Abstract
Much effort has focused recently on determining the mechanisms that control the allele-specific expression of genes subject to genomic imprinting, yet imprinting regulation is only one aspect of configuring appropriate expression of these genes. Imprinting control mechanisms must interact with those regulating the tissue-specific expression pattern of each imprinted gene in a cluster. Proper expression of the imprinted Delta-like 1 (Dlk1) - Maternally expressed gene 3 (Meg3) gene pair is required for normal fetal development in mammals, yet the mechanisms that control tissue-specific expression of these genes are unknown. We have used a combination of in vivo and in vitro expression assays to localize cis-regulatory elements that may regulate Dlk1 expression in the mouse embryo. A bacterial artificial chromosome transgene encompassing the Dlk1 gene and 77 kb of flanking sequence conferred expression in most endogenous Dlk1-expressing tissues. In combination with previous transgenic data, these experiments localize the majority of Dlk1 cis-regulatory elements to a 41 kb region upstream of the gene. Cross-species sequence conservation was used to further define potential regulatory elements, several of which functioned as enhancers in a luciferase expression assay. Two of these elements were able to drive expression of a lacZ reporter transgene in Dlk1-expressing tissues in the mouse embryo. The sequence proximal to Dlk1 therefore contains at least two discrete regions that may regulate tissue-specificity of Dlk1 expression.
Introduction
Multicellular organisms generate an enormous diversity of distinct cell types during embryonic development by the spatial and temporal regulation of gene expression. Much of this regulation occurs at the level of transcription, where positive and negative cis-regulatory elements are employed to achieve fine control over expression. Enhancers and silencers are DNA elements composed of binding sites for transcription factors that interact in cis with promoter-bound proteins to control gene expression, functioning to allow or prevent, respectively, interaction with the promoter-proximal transcriptional machinery. Many developmentally important genes are also regulated by genomic imprinting, the parental-specific expression of the two alleles of an autosomal gene [1]. Imprinted gene expression is controlled on multiple levels by DNA CpG methylation, histone protein modification, higher order chromatin packaging, and likely additional unknown processes. These imprinting regulatory mechanisms are believed to specify the on or off state of each allele of an imprinted gene, with the pattern and levels of expression of that gene determined by tissue-specific enhancers. The imprinting regulatory machinery must therefore coordinate with the transcriptional regulatory machinery, and understanding the control of imprinted gene expression requires full knowledge of both components.
The imprinted Dlk1-Meg3 locus on mouse chromosome 12 (human 14q32) contains maternally- and paternally-expressed genes, of which Dlk1 and Dio3 are the only members to produce unique protein products [2]–[4]. Dlk1 encodes a transmembrane protein containing six epidermal growth factor-like motifs with similarity to the Delta-Notch-Serrate family of signaling molecules [5], [6]. The full length Dlk1 protein is cleaved to produce membrane-bound and secreted/soluble forms of the protein by the tumor necrosis factor alpha converting enzyme (TACE) [7]. Conflicting data exist as to the roles of the membrane-bound and soluble forms of the Dlk1 protein, which appear to play complex and perhaps complementary roles in multiple differentiating cell types [6], [8], [9], [10]. Several studies have shown that Dlk1 can act as a Notch antagonist, a role consistent with its lack of the conserved DSL interaction domain found in other Notch ligands [11]–[14]. In cultured cells, soluble Dlk1 has been shown to inhibit the ability of Notch to transactivate target genes such as Hes1 [12], [13]. Additionally, Dlk1 appears to activate the insulin/IGF-I signaling pathway, leading to activation of the extracellular regulated kinase/mitogen-activated protein kinase (ERK/MAPK), but may also signal through non ERK-dependent mechanisms [15]–[17].
Dlk1-null mice display pre- and postnatal growth retardation, partially penetrant neonatal lethality, malformation of the ribs and sternum and altered lipid metabolism [18]. Overexpression of Dlk1 is also deleterious, as animals expressing a two-fold excess of Dlk1 showed overgrowth during embryogenesis and postnatal lethality possibly due to feeding defects [19]. Animals expressing a three-fold Dlk1 excess died during late gestation with severe edema, skeletal defects and underdeveloped lungs. These opposing animal models bear many similarities to the phenotype of human patients with either maternal or paternal uniparental disomy for chromosome 14, which results in a loss or increase of Dlk1 expression, respectively [20].
Functionally, Dlk1 is best understood as a regulator of the transition of preadipocyte cells to mature adipocytes, and it is downregulated upon the onset of differentiation in cultured preadipocyte cells and in differentiating mesenchymal stem cells [6], [21]–[23]. Dlk1 also plays a role in skeletal muscle development and regeneration, and mice with a myoblast-specific deletion of Dlk1 are smaller than normal, with decreased skeletal muscle mass [24]. These animals also show impaired regeneration of adult muscle after injury, when satellite cells must be stimulated first to proliferate and then to differentiate into new and engrafting myoblasts.
Dlk1 and Meg3 display complex patterns of tissue- and temporal-specific regulation. Dlk1 is expressed in the placenta and many embryonic tissues, and expression is generally concordant between mouse and human. During development, Dlk1 is expressed in fetal hepatocytes, lung epithelium, adrenal cortex, the chondroblasts of the developing skeleton, developing skeletal myoblasts, the ventral diencephalon and Rathke’s pouch, peripheral nerves prior to myelination, bone marrow and the thymic epithelium [25]–[31]. In the pancreas, Dlk1 is initially expressed in most cells of the pancreatic rudiment, becoming restricted first to the differentiating endocrine cells, and finally to the beta cells alone [32]. In the extraembryonic tissues of the mouse, Dlk1 is expressed in the fetal endothelium of the yolk sac and the placental labyrinth [27], [33]. Dlk1 expression declines after birth, and in the adult becomes restricted to the pancreatic beta cells, the pituitary somatotrophs, the bone marrow, and the zona glomerulosa of the adrenal gland [25], [28], [31], [34], [35]. Previous groups have identified general positive and negative transcriptional elements within the Dlk1 promoter-proximal region, but nothing is known about the distal elements that govern the complex tissue- and temporal-specific expression pattern of this gene [36]–[39].
Closely linked to Dlk1 is the maternally expressed Meg3 gene, which produces a long noncoding RNA that may function as a developmental regulator and/or tumor suppressor [2], [3], [40]–[42]. Meg3 is expressed in the preimplantation mouse embryo, and as differentiation proceeds it is found in the developing pancreas, the paraxial mesoderm and the resulting myoblasts, the ventral diencephalon and Rathke’s pouch, and in the dorsal root ganglia, spinal cord and forebrain [43], [44]. In mouse extraembryonic tissue, Meg3 is expressed in the yolk sac, the labyrinthine trophoblast, and some labyrinth endothelial cells [33], [43]. Comparison of Dlk1 and Meg3 expression shows a complex pattern of both shared and discrete cell type-specific expression. Both genes are strongly expressed in the myotome and migrating myoblasts, for example, and both are down-regulated in skeletal muscle postnatally [27], [33]. In the pituitary, however, Dlk1 and Meg3 show coordinate expression in the infundibulum and Rathke’s pouch during development, but Dlk1 subsequently becomes restricted to somatotrope cells, while Meg3 is expressed only in gonadotropes [27], [33].
Bacterial artificial chromosome (BAC) transgenesis in the mouse is often used to localize regulatory elements within the sequences surrounding a genomic locus [45]. Large BAC transgenes can recapitulate genomic imprinting as well as expression, making them ideal for studying imprinted gene regulation. Previous work from our laboratory showed that the Meg3 gene was expressed and properly imprinted from the 28G5 BAC transgene that spans the region from 3.5 kb upstream of Dlk1 to 69 kb downstream of Meg3, however, Dlk1 was not expressed from this transgene [46]. Another group subsequently showed that Dlk1 is expressed in a subset of its normal pattern from a BAC transgene carrying 49 kb of sequence upstream of Dlk1 (TgDlk1−70), but not from a BAC transgene carrying only 8 kb of upstream sequence (TgDlk1−31) [19]. In the current study we use additional transgenic and in vitro approaches to identify discrete regions of sequence proximal to Dlk1 that contain cis-regulatory elements capable of directing Dlk1-like expression in the mouse embryo.
