Mammalian Atg9 (mAtg9) is a multispanning membrane protein that resides in a novel compartment. mAtg9 interacts dynamically with phagophores and forming autophagosomes. It is proposed that mAtg9 function is required to initiate autophagosome formation and increase the number of autophagosomes.
Abstract
Autophagy is a catabolic process essential for cell homeostasis, at the core of which is the formation of double-membrane organelles called autophagosomes. Atg9 is the only known transmembrane protein required for autophagy and is proposed to deliver membrane to the preautophagosome structures and autophagosomes. We show here that mammalian Atg9 (mAtg9) is required for the formation of DFCP1-positive autophagosome precursors called phagophores. mAtg9 is recruited to phagophores independent of early autophagy proteins, such as ULK1 and WIPI2, but does not become a stable component of the autophagosome membrane. In fact, mAtg9-positive structures interact dynamically with phagophores and autophagosomes without being incorporated into them. The membrane compartment enriched in mAtg9 displays a unique sedimentation profile, which is unaltered upon starvation-induced autophagy. Correlative light electron microscopy reveals that mAtg9 is present on tubular–vesicular membranes emanating from vacuolar structures. We show that mAtg9 resides in a unique endosomal-like compartment and on endosomes, including recycling endosomes, where it interacts with the transferrin receptor. We propose that mAtg9 trafficking through multiple organelles, including recycling endosomes, is essential for the initiation and progression of autophagy; however, rather than acting as a structural component of the autophagosome, it is required for the expansion of the autophagosome precursor.
INTRODUCTION
To remain healthy, eukaryotic cells require a constant turnover and replacement of old components with functional new ones. Disposing of old organelles, often as big as mitochondria, is a challenging task that cells tackle using autophagy. Thus autophagy plays a major role in cell homeostasis (Mizushima, 2007). The key event in autophagy is the formation of a double-membrane structure called an autophagosome, which engulfs portions of cytosol and entire organelles. Autophagosomes fuse with the endolysosomal system, resulting in the degradation of their content by lysosomal enzymes (Orsi et al., 2010). Although it is active at basal levels under normal nutrient conditions, autophagy can be up-regulated in conditions of nutrient deprivation to maintain cytosolic pools of amino acids and macromolecules (Rabinowitz and White, 2010). Moreover, autophagy is used to clear protein aggregates (Tyedmers et al., 2010), as a defense against pathogens (Deretic and Levine, 2009), and to regulate immunity and inflammation (Levine et al., 2011). For these reasons, deregulation of autophagy has been implicated in several pathological conditions, including neurodegeneration, Crohn's disease, and cancer (Mizushima et al., 2008).
The process of autophagy is orchestrated by >30 proteins originally identified in yeast and collectively referred to as Atg proteins (Klionsky et al., 2003). Because autophagy is a multistep process, the different Atg proteins are used sequentially and ultimately result in the completion of the autophagosome. ULK1/2 (Atg1 in yeast), FIP200 (Atg17), and the phosphatidylinositol 3-kinase (PI3K) class III complex, including Vps34, Atg14, and Beclin 1 (Atg6), are involved in the induction and initiation phases (Yang and Klionsky, 2010). In yeast, the autophagosome originates at a precise and unique location in the cell called the preautophagosomal structure (PAS). In mammalian cells autophagosomes are formed at multiple locations from phagophores (Yang and Klionsky, 2010). Double FYVE–containing protein 1, (DFCP1), which is found on omegasomes, is a marker for these early phagophores (Axe et al., 2008; Hayashi-Nishino et al., 2009; Mari et al., 2011). In mammals, two ubiquitin-like conjugation systems produce the Atg5-12-16 complex and lipidated LC3 (LC3-II; Mizushima, 2007). LC3-II remains associated with the expanding autophagosome membrane and is sequestered inside the closed, completed autophagosome, providing a useful marker for the autophagosome. Subsequently, the autophagosome undergoes multiple fusion events with endosomes and lysosomes to become an autolysosome.
On amino acid withdrawal, autophagy is induced efficiently and green fluorescent protein (GFP)-LC3–positive autophagosomes are detected within minutes (Kochl et al., 2006). Autophagosome formation requires extensive membrane remodeling, but the source of the phagophore and the expanding autophagosome membrane is under investigation in many laboratories. Recent data suggest that multiple sources of membranes can contribute to the process (Tooze and Yoshimori, 2010), but a complete understanding is hampered by the fact that the association of nearly all Atg proteins with phagophores and autophagosomes is transient, and the majority of the Atg proteins, with the exception of Atg9, have no known integral membrane domains. In contrast, Atg9 has six conserved transmembrane domains (He et al., 2006; Young et al., 2006) and therefore should be the most useful Atg protein to track the origin of the autophagosome membrane.
In yeast, Atg9 is found on “Atg9 reservoirs,” which by electron microscopy appear as bundles of vesicles and tubules (Mari et al., 2010). On induction of autophagy by nutrient deprivation, these reservoirs move en bloc to the PAS, and this recruitment depends on Atg17 and requires Atg1 (Sekito et al., 2009). Because Atg9 has not been found on autophagic bodies in the yeast vacuole, it has been postulated that it recycles back to the PAS before autophagosomes fuse with the vacuole, and that the recycling process is dependent on Atg18–Atg2 (Reggiori et al., 2004a), which are the yeast homologues of the mammalian WIPI (1–4) proteins, and Atg2A and B. In mice, Atg9 (also known as Atg9a, Atg9L1, or mAtg9), like Atg5 or Atg7, is required for survival during the early neonatal starvation period, which depends on autophagy (Kuma et al., 2004; Komatsu et al., 2005), thus establishing an essential role for mAtg9 (Saitoh et al., 2009). In addition, mAtg9−/− mouse embryonic fibroblasts (MEFs) display reduced autophagic response, as revealed by fewer LC3-positive autophagosomes and reduced long-lived protein degradation (Saitoh et al., 2009). Atg9 is also essential for formation of a double-membrane around Salmonella in the Salmonella-containing vacuole and recruitment of the PI3K complex (Kageyama et al., 2011). In contrast to the yeast protein, mAtg9 localizes to both a juxtanuclear region, which corresponds to the trans-Golgi network (TGN), and a peripheral population, which was initially shown to partially colocalize with markers of late endosomes (LEs) such as Rab7 and Rab9 (Young et al., 2006). On amino acid deprivation, the tight juxtanuclear population redistributes in a ULK1-dependent manner, suggesting that mAtg9 is mobilized to support an increasing demand for autophagosomes. In support of this, it has been shown that manipulating mAtg9 relocalization to peripheral endosomes through activation of the p38α mitogen-activated protein (MAP) kinase pathway results in increased autophagy (Webber and Tooze, 2010a).
However, the exact function of Atg9 is elusive. Our current understanding of the localization of Atg9 and its role in autophagy is primarily derived from the yeast Saccharomyces cerevisiae (see earlier discussion) and correlation with relatively late events in mammalian cells (LC3 lipidation, LC3 spot formation, Rab7 and 9 colocalization, and long-lived protein degradation; Young et al., 2006; Saitoh et al., 2009). The current view that Atg9 may function to deliver membrane to growing phagophores and autophagosomes has not been formally proven (Orsi et al., 2010). Given the lack of amino acid conservation between the cytosolic domains of yeast and mammalian Atg9 (Figure 1A), which are among the domains likely to interact with cytosolic vesicle-sorting machinery, we addressed the function of mammalian Atg9 by determining its role in phagophore formation and expansion and the dynamics of how it interacts with these autophagosome precursors and autophagosomes. In addition, we characterized the membrane compartment where Atg9 is enriched, both morphologically and biochemically. We found that a population of mAtg9 localizes to several organelles, including recycling endosomes (REs) and early endosomes, in addition to TGN and LEs as previously described (Young et al., 2006). mAtg9 undergoes a dynamic interaction with phagophores and autophagosomes, without, however, becoming incorporated into their membrane and shows substantially less colocalization with autophagy markers than the organelle markers. By clarifying its relationship with other master regulators of autophagy such as ULK1 and WIPI2, we also show that mAtg9 acts at or on the phagophore and omegasome, revealing its position at the apex of the Atg protein hierarchy in mammalian cells.
FIGURE 1:
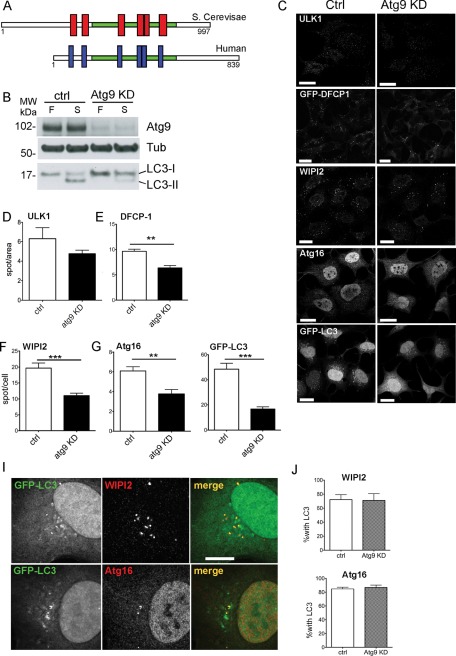
mAtg9 affects autophagy at an early omegasome stage. (A) Domain structure of yeast and human Atg9. Red and blue boxes represent conserved transmembrane domains. (B) HEK293 cells were treated with either control siRNA (ctrl) or siRNA against mAtg9 (Atg9 KD) and incubated in full (F) or starvation medium (S) for 2 h. Cell lysates were analyzed by Western blot using antibodies against mAtg9, β-tubulin, and LC3. (C) Confocal microscopy of HEK293/GFP-LC3 and HEK293/GFP-DFCP1 cells treated with RISC-Free (ctrl) or mAtg9 (Atg9 KD) siRNA and starved for 2 h. ULK1, WIPI2, and Atg16 were detected by indirect immunofluorescence using anti-ULK1, -WIPI2, and -Atg16 antibodies. Bars, 10 μm. (D–H) Quantification of ULK1, GFP-DFCP1, WIPI2, Atg16, or GFP-LC3 spots from B expressed as spots/cell area (ULK1, GFP-DFCP1) or spots/cell (WIPI2, Atg16, GFP-LC3). Error bars, SEM; **p < 0.01 ***p < 0.001 (n = 3, two-tailed unpaired t test). (I) Colocalization of GFP-LC3 with WIPI2 and Atg16 in HEK293/GFP-LC3 cells treated with mAtg9 siRNA, starved, and detected in C. (J) Quantification of colocalization of WIPI2 and Atg16 with GFP-LC3 as shown in (I). Error bars, SEM, n = 3.
