The association of Rad18 with Polη is crucial for efficient translesion synthesis and DNA damage tolerance. Rad18–Polη interactions and UV tolerance depend on JNK-dependent Rad18 phosphorylation. These results provide a new mechanism by which SAPK signaling promotes genome maintenance.
Abstract
The E3 ubiquitin ligase Rad18 chaperones DNA polymerase η (Polη) to sites of UV-induced DNA damage and monoubiquitinates proliferating cell nuclear antigen (PCNA), facilitating engagement of Polη with stalled replication forks and promoting translesion synthesis (TLS). It is unclear how Rad18 activities are coordinated with other elements of the DNA damage response. We show here that Ser-409 residing in the Polη-binding motif of Rad18 is phosphorylated in a checkpoint kinase 1–dependent manner in genotoxin-treated cells. Recombinant Rad18 was phosphorylated specifically at S409 by c-Jun N-terminal kinase (JNK) in vitro. In UV-treated cells, Rad18 S409 phosphorylation was inhibited by a pharmacological JNK inhibitor. Conversely, ectopic expression of JNK and its upstream kinase mitogen-activated protein kinase kinase 4 led to DNA damage–independent Rad18 S409 phosphorylation. These results identify Rad18 as a novel JNK substrate. A Rad18 mutant harboring a Ser → Ala substitution at S409 was compromised for Polη association and did not redistribute Polη to nuclear foci or promote Polη−PCNA interaction efficiently relative to wild-type Rad18. Rad18 S409A also failed to fully complement the UV sensitivity of Rad18-depleted cells. Taken together, these results show that Rad18 phosphorylation by JNK represents a novel mechanism for promoting TLS and DNA damage tolerance.
INTRODUCTION
Various environmental agents induce DNA lesions termed “adducts” that impede the progression of the DNA replication machinery. For example the chemical carcinogen benzo[a]pyrene generates genotoxic metabolites, including (+)-r-7,t-8-dihydroxy-t-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene (BPDE), which reacts primarily with the N2 amino group of guanine to form a BPDE–DNA (BPDE–N2-dG) adduct (Thakker et al., 1976; Conney, 1982; Dipple, 1995). Similar helix-distorting DNA lesions result from solar UV radiation, which generates cyclobutane pyrimidine dimers (CPDs; formed by linkage of two adjacent pyrimidines) and to a lesser extent pyrimidine (6-4) pyrimidone photoproducts (Pfeifer et al., 2005). BPDE adducts, CPDs, and many other DNA lesions block the progression of replicative DNA polymerases and cause stalling of active replication forks. Failure to preserve stalled replication forks can lead to fork collapse and acquisition of DNA double strand breaks (DSBs). Moreover, error-prone replicative bypass of damaged DNA can lead to mutations and cancer. Therefore appropriate replication and repair of damaged DNA is necessary to prevent genomic instability.
