Abstract
The present study demonstrates that multiple NoV genotypes belonging to genogroup II contributed to an acute gastroenteritis outbreak at a US military facility in Turkey that was associated with significant negative operational impact. Norovirus (NoV) is an important pathogen associated with acute gastroenteritis among military populations. We describe the genotypes of NoV outbreak occurred at a United States military facility in Turkey. Stool samples were collected from 37 out of 97 patients presenting to the clinic on base with acute gastroenteritis and evaluated for bacterial and viral pathogens. NoV genogroup II (GII) was identified by RT-PCR in 43% (16/37) stool samples. Phylogenetic analysis of a 260 base pair fragment of the NoV capsid gene from ten stool samples indicated the circulation of multiple and rare genotypes of GII NoV during the outbreak. We detected four GII.8 isolates, three GII.15, two GII.9 and a sole GII.10 NoV. Viral sequences could be grouped into four clusters, three of which have not been previously reported in Turkey. The fact that current NoV outbreak was caused by rare genotypes highlights the importance of norovirus strain typing. While NoV genogroup II is recognized as causative agent of outbreak, circulation of current genotypes has been rarely observed in large number of outbreaks.
Introduction
Human noroviruses (NoV) are the most common cause of outbreaks of acute gastroenteritis, worldwide [1]. Annually, NoV have been estimated to be implicated in 21 million cases of acute gastroenteritis in the United States [2], and is the most common cause of diarrhea in adults [3]. One in fourteen people will have a norovirus infection each year, and most people will have suffered several NoV infections within their lifetime [2]. Worldwide, it has been estimated that over 218,000 of pediatric deaths that are attributed to gastrointestinal disease each year from a total of 1.6 million can be directly attributed to NoV illness [3]. Most children will have at least one NoV infection within the first 5 years of life [4]. Sporadic NoV infection has been reported as second to rotavirus as an enteric pathogen associated with gastroenteritis among children [5].
Norovirus is recognized as a major cause of gastroenteritis outbreaks in military populations [6], and other semi-closed or closed institutions such as hospitals, elder care homes, cruise ships, and nurseries [7]. Norovirus strains were a major cause of outbreaks and sporadic cases of gastroenteritis among US ground troops during the Persian Gulf War in 1991, where NoV infection was in one report the most common cause of disability [6] as well as coalition forces in Gulf War II and the Afghanistan campaign beginning in 2002 [8], [9], [10], [11]. As many as 73% of enteric viral gastroenteritis cases among British troops deployed from 2002–2007 were associated with NoV [9]. In 2002, a NoV outbreak among British forces deployed to Afghanistan forced medical evacuations to European hospitals because of severe dehydration and other complications, resulting in closure of the field hospital, leaving the remaining military contingent without full medical services until the outbreak could be characterized [8]. Studies from an array of military units continue to demonstrate that NoV outbreaks have the capacity to impede force readiness and have even caused the stand down of the entire air wing of a battle group [12].
Norovirus belong to the family Caliciviridae and have a single-stranded, positive-sense RNA genome that is genetically highly variable, with five known genogroups [1], three (GI, GII, GIV) of which are associated with human disease [1]. The GII NoV are most commonly associated with human disease, with 19 known genotypes [13]; strains of genotype GII.4 in particular are recognized as a dominant circulating strain. Between 1995 and 2006, genotype GII.4 caused at least four pandemic seasons during which novel GII.4 variants rapidly emerged and displaced previously circulating strains [14]. Because of the dramatic increase in GII.4 NoV outbreaks in the past several years, much of the attention with respect to NoV control has focused on strains of this genotype. However, other NoV strains are always circulating, causing local and regional outbreaks [15].
In May 2009, a significant increase in acute gastroenteritis (AGE) cases was noted in the American health clinic at Incirlik Air Base (IAB) in Adana, Turkey. This increased rate of AGE led to discussions with local Turkish military public health authorities, which confirmed that the Turkish military community and the residents of Adana were also experiencing an anecdotal increase in AGE illnesses [16]. An epidemiologic investigation was launched to attempt to identify the cause and possible source of this AGE outbreak at IAB.