RESULTS
mAtg9 affects autophagy at an early stage after induction
To more precisely examine when mAtg9 acts, we measured the number of autophagic and preautophagic structures in HEK293 cells depleted or not of mAtg9. We used markers that are recruited to the phagophore and form autophagosome at different stages: 1) ULK1, proposed to be one of the earliest (Itakura and Mizushima, 2010); 2) DFCP1, a early marker that labels the omegasome, expanding phagophores (Axe et al., 2008); 3) WIPI2, which, like DFCP1, is recruited by PI3P to the phagophore, but remains associated longer with the autophagosome (Polson et al., 2010); 4) Atg16, which, together with the Atg12–Atg5 complex, is required for lipidation and recruitment of LC3 (Mizushima, 2007); and 5) LC3, which is present during the latter stages of autophagosome formation and maturation.
Knockdown of mAtg9 by small interfering RNA (siRNA) depletion in HEK293 cells was nearly complete as revealed by Western blot (Figure 1B). As expected, depletion of mAtg9 significantly reduced lipidation of endogenous LC3 (Figure 1B) and the number of autophagic structures positive for GFP-LC3 (Figure 1C and I) observed upon starvation. All other autophagic structures, that is, those positive for DFCP1, WIPI2, and Atg16, were also significantly reduced (Figure 1, C and E–H), suggesting that mAtg9 may act very early, at the same step as or even before DFCP1. ULK1-positive puncta were reduced after depletion of Atg9 but not significantly (Figure 1, C and D). These results are in agreement with results in yeast, in which Atg9 is one of the first Atg proteins to be recruited to the PAS (Suzuki et al., 2007).
To understand whether the few autophagosomal structures still forming in cells depleted of mAtg9 were detected because of the presence of residual mAtg9, we used MEFs derived from Atg9−/− mice (Saitoh et al., 2009). In these MEFs, LC3 lipidation (Supplemental Figure S1, A and B) and the number of LC3- and WIPI2-positive spots induced by starvation were markedly reduced in comparison to wild-type MEFs (Supplemental Figure S1, C and D). However, a few autophagosomes were still able to form in Atg9−/− MEFs, suggesting that mAtg9 is important for the formation of autophagosomes but might not be strictly required.
Atg9 is not a structural component of the autophagosomal membrane
We next investigated whether mAtg9 was required for the correct recruitment of Atg proteins to the forming autophagosome, and, in particular, we looked at the recruitment of LC3 to WIPI2- or Atg16-positive structures. In control starved cells, ∼72 and 85% of WIPI2 and Atg16 spots, respectively, were also positive for LC3 (Figure 1, I and J). In cells depleted of mAtg9, although the total number of these structures was greatly reduced (Figure 1, D–H), the percentage of WIPI2 and Atg16 spots containing LC3 was unaltered (Figure 1J). This indicates that in the absence of mAtg9, these phagophores and autophagosome membranes were able to recruit Atg proteins upstream of LC3 lipidation and conjugation. Thus we hypothesized that mAtg9 may be delivering membrane and/or other components to speed up the process of autophagosome formation.
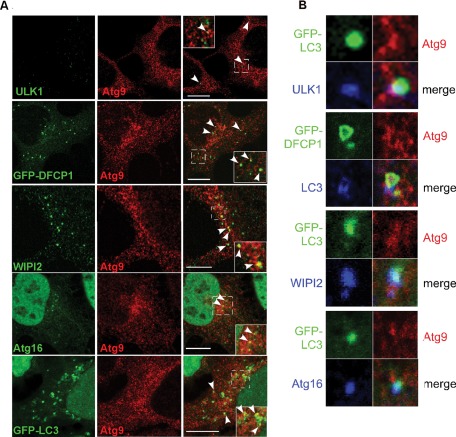
We examined the localization of mAtg9 during starvation and found that mAtg9 colocalizes, albeit to a limited extent, with several autophagosome markers. These include both early (ULK1, GFP-DFCP1, WIPI2) and later markers (Atg16, GFP-LC3; Figure 2A and Table 1). However, the vast majority of mAtg9 was not present on autophagosome structures (Figure 2A and Table 1). After amino acid starvation the percentage of the total population of mAtg9 that is found with autophagy proteins is ∼4%, which is barely above background of an irrelevant marker (Table 2). However, in the area of the phagophore and autophagosome-positive spots there is a higher percentage of Atg9, approaching 23% for DFCP1, colocalizing with the autophagosome marker (Table 1, local values). These values correlate with those previously reported for Atg9 and GFP-LC3 (Young et al., 2006). As expected, when we asked how many ULK1, DFCP1, WIPI2, Atg16, or LC3 spots contain Atg9 we found a range between 10 and 23% (Table 1). Moreover, when mAtg9 colocalizes with one of the autophagic markers in Figure 2, the colocalization is often only partial, suggesting that mAtg9 is present on an adjacent but distinct structure. Autophagosome markers ULK1, WIPI2, and Atg16 overlap perfectly with GFP-LC3 (Figure 2B); mAtg9 is found in the vicinity of ULK1 and LC3 but not completely overlapping, and the shape of the mAtg9-positive structure does not coincide with that of the other markers (Figure 2B). In the same way, mAtg9 partially colocalizes with DFCP1 (Figure 2, A and B). As expected, GFP-DFCP1 and LC3 are close but not overlapping, in agreement with previous data showing that LC3-positive structures emerge from DFCP1-positive omegasomes (Axe et al., 2008).
FIGURE 2:
mAtg9 colocalizes with both early and late autophagosome autophagy markers. (A) Confocal microscopy of mAtg9 (red) in HEK293/GFP-LC3 and HEK293/GFP-DFCP1 cells. GFP-DFCP1, ULK1, WIPI2, Atg16, or GFP-LC3 in green as indicated. ULK1, WIPI2, Atg16, and mAtg9 were detected using antibodies as in Figure 1. Arrows in merge show mAtg9 colocalization with markers. Bars, 10 μm. (B) Confocal microscopy of HEK293/GFP-LC3 cells, GFP-LC3 (green), ULK1 (blue), mAtg9 (red); HEK293/GFP-DFCP1 cells with mAtg9 (red), LC3 (blue); HEK293/GFP-LC3 cells, GFP-LC3 (green), Atg9 (red), and WIPI2 and Atg16 (blue). A low and partial colocalization of ULK1 and Atg9 is shown and is in contrast to the colocalization of ULK1 with LC3. LC3 emerges from the GFP-DFCP1 structure, and mAtg9 coincides partially with both structures. Bottom, WIPI2, Atg16, and GFP-LC3 signals overlap, whereas mAtg9 is found in close proximity with WIPI2, Atg16, and GFP-LC3. Quantification of the percentage of Atg9 found with ULK1, DFCP1, WIPI2, Atg16, and LC3 is shown in Table 1.
TABLE 1:
Colocalization of mAtg9 with phagophore and autophagosome markers.
| Percentage of Atg9 with phagophore and autophagosome markers | ||||
|---|---|---|---|---|
| Total Atg9 | SEM | Local Atg9 | SEM | |
| Ulk1 | 0.52 | 0.37, n = 2 | 15.03 | 1.01 n = 2 |
| DFCP1 | 4.00 | 0.92, n = 3 | 23.20 | 4.02 n = 3 |
| WIPI2 | 1.73 | 0.16, n = 3 | 15.60 | 1.26 n = 2 |
| Atg16 | 1.08 | 0.89, n = 3 | 9.77 | 2.48 n = 3 |
| LC3 | 4.50 | 0.64, n = 3 | 18.42 | 2.48 n = 3 |
| Phagophore and autophagosome markers | ||||
| Percentage with Atg9 | SEM | |||
| ULK1 | 19.01 | 5.82, n = 2 | ||
| DFCP1 | 15.71 | 3.71, n = 3 | ||
| WIPI2 | 10.44 | 2.73, n = 2 | ||
| Atg16 | 22.39 | 6.17, n = 2 | ||
| LC3 | 16.31 | 5.81, n = 3 | ||
All quantifications were performed with Imaris software, setting an appropriate threshold, which was kept constant during the whole analysis. Images for quantification were taken under the same conditions, and about five images per experiment were quantified. The values of all images from a single experiment were averaged together, and this value was used as n = 1 for statistical analysis with Excel and Prism software.
For “total Atg9,” the signal of Atg9 and the relative marker in the whole cell were compared. For “local” colocalization, at least 20 spots that were positive for the marker of interest were picked in blind per experiment. The colocalization between Atg9 and the marker was then evaluated only in a squared area of approximately twice the diameter around each spot.
TABLE 2:
Percentage of Atg9 colocalizing with organelle markers.
| Percentage Atg9 | SEM | |
|---|---|---|
| EEA1 | 18.90, n = 2 | 10.90 |
| TfR | 24.10, n = 2 | 7.43 |
| TGN46 | 26.30, n = 3 | 4.72 |
| Rab11 | 9.45, n = 2 | 2.20 |
| COPI | 6.40, n = 1 | ND |
| PDI | 8.91, n = 1 | ND |
| Tubulin | 11.24, n = 1 | ND |
Colocalization analyses were performed with Imaris software as described for Table 1. The whole Atg9 signal and that of the relative organelle marker were analyzed. TGN46 was included as positive control (as described in Young et al. 2006), whereas tubulin, COPI and PDI were taken as “irrelevant” markers with which Atg9 may appear to colocalize due to its diffuse distribution.