Materials and Methods
Generation of 127H5 BAC Transgenic Mice
The 127H5 BAC clone was isolated from the CITB 129/Sv mouse genomic library (Children’s Hospital Oakland Research Institute). The clone was end-sequenced and found to span 84 kb, from 41 kb upstream to 36 kb downstream of the Dlk1 gene. The BAC clone was linearized at a unique ClaI site 10.5 kb upstream of Dlk1, and injected into fertilized FVB/N embryos as described previously [46]. The resulting offspring were genotyped by PCR using the primers OL477, 5′-GCTTGAGTATTCTATAGTGTCA-3′, and OL478, 5′-CAACGCAATTAATGTGAGTTAG-3′, which amplify a 170 bp fragment of the BAC vector. Founder animals were bred to wild type FVB/N mice to establish independent transgenic lines, and transgene copy number was calculated by Southern blotting as described [46].
Expression and Imprinting Analysis
Dlk1 expression was analyzed by Northern blotting, and imprinting analysis was performed using RT-PCR and direct sequencing as described [2]. Embryos and placentae were recovered between e12.5 and e14.5 and adult tissues were obtained from 3–4 week old animals. Total RNA was purified using LiCl-urea precipitation [47]. For Northern analysis, 10 µg of total RNA was separated on 1% formaldehyde agarose gels and transferred to Hybond N+ membranes. Membranes were hybridized with Dlk1 and β-actin probes; the Dlk1 probe is a 735 bp fragment corresponding to nucleotides 685 to 1420, and the β-actin probe is a 1200 bp fragment spanning exons 3–7. Hybridization was carried out using Express-Hyb (Clontech) overnight at 65 oC, and the membrane was washed three times for 15 min at 50°C with Northern wash I (2×SSC and 0.05% SDS), and twice for 15 min at 50°C with Northern wash II (0.1×SSC and 0.1% SDS). The membrane was exposed to a phosphorimager screen or film for analysis and Dlk1 signal intensity was normalized to β-actin expression. A student’s two-tailed t-test was used to determine statistical significance. For RT-PCR, 2 µg total RNA was reverse transcribed using Superscript III (Invitrogen) and oligo-dT, RT reactions were diluted 1∶10 and 2 µl was used for all PCR analyses.
Identification and Cloning of Conserved Elements Upstream of Dlk1
Sequence conservation was analyzed using the University of California Santa Cruz Genome Browser (http://genome.ucsc.edu/) to compare the genomes of Mus musculus (July 2007 assembly), Rattus norvegicus (Nov. 2004), Homo sapiens (Feb. 2009), Pan troglodytes (Oct. 2010), Macaca mulatta (Jan. 2006), Pongo abelii (July 2007), Callithrix jacchus (Mar. 2009), Otolemur garnettii (Dec. 2006), Canis familiaris (May 2005), Felis catus (Dec. 2008), Equus caballus (Sept. 2007), Bos taurus (Oct. 2007), Dasypus novemcinctus (May 2005) and Loxodonta africana (July 2009). Vista browser (http://genome.lbl.gov/vista) was also used to align Mus musculus (July 2007) and Homo sapiens (Feb. 2009) sequences downloaded from Ensembl (http://ensenmbl.org). Nine elements upstream of Dlk1 (CE1–CE9) were selected that contained ≥50 bp stretches of DNA with ≥70% sequence homology. The Dlk1 promoter [36], [37], [48] was isolated as a 256 bp XhoI-RsrII restriction fragment and cloned into the XhoI site of the pGL3-B plasmid (Promega), generating plasmid pGL3-Dlk1P. The conserved elements were amplified by PCR from 127H5 DNA using primers listed in Table 1; PCR conditions for all primers were 94°C, 30 sec, 60°C, 1 min and 72°C, 1 min, for 35 cycles. The PCR products were cloned into pCRII-TOPO, then excised using EcoRI, blunt ended and inserted into the SmaI site of pGL3-Dlk1P. These plasmids were termed pGL3-CE, where CE stands for the conserved element.
Table 1. Primers used to amplify Dlk1 upstream conserved elements.
| Conserved element | Primer name | Primer sequence |
| CE1 | OL1152 | 5′-TTAAGGAGGAGAGCCACTCACTGT-3′ |
| OL1153 | 5′-TCAGGCAAAGGCCAGAGACAGAAA-3′ | |
| CE2 | OL1154 | 5′-CTGTTGCGATGTAACTAGGTGGGA-3′ |
| OL1155 | 5′-AAAGGCCAAGGACACCAGATCAGA-3′ | |
| CE3 | OL1156 | 5′-TGGTTTGCCCTGCCTTCCTAGTAT-3′ |
| OL1157 | 5′-ATCTGAAGGGTGCCACAACTGTCT-3′ | |
| CE4 | OL1158 | 5′-GCAGGGCTTCGTTCTTTCCATGTT-3′ |
| OL1159 | 5′-TCCATACATGGCCGGATGTGGTTA-3′ | |
| CE5 | OL1101 | 5′-TCATCACCCAGCAAGAAGACAGGT-3′ |
| OL1102 | 5′-TCTGCTCAACCAGCCTAGCTTACT-3′ | |
| CE6 | OL1090 | 5′-TAAGGCACTACAGCAAGGAAGCCA-3′ |
| OL1100 | 5′-TCCTGGGCATCAAACATGACCACT-3′ | |
| CE7 | OL1093 | 5′-ACCTGAGGACGCCATTTGACCATA-3′ |
| OL1094 | 5′-GCTCGCCAGCCAGAAGTAGAATTT-3′ | |
| CE8 | OL1095 | 5′-GCTCTGTGTGCAATCTGCTTTCCA-3′ |
| OL1096 | 5′-CAATGCCTGCAGCTTACCACACTT-3′ | |
| CE9 | OL1097 | 5′-CCACACAACCTTCACCCAACCATT-3′ |
| OL1098 | 5′-TTACTGGCTAGGCTCACAGAGCAA-3′ |
Cell Culture and Transfection
The C2C12 cells were maintained in high glucose Dulbecco’s Modified Eagle Medium (DMEM) (Invitrogen) with 10% fetal bovine serum (FBS) and 4 mM L-glutamine. NIH-3T3 cells were maintained in high glucose DMEM with 10% calf serum and 4 mM L-glutamine. SVR cells were maintained in high-glucose DMEM with 5% FBS and 4 mM L-glutamine. Y-1 cells were maintained in F-12K medium (Invitrogen) with 15% horse serum, 2.5% FBS and 2 mM L-glutamine. All cell lines were obtained from ATCC. All media contained 100 µg/ml penicillin/streptomycin and the cells were maintained in a humidified incubator at 37°C with 5% CO2.
For transfection, C2C12, NIH-3T3 and SVR cells were plated in 24-well plates at a density of 50,000 cells/well; Y-1 cells were plated at a density of 150,000 cells/well. Cell lines were transfected with 200 µl of serum-free media per well containing Transfast reagent at a 1∶1 ratio, 0.7 µg of the appropriate pGL3 DNA, and 0.3 µg of the pSV-βgal control vector. Growth media was replaced after 1 hour, and the cells were lysed 48 hours post-transfection with 150 µl of 1× Glo Lysis Buffer (Promega). Luciferase production was assayed using a Clarity Microplate Luminometer (Bio-Tek Instruments Inc.) by adding 50 µl of cell lysate to 50 µl of Steady-Glo Luciferase Assay Substrate (Promega). Expression of lacZ as β-galactosidase activity was measured by incubating 50 µl of cell lysate with 143 µl of Z-Buffer (0.06 M Na2HPO4, 0.04 M NaH2PO4, 0.01 M KCl, 0.001 M MgSO4), 36 µl ONPG (4 mg/ml in diH20) and 1 µl 2–mercaptoethanol reagent (Fisher Scientific) at 37°C for 30 min. After incubation, 300 µl of diH2O was added to the reactions and the absorbance was measured at 414 nm. Luciferase expression was normalized to β-galactosidase activity for each well. A student’s two-tailed t-test was used to determine statistical significance.