Materials and Methods
Clinical and Epidemiologic Methods
The current investigation describes a gastrointestinal outbreak which occurred on a US military base in Turkey. The main objective was to identify the etiological agent(s) responsible for the outbreak as a public health response to formulate containment strategy and provide data that would support an appropriate intervention control strategy. The project was conducted as an activity not involving human research, therefore IRB review was not required. Participants voluntarily agreed to provide stool samples and complete the questionnaire information.
IAB is located in the village of Incirlik, about six miles from the city of Adana in southeastern Turkey. It is a Turkish military complex, composed of both Turkish and American personnel. The American component has approximately 1250 active duty personnel and 1300 civilians and dependents. The vast majority of U.S. military and their dependents live and work on the base, but travel within and outside Turkey is common for base personnel and their families.
Due to a rapid increased report of AGE in the local community and anecdotal cases reported among US personnel at IAB in May, 2009, a clinic case-based investigation was performed. The goal of this investigation was to identify whether an outbreak was underway and if so, to determine the scope of the outbreak, the etiologic agent and its source, in order to mitigate risk and minimize impact as necessary. Patients presenting with diarrhea were requested to fill in a brief self-completed survey starting on 18 May, 2009. The survey collected demographic information, clinical signs and symptoms, and risk factors. Patients were asked to voluntarily submit a diarrheal stool sample for laboratory analysis. The case-based investigation ended on 18 June, 2009 as gastroenteritis cases dropped to a frequency typical for summer.
To identify the frequency of patients presenting with AGE just prior to the beginning of the presumed outbreak and continuing over time (from May 17–June 27, 2009), data from the clinic’s electronic medical record was utilized by public health officials. This was done through identification of cases with a disease non-battle injury (DNI) code of ‘gastrointestinal, infectious’ which encompasses AGE, to characterize the likely onset and duration of the outbreak.
Microbiological Analysis
Stool samples were collected in clean stool hats and transferred into a sample cup and stored at 4°C until initial testing. An aliquot of each sample was tested locally using an ELISA for NoV (IDEIA Oxoid (Ely) Ltd, Denmark House, Angel Drove, UK). An additional aliquot of each specimen was also tested by EIA for Cryptosporidium parvum, Entamoeba histolytica, and Giardia lamblia (Techlab Inc., Blacksburg, VA, USA). The remaining sample was stored in Cary-Blair transport medium until transferred to the enteric disease research laboratory at US Naval Medical Research Unit No.3, Cairo, Egypt. In Cairo, standard bacterial culture investigation was performed to detect Campylobacter spp., Salmonella spp., Shigella spp., and Vibrio spp. Bacterial isolates were identified and confirmed using standard biochemical methods, and antibiotic sensitivity was determined via disk-diffusion (Becton Dickinson, Cockeysville, MD, USA), and/or E-tests (AB- Biodisk, Sölna, Sweden) as guided by the Clinical Laboratory Standards Institute (CLSI) [17]. EIA was also performed to test for astrovirus, rotavirus, adenovirus, and repeated for NoV (IDEIA, Oxoid, UK) according to the manufacturer’s instructions. DNA was extracted from Escherichia coli-like colonies grown on MacConkey-lactose medium and subsequently tested for the enterotoxin genes eltB and estA [18].
RNA Extraction, RT-PCR and cDNA Sequencing
Viral RNA was extracted from 140 µl of a 10% fecal suspension in vertrel/water solution using a QIAamp Viral RNA Mini Kit (QIAGEN, Valencia, CA, USA) according to the manufacturer’s protocol. Identification of NoV was performed by RT-PCR as previously described [19]. Genogroup II NoV was identified using primers targeting GII-specific regions in the 5′ capsid gene as described [20]. PCR amplicons from NoV-positive specimens were sequenced using the ABI Prism Big Dye Terminator cycle sequencing kit and an ABI Prism 3100 DNA sequence (Applied Biosystems, Foster City, CA, USA). Sequences were aligned against NoV capsid sequences obtained from Genbank using BioEdit software, version 5.1. Phylogenetic analysis was carried out using MEGA 5.0 [21] and a dendrogram was constructed using the neighbor-joining method with genetic distance modeled using the Kimura 2-parameter model [22]. Statistical confidence for the phylogenetic branches was assessed by bootstrap analysis (2,000 replicates) [23]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree [24].