Atg9 interacts with autophagosomes transiently
To reconcile these data with the role of mAtg9 in autophagy, we hypothesized that mAtg9 may interact with phagophores and autophagosomes transiently and not remain on the autophagosome membrane. To visualize the movement of mAtg9 in live cells, we used time-lapse widefield microscopy, allowing us to track the fluorescent vesicles continuously without the risk of vesicles moving out of the focal plane. For these experiments, we generated stable cell lines expressing monomeric red fluorescent protein (mRFP)–Atg9 alone and in combination with GFP-LC3 (Supplemental Figure S2). Unfortunately, we could not establish a stable cell line expressing both mRFP-Atg9 and GFP-DFCP1 and had to rely on transient transfection of mRFP-Atg9 in GFP-DFCP1–expressing 2O1 cells. When stably expressed at low levels, mRFP-Atg9 showed an identical behavior and localization profile to that of endogenous mAtg9 (Supplemental Figure S2A). In stable cell lines, LC3 lipidation and GFP-LC3 spot formation were comparable with parental lines (Figure S2, B and C), showing that they undergo normal autophagy.
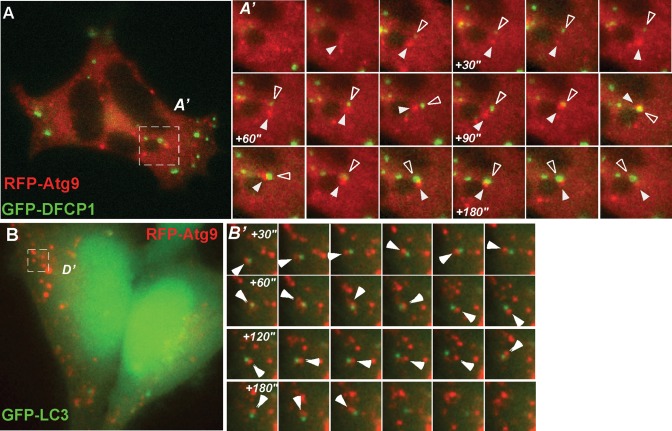
In cells expressing mRFP-Atg9 and GFP-DFCP1, GFP-DFCP1 spots form within 5 min of incubation in starvation medium, as in cells expressing GFP-DFCP1 alone, and in both we confirmed that GFP-DFCP1–positive omegasomes are relatively immobile (Axe et al., 2008). mRFP-Atg9 vesicles, however, appear to be very mobile and dynamic and transiently interact with GFP-DFCP1 structures (Supplementary Video 1 and Figure 3A′). Occasionally a GFP-DFCP1–positive structure emerges from a mAtg9 spot (Supplementary Video 2 and Figure 3A), whereas in other areas of the cell the GFP-DFCP1 structure encounters mAtg9 structures after it has formed. We observed both brief (<10 s) and prolonged (≤2 min) contacts but never observed mRFP-Atg9 fusing to GFP-DFPC1–positive structures. Instead, mAtg9 appears to orbit around GFP-DFCP1 structures (Figure 3A). In GFP-LC3/mRFP-Atg9 cells, GFP-LC3 structures are more dynamic than GFP-DFCP1 structures and less restricted to the juxtanuclear region (Figure 3B). Again, mAtg9, which is as mobile as LC3, orbits GFP-LC3–positive structures without fusing with them (Supplementary Video 2 and Figure 3B′). Frequently we observed GFP-LC3 spots interacting with more than one mAtg9 structure.
FIGURE 3:
(A) Live-cell imaging of HEK293/GFP-DFCP1 cell transfected with mRFP-Atg9 using widefield fluorescence microscopy. (A′) Magnified frames from the boxed region. Frames are taken from Supplementary Video S1, which started after 5 min of incubation in starvation medium. Images were captured every 5 s, and every other frame is shown. White arrowhead, mRFP-Atg9 vesicle; empty arrowhead, GFP-DFCP1. Note how the two structures orbit each other without significant overlap except at +110 s, immediately after which they separate. (B) Live-cell imaging of HEK293/GFP-LC3/mRFP-Atg9 cells by widefield fluorescence microscopy. (B′) Magnified boxed regions. Images are extracted from Supplementary Video S2. Frames were taken every 5 s, and every other frame is shown. Note how the two structures orbit each other without significant overlap except at +60 s, immediately after which they separate. White arrowhead, mRFP-Atg9 vesicle.
Previous data in yeast demonstrated that Atg9 was not found on autophagosomes or in autophagosomes fused with the vacuole (Lang et al., 2000; Noda et al., 2000; Kim et al., 2002). Our observations support the conclusion that mAtg9 interacts dynamically with phagophores and early and late autophagosomes but does not at any stage become a stable part of the autophagosome membrane. Due to their high mobility, it is also unlikely that mAtg9-positive vesicles or membranes act as a stable platform for recruitment of other Atg proteins.
mAtg9 is not found on the autophagosome membrane
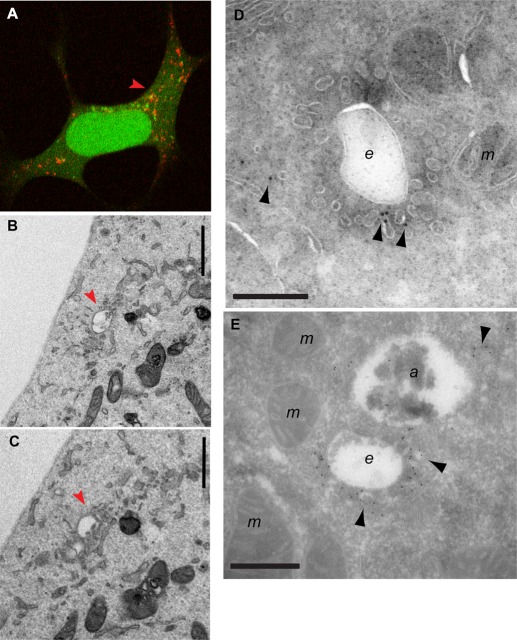
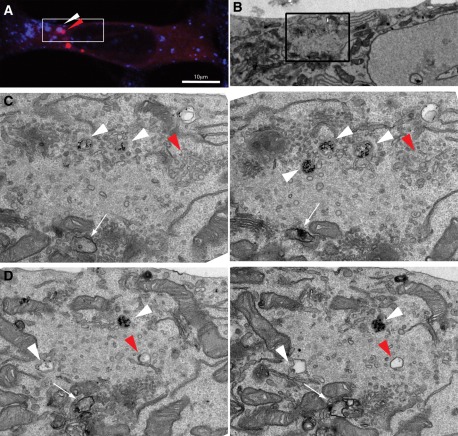
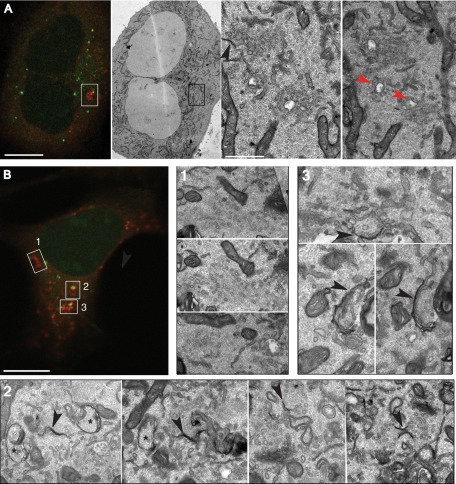
To more clearly visualize the mAtg9 compartment and to understand how it relates to phagophores and autophagosomes, we performed correlative light electron microscopy (CLEM) using cells expressing GFP-LC3 and mRFP-Atg9 (Figure 4 and Supplemental Figure S3A). Structures containing GFP and mRFP were identified by confocal microscopy in fixed cells and then relocated after Epon embedding for transmission electron microscopy (TEM) and imaged by TEM. These GFP-LC3-mRFP-Atg9 structures appear to be very complex, as can be appreciated in sequential serial sections (Figure 4, B and C). We observed clusters of tubules and vesicles emanating from endosome- or MVB-like structures corresponding to the mRFP-Atg9 signal (Figure 4, red arrows), in areas where GFP-LC3 was detected by fluorescence microscopy (Figure 4A).
FIGURE 4:
CLEM and cryoimmuno-EM demonstrates mAtg9 is present on tubulovesicular membranes surrounding autophagosomes. (A) HEK293/GFP-LC3/mRFP-Atg9 cells starved for 2 h were fixed, and GFP-LC3 (green) or mAtg9 (red) fluorescent structures were identified by confocal microscopy and subsequently in thin sections prepared for TEM. Low-magnification TEM of the whole cell is shown in Supplemental Figure S3A. (B, C) two 70- to 100-nm serial sections of the boxed area in Supplemental Figure S3A. The mRFP-Atg9–positive membranes (red arrows) corresponding to vesicular clusters and tubules emerging from multivesicular structures (red arrows). Bar, 500 nm. (D, E) Cryoimmuno-EM of HEK293/GFP-LC3/mRFP-Atg9 cells (D) or primary rat hepatocytes (E), transduced with mRFP-Atg9, starved for 2 h, and labeled for mAtg9 (10- or 5-nm gold, respectively). mAtg9 (arrowheads) is found on vesicles and tubules, often surrounding large endosomal-like structures. In E, note that mAtg9 is not found on the autolysosome membrane, although some signal is present inside that could represent trapped material. Bar, 500 nm. a, autolysosome; e, endosome; m, mitochondria. Bars, 500 nm (B, C), 1 μm (D, E).