Generation of CE-lacZ Transgenic Mice
A 2.4 kb fragment containing two copies of the chicken β-globin insulator, 5′HS4 [49], was excised from plasmid pJC13-1 using EcoRI and BamHI and cloned in duplicate into the BglII and HpaI sites of the pSL1180 shuttle vector, generating plasmid pSL-I. An XbaI site in pNASS-β was destroyed to allow subsequent cloning, and the lacZ gene was excised from this plasmid using EcoRI and HindIII and inserted into the EcoRV site of pSL-I, generating pSL-IL. The Dlk1 promoter was excised from pGL3-Dlk1P using XhoI and NheI and cloned into the NheI site of pSL-IL to generate pSL-ILP. The CE4, CE8 and CE9 fragments were excised from pCRII-TOPO using EcoRI and cloned into the XbaI site of pSL-ILP to generate CE4-lacZ, CE8-lacZ and CE9-lacZ, respectively. CE4-lacZ and CE8-lacZ were linearized using NcoI and SnaBI, CE9-lacZ was linearized using BstBI and SnaBI, and all transgenes were purified with QIAEX II (Qiagen). Linearized DNA was injected into FVB/N embryos by the UIC Transgenic Production Service, and the embryos implanted in pseudopregnant ICR females. Embryos were collected at e13.5 and fixed in 4% paraformaldehyde for 90 min on ice, washed 3 times at room temperature (RT) for 30 min (2 mM MgCl2, 0.01% deoxycholic acid, 0.02% NP-40 in 1× PBS), and stained overnight in the dark at RT (1 mg/ml X-gal, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 2 mM MgCl2, 0.01% deoxycholic acid, 0.02% NP-40). The reaction was stopped by washing the embryos 3 times in 1× PBS at RT. Embryos were genotyped from yolk sac DNA using primers OL410, 5′-GGGGACGACAGTACGAAAAGGC-3′, and OL411, 5′-GTATCGGCCTCAGGAAGATCGC-3′ to generate a 789 bp lacZ product. PCR conditions were 94°C, 30 sec, 60°C, 1 min and 72°C, 1 min, for 35 cycles. Transgenic embryos were embedded in paraffin, and 8-micron sagittal sections were cut and counterstained with eosin. Low power images were collected on a Leica MZFLIII dissecting microscope using a Leica DFC320 camera. High power images were collected on a Zeiss Axiovert 200 M microscope using a Zeiss AxioCam MRc5 camera.
Results
Dlk1 is Expressed but not Imprinted from the BAC 127H5 Transgene
Previous work showed that Dlk1 was not expressed from the 28G5 transgene spanning the mouse Dlk1-Meg3 region, suggesting that elements outside of this BAC are required for Dlk1 expression (Fig. 1A) [46]. To expand the search for Dlk1 regulatory sequences, transgenic mice were produced using the overlapping BAC 127H5, which extends 41 kb upstream and 36 kb downstream of the mouse Dlk1 gene (Fig. 1A). The BAC was linearized at a unique ClaI site at −10.5 kb relative to the Dlk1 transcriptional initiation site. While this places much of the upstream sequence in a position downstream of Dlk1 in the transgene, this is not likely to alter the function of tissue-specific enhancers. Enhancers are by definition position- and orientation-independent, a fact that has been exploited in a wide variety of heterologous in vivo expression vectors. We have also shown that enhancers directing tissue-specific expression of the linked Meg3 gene function appropriately even when located on a separate, cointegrated DNA fragment [46].
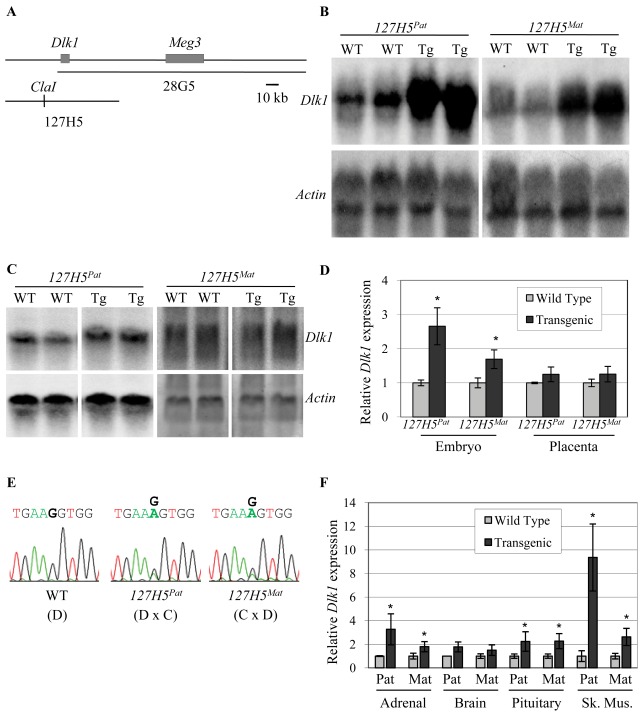
Figure 1. Dlk1 is expressed but not imprinted from the 127H5 transgene.
(A) Schematic of the Dlk1-Meg3 BAC clones used to generate transgenic mice; the 28G5 transgene was described previously [46]. The 127H5 BAC was linearized at a unique ClaI site. (B, C) Representative Northern blots for Dlk1 mRNA in midgestation wild type (WT) and heterozygous transgenic (Tg) embryo (B) and placenta (C), after paternal (127H5Pat) and maternal (127H5Mat) inheritance. The mouse β-actin gene was used as a loading control. (D) Quantitative Northern data for blots shown in B & C; expression is normalized to β-actin. Gray bars represent wild type samples and black bars represent 127H5 transgenic samples upon paternal (127H5Pat) or maternal (127H5Mat) transmission in crosses to Cg12. (E) Direct sequencing assay for Dlk1 imprinting in wild type (WT) and heterozygous transgenic (Tg) F1 embryos after paternal (127H5Pat) and maternal (127H5Mat) inheritance. D indicates wild type animals carrying only the M. domesticus allele, while D×C or C×D indicates offspring of crosses to the Cg12 line carrying a M. castaneus allele, with the female genotype listed first. (F) Quantitative Northern blot analysis for Dlk1 mRNA in 3–4 week old 127H5 tissues. Expression is normalized to β-actin, and each bar represents 8–10 animals. Gray bars represent wild type samples and the black bars represent 127H5 transgenic samples upon paternal (Pat) or maternal (Mat) transmission. In all figures asterisks indicate p≤0.05.
Three founder animals were obtained that carried 8, 2, and 2 copies of the BAC (data not shown). Individual transgenic lines were established by breeding to FVB/N mice, and Dlk1 expression levels were analyzed in whole embryo at embryonic day 12.5 (e12.5). Since the Dlk1 gene is subject to genomic imprinting, expression levels were assayed in heterozygous embryos after both maternal and paternal transmission of the transgene. Dlk1 expression from two of the lines was equal to wild type littermates by Northern blotting, suggesting that Dlk1 was not expressed from these transgenes (data not shown). In our experience, the lack of expression from two of the three lines is not unusual for transgenic animals, in which integrated sequences may be silenced by virtue of their integration site, concatenated structure or bacterially-derived vector sequences. The third line, which carries 2 copies of the BAC, showed a 1.8 and 2.5 fold increase in Dlk1 expression upon maternal and paternal transmission of the transgene, respectively (Fig. 1B). These data show that Dlk1 is expressed from the transgene in this 127H5 line, but suggest that it is not imprinted (Fig. 1B, D). Dlk1 is normally expressed in the mouse placenta at e12.5, but none of the 127H5 lines showed increased levels of Dlk1 in placenta when compared to wild type (Fig. 1C, D and data not shown).
To more directly analyze imprinting of the 127H5 transgene, we took advantage of a congenic mouse line (Cg12) that carries the Dlk1-Meg3 region of chromosome 12 from Mus musculus castaneus (C) on a Mus musculus domesticus (D) background [50]. The Cg12 mice allow the use of sequence polymorphisms between C and D animals for imprinting analysis in F1 offspring. Analysis of embryos derived from a cross between 127H5 mice carrying D alleles, and Cg12 mice carrying C alleles, showed that both parental alleles were expressed, and that Dlk1 is not imprinted on the 127H5 transgene (Fig. 1E).