Results
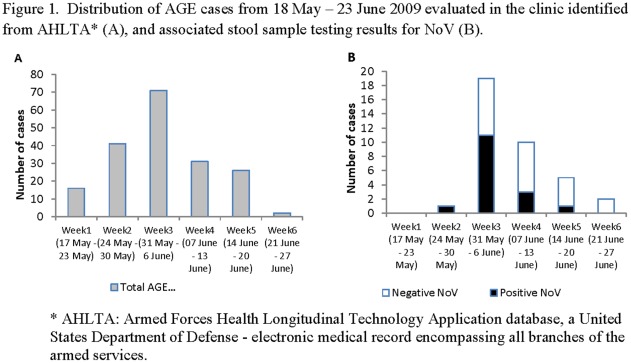
From the electronic medical record review from 18 May to 19 Jun 2009, 187 patients presented to the IAB clinic with AGE. The peak incidence of AGE cases was during the week of 31 May –06 June with a total of 71 patients seeking medical care at the clinic (Figure 1), with the outbreak occurring from 26 May to 15 June, 2009. Of the 187, 82 patients completed the case survey, 79% reported diarrhea, 46% reported vomiting, and 29% reported fever (Table 1). The median number of days between symptom onset and clinic visit was 2 days (interquartile range 1, 5). During the 7 days prior to symptoms, 73% of respondents reported travelling off base, 56% reported eating off base, and 24% reported using an outdoor pool.
Figure 1. Distribution of AGE cases from 18 May –23 June 2009 evaluated in the clinic identified from AHLTA* (A), and associated stool sample testing results for NoV (B).
Table 1. Characteristics and activities of gastroenteritis cases reported by case-based surveillance survey responders.
| Characteristics | Total; n = 82 |
| Age: Median (IQR) | 27(23–35) |
| Gender: Male, n (%) | 51(62) |
| Symptoms, n (%) | |
| Watery diarrhea | 65(79) |
| Nausea | 52(63) |
| Vomiting | 38(46) |
| Fever | 24(29) |
| Headache | 42(51) |
| Chills | 31(38) |
| Stomach and/or abdominal pain | 63(77) |
| Base facilities, n (%) | |
| Consumption of vegetables, fruits, meat, milk off base | 46(56) |
| Consumption of bottled water | 56(68) |
| Travel off base | 60(73) |
| Self-report of an ill family member | 22(27) |
| Outdoor Pool | 20(24) |
Stool samples were collected from 37 patients; only 22 of these patients completed the survey questionnaire. Of the 22 stool samples collected along with a completed survey form, 9 were positive for NoV and we could identify their clinical characteristics. Of the NoV positive cases, 44% (4/9) suffered from nausea, 78% (7/9) had watery diarrhea and 67% (6/9) experienced vomiting. No significant difference was noticed between cases positive for NoV as opposed to negative cases in regard to consumption of bottled water, eating and travelling off base, pool usage and presence of a family member with a similar symptom.
Norovirus genogroup II was detected in 43% (16/37) stool samples by RT-PCR. Three of the 16 NoV positive cases were identified with a second enteropathogen: C. jejuni, C. parvum, or ETEC whereas enteropathogens were recovered from 59% (22/37) of remaining cases (Table 2). With regards to identification of other sole-pathogen infections, 5% (2/37) were positive for Salmonella group B, 3% (1/37) were positive for ETEC; 3% (1/37) were positive for Cryptosporidium, and 5% (2/37) were positive by EIA for rotavirus; 41% (15/37) of the samples were negative for all pathogens tested (Table 2).