To confirm the presence of mAtg9 on these structures, we performed cryoimmuno-EM of HEK293 cells or primary rat hepatocytes expressing mRFP-Atg9 (Figure 4, D and E, and Supplemental Figure S3, B–E). A low but specific signal for mAtg9 was mostly found on clusters of small vesicles and tubular structures scattered throughout the cytosol, similar to the level of signal detected on the Golgi (Young et al., 2006). These vesicular–tubular structures are reminiscent of what has been described as “Atg9 reservoirs” in yeast (Mari et al., 2010). Very often we observed large, endosome-like structures surrounded by small, mAtg9-positive vesicles/tubules, similar to what we observed by CLEM (Figure 4, A–C, and Supplemental Figure S3, B and E). However, mAtg9 could not be found on the membrane of morphologically identifiable autophagosomes (unpublished data) or autolysosomes (Supplemental Figure S3, C and D). Occasionally we detected gold particles labeling mRFP-Atg9 inside autophagosomes (unpublished data) or in autolysosomes (Figure 4E), which could represent mAtg9-positive entrapped membranes. In summary, these data show that mAtg9 resides on a tubular/vesicular compartment, in proximity to endosomes and autophagosomes, but apparently not on the autophagosome membrane itself.
mAtg9 is found in a transferrin receptor–positive compartment
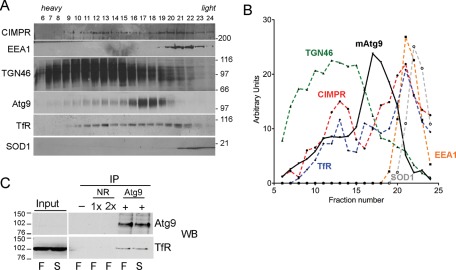
To further understand the mAtg9 compartment, we used subcellular fractionation. On the basis of the foregoing EM analysis and our results demonstrating that early and late endosomes cofractionated with mAtg9 (Young et al., 2006), we followed the RE marker transferrin receptor (TfR) and compared it to the behavior of mAtg9. Cell homogenates from HEK293 cells were fractionated on a Ficoll gradient, and the fractions were resolved by SDS–PAGE and probed for markers of cellular compartments. As expected, mAtg9 partially overlaps with TGN (TGN46) and late endosome (cation-independent mannose-6-phosphate receptor [CI-MPR]) markers (Figure 5, A and B). The overall profile of mAtg9 is different from the markers tested; however, a small amount of mAtg9 is found with EEA1 (fractions 19 and 20), and a more significant overlapping peak of TfR and mAtg9 is detected in fractions 16–18.
FIGURE 5:
mAtg9 is on recycling endosomes. (A) Homogenates from HEK293 cells were separated on 1–22% Ficoll gradients. Gradient fractions were loaded from left (heavy) to right (light) and then analyzed by Western blot using antibodies against CI-MPR (marker for late endosomes/TGN), EEA1 (early endosomes), TGN46 (TGN), mAtg9, TfR (RE), and SOD1 (cytosol). (B) Distribution of marker proteins. The intensity of each band (in arbitrary units) is plotted on the y-axis and the fraction number on the x axis. mAtg9, black solid line; all other markers are dashed lines: red, CI-MPR; yellow, EEA1; green, TGN46; blue, TfR; gray, SOD1. (C) Lysates from HEK293 cells incubated in full medium (F) or starved for 2 h (S) were immunoprecipitated using beads alone (–), a nonrelevant antibody (NR) at 1× and 2× immunoglobulin G (IgG), and anti-mAtg9 IgG and then analyzed by Western blot using antibodies against mAtg9 and TfR. Input, 5% of total lysate before immunoprecipitates (IP). Data are representative of three independent experiments.
Because the mAtg9-positive membranes appear to be distinct, we obtained more information about this compartment by screening for potential interactors using immunoprecipitation followed by SDS–PAGE and mass spectroscopy analysis. Hemagglutinin (HA)-tagged mAtg9 was expressed in HEK293A cells and immunoprecipitated using anti-HA antibodies (Supplemental Figure S4A). In two independent determinations (Supplemental Figure S4, B and C) we identified CI-MPR and TfR as specific proteins that coimmunoprecipitate with HA-mAtg9. The analysis was performed in both basal (fed) and starved (amino acid depleted) conditions, but we did not observe any differences in the peptide numbers detected. These results further support the presence of mAtg9 in endosomal compartments, including the RE.
We previously found a colocalization of mAtg9 with CI-MPR both by indirect immunofluorescence and cryoimmunogold labeling of mAtg9 and CI-MPR on late endosomes (Young et al., 2006). We were able to confirm the coimmunoprecipitation of CI-MPR with HA-mAtg9, but depletion of CI-MPR had no effect on mAtg9 distribution or autophagy (Robinson, unpublished results). We confirmed the interaction between mAtg9 and TfR by immunoprecipitation using both mRFP- and HA-tagged mAtg9 (Supplemental Figure S4D) and coimmunoprecipitation of endogenous mAtg9 with TfR (Figure 5C). We were able to demonstrate that another RE membrane protein, syntaxin-13 (Prekeris et al., 1998), did not coimmunoprecipitate with endogenous Atg9 (Supplemental Figure S4E). However we cannot formally exclude the possibility that the coprecipitation of Atg9 and TFR is indirect due to the mild solubilization conditions (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate [CHAPS] detergent lysis). Furthermore, we tested the effect of TfR depletion on mAtg9 trafficking and LC3 lipidation using previously published siRNA duplexes (Herbison et al., 2009) but obtained conflicting results and were unable to draw any conclusions about the role of TfR in autophagy. This may be due to the potentially toxic and stressful effects in our cell lines following loss of the TfR combined with amino acid depletion. However, knockdown of mAtg9 did not affect TfR or CI-MPR localization (unpublished data), suggesting that these proteins are found in the same membrane compartment but that mAtg9 does not regulate the localization of either protein.
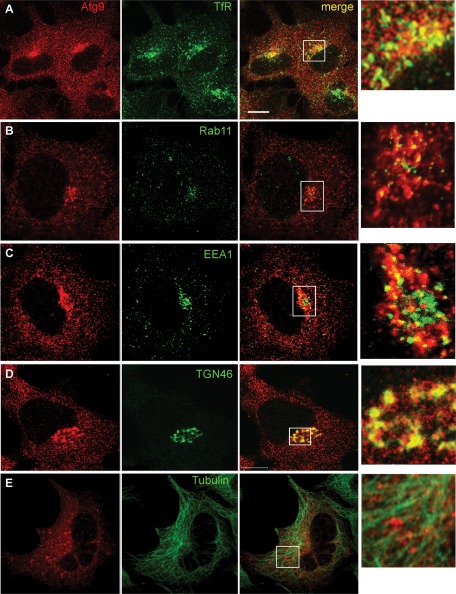
We next asked whether mAtg9 colocalized with TfR in fed and starved cells. We could detect a juxtanuclear population of mAtg9 that colocalized with TfR (Figure 6A). This colocalization was unchanged in amino acid starvation (unpublished data). We also examined other markers of RE, including Rab11, and early endosomes, EEA1, and found that they both showed some degree of colocalization (Figure 6, B and C). Because mAtg9 has a very widespread distribution in the cell, we quantified the percentage of Atg9 that colocalized with TfR, Rab11, and EEA1 and compared this to our positive control TGN46 (Figure 6D) and markers including tubulin (shown in Figure 6E), COPI, and protein disulfide isomerase (PDI), which should not colocalize to a large extent with mAtg9. As shown in Table 2, 26% of total mAtg9 colocalized with TGN46, followed by TfR, with 24% colocalization of total Atg9. Colocalization with Rab11 was not substantially higher than that of the irrelevant markers, whereas EEA1 showed a reasonable degree of colocalization.
FIGURE 6:
mAtg9 colocalizes with endosomes and TGN. Confocal microscopy of HEK293/GFP-LC3 cells. mAtg9 (red) and (A) TfR, (B) Rab11, (C) EEA1, (D) TGN46, and (E) tubulin were detected by indirect immunofluorescence with antibodies to indicated proteins (green). TfR and TGN46 show the highest degree of partial colocalization with mAtg9. Insets in the merge panel are shown on the right. Quantification is shown in Table 2. Bar, 10 μm.
Our data show that mAtg9 resides in a distinct membrane compartment with characteristics that partially overlap with those of the Golgi, early and late endosomes, and REs. Moreover, mAtg9 interacts and colocalizes with a population of TfR. These conclusions are supported by the EM analysis, which identified mAtg9 juxtaposed to and labeling tubulovesicular structures with a morphology resembling that of REs. Note that these structures were not close to stacked Golgi complex membranes, making it unlikely that they were Golgi derived (Figure 4).
To further study the relationship between the mAtg9-positive membranes and TfR-positive REs, we performed an uptake experiment using fluorescently labeled Tfn and gold-conjugated anti-TfR antibody in HEK293/mRFP-Atg9 cells for 2 h under starvation conditions, followed by CLEM (Figure 7). This approach enabled us to visualize the entire RE pathway using TEM. We focused on mAtg9-positive structures that contained the fluorescent Tfn ligand (Figure 7A) and then located them by TEM (Figure 7B-D). We found heavily labeled endosomes containing anti–TfR-gold where the fluorescent ligand was present (Figure 7, C and D, white arrows), and a tubular vesicular network of membrane where mRFP-Atg9 was localized (red arrowheads). In addition, within this tubular network were vacuolar endosome-like structures that contained a small amount of anti–TfR-gold (Figure 7D, white arrowheads). Of importance, the mRFP-Atg9 and sparsely labeled endosomes in Figure 7D were near forming and newly formed autophagosomes (Figure 7, C and D, white arrows). These results demonstrate the close interconnection between the unique Atg9 membranes and the TfR-containing endosomes.