To determine the tissue-specific profile of 127H5 Dlk1 expression, three-week old 127H5 mice were analyzed for Dlk1 mRNA levels in isolated adrenal gland, brain, pituitary gland and skeletal muscle (Fig. 1F). A significant increase in Dlk1 expression over wild type was observed by Northern blotting in the adrenal and pituitary glands and skeletal muscle of 127H5 mice, but no increase in Dlk1 expression was observed in brain (Fig. 1F). The levels of 127H5-derived Dlk1 expression were independent of the direction of parental transmission, consistent with the lack of imprinting observed in the embryo (Fig. 1E). Levels of Dlk1 in skeletal muscle were increased to a greater degree than those seen in other tissues, indicating a strong skeletal muscle enhancer may lie within 127H5, and/or that a silencer that modifies Dlk1 expression is not present on the BAC. Skeletal muscle Dlk1 levels were also higher in animals inheriting the transgene paternally than in those inheriting it maternally, despite the lack of overt imprinting of the transgene as a whole. While the reason for this expression difference is unclear, it is possible the transgene carries some component of the imprinting machinery that, in isolation, remains capable of driving increased paternal expression in certain tissues or in highly-expressing tissues. These data suggest that 127H5 contains the tissue specific elements required for Dlk1 expression in some, but not all, sites of endogenous Dlk1 expression.
These results are consistent with work from da Rocha et al, which showed that a BAC transgene carrying 49 kb of sequence upstream of Dlk1 (extending 8 kb beyond the 5′ end of 127H5) is active in many sites of endogenous Dlk1 expression, including kidney, skeletal muscle and liver, but is not expressed in placenta [19]. Similarly to the data shown here, the Dlk1 gene was not imprinted in the context of this transgene.
Conserved Elements Upstream of Dlk1 Function as Enhancers in Cell Culture
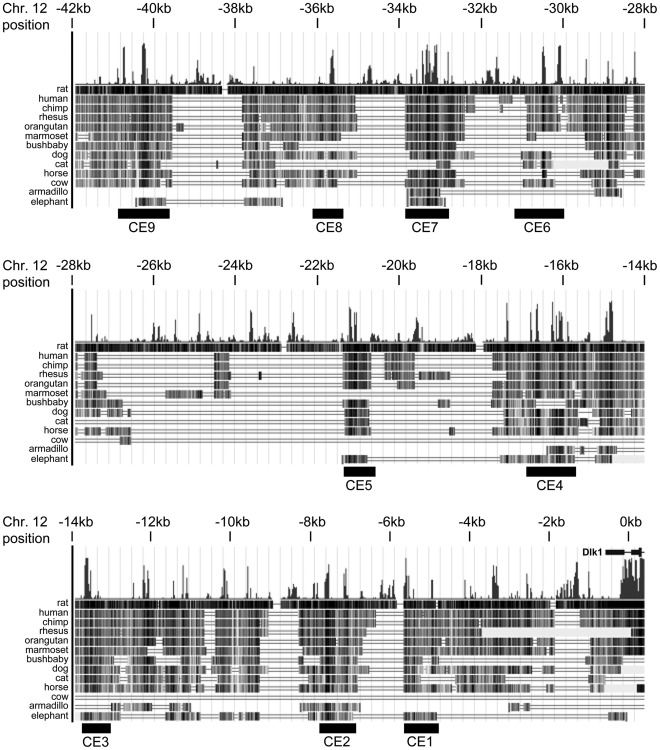
The data obtained from the 28G5 and 127H5 transgenes suggest that elements located in the non-overlapping sequence (−3.5 to −41 kb upstream of Dlk1) are capable of directing gene expression in multiple Dlk1-expressing tissues. The sequence upstream of Dlk1 was analyzed for conserved regions across species using the University of California Santa Cruz (UCSC) Genome Browser (http://genome.ucsc.edu/) and the Vista browser (http://genome.lbl.gov/vista) (Fig. 2 and data not shown). The UCSC browser measures evolutionary conservation between multiple vertebrate species using phastCons and phyloP. Vista uses a combination of Shuffle-LAGAN global alignments and local BLAT alignments to construct genome-wide pairwise DNA alignments between two species. The UCSC Genome Browser alignment shown in Figure 2 makes use of all currently available genome sequence, and with one exception supports the original alignments performed in 2006. The highly conserved region at approximately −15 kb was not present in comparisons performed when fewer species were available, and awaits future functional analysis.
Figure 2. Sequence conservation in the Dlk1 upstream region.
The sequence upstream of Dlk1 was analyzed for regions of conservation across multiple species. The conserved elements chosen for further analysis are numbered CE1 to CE9. The region being displayed corresponds to the July 2007 mouse genome assembly; assembly dates for other species are given in the Methods (adapted from the UCSC Genome Browser).
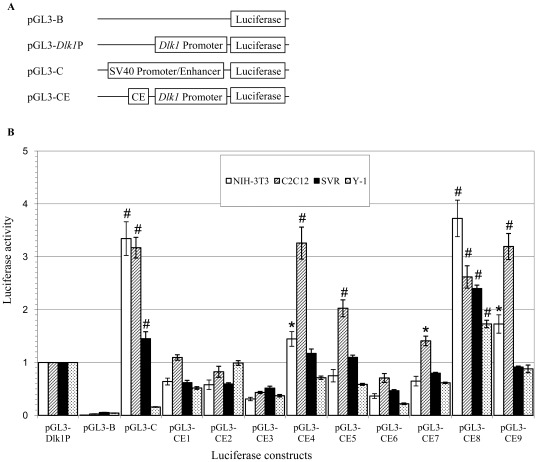
Nine conserved elements of approximately 1 kb (CE1–9), that contained ≥50 bp stretches of DNA with ≥70% sequence homology, were chosen for further analysis (Fig. 2 and data not shown). A luciferase reporter assay was used to screen the CEs for regulatory activity; CEs were cloned into a luciferase expression vector driven by the Dlk1 promoter, generating plasmids pGL3-CE1 through pGL3-CE9 (Fig. 3A). Established cell lines selected from tissues that express the endogenous Dlk1 gene: NIH-3T3 (mouse embryonic fibroblast), C2C12 (mouse myoblast), SVR (mouse pancreatic islet) and Y-1 (mouse adrenal tumor), were transiently transfected with the plasmids pGL3-B, pGL3-C, pGL3-Dlk1P, or the individual pGL3-CEs. Luciferase activity was measured relative to that driven by the Dlk1 promoter alone, and all values were normalized to a transfection control (Fig. 3B). Multiple CEs functioned as enhancers in the luciferase assay, with CE4, 5, 7 and 9 showing higher levels of luciferase expression in C2C12 cells than in other cell types, suggesting that these elements might contain muscle specific enhancers (Fig. 3B). CE8 directed high levels of luciferase expression in multiple cell lines, suggesting that CE8 contains multiple enhancers or an enhancer that can function in multiple tissues (Fig. 3B). Expression plasmids carrying CE1, 2, 3 and 6 produced little or no luciferase activity over the Dlk1 promoter alone.
Figure 3. Transcriptional activation by conserved elements is cell line-dependent.
(A) Luciferase expression plasmids used to assay enhancer function in cell culture. “Dlk1 Promoter” signifies the Dlk1 basal promoter and CE signifies the individual conserved elements tested. Plasmid pGL3-B contains the promoterless luciferase gene, pGL3-Dlk1P contains the endogenous Dlk1 promoter upstream of luciferase and pGL3-C contains the SV40 promoter/enhancer upstream of luciferase. (B) Enhancer activity observed in the NIH-3T3, C2C12, SVR and Y-1 cell lines. Expression is normalized to pGL3-Dlk1P for each cell type; the results are presented as mean ± SEM. P-values relative to pGL3-Dlk1P expression are indicated by *, p≤0.05; #, p≤0.01 (n = 3) (Student’s t-test).
