and LSK cells in the spleen (middle panel), and total thymic cells, CD4+CD8- cells, CD4-CD8+ cells, and CD4+CD8+ cells in the thymus (lower panels) are shown as the mean ± S.D. (Tie2-Cre; n = 8, Tie2-Cre;R26StopFLBmi1; n = 7). (B) Cell cycle status of CD34-LSK cells examined by Pyronin Y incorporation. Proportion of CD34-LSK cells in the G0 phase of the cell cycle (Pyronin Y-) was shown as the mean ± S.D. (n = 4) (left panel). Representative flow cytometric profiles are also depicted (right panel).
(EPS)
Apoptosis and cell cycle status of Tie2-Cre ; R26StopFLBmi1 LSK cells in culture. (A) The proportion of apoptotic cells in the LSK fraction in culture. CD34-LSK cells from Tie2-Cre (Control) and Tie2-Cre;R26StopFLBmi1 (Bmi1) mice were cultured in the SF-O3 serum-free medium supplemented with 50 ng/ml SCF and TPO. At day 10 of culture, apoptotic cells were detected by staining culture cells with anti-Annexin V and propidium iodide (PI). The percentage of Annexin V+PI- apoptotic cells in the LSK fraction is shown as the mean ± S.D. (n = 5). (B) The cell cycle status of LSK cells overexpressing Bmi1. CD34-LSK cells from Tie2-Cre (Control) and Tie2-Cre;R26StopFLBmi1 (Bmi1) mice were cultured in the SF-O3 serum-free medium supplemented with 50 ng/ml SCF and TPO. At day 10 of culture, the cells were incubated with 10 µM BrdU at 37°C for 30 min and then analyzed using a BrdU Flow Kit. Data are shown as the mean ± SD (n = 4).
(EPS)
Hematopoietic recovery in recipients of Tie2-Cre;R26StopFLBmi1 HSCs after irradiation. Fresh BM cells from Tie2-Cre and Tie2-Cre;R26StopFLBmi1 mice (1×107, CD45.2) were transplanted into 8-week-old CD45.1 mice irradiated at a dose of 9.5 Gy without competitor cells. Four months later, the recipient mice were irradiated at a dose of 5 Gy. Changes in the PB cell count were monitored for 4 weeks (A) and the absolute number of BM LSK cells in bilateral femurs and tibiae was examined at 4 weeks post-irradiation (B). Data are shown as the mean ± SD (n = 5).
(EPS)
ROS levels in Tie2-Cre;R26StopFLBmi1 cells in culture. Levels of ROS in cells overexpressing Bmi1 in culture. CD34-LSK cells from Tie2-Cre (Control) and Tie2-Cre;R26StopFLBmi1 (Bmi1) mice were cultured in the SF-O3 serum-free medium supplemented with 50 ng/ml SCF and TPO. Cells from day 11 or 12 of culture were further cultured for 2 days in the presence of 0.2 mM BSO, then levels of ROS in Lin-Sca-1+c-Kit+ cells and Lin-Sca-1low/−c-Kit+ cells were analyzed using CellROXTM Deep Red Reagent. Data are shown as dots and the mean values are indicated by bars (n = 4).
(EPS)