Table 2. Distribution of pathogens identified from stool samples (n = 37).
| Pathogen detected (Method) | Sole, n (%) | Mixed, n (%) |
| Cryptosporidium (EIA) | 1(3) | 1(3) |
| ETEC (PCR) | 2(5) | 1(3) |
| Norovirus (PCR) | 13(35) | 3(8) |
| Rotavirus (EIA) | 2(6) | 0(0) |
| Campylobacter (culture) | 0(0) | 1(3) |
| Salmonella (culture) | 2(5) | 0(0) |
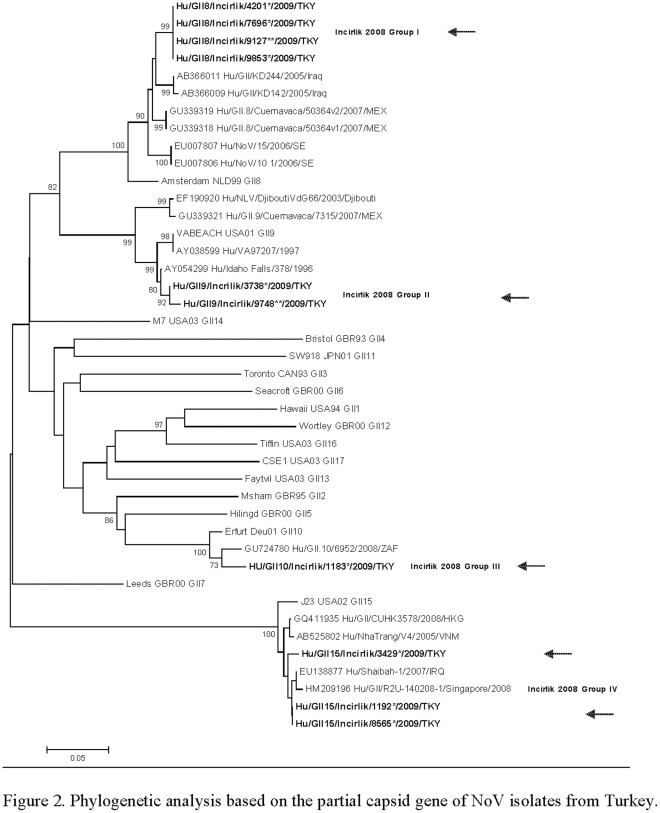
We were able to amplify a fragment of the gene encoding NoV capsid protein from 10 of the 16 NoV-positive stool samples; a phylogenetic analysis based on sequence comparison of these products indicated that multiple and rare genotypes of GII NoV were circulating during the outbreak (Figure 2). Four indistinguishable strains (Group I; accession number: JF895615, JF89616, JF973385, JF895613) could be typed as GII.8. A second group of two strains (Group II; accession number: JF895614, JF973386) could be typed as GII.9 and had 99% nucleotide sequence similarity with prototype strain Idaho Falls (accession number AY054299). A third group included one strain that could be typed as GII.10 (Group III; accession number JF895609). Yet another three strains belonged to genotype II.15 (Group IV; accession number: JF895612, JF895610, JF8956111) which had 98–99% nucleotide sequence similarity with other GII.15 strains in the GenBank database.
Figure 2. The evolutionary history was inferred using the Neighbor-Joining method.
The optimal tree with the sum of branch length = 2.38 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (2000 replicates) are shown next to the branches. Boot strap values below 70% were considered insignificant and are not shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura 2-parameter method and are in the units of the number of base substitutions per site. The analysis involved 42 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated. There were a total of 260 positions in the final dataset. Bold sequences represent strains from current study.
Discussion
This report summarizes the results of an AGE outbreak investigation at a US military facility in Turkey that was associated with significant negative operational impact, degrading mission readiness with nearly 20% of the American population in a one month period affected. Initiation of a clinic case-based investigation yielded 37 stool samples in which NoV was detected in 43% (16/37) of the samples with 81% (13/16) of the positive NoV samples identified without a copathogen.