Conserved Elements can Direct lacZ Expression in vivo
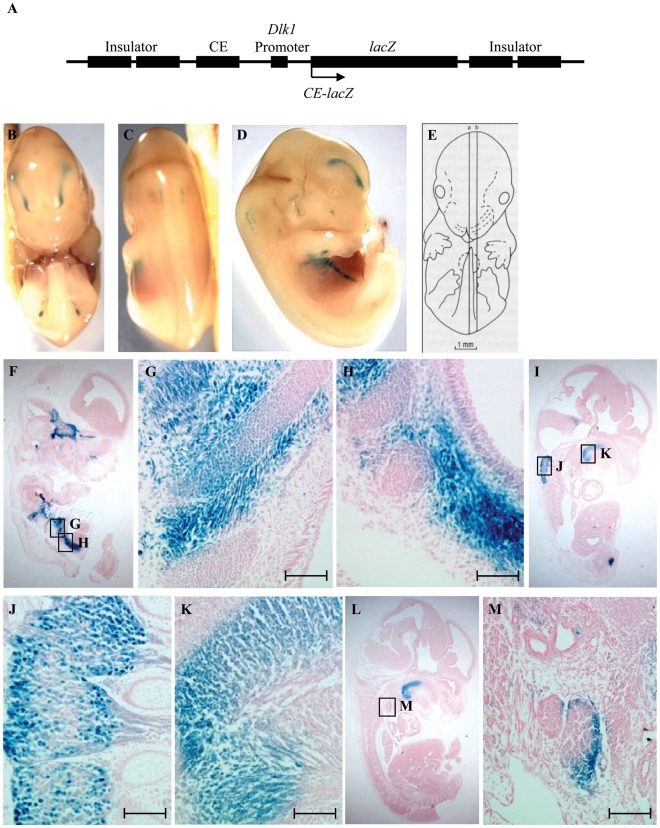
As CE4, CE8 and CE9 conferred the highest level of enhancer activity in the reporter assay, these three regions were selected for in vivo analysis. Reporter transgenes were generated carrying each conserved element, and the Dlk1 promoter, upstream of a lacZ cassette (CE4-lacZ, CE8-lacZ, and CE9-lacZ), and the transgenes were flanked by insulator elements to reduce position effects (Fig. 4A) [49]. The transgenes were injected into FVB/N fertilized eggs, and F0 embryos were collected at e13.5 for whole-mount analysis of lacZ expression. A total of 51 embryos were obtained from the CE4-lacZ injections, of which 5 carried the transgene and 4 showed lacZ expression. A total of 77 embryos were obtained from the CE8-lacZ injections, of which 16 embryos carried the transgene and 9 showed lacZ expression. A total of 45 embryos were obtained from the CE9-lacZ injections, of which 7 carried the transgene and 5 showed lacZ expression. Positively staining embryos were embedded and sectioned for histological analysis.
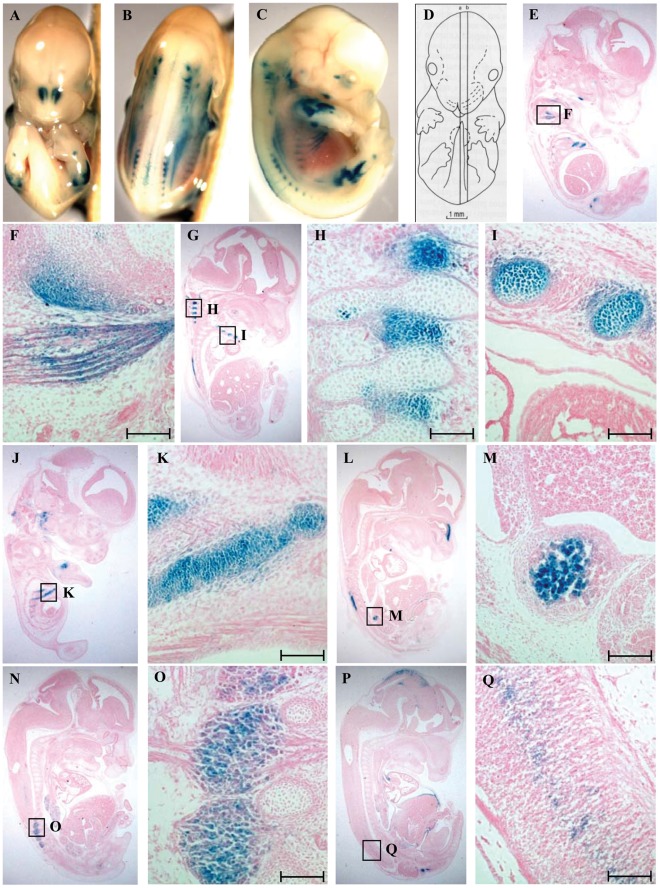
Figure 4. CE4-lacZ displays expression in a subset of Dlk1-expressing tissues.
(A) Diagram of lacZ expression constructs used to produce CE4-lacZ and CE8-lacZ transgenic embryos. The arrow represents the direction of transcription, and CE stands for conserved element 4 or 8. (B–D) Ventral, dorsal and lateral views, respectively, of a representative whole mount transgenic embryo at e13.5. (E) Schematic representation of sagittal section planes used in these images [64]. (F–M) Sagittal sections of embryos under low magnification (F, I, L) and high magnification (G, H, J, K, M). All sections are oriented with anterior on top and dorsal to the left. Expression was seen in (F–H) intercostal muscle, body wall muscle, and ribs; (I, J) dorsal root ganglia; (I, K) intrinsic tongue muscle; (L, M) thymus. The embryo shown in (F) displays lacZ expression in the pituitary gland and trigeminal ganglion, both sites of endogenous Dlk1 expression, but this pattern was not seen in other embryos carrying CE4-lacZ. Scale bars represent 100 µm.
The chicken β-globin insulators functioned well in this system, as most embryos carrying a particular transgene showed similar patterns of expression despite their unique integration sites. In previous work from our laboratory, the β-globin insulators were shown to block the effects of virtually all external regulatory elements, and to confer no activity on their own (Steshina & Schmidt, unpublished). A particular tissue was scored as positive for lacZ expression only if it was found in 3 or more embryos for a given transgene. Embryos inheriting the CE4-lacZ transgene displayed lacZ expression in multiple regions of the musculoskeletal system and in some neuronal tissues (Fig. 4) (Table 2). CE4-lacZ expression was seen in the intercostal muscles, the epiphyses of the ribs and the body wall muscles of the abdomen (Fig. 4B–D, F–H). Expression was also found in the intrinsic muscles of the tongue (Fig. 4I, K). The expression seen in the dorsal root ganglia was present in the cell bodies as well as the axons (Fig. 4I, J). CE4-lacZ expression was also seen in the cortical region of the thymus (Fig. 4L, M).
Table 2. Embryonic expression of CE4-lacZ, CE8-lacZ and Dlk1.
| Tissue/organ | CE4-lacZ | CE8-lacZ | Dlk1 |
| Tongue muscle | + | – | + |
| Intercostal muscle | + | – | + |
| Body wall muscle | + | – | + |
| Limb muscle | – | + | + |
| Intervertebral cartilage | – | + | + |
| Ribs | + | + | + |
| Thymus | + | – | + |
| Pituitary gland | – | – | + |
| Adrenal gland | – | + | + |
| Liver | – | – | + |
| Pancreas | – | – | + |
| Lung | – | – | + |
| Spinal cord | – | + | – |
| Dorsal root ganglia | + | + | – |
+ indicates expression; – indicates no expression.
Embryos inheriting the CE8-lacZ transgene showed expression in the migrating skeletal muscle of the limbs (Fig. 5A–F), and in the developing skeleton including the ribs, costosternal junctions, intervertebral cartilage and vertebrae (Fig. 5B, C, G–K) (Table 2). CE8-lacZ expression was also observed in a subset of neuronal and neuroendocrine tissues, including the chromaffin cells of the adrenal medulla (Fig. 5L, M), the dorsal root ganglia (Fig. 5N, O), and the spinal cord (Fig. 5P, Q). No consistent pattern of expression was seen among the CE9-lacZ embryos, and so this element was not analyzed further. Overall, expression in the musculoskeletal system predominated from both the CE4 and CE8 elements, consistent with the high-level expression seen in muscle in the 127H5 transgenic mice, and the high C2C12 activity in the luciferase expression system. The pattern of expression from the CE4-lacZ and CE8-lacZ transgenes overlaps significantly with that of the endogenous Dlk1 gene [27], [33], suggesting Dlk1-specific cis-regulatory elements are contained within CE4 and CE8. Despite this significant muscle expression, not all developing muscle is positive for lacZ, suggesting the presence of elements that direct Dlk1 expression in a specific subset of muscle cells or at a specific stage of muscle development.
Figure 5. CE8-lacZ displays expression in a subset of Dlk1-expressing tissues.
. (A–C) Ventral, dorsal and lateral views, respectively, of a representative whole mount transgenic embryo at e13.5. (D) Schematic representation of sagittal section planes used in these images [64]. (E–Q) Sagittal sections of embryos under low magnification (E, G, J, L, N, P) and high magnification (F, H, I, K, M, O, Q). All sections are oriented with anterior on top and dorsal to the left. Expression was seen in (E, F) skeletal muscle of the limb, (G, H) intervertebral cartilage; (G, I–K) ribs and costosternal junctions; (L, M) chromaffin cells of the adrenal gland; (N, O) dorsal root ganglia; (P, Q) spinal cord. Scale bars represent 100 µm.