Cell cycle checkpoints and translesion synthesis are two ubiquitous responses to DNA damage and replication fork stalling. Both processes are hypothesized to play important roles in maintaining genome stability and preventing cancer. Checkpoints are DNA damage–induced signal transduction pathways that delay cell cycle phase transitions and promote repair mechanisms, thereby integrating cell cycle progression with DNA repair (Zhou and Elledge, 2000; Sancar et al., 2004; Cimprich and Cortez, 2008). Many cell cycle checkpoint mechanisms have been described, and these respond to different forms of DNA damage (Zhou and Elledge, 2000; Sancar et al., 2004; Cimprich and Cortez, 2008). The S-phase checkpoint induced by bulky lesions such as BPDE or CPD is mediated via the ATR and Chk1 protein kinases (Guo et al., 2002; Heffernan et al., 2002; Liu et al., 2006; Barkley et al., 2007; Heffernan et al., 2007). ATR/Chk1-mediated S-phase checkpoint signaling is triggered by uncoupling of replicative DNA helicase (comprising Cdc45, Mcm2-7, and the GINS complex) and DNA polymerase activities (Byun et al., 2005; Cimprich and Cortez, 2008). When replicative DNA polymerases are blocked by DNA lesions (CPD and BPDE–DNA adducts), helicase activity continues to unwind DNA ahead of the stalled polymerase (Byun et al., 2005; Cimprich and Cortez, 2008). Replicative helicase–polymerase uncoupling results in accumulation of single-stranded DNA (ssDNA), which is coated by replication protein A (RPA; Zou and Elledge, 2003; Zou et al., 2003). RPA-coated ssDNA helps to recruit and activate the ATR kinase (Burrows and Elledge, 2008; Mordes and Cortez, 2008; Mordes et al., 2008), leading to phosphorylation and activation of the checkpoint kinase Chk1 (a key effector of the ATR pathway). Chk1 signaling inhibits DNA synthesis at unfired origins, stabilizes stalled replication forks, and inhibits entry into mitosis in the presence of unreplicated DNA (Sancar et al., 2004; Cimprich and Cortez, 2008). The precise mechanisms by which stalled forks are stabilized and the mechanisms by which late origins are inhibited by Chk1 in mammalian cells are not well understood. Nevertheless, fork stabilization and repression of origin firing by Chk1 are believed to play key roles in cell cycle progression and maintenance of genomic stability even in cells that do not acquire DNA damage from exogenous sources (Sorensen et al., 2004).
Concomitant with activation of ATR/Chk1 signaling, translesion synthesis (TLS) DNA polymerases are recruited to stalled replication forks (Bi et al., 2005, 2006; Bomgarden et al., 2006). In contrast to the DNA polymerases that perform bulk replicative DNA synthesis during S phase, “Y-family” TLS polymerases have low fidelity and processivity on undamaged DNA (Ohmori et al., 2001; Prakash et al., 2005). However, because of active sites that can accommodate helix-distorting lesions, TLS enzymes can bypass various forms of DNA damage with relative accuracy compared with replicative polymerases (Prakash et al., 2005). In eukaryotes, the main TLS polymerases are DNA polymerase η (Polη), DNA polymerase κ (Polκ), DNA polymerase ι (Polι), REV1, and DNA polymerase ζ (Polζ). Collectively, these enzymes are responsible for translesion synthesis of various DNA adducts.
In a process termed polymerase switching, replicative DNA polymerases that encounter DNA lesions are replaced by TLS enzymes. The molecular basis of the DNA polymerase switch is best understood for Saccharomyces cerevisiae Rad30 and its mammalian homologue Polη (Kannouche and Lehmann, 2004; Ulrich, 2004). When replication forks stall at sites of DNA damage, proliferating cell nuclear antigen (PCNA) is monoubiquitinated on lysine 164 (Ohmori et al., 2001; Prakash et al., 2005). PCNA K164 monoubiquitination is mediated mainly by Rad18 (an E3 ubiquitin ligase) and Rad6 (an E2 ubiquitin–conjugating enzyme). PCNA ubiquitination results in part from DNA damage–induced recruitment of Rad18 to stalled replication forks. The Ulrich and Tateishi laboratories proposed that Rad18 associates with stalled replication forks via direct interactions with RPA-coated ssDNA (Davies et al., 2008; Huttner and Ulrich, 2008; Tsuji et al., 2008), and the p95/NBS protein may facilitate RPA-mediated Rad18 recruitment (Yanagihara et al., 2011). DNA damage–induced degradation of the PCNA deubiquitinating enzyme USP1 may also contribute to accumulation of ubiquitinated PCNA in genotoxin-treated cells (Huang et al., 2006). Monoubiquitinated PCNA is considered to promote recruitment of TLS polymerases to the replication fork. Specialized ubiquitin (Ub)-binding domains (termed UBZ or UBM motifs) facilitate interactions between TLS polymerases and monoubiquitinated PCNA (Bienko et al., 2005). After lesion bypass, another switch is presumed to restore the replicative DNA polymerase to the fork.