DNA sequencing data demonstrated that several relatively rare genotypes of NoV contributed to this outbreak. Among the 16 NoV positive samples identified, 4 different genotypes were identified. Two of the NoV strains (Shaibah-like, and KD244-like) were previously reported in Iraq and only from deployed troops, while the other two genotypes were reported in South Africa and in the US [25], [26]. In Turkey little systematic data on circulating NoV genotypes exist. However, GIIb/GII.4 strains have been frequently identified in Turkish children with gastroenteritis; strains belonging to this genotype have been found in Europe and mainly in children [27]. Our phylogenetic analysis, although based only on partial sequences from the capsid gene, showed unique strains not previously reported in Turkey. The capsid region of the NoV genome demonstrates great genetic diversity with 20% amino acid differences across capsid sequences [28]. The gene encoding the capsid region is widely used for classification of NoV and provides a reliable marker for genotyping NoV [29]. Homologous recombination, in particular at the junction between ORF1 and ORF2, but also within ORF2 contributes significantly to this high degree of variation [30]. Full capsid sequencing analyses is the gold standard for genotyping of NoV strains, however, for clinical samples with low copy numbers, amplifying and sequence analysis of partial capsid sequences is more practical and is only slightly less discriminatory than full capsid sequencing [31]. In addition, full capsid sequencing has been used to distinguish differences between closely related GII.4 variants [32]; this genotype was not the predominant NoV in our study and partial capsid sequencing sufficiently demonstrated viral relationships.
Previous reports from British troops deployed to Iraq indicated that two NoV strains were responsible for cases of gastroenteritis. Similar mixed NoV outbreaks have been previously observed and are often attributed to systematic failure of cooking/cleaning/drinking water supplies [33]. One limitation of this investigation was that the survey was not used to capture data from a control group (persons without recent AGE); thus we were unable to perform a risk factor analysis. Another limitation was the lack of of environmental samples that could be tested for NoV in order to track the source of outbreak.
To our knowledge a formal outbreak investigation in the Turkish population was never performed. With the reported lack of sensitivity of commercial norovirus EIAs [34], it is important to use molecular diagnostics such as real-time RT-PCR for detection of NoV [35], [36]. Norovirus strain typing of samples collected during outbreaks of AGE in the military is infrequently done. However, with NoV being the most important cause of foodborne illness in the US [2], a full epidemiologic investigation of AGE illness in the military including strain typing of stool samples may be able to limit the magnitude of the outbreak, link unrelated cases to a single contaminated food or water source, and better identify future risks of NoV illness in the military. As a part of the efforts to reduce the risk of outbreaks of NoV gastroenteritis, the US Department of Defense research community is working with US Centers for Disease Control and Prevention to join the electronic NoV outbreak surveillance network CaliciNet [36]. This NoV surveillance network provides centralized reporting of causative NoV strains in the US, epidemiologic information such as likely transmission route, and viral typing information. By combining this existing information new observations, such as seasonality of outbreaks or patterns of emergence of new variants or frequency of food- and waterborne exposure, can be more broadly studied.
NoVs are an important cause of both outbreaks and sporadic disease among military service members. Although usually self-limited, NoV has been associated with significant operational impact. The global significance of the emergence of NoV in recent years as well as its association with military operations, and its divergent molecular epidemiology highlight the need to perform surveillance of circulating NoV infections to characterize and detect evolving strains. This is particularly important now that efforts are underway to develop a NoV vaccine.
Acknowledgments
We are grateful to Col Johnson and Lt Col Green for their support with the outbreak investigation. We would also like to thank MSgt Christopher Duffy, TSgt Angelita Pfeiffer, SSgt Tiffany (Woods) Mitchell and SSgt Christa (Lewis) Premo for their technical assistance in the laboratory at IAB. The laboratory support of Ms.Mireille Kamel is gratefully acknowledged.
We are indebted to Dr J.Vinjé for his review and critique of this manuscript.
Author’s disclaimer. The views expressed in this article are those of the author and do not necessarily reflect the official policy or the opinions of the Department of the Navy, Centers for Disease Control and Prevention.