FIGURE 7:
CLEM of transferrin-positive compartments and mAtg9 compartments. HEK293/mRFP-Atg9 cells were incubated for 2 h with anti-TfR antibody conjugated to 10-nm gold and Alexa 647–conjugated Tfn. Cells were fixed, and (A) Tfn-Alexa 647 (blue) or mRFP-Atg9 (red) fluorescent structures were identified by confocal microscopy and subsequently in thin sections prepared for EM. (B) Low-magnification TEM of the top of the cell from A. (C, D) two sets of 70- to 100-nm serial sections from the boxed area in B. TfR-gold–positive vacuoles were identified in TEM (white arrowheads) near the Atg9 compartment: mRFP-Atg9–positive membranes (red arrowheads) corresponding to vesicular clusters and tubules, and vacuoles, some of which contain small amounts of TfR-gold. White arrows show phagophore and autophagosome membranes. Bar (A), 10 μM.
Retromer is an important regulator of retrograde transport from the endosome and has been implicated in the traffic of both CI-MPR and TfR (Seaman, 2004; Popoff et al., 2007). Thus we tested whether retromer likewise controlled the traffic of mAtg9 or had a role in autophagy. We found a partial colocalization between mAtg9 and Vps26 (Supplemental Figure S5A), an essential subunit of the retromer complex. However, in cells depleted for Vps26 (Supplemental Figure S5A, bottom), we detected no difference either in starvation-dependent mAtg9 redistribution or in the reestablishment of mAtg9 juxtanuclear population after 30 min of recovery from nutrient depletion (Supplemental Figure S5, B and C). LC3 lipidation and LC3-II turnover were also unaffected (Supplemental Figure S5, D and E), showing that retromer activity is dispensable for autophagy.
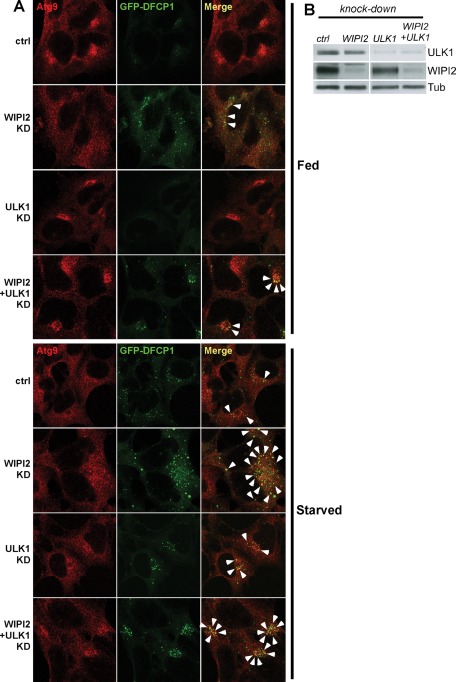
mAtg9 localization is dependent on WIPI2
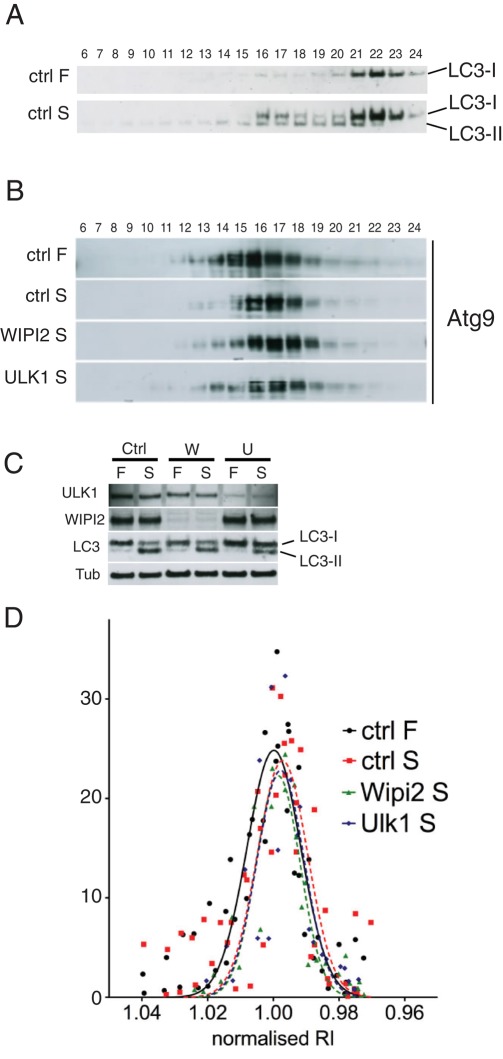
In yeast, Atg9 cycles between the “Atg9 reservoir” and the PAS. This traffic largely depends on Atg1 and Atg18, and in the absence of Atg18, Atg9 remains on the PAS (Reggiori et al., 2004a; Mari et al., 2010). Recent data in mammalian cells suggest that DFCP1-positive omegasomes may be considered a mammalian equivalent of the PAS (Axe et al., 2008). Depletion of WIPI2, one of the mammalian homologues of Atg18, causes an accumulation of GFP-DFCP1–positive omegasomes in both fed and starved cells. These structures are unable to mature, as they do not recruit LC3 (Polson et al., 2010). Thus we asked whether WIPI2 was required for mAtg9 trafficking. After depletion of WIPI2, we observed that mAtg9 is found in the regions of these stalled omegasomes, suggesting that mAtg9 can move to these structures in the absence of WIPI2 but is unable to be removed (Figure 8A). Depletion of ULK1, the mammalian homologue of Atg1, was previously shown to cause a concentration of mAtg9 around the juxtanuclear region (Young et al., 2006). Under these conditions, the omegasome structures cluster in a juxtanuclear region; however, the extent of DFCP1-mAtg9 colocalization is unaltered, suggesting that ULK1 is required neither for mAtg9 recruitment to the omegasomes nor for its retrieval (Figure 8A). Double knockdown of WIPI2 and ULK1 (Figure 8, A and B) recapitulates both phenotypes: an increased number of DFCP1 structures (especially in full medium) with increased mAtg9-DFCP1 colocalization (WIPI2 phenotype) and clustering of these structures around the nucleus (ULK1 phenotype). Thus we propose that mAtg9 is able to traffic to DFCP1-positive omegasomes independent of WIPI2 and ULK1, whereas its retrieval depends on WIPI2. ULK1 instead appears to regulate the distribution of mAtg9 in peripheral structures and the areas where new omegasomes can form.
FIGURE 8:
Regulation of mAtg9 traffic by ULK1 and WIPI2. (A) Confocal microscopy of HEK293/GFP-DFCP1 cells treated with siRNAs against RISC-free (ctrl), ULK1, WIPI2, or WIPI2 and ULK1. Cells were incubated in full medium or starved for 2 h before fixation. mAtg9 was detected by indirect immunofluorescence. Arrows indicate areas of colocalization between GFP-DFCP1 and mAtg9. In WIPI2 KD virtually every DFCP1 spot colocalizes with mAtg9. (B) The efficiency of the knockdowns in A was confirmed by Western blot using antibodies against ULK1, WIPI2, and β-tubulin. One representative experiment of three is shown.
The mAtg9 compartment retains its distinct properties after starvation and loss of WIPI2 and ULK1
Our data so far suggest that mAtg9, despite being essential for the formation and expansion of autophagosomes, does not actually become a stable component of their membranes at any stage. Thus we hypothesized that the characteristics of the mAtg9 compartment may not change upon induction of autophagy. Using subcellular fractionation in fed and starved conditions, we detected the formation of autophagosomes using lipidated LC3. In starved conditions LC3-II appears in the heavy fractions, which correspond to autophagosomes (Figure 9A). However, we could not detect any major difference in the distribution of endogenous mAtg9 between full-medium and starved conditions (Figure 9, B and D, and Supplemental Figure S6C).
FIGURE 9:
The mAtg9 compartment is unaltered after starvation. (A) HEK293 cells were incubated in full (F) or starvation medium for 2 h (S) before homogenization and separation on a 1–22% Ficoll gradient as in Figure 6A. Fractions were loaded from left (heavy) to right (light) and analyzed by Western blot using an antibody against LC3. Membrane-associated LC3-II is found in fractions 13–21. (B) HEK293 cells were treated with RISC-Free (ctrl) siRNA or siRNA against WIPI2 or ULK1 and then incubated in full medium (F) or starved (S) for 2 h before homogenization and fractionation as in A, followed by Western blot for endogenous mAtg9. (C) The efficiency of the knockdown in B was evaluated using antibodies against ULK1, WIPI2, and β-tubulin and confirmed by inhibition of LC3 lipidation after both WIPI2 and ULK1 KD. (D) The intensity of mAtg9 signal was quantified (in arbitrary units) and plotted against the refractive index of each fraction (see example in Supplemental Figure S5D and Materials and Methods). To allow comparison of experiments, a Gaussian curve was fitted for each profile. For each experiment the values were normalized to the median value of the ctrl-F sample, and a Gaussian curve was fitted for each value. No significant shift from the median value was detected. Representative experiments are shown. Ctrl-F and ctrl-S, n = 4; WIPI2 and ULK1, n = 2.
We took advantage of the ULK1- and WIPI2-knockdown phenotypes to provide a way to substantially alter the mAtg9 compartment. We predicted that in starved cells devoid of ULK1, mAtg9 would be predominantly in its nonautophagosomal locations, at the TGN and endosomes, whereas after WIPI2 knockdown mAtg9 would accumulate at the stalled preautophagosome structures. However, surprisingly, even in these knockdown conditions we were unable to detect any significant difference in the sedimentation behavior of endogenous mAtg9 (Figure 9, B–D). This suggests that either the bulk of mAtg9 is not on the stalled or clustered omegasomes or the apparent colocalization results from close juxtaposition of mAtg9 and GFP-DFCP1–positive membranes. Our quantification, shown in Table 1, also supports the conclusion that the bulk of mAtg9 is not present on autophagosomes. In light of these data, we decided to investigate the localization of mAtg9 in WIPI2-depleted cells and performed CLEM analysis on starved GFP-DFCP1 HEK293 cells expressing mRFP-Atg9 treated with control or WIPI2-specific siRNA (Figure 10). In WIPI2-depleted cells (Figure 10A), we examined the regions positive for both GFP-DFCP1 and mRFP-Atg9 using TEM and again observed tubular–vacuolar structures and clusters of vesicles (Figure 10A, red arrowheads) that were very similar to those structures seen in starved HEK293 cells and primary rat hepatocytes (Figures 4 and 7). In the control siRNA-treated GFP-DFCP1 cells expressing mRFP-Atg9 we were also able to identify the mRFP-Atg9–positive membranes in the vicinity of phagophores and forming autophagosomes (Figure 10B, black arrowheads). These biochemical and morphological data indicate that the characteristics of the mAtg9 compartment do not depend on ULK1 or WIPI2, and they provide further evidence that mAtg9 is not incorporated into precursor autophagosomes, phagophores, or autophagosome membranes but instead are found in a compartment that equates to the yeast Atg9 reservoir.