TLS can be error-free or error-prone. Whereas error-free TLS suppresses mutagenesis and promotes DNA damage tolerance, error-prone TLS is mutagenic. TLS polymerases display specificity for bypass of different lesions in vitro. For example, Polη correctly inserts A-A across T-T dimers, whereas other TLS polymerases (likely Polι and Polκ) perform error-prone insertion across CPD (Ziv et al., 2009). Therefore Polη is crucial for error-free TLS, maintenance of ongoing DNA replication, and DNA damage tolerance in UV-treated cells. Indeed, UV-sensitivity and mutagenesis are hallmarks of xeroderma pigmentosum variant cells that lack Polη (Maher et al., 1976; Johnson et al., 1999; Masutani et al., 1999). Clearly, selection of the appropriate TLS polymerase for a particular lesion is crucial for maintaining genomic stability. However, the molecular mechanism for recruiting appropriate TLS polymerase(s) and ensuring error-free lesion bypass is not fully understood. Polη is a versatile enzyme and may be the preferred TLS polymerase and the default enzyme selected for bypass of most lesions. Rad18 interacts directly with and helps to guide Polη to sites of replication fork stalling (Watanabe et al., 2004; Day et al., 2010), leading to stable engagement of Polη with PCNA. In part, the preferential selection of Polη over other TLS polymerases might result from specific and direct associations between Polη and Rad18.
Although S-phase checkpoint signaling and TLS are initiated independently via a common damage-induced DNA replication intermediate (RPA-coated ssDNA; Chang et al., 2006a), there are some indications that TLS and checkpoint signaling are coupled in a manner that is necessary for appropriate responses to fork-stalling DNA lesions. For example, defective TLS results in elevated ATR/Chk1 signaling, most likely due to persistence of the postreplicative ssDNA gaps generated by polymerase/helicase uncoupling (Bi et al., 2005, 2006; Bomgarden et al., 2006). Conversely, certain aspects of TLS may be facilitated by checkpoint signaling. For example, we showed that PCNA monoubiquitylation and its association with Polκ are partially dependent on ATR/Chk1 signaling (Bi et al., 2006). Zou and colleagues also showed that DNA damage–inducible PCNA ubiquitination is ablated in Chk1-depleted cells (Yang et al., 2008; Yang and Zou, 2009). Lehmann and colleagues found that Polη is an ATR substrate whose genotoxin-inducible phosphorylation modulates checkpoint signaling (Gohler et al., 2011). Nevertheless, the mechanisms that integrate checkpoint signaling with TLS are incompletely understood.
Because checkpoints are mediated by protein kinases, we hypothesized that Chk1-dependent phosphorylation of TLS proteins might provide a potential mechanism for linking S-phase checkpoint signaling with TLS, thereby coordinating distinct branches of DNA damage response. Rad18 plays a central role in TLS and additional DNA repair pathways (Huang et al., 2009; Watanabe et al., 2009). Therefore we hypothesized that Rad18 represented a likely target of Chk1 signaling. Consistent with a hypothetical link between Chk1 and Rad18, we show here that Rad18 is phosphorylated inducibly (and in a Chk1-dependent manner) in response to DNA damage. We identified Ser-409 as a DNA damage–induced phosphorylation site that resides within the Polη-binding motif of Rad18. We demonstrate that Rad18 phosphorylation at S409 specifically promotes Rad18–Polη associations, facilitates efficient recruitment of Polη to the replication machinery, and confers DNA damage tolerance. Surprisingly, however, we show that Rad18 S409 is not phosphorylated directly by checkpoint kinases. Instead we identify c-Jun N-terminal kinase (JNK; a member of the stress-activated protein kinase [SAPK] family) as the Rad18 S409 kinase. It is well established that JNK and other SAPKs mediate transcriptional events that modulate survival and apoptotic signaling in response to various cellular stresses, including UV radiation (Davis, 2000). There are recent indications that SAPKs may also participate in checkpoint control when the genome is compromised (Reinhardt et al., 2007; Reinhardt and Yaffe, 2009). However, a relationship between SAPK signaling and TLS had not previously been described. Therefore our results demonstrate novel and unanticipated integration of checkpoint kinase signaling, SAPK cascades, and TLS, three major effector branches of the cellular response to UV irradiation.