Copyright Statement. I am an employee of the U.S. Government. This work was prepared as part of my official duties. Title 17 U.S.C. §105 provides that ‘Copyright protection under this title is not available for any work of the United States Government.’ Title 17 U.S.C. §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person’s official duties.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Military Infectious Disease Research Program (MIDRP) proposal D0105_09_N3 - The Epidemiology and Management of Diarrhea Among Deployed US Forces to CENTCOM and EUCOM theaters of operation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Patel MM, Hall AJ, Vinje J, Parashar UD. Norovirus: a comprehensive review. J Clin Virol. 2009;44:1–8. doi: 10.1016/j.jcv.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 2.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, et al. Foodborne illness acquired in the United States–major pathogens. Emerg Infect. 2011;Dis17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koo HL, Ajami N, Atmar RL, DuPont HL. Noroviruses: The leading cause of gastroenteritis worldwide. Discov. 2010;Med10:61–70. [PMC free article] [PubMed] [Google Scholar]
- 4.Lopman B, Armstrong B, Atchison C, Gray JJ. Host, weather and virological factors drive norovirus epidemiology: time-series analysis of laboratory surveillance data in England and Wales. PLoS One. 2009;4:e6671. doi: 10.1371/journal.pone.0006671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel MM, Widdowson MA, Glass RI, Akazawa K, Vinjé J, et al. Parashar UD. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg Infect Dis. 2008;14:1224–31. doi: 10.3201/eid1408.071114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCarthy M, Estes MK, Hyams KC. Norwalk-like virus infection in military forces: epidemic potential, sporadic disease, and the future direction of prevention and control efforts. Systematic literature review of role of Noroviruses in sporadic gastroenteritis. J Infect. 2000;is181:S387–91. doi: 10.1086/315582. [DOI] [PubMed] [Google Scholar]
- 7.Gallimore CI, Richards AF, Gray JJ. Molecular diversity of noroviruses associated with outbreaks on cruise ships: comparison with strains circulating within the UK. Commun Dis Public Health. 2003;6(4):285–93. [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention (CDC) Outbreak of acute gastroenteritis associated with Norwalk-like viruses among British military personnel Afghanistan, May 2002 MMWR Morb Mortal Wkly Rep. 2002;51:477–9. [PubMed] [Google Scholar]
- 9.Bailey MS, Gallimore CI, Lines LD, Green AD, Lopman BA, et al. Viral gastroenteritis outbreaks in deployed British troops during 2002–7. J R Army Med Corps. 2008;154:156–9. doi: 10.1136/jramc-154-03-04. [DOI] [PubMed] [Google Scholar]
- 10.Goodgame R. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Curr Infect Dis Rep. 2007;9:102–9. [Google Scholar]
- 11.Thornton SA, Sherman SS, Farkas T, Zhong W, Torres P, et al. Gastroenteritis in US Marines during Operation Iraqi Freedom. Clin Infect Dis. 2005;40:519–25. doi: 10.1086/427501. [DOI] [PubMed] [Google Scholar]
- 12.Matson DO. Norovirus gastroenteritis in US marines in Iraq. Clin Infect Dis. 2005;40:526–7. doi: 10.1086/427508. [DOI] [PubMed] [Google Scholar]
- 13.Siebenga JJ, Vennema H, Zheng DP, Vinjé J, Lee BE, et al. Norovirus illness is a global problem: emergence and spread of Norovirus GII.4 variants, 2001–2007. J Infect Dis. 2009;200:802–12. doi: 10.1086/605127. [DOI] [PubMed] [Google Scholar]
- 14.Siebenga JJ, Vennema H, Renckens B, de Bruin E, van der Veer B, et al. Epochal evolution of GGII.4 Norovirus capsid proteins from 1995 to 2006. J Virol. 2007;9:932–41. doi: 10.1128/JVI.00674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroneman A, Verhoef L, Harris J, Vennema H, Duizer E, et al. Analysis of integrated virological and epidemiological reports of Norovirus outbreaks collected within the Foodborne viruses in Europe network from 1 Jul 2001 to 30 June 2006. J Clin Microbiol. 2008;46:2959–65. doi: 10.1128/JCM.00499-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Real-World Spring 2009 Disease outbreak After Action report–Incirlik AB, Turkey.