FIGURE 10:
CLEM analysis of mRFP-Atg9 in WIPI2-depleted GFP-DFCP1 cells. (A) CLEM of WIPI2-depleted starved or (B) RF starved cells. (A) Left, GFP-DFCP1/mRFP-Atg9–expressing cell. Right, boxed area in low-magnification image and two serial sections from this region. Red arrowheads indicate tubular and vacuolar Atg9-positive membranes. Black arrowhead indicates double-membrane phagophore. (B) Top left, confocal microscopy of mRFP-Atg9 and GFP-DFCP1–positive structures (boxes 1–3). These structures were then identified in low-magnification TEM and then in higher-magnification images, and sections from three different areas (1–3) are shown. Black arrowhead indicates double-membrane phagophores. Asterisk indicates autophagosome. Bars, 50 μm (A, B), 1 μm (A, middle panel).
DISCUSSION
As the only transmembrane Atg protein, Atg9 has been regarded as key to one of the mysteries of autophagy: where does the autophagosome membrane come from? Despite the fact that Atg9−/− mice are deficient in autophagy (Saitoh et al., 2009), it is evident that low numbers of autophagosomes are still able to form without mAtg9. This suggests that mAtg9 is not strictly required for the process of autophagosome formation. We propose that mAtg9's function is not essential in basal conditions (when few autophagosomes form) but might become essential when autophagy is induced and the demand for new autophagosome membranes increases. This proposal is in line with the current hypothesis that Atg9 delivers lipids required for the initiation and expansion of autophagosomes (Reggiori et al., 2004a; Longatti and Tooze, 2009), but it may also be possible that Atg9 delivers regulators to the growing phagophore.
Yeast, as well as mammalian, Atg9 is translocated into the secretory pathway at the level of the ER, where it is glycosylated and then trafficked through the Golgi to a post-Golgi compartment (Young et al., 2006; Mari et al., 2010; van der Vaart et al., 2010). Moreover, it appears that Atg9 must traffic through and exit the Golgi to exert its function in autophagy (Ohashi and Munro, 2010). The post-Golgi pool of yeast Atg9 is found in a novel organelle—the “Atg9 reservoir” (Mari et al., 2010)—and is retrieved from the autophagosome before fusion with the vacuole (Lang et al., 2000; Noda et al., 2000; Kim et al., 2002). Furthermore, we showed that Atg9 colocalizes with CI-MPR and Rab7- and Rab9-positive endosomes (Young et al., 2006). Here we show that mammalian Atg9 is on a distinct, vesicular compartment: the colocalization of mAtg9 with phagophores and autophagosome markers is partial and asymmetric, and mAtg9 is not detectable on autophagosome membranes. Furthermore, the regulation of Atg9 trafficking originally described in yeast appears to be conserved between yeast and mammals. Given the complexity of the mammalian endosomal pathway and autophagosome maturation, this may be surprising; however, our data demonstrate that mAtg9 is present on a unique tubular vesicular compartment that has many of the same properties as the yeast Atg9 reservoirs.
Our biochemical analysis revealed that the characteristics of the mAtg9 compartment are unaffected by starvation-induced autophagy or by disruption of its starvation-stimulated trafficking, as controlled by Ulk1 and WIPI2. Similarly, Atg9 reservoirs in yeast display the same sedimentation properties in both atg1∆ and atg11∆ mutants (Mari et al., 2010), underscoring the unique behavior of Atg9-positive membranes. Using CLEM and immuno-EM, we show that mAtg9 is most often found on clusters of vesicles or tubules, occasionally surrounding endosome-like vacuolar structures in the vicinity of double-membrane phagophores. Our results may also suggest that the majority of mAtg9 is found on a completely independent compartment, which neither buds from nor fuses with other organelles. This is unlikely, since we report here that mAtg9 physically interacts with endosomal markers, in particular TfR. We favor the hypothesis that mAtg9 traffics by tubulation from an existing compartment and in doing so is able to maintain the same density as the donor compartment. Thus we propose that mammalian equivalents of yeast “Atg9 reservoirs” continuously emanate from vacuolar RE-like compartment, help promote autophagosomes formation, and then rapidly return to the donor compartment.
Recently, mAtg9-positive Golgi membranes, after amino acid starvation, were shown to undergo tubulation and fragmentation to produce cytoplasmic punctate structures dependent upon the N-BAR domain of Bif-1 (Takahashi et al., 2011). Vesicular traffic of mAtg9 may be occurring through these tubulated membranes. Given that REs are known to be highly tubulated (Bonifacino and Rojas, 2006), it would be interesting to test whether Bif-1 was also acting on the RE-like compartment we identified. Retromer mediates the tubulation of the RE and the sorting of CI-MPR and TnR from the REs (Seaman, 2004; Bonifacino and Rojas, 2006; Popoff et al., 2007), and recent experiments suggested that Atg9 trafficking from a Golgi–endosome trafficking pathway in yeast contributes to autophagy (Ohashi and Munro, 2010). In agreement with previous results in yeast (Reggiori et al., 2004b), we did not detect any role for retromer in mammalian autophagy.
Evidence from several groups suggests that autophagosomes can obtain membranes from multiple sources, including the ER (Hayashi-Nishino et al., 2009; Yla-Anttila et al., 2009), Golgi (Geng et al., 2010; van der Vaart et al., 2010; Yen et al., 2010), mitochondria (Hailey et al., 2010), and the plasma membrane (Ravikumar et al., 2010). We speculate that due to its widespread distribution, mAtg9 is the perfect candidate to relay membranes from different sources. In this context, mAtg9 may be strategically placed at the RE, a compartment that regulates endocytotic trafficking and connects the endocytic to the exocytic pathway (Bonifacino and Rojas, 2006; van Ijzendoorn, 2006). Further support for our hypothesis comes from data showing a functional connection between REs and the ER–Golgi intermediate compartment (Marie et al., 2009). It is tempting to speculate that mAtg9 traffic through the RE is essential for autophagy, and further experiments are required to directly test this hypothesis.
Atg9 is one of the first proteins to be recruited to the PAS in yeast (Suzuki et al., 2007). We show here for the first time that in starved mammalian cells mAtg9 acts at an early stage, as depletion of mAtg9 inhibits the formation of early ULK1-, DFCP1-, and WIPI2-positive phagophores. Similar findings on the role of Atg9 early in membrane formation, in particular recruitment of Atg14, was found in Salmonella-infected cells (Kageyama et al., 2011). Conversely, ULK1 and Atg13 knockdown inhibit mAtg9 trafficking, and mAtg9 localizes to a juxtanuclear region (Young et al., 2006; Chan et al., 2009), and early DFCP1-positive omegasomes are also restricted to this area and are unable to mature into LC3-positive autophagosomes. When WIPI2 is depleted, mAtg9 accumulates on DFCP1-positive omegasomes that are unable to mature. A similar result was recently obtained in Caenorhabditis elegans, in which an accumulation of large, Atg9-positive structures was seen after loss of EPG-6 (WIPI4), which had a unique distribution compared with Unc-51 and EPG-1 (Atg1 and Atg13 homologues in C. elegans) mutants (Lu et al., 2011). The ULK1/2 complex, in particular FIP200, has been shown to be the most upstream protein to be recruited to ER-localized phagophores and is required for localization of the class III PI3K complex (Itakura and Mizushima, 2010), followed by recruitment of WIPI2 (Polson et al., 2010). Because the combined knockdown of WIPI2 and ULK1 still allows mAtg9 to reach DFCP1-positive membranes, the initial recruitment of mAtg9 to the omegasome is independent of these two proteins (ULK1 and WIPI2), again placing mAtg9 high in the hierarchy of Atg proteins.
Finally, despite its essential role in autophagy, mAtg9 it is not a component of autophagosomes. In fact, mAtg9 resides on a separate compartment, acts dynamically and independently on the phagophore, and remains distinct from the autophagosome membrane. Our observations that phagophores and omegasomes form mainly in Atg9-positive regions lead us to propose that the widespread distribution of mAtg9 facilitates formation of autophagosomes throughout the cell. Our previous experiments revealed a role for p38 MAP kinase (Webber and Tooze, 2010b) and moreover showed that when the ability of mAtg9 to traffic to dispersed peripheral sites is restricted, autophagy is compromised. Further work is required to elucidate the signals in mammalian cells controlling Atg9 trafficking from the unique Atg9 compartment.