RESULTS
Rad18 is phosphorylated in response to DNA damage
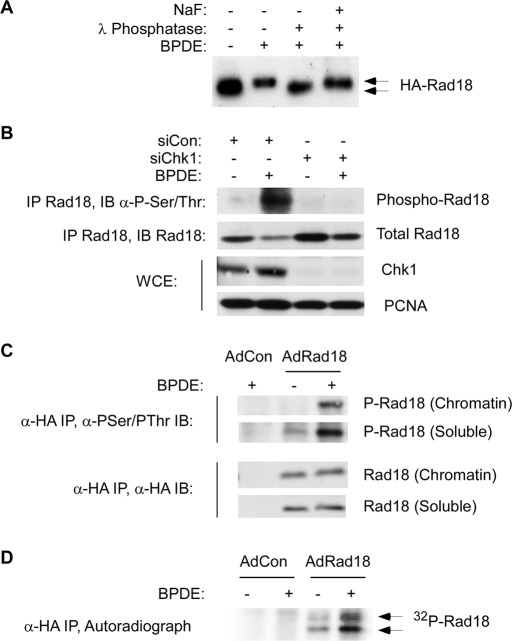
During immunoblot analyses we noticed that Rad18 in extracts from genotoxin-treated cells migrated with retarded electrophoretic mobility relative to Rad18 in control (untreated) cell extracts (Figure 1A). Moreover, the mobility shift of Rad18 was reversed by incubation with λ phosphatase (Figure 1A), indicating that Rad18 was phosphorylated inducibly in response to DNA damage. In immunoblotting experiments anti–phosphoserine/phosphothreonine antibodies preferentially detected Rad18 that was immunoprecipitated from genotoxin-treated cells (Figure 1B). Moreover, the DNA damage–induced phosphorylation of Rad18 was ablated in Chk1-depleted cells (Figure 1B), further consistent with DNA damage–inducible Rad18 phosphorylation. The genotoxin-inducible electrophoretic mobility shifts and anti–phosphoserine/threonine immunoreactivity were evident in both soluble and chromatin-associated pools of Rad18 (Figure 1C). In metabolic labeling experiments with radiolabeled orthophosphate, immunoprecipitated Rad18 from genotoxin-treated cells contained basal levels of incorporated 32P, which were further increased following acquisition of DNA damage (Figure 1D). We conclude that Rad18 is phosphorylated basally and in a DNA damage–inducible manner.
FIGURE 1:
Rad18 phosphorylation is genotoxin inducible and requires Chk1. (A) HA-Rad18–expressing H1299 cells were treated with 600 nM BPDE for 4 h or were left untreated for controls. CSK extracts from the cells were incubated with λ phosphatase (1 U/μl) in the presence or absence of 50 mM NaF at 37 C for 30 min. The resulting extracts were analyzed by immunoblotting with anti-HA antibodies. BPDE-induced electrophoretic mobility shifts are indicated by the arrows. (B) Control or Chk1-depleted HA-Rad18–expressing H1299 cells were treated with 600 nM BPDE for 4 h or were left untreated for controls. Anti-HA immune complexes were analyzed by SDS–PAGE and immunoblotting sequentially with anti–phosphoserine/phosphothreonine and anti-HA antibodies. Whole-cell extracts were analyzed for Chk1 expression, and PCNA was used as a loading control. (C) AdCon or AdHA-Rad18–infected H1299 cells were treated with 600 nM BPDE for 4 h or were left untreated for controls. Chromatin and soluble fractions from the cells were immunoprecipitated with anti-HA antibodies, and the resulting immunoprecipitates were analyzed by SDS–PAGE and immunoblotting with anti–phosphoserine/phosphothreonine and anti-HA antibodies. (D) AdCon or AdHA-Rad18–infected H1299 cells were transferred to phosphate-free DMEM and incubated for 4 h in the presence of 0.2 mCi/ml of 32P-orthophosphate. The 32P-orthophosphate–labeled cells were treated with 600 nM BPDE for 4 h or were left untreated for controls. Whole-cell extracts from the cells were immunoprecipitated with anti-HA antibodies, and the resulting immunoprecipitates were resolved by SDS–PAGE. The resulting gels were washed extensively in 40% methanol/10% acetic acid. The fixed gel was dried, and radioactive proteins were detected by autoradiography.