- 17.CLSI . Performance standards for antimicrobial susceptibility testing, 19 information supplement. M100-S19.9 (3). Wayne, Pennsylvania (USA); 2009. Clinical Laboratory Standars Institute. [Google Scholar]
- 18.Nada RA, Shaheen HI, Touni I, Fahmy D, Armstrong AW, et al. Design and validation of a multiplex polymerase chain reaction for the identification of enterotoxigenic Escherichia coli and associated colonization factor antigens. Diag Microbiol Infect Dis. 2010;67:134–42. doi: 10.1016/j.diagmicrobio.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 19.Kojima S, Kageyama T, Fukushi S, Hoshino FB, Shinohara M, et al. Genogroup-specific PCR primers for detection of Norwalk-like viruses. Journal of Virological Methods. 2002;100:107–114. doi: 10.1016/s0166-0934(01)00404-9. [DOI] [PubMed] [Google Scholar]
- 20.KageyamaT, Shinohara M, Uchida K, Fukushi S, Hoshino FB, et al. Coexistence of multiple genotypes, including newly identified genotypes, in outbreaks of gastroenteritis due to Norovirus in Japan. J Clin Microbiol. 2004;42:2988–95. doi: 10.1128/JCM.42.7.2988-2995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–39. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 23.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 24.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–91. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 25.Mans J, de Villiers JC, du Plessis NM, Avenant T, Taylor MB. Emerging norovirus GII.4 2008 variant detected in hospitalised paediatric patients in South Africa. J Clin Virol. 2010;49(4):258–64. doi: 10.1016/j.jcv.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 26.Fankhauser RL, Noel JS, Monroe SS, Ando T, Glass RI. Molecular epidemiology of "Norwalk-like viruses" in outbreaks of gastroenteritis in the United States. J Infect Dis. 1998;178(6):1571–8. doi: 10.1086/314525. [DOI] [PubMed] [Google Scholar]
- 27.Altindis M, Bányai K, Kalayci R, Gulamber C, Koken R, et al. Frequency of norovirus in stool samples from hospitalized children due to acute gastroenteritis in Anatolia, Turkey, 2006–2007. Scand J Infect Dis. 2009;41:685–8. doi: 10.1080/00365540903071342. [DOI] [PubMed] [Google Scholar]
- 28.Vinjé J, Green J, Lewis DC, Gallimore CI, Brown DW, et al. Genetic polymorphism across regions of the three open reading frames of "Norwalk-like viruses". Arch Virol. 2000;145(2):223–41. doi: 10.1007/s007050050020. [DOI] [PubMed] [Google Scholar]
- 29.Gallimore CI, Iturriza-Gomara M, Xerry J, Adigwe J, Gray JJ. Inter-seasonal diversity of norovirus genotypes: emergence and selection of virus variants. Arch Virol. 2007;52(7):1295–303. doi: 10.1007/s00705-007-0954-9. [DOI] [PubMed] [Google Scholar]
- 30.Allen DJ, Gray JJ, Gallimore CI, Xerry J, Iturriza-Gómara M. Analysis of amino acid variation in the P2 domain of the GII-4 norovirus VP1 protein reveals putative variant-specific epitopes. PLoS One. 23; 2008;3(1):e1485. doi: 10.1371/journal.pone.0001485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vinjé J, Hamidjaja RA, Sobsey MD. Development and application of a capsid VP1 (region D) based reverse transcription PCR assay for genotyping of genogroup I and II noroviruses. J Virol Methods. 2004;116(2):109–17. doi: 10.1016/j.jviromet.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 32.MMWR Recomm Rep. Updated norovirus outbreak management and disease prevention guidelines. 2011;60(RR-3):1–18. [PubMed] [Google Scholar]
- 33.O'Reilly CE, Bowen AB, Perez NE, Sarisky JP, Shepherd CA, et al. A waterborne outbreak of gastroenteritis with multiple etiologies among resort island visitors and residents: Ohio, 2004. Clin Infect Dis. 2007;44:506–12. doi: 10.1086/511043. [DOI] [PubMed] [Google Scholar]
- 34.Costantini V, Grenz L, Fritzinger A, Lewis D, Biggs C, et al. Diagnostic accuracy and analytical sensitivity of IDEIA Norovirus assay for routine screening of human norovirus. J Clin Microbiol. 2010;48(8):2770–8. doi: 10.1128/JCM.00654-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mattison K, Grudeski E, Auk B, Charest H, Drews SJ, et al. Multicenter comparison of two norovirus ORF2-based genotyping protocols. J Clin Microbiol. 2009;47(12):3927–32. doi: 10.1128/JCM.00497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Everardo Vega, Jan Vinjé. Novel GII.12 Norovirus Strain, United States, 2009–2010. Emerg Infect Dis. 2011;17(8):1516–8. doi: 10.3201/eid1708.110025. [DOI] [PMC free article] [PubMed] [Google Scholar]