Discussion
The 127H5 Transgene Recapitulates Endogenous Dlk1 Expression
Previous work from our laboratory showed that the mouse Meg3 gene was expressed in a subset of its endogenous pattern from the 28G5 transgene, and was imprinted in those tissues where it was expressed [46]. Dlk1 was not expressed from 28G5 in any tissues, suggesting that cis-regulatory elements beyond the extent of 28G5 are required for Dlk1 expression. Consistent with this idea, a subsequent study showed that the TgDlk1−70 BAC transgene containing 49 kb of sequence upstream of Dlk1 could direct Dlk1 expression in a subset of endogenous Dlk1-expressing tissues [19] . The more proximal 127H5 transgene described here expands and refines this work, localizing multiple cis-regulatory elements that may control Dlk1 in developing skeletal muscle, and in neuroendocrine tissues such as the pituitary and adrenal glands, to a 41 kb region upstream of Dlk1 (Fig. 1). The similarities in expression between the 127H5 and TgDlk1−70 BAC transgenes, and the lack of expression from TgDlk1−31, suggest most upstream regulatory elements lie within the −41 kb to −8 kb region.
Dlk1 is not imprinted in the context of the 127H5 transgene; this was not surprising, as this transgene lacks both the intergenic and Meg3 differentially methylated regions shown to be required for proper imprinting of the Dlk1-Meg3 locus (Fig. 1E) [51]–[53]. In addition to expression in the embryo, placental expression of Dlk1-Meg3 is also of interest, as many imprinted genes appear to play important roles in placental development and function [54]. Meg3 is expressed in the placenta from the 28G5 transgene, but Dlk1 does not show placental expression from any of the BAC transgenes, indicating yet more distal control elements exist for Dlk1 extraembryonic expression [46] (Fig. 1C, D). We showed previously that 28G5 carries some Meg3 control elements, and have now shown that 127H5 carries some Dlk1 control elements; this gene pair therefore has a complex regulatory domain spanning more than 200 kb of sequence.
Coordinate versus Independent Regulation of Dlk1 and Meg3
Dlk1 and Meg3 demonstrate a significant degree of overlap in tissue-specific expression during development, but within a single tissue these two genes are often expressed in dynamic and unique patterns in different cell types. Both genes are highly expressed in developing skeletal muscle, and they are coexpressed in fetal hepatocytes and in the early pancreas. During later pancreas development, however, Meg3 expression is downregulated, while Dlk1 retains high level expression but becomes restricted to the insulin-producing beta cells. Both genes are expressed in the developing pituitary gland, initially in the infundibulum that gives rise to the posterior lobe, and in Rathke’s pouch that gives rise to the anterior lobe. In adults, however, Dlk1 expression is restricted to the anterior somatotrophs, while Meg3 is expressed primarily in gonadotrophs [27], [33]. Dlk1 is highly expressed in the lung, where Meg3 is not expressed, and Meg3 exhibits widespread expression in the central nervous system, while Dlk1 is localized to a few brain regions where Meg3 is absent [33]. While other imprinted gene pairs such as H19-Igf2 share common regulatory elements, the limited overlap in cellular expression of Dlk1-Meg3 suggests that the many of their cis-regulatory elements are unique to each gene. Data from the 28G5 and 127H5 transgenes largely support this hypothesis, with Meg3 enhancers within 28G5 failing to activate Dlk1, while most Dlk1 enhancers lie within the nonoverlapping region of 127H5. Interestingly, the CE4-lacZ and CE8-lacZ transgenes directed expression in a subset of neuronal tissues where Meg3, but not Dlk1, is expressed, including the dorsal root ganglia (Fig. 4I–J, 5N–O) and the spinal cord (Fig. 5P–Q). Some Meg3-specific enhancers may therefore lie upstream of Dlk1, but are prevented in vivo from activating expression of Dlk1.
Dissecting the Dlk1 Upstream Region
Multi-species comparison of the Dlk1 upstream region not surprisingly demonstrated stronger conservation among more closely related mammalian species, and less conservation with non-mammalian vertebrates (Fig. 2 and data not shown). Although the long-standing dogma has been that key regulatory elements will show deep evolutionary conservation, recent analyses have often refuted this idea. While the recognition sites for some DNA binding factors are well-conserved across species [55], many are poorly conserved even within mammals, and less so in lower vertebrates [56], [57]. Genomic imprinting is a uniquely mammalian phenomenon, however, and while Dlk1 is present in many non-mammalian vertebrates, it is not imprinted in these species. For genes subject to genomic imprinting, it is possible that constraint against harmful loss of imprinting would drive stronger conservation within mammals.
Within the 41 kb Dlk1 upstream region of 127H5, five highly conserved elements were able to function as enhancers in cell culture assays (Fig. 3), and two of these elements (CE4 and CE8) directed reproducible expression of a lacZ reporter transgene in a subset of Dlk1-expressing tissues in the mouse embryo (Fig. 4, 5). These data suggest that CE4 and CE8 contain cis-regulatory elements that direct Dlk1 expression in vivo. CE4-lacZ and CE8-lacZ were both expressed in regions of the developing skeletal muscle, and the developing cartilage of the axial skeleton, particularly in the ribs and intervertebral discs (Fig. 4, 5). These patterns resemble the high level expression of endogenous Dlk1 found in the developing skeletal muscle and the ossifying skeleton [27], [33]. Endogenous Dlk1 is also expressed at high levels in many neuroendocrine cell types, including the pituitary somatotrophs, the chromaffin cells of the adrenal medulla, and the beta cells of the pancreas. The 127H5 transgene directed pituitary and adrenal expression (pancreas was not analyzed), and CE8-lacZ expressed lacZ in the chromaffin cells of the adrenal gland (Fig. 1, 5). Neither CE4-lacZ nor CE8-lacZ conferred expression in the pituitary, however, and no expression was seen from the lacZ transgenes in the developing pancreas. As 127H5 did not fully recapitulate endogenous Dlk1 expression, and the sum of CE4-lacZ and CE8-lacZ did not fully recapitulate 127H5 expression, there remain Dlk1 regulatory elements to be identified both within the span of 127H5, and in sequences outside this region.
Multiple Elements Regulate Dlk1 Expression in Developing Skeletal Muscle
In the late gestation embryo, Dlk1 is most strongly expressed in developing skeletal muscle, but the gene is silenced in muscle shortly after birth. Sheep carrying the naturally-occurring Callipyge (Clpg) mutation fail to down-regulate Dlk1 in specific muscle groups, resulting in increased skeletal muscle mass [58]–[61]. Clpg has been mapped to a point mutation in the Dlk1-Meg3 intergenic region that is proposed to function as a cis-acting silencer in postnatal tissues [62]. In myoblasts, as in many other developing tissues, Dlk1 may function to regulate the switch from a proliferating, immature cell population to a differentiating, mature one. In adult mice, Dlk1 is reactivated in regenerating skeletal muscle, where it may be involved in satellite cell activation and/or proliferation [24].
MyoD is a key regulator of skeletal muscle development, where it transactivates muscle-specific genes through cis-acting E-box elements [63]. The loss of Dlk1 in skeletal muscle-specific knockout mice is associated with reduced MyoD expression, perhaps through activation of the NF-kappa B pathway, providing support for an interaction between Dlk1 and MyoD in skeletal muscle [24]. The CE4-lacZ and CE8-lacZ transgenes showed strong myoblast expression (Fig. 4, 5), indicating that muscle-specific transcription factors may recognize the CE4 and CE8 elements. This work significantly narrows the regions to be investigated for possible interaction with MyoD and other, yet unknown, regulators of Dlk1 tissue-expression in the mouse embryo.
Acknowledgments
The authors thank Gary Felsenfeld for the pJC13-1 plasmid, the UIC Transgenic Production Service and the CBC/UIC RRC Proteomics and Informatics Services Facility. All animals used in these experiments were maintained in compliance with the National Institutes of Health’s Guide for Care and Use of Laboratory Animals and the University of Illinois at Chicago Animal Care Committee guidelines, which specifically approved this study (Protocols 06–159 and 09–115).