MATERIALS AND METHODS
Cell culture and transfection
HEK293A cells and derivatives were maintained in full medium (FM): DMEM with 10% fetal calf serum (FCS) as previously described (Kochl et al., 2006; Chan et al., 2007). To induce autophagy, cells were incubated for 2 h in starvation medium: Earle's balanced salt solution (EBSS). When indicated, bafilomycin A1 (Calbiochem, La Jolla, CA) in dimethyl sulfoxide was used at 100 nM. The 293/GFP-LC3 cells were previously described (Kochl et al., 2006). The 293/GFP-DFCP1 (clone 2O1; Axe et al., 2008) was a kind gift from N. T. Ktistakis (Brabaham Institute, Cambridge, United Kingdom) and was were maintained in the presence of G418 at 400 μg/ml. The Atg9−/− and matched wild-type immortalized MEFs (Saitoh et al., 2009) were a kind gift from T. Saitoh and S. Akira (Osaka University, Osaka, Japan) and were maintained in FM. The 293/GFP-LC3/mRFP-Atg9 and 293/mRFP-Atg9 were generated by transfection of 293/GFP-LC3 and HEK293 cells with pCDNA4-TO-mRFP-Atg9 or pEGFP-mRFP-Atg9 plasmids and selected with 750 μg/ml of Zeocin (Invitrogen, Carlsbad, CA) or 800 μg/ml of G418 (Life Technologies, Carlsbad, CA), respectively, followed by clonal dilution. Clones were screened by Western blot and confocal microscopy for low-level expression, in which mRFP-Atg9 distribution resembled that of endogenous mAtg9. The 293/GFP-LC3/mRFP-Atg9 clone 9B9 was maintained in the presence of Zeocin at 400 μg/ml. The 293/mRFP-Atg9 clone 1F10 was maintained in the presence of G418 at 400 μg/ml.
Transfection
For RNA interference, cells were transfected using Oligofectamine (Invitrogen), followed by a second transfection 24 h later with Lipofectamine 2000 (Invitrogen). Cells were analyzed 72 h after the first transfection. The final concentration of siRNA oligos ranged between 20 and 50 μM, depending on the target protein: D-001220-01 (RISC-Free, control), D-014294-02 or -04 (Atg9), D-020521-09 or -12 (WIPI2), or D-005049-04 (ULK1; Dharmacon, Lafayette, CO). Custom siRNA oligo UGA UGG GGA AAC CAG GAA AUU was used to knock down Vps26. Transferrin receptor was knocked down using siRNA oligos (pools of two) as previously described (Herbison et al., 2009).
Lipofectamine 2000 was used for transient transfection of DNA. For biochemical analysis, 1 μg of plasmid per milliliter of transfection mix was used, whereas for immunofluorescence (IF) and live-cell imaging the plasmid was diluted 1:5 with a carrier vector.
Antibodies
The following primary antibodies were used: rabbit anti-mAtg9 (STO215, STO219; Young et al., 2006); Armenian hamster anti-mAtg9 (clone 14F2; Webber and Tooze, 2010a); mouse anti-Atg16L (TMD-PH-AT16; CosmoBio, Carlsbad, CA); rabbit anti–β-tubulin (Abcam, Cambridge, MA); mouse anti-LC3 (clone 5F10; 0231-100; NanoTools, Teningen, Germany); rabbit anti-LC3 (ab48394; Abcam); rabbit anti-WIPI2 (Polson et al., 2010); mouse anti-TfR (13-6800; Invitrogen); sheep anti-TGN46 (AHP00G; AbD Serotec, Raleigh, NC); rabbit anti-CIMPR (STO52; Dittie et al., 1999); mouse anti-EEA1 (610457; BD Biosciences PharMingen, San Diego, CA); polyclonal anti-ULK1 (sc-33182; Santa Cruz Biotechnology, Santa Cruz, CA); rabbit anti-SOD1 (16831; Abcam); mouse anti-GFP (clone 3E1; Cancer Research UK, London, United Kingdom); mouse anti-Rab11 (610657; BD Biosciences PharMingen); rabbit anti-PDI (SPA-891; Bioquote, York, United Kingdom); mouse anti-tubulin (7291, Abcam); rabbit anti–syntaxin-13 (110 132; Synaptic Systems, Göttingen, Germany); rabbit anti-Vps26 was a kind gift of M. Seaman (Cambridge Institute for Medical Research, Cambridge, United Kingdom); COPI was a kind gift of F. Wieland (University of Heidelberg, Heidelberg, Germany). The gold-conjugated anti-TfR antibody was a kind gift of Clare Futter (Institute of Ophthalmology, London, United Kingdom).
Light microscopy
Indirect IF was performed as previously described (Young et al., 2006). For visualization of endogenous LC3, cells were either fixed in 3% paraformaldehyde and permeabilized in methanol and blocked in 5% bovine serum albumin. Secondary antibodies were all Alexa Fluor conjugated (488, 555, or 647) from Invitrogen, with the exception of Cy3-conjugated goat anti–Armenian hamster (Jackson ImmunoResearch Laboratories, West Grove, PA).
Cells were imaged with either an LSM 510 or an LSM 710 laser scanning confocal microscope equipped with a 63×, 1.4 numerical aperture (NA), Plan Apochromat oil immersion objective (Carl Zeiss MicroImaging, Jena, Germany). Confocal sections for colocalization analysis were 0.8 μm thick. Images were processed using LSM 510 and Zen software.
Quantification of Atg9
All quantifications were performed with Imaris software (Bitplane, Zurich, Switzerland), setting an appropriate threshold, which was kept constant during the whole analysis. Images for quantification were taken under the same conditions, and about five images per experiment were quantified. The values of all images from a single experiment were averaged together, and this value was used as n = 1 for statistical analysis with Excel (Microsoft, Redmond, WA) and Prism (GraphPad Software, La Jolla, CA) software.
For “total Atg9” the signal of Atg9 and the relative marker in the whole cell were compared. For “local” colocalization, at least 20 spots that were positive for the marker of interest were picked in blind per experiment. The colocalization between Atg9 and the marker was then evaluated only in a squared area of approximately twice the diameter around each spot.
Live-cell imaging
For live-cell imaging, cells were grown on MatTek dishes (MatTek Corporation, Ashland, MA). Full-medium imaging medium consisted of DMEM, 10% FCS, 30 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.4, low bicarbonate, minus phenol red, minus riboflavin, and minus folic acid. Starvation imaging medium was EBSS and 30 mM HEPES, pH 7.4.
Cells were imaged at 37°C using a widefield inverted Nikon Eclipse TE2000-E microscope and a 60×, 1.40 NA, oil immersion objective (Nikon, Melville, NY). Image sequences were then processed using MetaMorph (Molecular Devices, Sunnyvale, CA) and exported as .avi files (Microsoft Video 1 compression).
Electron microscopy
For CLEM, GFP-LC3/mRFP-Atg9 cells were grown on photoetched gridded coverslips and incubated for 2 h in starvation medium. For the CLEM experiments with Tfn and TnR, cells were starved for 2 h in EBSS and the Tnf-Alexa 647 ligand and anti-TnR gold conjugate (10-nm gold) was added to the EBSS. Cells were then fixed in 8% formaldehyde/0.2 M HEPES, pH 7.2, added directly to the cell medium for 10 min, followed by fixation in 4% formaldehyde/0.2 M HEPES, pH 7.2, for 1 h. Cells of interest were identified and imaged by phase contrast and fluorescence microscopy, using a 63× water objective and taking 0.8-μm-thick z-stacks at 0.4-μm intervals. Cells were then fixed in 2.5% glutaraldehyde/4% formaldehyde in 0.1 M phosphate buffer (PB) for 1 h. The samples were postfixed in reduced osmium tetroxide, stained with tannic acid, dehydrated stepwise to 100% ethanol, and embedded in Epon. Serial sections (∼70 nm) were cut using an Ultracut UCT ultramicrotome (Leica Microsystems UK, Milton Keynes, United Kingdom), collected on formvar-coated slot grids, and poststained with lead citrate.
EM immunolocalization of Atg9 was performed by cryosectioning and immunolabeling (Tokuyasu, 1973). Briefly, cells were fixed in 8% formaldehyde/0.4% gluteraldehyde/0.1 M PB, added directly to the cell medium at 37°C for 10 min, and followed by fixation in 4% formaldehyde/0.2% gluteraldehyde/0.1 M PB for 30 min at room temperature. The cells were embedded in 2% gelatin, cryoprotected in 2.3 M sucrose, mounted onto pins, and plunge-frozen in liquid nitrogen. Ultrathin cryosections were cut using an FC6 cryo-ultramicrotome (Leica Microsystems UK). Cryosections were immunolabeled with rabbit polyclonal anti-Atg9 (1:10; Young et al., 2006) and protein A conjugated to 10-nm gold (Cell Microscopy Center, University Medical Center Utrecht, Utrecht, Netherlands).
Sections were viewed using a Tecnai G2 Spirit 120-kV transmission electron microscope (FEI Company, Eindhoven, Netherlands) with either an Orius or an Ultrascan 1000 charge-coupled device camera (Gatan UK, Abingdon, United Kingdom).
Rat hepatocytes were prepared and infected with mRFP-Atg9 and GFP-LC3 viruses as previously described (Kochl et al., 2006; Young et al., 2006) before immuno-EM immunolocalization as described.
Western blot
Cells were lysed in ice-cold TNTE buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 0.3% wt/vol Triton X-100, 5 mM EDTA) containing EDTA-free Complete Protease Inhibitor cocktail (Roche, Indianapolis, IN). Lysates were cleared by centrifugation and resolved by SDS–PAGE. For LC3 lipidation assays NuPage 4–12% Bis-Tris gels in 2-(N-morpholino)ethanesulfonic acid buffer were used (Invitrogen). For gradient fractions 20 × 20 cm format 8% Laemmli SDS–PAGE was performed. Gels were transferred onto polyvinylidene fluoride membrane using Bio-Rad Semidry apparatus (Bio-RAD, Hercules, CA). Blots were revealed by horseradish peroxidase–conjugated secondary antibodies, followed by enhanced chemiluminescence (GE Healthcare, Piscataway, NJ). When needed, membranes were stripped using Restore PLUS (Thermo Scientific, Waltham, MA).