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by grant HD042013 from the National Institutes of Health awarded to JVS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Morison IM, Paton CJ, Cleverley SD. The imprinted gene and parent-of-origin effect database. Nucleic Acids Res. 2001;29:275–276. doi: 10.1093/nar/29.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmidt JV, Matteson PG, Jones BK, Guan XJ, Tilghman SM. The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes Dev. 2000;14:1997–2002. [PMC free article] [PubMed] [Google Scholar]
- 3.Takada S, Tevendale M, Baker J, Georgiades P, Campbell E, et al. Delta-like and gtl2 are reciprocally expressed, differentially methylated linked imprinted genes on mouse chromosome 12. Curr Biol. 2000;10:1135–1138. doi: 10.1016/s0960-9822(00)00704-1. [DOI] [PubMed] [Google Scholar]
- 4.Hernandez A, Fiering S, Martinez E, Galton VA, St Germain D. The gene locus encoding iodothyronine deiodinase type 3 (Dio3) is imprinted in the fetus and expresses antisense transcripts. Endocrinology. 2002;143:4483–4486. doi: 10.1210/en.2002-220800. [DOI] [PubMed] [Google Scholar]
- 5.Laborda J, Sausville EA, Hoffman T, Notario V. dlk, a putative mammalian homeotic gene differentially expressed in small cell lung carcinoma and neuroendocrine tumor cell line. J Biol Chem. 1993;268:3817–3820. [PubMed] [Google Scholar]
- 6.Smas CM, Sul HS. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell. 1993;73:725–734. doi: 10.1016/0092-8674(93)90252-l. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Sul HS. Ectodomain shedding of preadipocyte factor 1 (Pref-1) by tumor necrosis factor alpha converting enzyme (TACE) and inhibition of adipocyte differentiation. Mol Cell Biol. 2006;26:5421–5435. doi: 10.1128/MCB.02437-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garces C, Ruiz-Hidalgo MJ, Bonvini E, Goldstein J, Laborda J. Adipocyte differentiation is modulated by secreted delta-like (dlk) variants and requires the expression of membrane-associated dlk. Differentiation. 1999;64:103–114. doi: 10.1046/j.1432-0436.1999.6420103.x. [DOI] [PubMed] [Google Scholar]
- 9.Mei B, Zhao L, Chen L, Sul HS. Only the large soluble form of preadipocyte factor-1 (Pref-1), but not the small soluble and membrane forms, inhibits adipocyte differentiation: role of alternative splicing. Biochem J. 2002;364:137–144. doi: 10.1042/bj3640137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nueda ML, Garcia-Ramirez JJ, Laborda J, Baladron V. dlk1 specifically interacts with insulin-like growth factor binding protein 1 to modulate adipogenesis of 3T3-L1 cells. J Mol Biol. 2008;379:428–442. doi: 10.1016/j.jmb.2008.03.070. [DOI] [PubMed] [Google Scholar]
- 11.Ross DA, Rao PK, Kadesch T. Dual roles for the Notch target gene Hes-1 in the differentiation of 3T3-L1 preadipocytes. Mol Cell Biol. 2004;24:3505–3513. doi: 10.1128/MCB.24.8.3505-3513.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baladron V, Ruiz-Hidalgo MJ, Nueda ML, Diaz-Guerra MJ, Garcia-Ramirez JJ, et al. dlk acts as a negative regulator of Notch1 activation through interactions with specific EGF-like repeats. Exp Cell Res. 2005;303:343–359. doi: 10.1016/j.yexcr.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Nueda ML, Baladron V, Sanchez-Solana B, Ballesteros MA, Laborda J. The EGF-like protein dlk1 inhibits notch signaling and potentiates adipogenesis of mesenchymal cells. J Mol Biol. 2007;367:1281–1293. doi: 10.1016/j.jmb.2006.10.043. [DOI] [PubMed] [Google Scholar]
- 14.Bray SJ, Takada S, Harrison E, Shen SC, Ferguson-Smith AC. The atypical mammalian ligand Delta-like homologue 1 (Dlk1) can regulate Notch signalling in Drosophila. BMC Dev Biol. 2008;8:11. doi: 10.1186/1471-213X-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruiz-Hidalgo MJ, Gubina E, Tull L, Baladron V, Laborda J. dlk modulates mitogen-activated protein kinase signaling to allow or prevent differentiation. Exp Cell Res. 2002;274:178–188. doi: 10.1006/excr.2001.5464. [DOI] [PubMed] [Google Scholar]
- 16.Kim KA, Kim JH, Wang Y, Sul HS. Pref-1 (preadipocyte factor 1) activates the MEK/extracellular signal-regulated kinase pathway to inhibit adipocyte differentiation. Mol Cell Biol. 2007;27:2294–2308. doi: 10.1128/MCB.02207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L, Qanie D, Jafari A, Taipaleenmaki H, Jensen CH, et al. Delta-like 1/fetal antigen-1 (Dlk1/FA1) is a novel regulator of chondrogenic cell differentiation via inhibition of the Akt kinase-dependent pathway. J Biol Chem. 2011;286:32140–9. doi: 10.1074/jbc.M111.230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moon YS, Smas CM, Lee K, Villena JA, Kim KH, et al. Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol Cell Biol. 2002;22:5585–5592. doi: 10.1128/MCB.22.15.5585-5592.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.da Rocha ST, Charalambous M, Lin SP, Gutteridge I, Ito Y, et al. Gene dosage effects of the imprinted delta-like homologue 1 (dlk1/pref1) in development: implications for the evolution of imprinting. PLoS Genet. 2009;5:e1000392. doi: 10.1371/journal.pgen.1000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kagami M, Sekita Y, Nishimura G, Irie M, Kato F, et al. Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat Genet. 2008;40:237–242. doi: 10.1038/ng.2007.56. [DOI] [PubMed] [Google Scholar]
- 21.Smas CM, Sul HS. Molecular mechanisms of adipocyte differentiation and inhibitory action of pref-1. Crit Rev Eukaryot Gene Expr. 1997;7:281–298. doi: 10.1615/critreveukargeneexpr.v7.i4.10. [DOI] [PubMed] [Google Scholar]
- 22.Abdallah BM, Jensen CH, Gutierrez G, Leslie RG, Jensen TG, et al. Regulation of human skeletal stem cells differentiation by Dlk1/Pref-1. J Bone Miner Res. 2004;19:841–852. doi: 10.1359/JBMR.040118. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Sul HS. Pref-1 regulates mesenchymal cell commitment and differentiation through Sox9. Cell Metab. 2009;9:287–302. doi: 10.1016/j.cmet.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waddell JN, Zhang P, Wen Y, Gupta SK, Yevtodiyenko A, et al. Dlk1 is necessary for proper skeletal muscle development and regeneration. PLoS One. 2010;5:e15055. doi: 10.1371/journal.pone.0015055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen CH, Teisner B, Hojrup P, Rasmussen HB, Madsen OD, et al. Studies on the isolation, structural analysis and tissue localization of fetal antigen 1 and its relation to a human adrenal-specific cDNA, pG2. Hum Reprod. 1993;8:635–641. doi: 10.1093/oxfordjournals.humrep.a138110. [DOI] [PubMed] [Google Scholar]
- 26.Floridon C, Jensen CH, Thorsen P, Nielsen O, Sunde L, et al. Does fetal antigen 1 (FA1) identify cells with regenerative, endocrine and neuroendocrine potentials? A study of FA1 in embryonic, fetal, and placental tissue and in maternal circulation. Differentiation. 2000;66:49–59. doi: 10.1046/j.1432-0436.2000.066001049.x. [DOI] [PubMed] [Google Scholar]
- 27.Yevtodiyenko A, Schmidt JV. Dlk1 expression marks developing endothelium and sites of branching morphogenesis in the mouse embryo and placenta. Dev Dyn. 2006;235:1115–1123. doi: 10.1002/dvdy.20705. [DOI] [PubMed] [Google Scholar]
- 28.Okamoto M, Takemori H, Halder SK, Nonaka Y, Hatano O. Implication of ZOG protein (zona glomerulosa-specific protein) in zone development of the adrenal cortex. Endocr Res. 1998;24:515–520. doi: 10.3109/07435809809032640. [DOI] [PubMed] [Google Scholar]
- 29.Costaglioli P, Come C, Knoll-Gellida A, Salles J, Cassagne C, et al. The homeotic protein dlk is expressed during peripheral nerve development. FEBS Lett. 2001;509:413–416. doi: 10.1016/s0014-5793(01)03205-7. [DOI] [PubMed] [Google Scholar]
- 30.Kaneta M, Osawa M, Sudo K, Nakauchi H, Farr AG, et al. A role for pref-1 and HES-1 in thymocyte development. J Immunol. 2000;164:256–264. doi: 10.4049/jimmunol.164.1.256. [DOI] [PubMed] [Google Scholar]
- 31.Moore KA, Pytowski B, Witte L, Hicklin D, Lemischka IR. Hematopoietic activity of a stromal cell transmembrane protein containing epidermal growth factor-like repeat motifs. Proc Natl Acad Sci U S A. 1997;94:4011–4016. doi: 10.1073/pnas.94.8.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tornehave D, Jensen CH, Teisner B, Larsson LI. FA1 immunoreactivity in endocrine tumours and during development of the human fetal pancreas; negative correlation with glucagon expression. Histochem Cell Biol. 1996;106:535–542. doi: 10.1007/BF02473268. [DOI] [PubMed] [Google Scholar]
- 33.da Rocha ST, Tevendale M, Knowles E, Takada S, Watkins M, et al. Restricted co-expression of Dlk1 and the reciprocally imprinted non-coding RNA, Gtl2: implications for cis-acting control. Dev Biol. 2007;306:810–823. doi: 10.1016/j.ydbio.2007.02.043. [DOI] [PubMed] [Google Scholar]
- 34.Bachmann E, Krogh TN, Hojrup P, Skjodt K, Teisner B. Mouse fetal antigen 1 (mFA1), the circulating gene product of mdlk, pref-1 and SCP-1: isolation, characterization and biology. J Reprod Fertil. 1996;107:279–285. doi: 10.1530/jrf.0.1070279. [DOI] [PubMed] [Google Scholar]
- 35.Larsen JB, Jensen CH, Schroder HD, Teisner B, Bjerre P, et al. Fetal antigen 1 and growth hormone in pituitary somatotroph cells. Lancet. 1996;347:191. doi: 10.1016/s0140-6736(96)90374-8. [DOI] [PubMed] [Google Scholar]
- 36.Smas CM, Kachinskas D, Liu CM, Xie X, Dircks LK, et al. Transcriptional control of the pref-1 gene in 3T3-L1 adipocyte differentiation. Sequence requirement for differentiation-dependent suppression. J Biol Chem. 1998;273:31751–31758. doi: 10.1074/jbc.273.48.31751. [DOI] [PubMed] [Google Scholar]
- 37.Takemori H, Doi J, Katoh Y, Halder SK, Lin XZ, et al. Characterization of a proximal element in the rat preadipocyte factor-1 (Pref-1) gene promoter. Eur J Biochem. 2001;268:205–217. doi: 10.1046/j.1432-1033.2001.01847.x. [DOI] [PubMed] [Google Scholar]
- 38.Shen YN, Kim YM, Yun CH, Moon YS, Kim SH. Transcriptional activation of pref-1 by E2F1 in 3T3 L1 cells. BMB Rep. 2009;42:691–696. doi: 10.5483/bmbrep.2009.42.10.691. [DOI] [PubMed] [Google Scholar]
- 39.Couture JP, Blouin R. The DLK gene is a transcriptional target of PPARgamma. Biochem J. 2011;438:93–101. doi: 10.1042/BJ20101840. [DOI] [PubMed] [Google Scholar]
- 40.Zhou Y, Cheunsuchon P, Nakayama Y, Lawlor MW, Zhong Y, et al. Activation of paternally expressed genes and perinatal death caused by deletion of the Gtl2 gene. Development. 2010;137:2643–2652. doi: 10.1242/dev.045724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi N, Okamoto A, Kobayashi R, Shirai M, Obata Y, et al. Deletion of Gtl2, imprinted non-coding RNA, with its differentially methylated region induces lethal parent-origin-dependent defects in mice. Hum Mol Genet. 2009;18:1879–1888. doi: 10.1093/hmg/ddp108. [DOI] [PubMed] [Google Scholar]
- 42.Zhou Y, Zhang X, Klibanski A. J Mol Endocrinol Online before print; 2012. MEG3 non-coding RNA: a tumor suppressor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuster-Gossler K, Bilinski P, Sado T, Ferguson-Smith A, Gossler A. The mouse Gtl2 gene is differentially expressed during embryonic development, encodes multiple alternatively spliced transcripts, and may act as an RNA. Dev Dyn. 1998;212:214–228. doi: 10.1002/(SICI)1097-0177(199806)212:2<214::AID-AJA6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 44.McLaughlin D, Vidaki M, Renieri E, Karagogeos D. Expression pattern of the maternally imprinted gene Gtl2 in the forebrain during embryonic development and adulthood. Gene Expr Patterns. 2006;6:394–399. doi: 10.1016/j.modgep.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 45.Tunster SJ, Van De Pette M, John RM. J Biomed Biotechnol 2011: 283013; 2011. BACs as tools for the study of genomic imprinting. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yevtodiyenko A, Steshina EY, Farner SC, Levorse JM, Schmidt JV. A 178-kb BAC transgene imprints the mouse Gtl2 gene and localizes tissue-specific regulatory elements. Genomics. 2004;84:277–287. doi: 10.1016/j.ygeno.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 47.Auffray C, Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980;107:303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- 48.Smas CM, Green D, Sul HS. Structural characterization and alternate splicing of the gene encoding the preadipocyte EGF-like protein pref-1. Biochemistry. 1994;33:9257–9265. doi: 10.1021/bi00197a029. [DOI] [PubMed] [Google Scholar]
- 49.Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 50.Yevtodiyenko A, Carr MS, Patel N, Schmidt JV. Analysis of candidate imprinted genes linked to Dlk1-Gtl2 using a congenic mouse line. Mamm Genome. 2002;13:633–638. doi: 10.1007/s00335-002-2208-1. [DOI] [PubMed] [Google Scholar]
- 51.Lin SP, Youngson N, Takada S, Seitz H, Reik W, et al. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet. 2003;35:97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- 52.Steshina EY, Carr MS, Glick EA, Yevtodiyenko A, Appelbe OK, et al. Loss of imprinting at the Dlk1-Gtl2 locus caused by insertional mutagenesis in the Gtl2 5′ region. BMC Genet. 2006;7:44. doi: 10.1186/1471-2156-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sekita Y, Wagatsuma H, Irie M, Kobayashi S, Kohda T, et al. Aberrant regulation of imprinted gene expression in Gtl2lacZ mice. Cytogenet Genome Res. 2006;113:223–229. doi: 10.1159/000090836. [DOI] [PubMed] [Google Scholar]
- 54.Fowden AL, Coan PM, Angiolini E, Burton GJ, Constancia M. Imprinted genes and the epigenetic regulation of placental phenotype. Prog Biophys Mol Biol. 2011;106:281–288. doi: 10.1016/j.pbiomolbio.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 55.Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 56.Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, et al. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet. 2010;42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cockett NE, Jackson SP, Shay TL, Farnir F, Berghmans S, et al. Polar overdominance at the ovine callipyge locus. Science. 1996;273:236–238. doi: 10.1126/science.273.5272.236. [DOI] [PubMed] [Google Scholar]
- 59.Charlier C, Segers K, Karim L, Shay T, Gyapay G, et al. The callipyge mutation enhances the expression of coregulated imprinted genes in cis without affecting their imprinting status. Nat Genet. 2001;27:367–369. doi: 10.1038/86856. [DOI] [PubMed] [Google Scholar]
- 60.Davis E, Jensen CH, Schroder HD, Farnir F, Shay-Hadfield T, et al. Ectopic expression of DLK1 protein in skeletal muscle of padumnal heterozygotes causes the callipyge phenotype. Curr Biol. 2004;14:1858–1862. doi: 10.1016/j.cub.2004.09.079. [DOI] [PubMed] [Google Scholar]
- 61.Murphy SK, Freking BA, Smith TP, Leymaster K, Nolan CM, et al. Abnormal postnatal maintenance of elevated DLK1 transcript levels in callipyge sheep. Mamm Genome. 2005;16:171–183. doi: 10.1007/s00335-004-2421-1. [DOI] [PubMed] [Google Scholar]
- 62.Freking BA, Murphy SK, Wylie AA, Rhodes SJ, Keele JW, et al. Identification of the single base change causing the callipyge muscle hypertrophy phenotype, the only known example of polar overdominance in mammals. Genome Res. 2002;12:1496–1506. doi: 10.1101/gr.571002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 64.Kaufman MH. San Diego: Academic Press. 525 p; 1999. The Atlas of Mouse Development. [Google Scholar]