Immunoprecipitation and mass spectrometry analysis
Cells were lysed in ice-cold CHAPS buffer (1% CHAPS, 150 mM NaCl, 20 mM Tris-HCl, pH 7.5, 5 mM EDTA) containing Complete Protease Inhibitor (Roche). Lysates cleared by centrifugation were incubated with protein G– or protein A–Sepharose beads (Sigma-Aldrich, St. Louis, MO) coupled to one of the following antibodies: mouse anti-HA (clone 12CA5; Cancer Research UK), rat anti-HA (11 867 423 001; Roche), mouse anti-GFP (clone 4E1; Cancer Research UK), rabbit affinity-purified anti-Atg9 (STO215), and hamster anti-Atg9 (14F2). After three washes in TNTE, proteins were eluted using 5× SDS sample buffer and heated to 65°C for 5 min.
For mass spectroscopy determination of mAtg9 interactors, HEK293 cells were transfected with either HA-tagged mAtg9 or empty vector. Cell lysates were immunoprecipitated as described. To reduce the risk of nonspecific binding, we performed two independent determinations using antibodies from different species: mouse or rat anti-HA antibodies. Proteins were resolved on NuPage 4–12% Bis-Tris gels and stained with GelCode Blue (Pierce, Thermo Fisher Scientific, Rockford, IL). Entire lanes were analyzed. Interactors were considered significant if they were present in both experiments.
Gradients
Cells were homogenized in basic homogenization buffer (HBB; 250 mM sucrose, 25 mM HEPES, pH 7.2; Figure 5), or rich homogenization buffer (HRB: 250 mM sucrose, 25 mM HEPES, pH 7.2, 25 mM KCl, 2.5 mM Mg acetate), both containing Complete Protease Inhibitor. Postnuclear supernatants were loaded on 1–22% Ficoll gradients, 20 mM TES (N-Tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid), pH 7.4, and 250 mM sucrose, on a 45% Nycodenz cushion and prepared in a 12.5 Quick-Seal tube (342413; Beckman Coulter, Brea, CA). Samples were centrifuged for 36 min at 50,000 rpm, using a VTi65.1 rotor in a Beckman Coulter Optima L-100 XP centrifuge.
We collected 500-μl fractions (∼24 or 25 per sample), and the refractive index of each fraction was determined using a refractometer (Bellingham + Stanley Ltd., Tunbridge Wells, United Kingdom). The fractions in the linear range (usually numbers 6–24) were analyzed by SDS–PAGE.
To determine the distribution profile of the different markers, Western blots were quantified using ImageJ (National Institutes of Health, Bethesda, MA) and plotted against the refractive index of the corresponding fraction. To compare the data across experiments, each profile was fitted to a Gaussian curve (least square fit) using GraphPad Prism, version 5.0d, determining the mean value of the control full-medium sample, to which all other values were compared.
Data analysis
Spots (GFP-DFCP1, GFP-LC3, WIPI2, Atg16) were counted using Imaris and expressed as spots/cell or spots/area unit. LC3-I and LC3-II blots were quantified using ImageJ. A two-tailed unpaired t test or one-way analysis of variance followed by a Tukey posttest was performed using GraphPad Prism, version 5. To measure mAtg9 dispersion, images were blinded and scored by two operators. From 50 to 90 cells were counted per condition in two independent experiments.
Supplementary Material
Acknowledgments
We thank F. E. McAlpine, N. C. McKnight, and G. Schiavo for critically reading the manuscript and the Secretory Pathways Laboratory for discussion and support. We thank N. T. Ktistakis for the HEK293/GFP-DFCP1 cell line, M. N. Seaman for retromer reagents, and T. Saitoh and S. Akira for the Atg9−/− MEFs.
Abbreviations used:
- CLEM
correlative light and electron microscopy
- DFCP1
double FYVE containing protein 1
- LE
late endosomes
- PI3K
phosphatidylinositol 3-kinase
- RE
recycling endosomes
- Tfn
transferrin
- TfR
transferrin receptor
- ULK1
unc-51–like kinase 1
- WIPI
WD-40 repeat containing protein that interacts with PtdIns
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-09-0746) on March 28, 2012.
REFERENCES
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Rojas R. Retrograde transport from endosomes to the trans-Golgi network. Nat Rev Mol Cell Biol. 2006;7:568–579. doi: 10.1038/nrm1985. [DOI] [PubMed] [Google Scholar]
- Chan EY, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem. 2007;282:25464–25474. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- Chan EY, Longatti A, McKnight NC, Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol. 2009;29:157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittie AS, Klumperman J, Tooze SA. Differential distribution of mannose-6-phosphate receptors and furin in immature secretory granules. J Cell Sci. 1999;112:3955–3966. doi: 10.1242/jcs.112.22.3955. [DOI] [PubMed] [Google Scholar]
- Geng J, Nair U, Yasumura-Yorimitsu K, Klionsky DJ. Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2257–2269. doi: 10.1091/mbc.E09-11-0969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- He C, Song H, Yorimitsu T, Monastyrska I, Yen WL, Legakis JE, Klionsky DJ. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J Cell Biol. 2006;175:925–935. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbison CE, Thorstensen K, Chua AC, Graham RM, Leedman P, Olynyk JK, Trinder D. The role of transferrin receptor 1 and 2 in transferrin-bound iron uptake in human hepatoma cells. Am J Physiol Cell Physiol. 2009;297:C1567–C1575. doi: 10.1152/ajpcell.00649.2008. [DOI] [PubMed] [Google Scholar]
- Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–776. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama S, Omori H, Saitoh T, Sone T, Guan JL, Akira S, Imamoto F, Noda T, Yoshimori T. The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella. Mol Biol Cell. 2011;22:2290–2300. doi: 10.1091/mbc.E10-11-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Huang WP, Stromhaug PE, Klionsky DJ. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J Biol Chem. 2002;277:763–773. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Kochl R, Hu XW, Chan EY, Tooze SA. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic. 2006;7:129–145. doi: 10.1111/j.1600-0854.2005.00368.x. [DOI] [PubMed] [Google Scholar]
- Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Lang T, Reiche S, Straub M, Bredschneider M, Thumm M. Autophagy and the cvt pathway both depend on AUT9. J Bacteriol. 2000;182:2125–2133. doi: 10.1128/jb.182.8.2125-2133.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longatti A, Tooze SA. Vesicular trafficking and autophagosome formation. Cell Death Differ. 2009;16:956–965. doi: 10.1038/cdd.2009.39. [DOI] [PubMed] [Google Scholar]
- Lu Q, Yang P, Huang X, Hu W, Guo B, Wu F, Lin L, Kovacs AL, Yu L, Zhang H. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev Cell. 2011;21:343–357. doi: 10.1016/j.devcel.2011.06.024. [DOI] [PubMed] [Google Scholar]
- Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari M, Tooze SA, Reggiori F. The puzzling origin of the autophagosomal membrane. F1000 Biol Rep. 2011;3:25. doi: 10.3410/B3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie M, Dale HA, Sannerud R, Saraste J. The function of the intermediate compartment in pre-Golgi trafficking involves its stable connection with the centrosome. Mol Biol Cell. 2009;20:4458–4470. doi: 10.1091/mbc.E08-12-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Kim J, Huang WP, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol. 2000;148:465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y, Munro S. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol Biol Cell. 2010;21:3998–4008. doi: 10.1091/mbc.E10-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi A, Polson HE, Tooze SA. Membrane trafficking events that partake in autophagy. Curr Opin Cell Biol. 2010;22:150–156. doi: 10.1016/j.ceb.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, Tooze SA. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:506–522. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- Popoff V, Mardones GA, Tenza D, Rojas R, Lamaze C, Bonifacino JS, Raposo G, Johannes L. The retromer complex and clathrin define an early endosomal retrograde exit site. J Cell Sci. 2007;120:2022–2031. doi: 10.1242/jcs.003020. [DOI] [PubMed] [Google Scholar]
- Prekeris R, Klumperman J, Chen YA, Scheller RH. Syntaxin 13 mediates cycling of plasma membrane proteins via tubulovesicular recycling endosomes. J Cell Biol. 1998;143:957–971. doi: 10.1083/jcb.143.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004a;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Wang CW, Nair U, Shintani T, Abeliovich H, Klionsky DJ. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol Biol Cell. 2004b;15:2189–2204. doi: 10.1091/mbc.E03-07-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci USA. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MNJ. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J Cell Biol. 2004;165:111–122. doi: 10.1083/jcb.200312034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekito T, Kawamata T, Ichikawa R, Suzuki K, Ohsumi Y. Atg17 recruits Atg9 to organize the pre-autophagosomal structure. Genes Cells. 2009;14:525–538. doi: 10.1111/j.1365-2443.2009.01299.x. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Meyerkord CL, Hori T, Runkle K, Fox TE, Kester M, Loughran TP, Wang HG. Bif-1 regulates Atg9 trafficking by mediating the fission of Golgi membranes during autophagy. Autophagy. 2011;7:61–73. doi: 10.4161/auto.7.1.14015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu KT. A technique for ultracryotomy of cell suspensions and tissues. J Cell Biol. 1973;57:551–565. doi: 10.1083/jcb.57.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol. 2010;11:777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- van der Vaart A, Griffith J, Reggiori F. Exit from the Golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2270–2284. doi: 10.1091/mbc.E09-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ijzendoorn SC. Recycling endosomes. J Cell Sci. 2006;119:1679–1681. doi: 10.1242/jcs.02948. [DOI] [PubMed] [Google Scholar]
- Webber JL, Tooze SA. Coordinated regulation of autophagy by p38alpha MAPK through mAtg9 and p38IP. EMBO J. 2010a;29:27–40. doi: 10.1038/emboj.2009.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber JL, Tooze SA. New insights into the function of Atg9. FEBS Lett. 2010b;584:1319–1326. doi: 10.1016/j.febslet.2010.01.020. [DOI] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen WL, Shintani T, Nair U, Cao Y, Richardson BC, Li Z, Hughson FM, Baba M, Klionsky DJ. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J Cell Biol. 2010;188:101–114. doi: 10.1083/jcb.200904075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- Young AR, Chan EY, Hu XW, Kochl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.