JNK phosphorylates Rad18 at S409
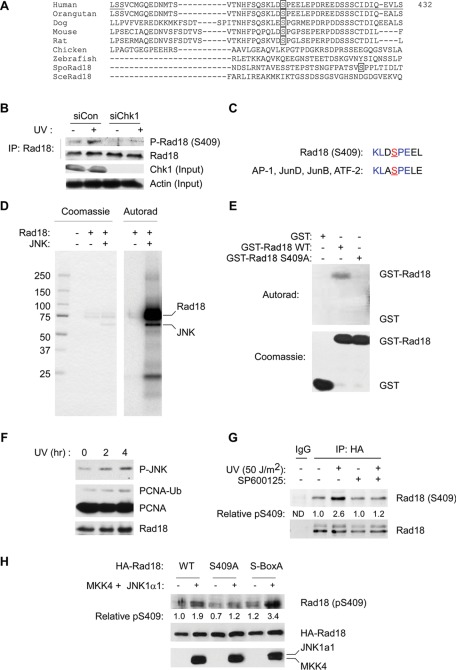
In a global screen for phosphoproteins, Nousiainen et al. (2006) detected phosphorylation of Rad18 at serine 409, although they did not explore the mechanism or significance of this phosphorylation event. We considered the possibility that S409 phosphorylation might contribute to the DNA damage–inducible changes in total Rad18 phosphorylation detected in Figure 1. Therefore we raised phosphospecific antiserum against the phosphopeptide CFSQSKLD[pS]PEEL, corresponding to residues 398–413 of hRad18 (see Materials and Methods). As shown in Figure 2B, the anti-Rad18 pS409 antibody detected a UV-inducible species in anti-Rad18 immunoprecipitates. The S409-reactive protein was not detected in Rad18 immunoprecipitates from Chk1-depleted cells (Figure 2B). Of importance, the anti-Rad18 pS409 antibody did not react with a Rad18 mutant harboring an S → A substitution at residue 409 (see later discussion of Figure 4), demonstrating phosphospecificity. We conclude that S409 is a DNA damage–inducible Rad18 phosphorylation site.
FIGURE 2:
Rad18 S409 is phosphorylated by JNK. (A) Amino acid sequence alignment showing conservation of S409 among different species. (B) Control or Chk1-depleted HA-Rad18–expressing H1299 cells were treated with UV (20 J/m2) or left untreated for controls. After 4 h, anti-HA immune complexes were prepared from chromatin fractions and analyzed by SDS–PAGE and immunoblotting with anti–phospho-Ser-409 and anti-HA antibodies. Soluble extracts were analyzed for Chk1 expression to confirm efficient siRNA-mediated knockdown. Immunoblotting with an anti-actin antibody was used to verify equivalent protein loading between lanes. (C) Comparison of sequences flanking Rad18 S409 (underlined) with JNK1 consensus phosphorylation sites. (D) Recombinant Rad18–Rad6 complex was subject to in vitro phosphorylation by JNK. The reaction contained 0.4 μg (4 pmol) of Rad18–Rad6, and half (0.2 μg; 2 pmol) was resolved on SDS–PAGE. The phosphorylated species contained 4000 cpm (determined by scintillation counting of the excised gel slice), which is equivalent to 2 pmol of incorporated ATP incorporation. (E) Recombinant GST-Rad18 398-495 (WT), GST-Rad18 S409A, and GST (control) were tested as JNK substrates using in vitro kinase assays. Reaction products were resolved on SDS–PAGE. The resulting gel was fixed, stained, and dried. Top, an autoradiogram; bottom, the Coomassie-stained proteins. (F) Exponentially growing H1299 cells were irradiated with UVC (20 J/m2) or sham irradiated for controls. Cells were harvested 2 or 4 h after UVC treatment and lysates were analyzed for pJNK and PCNA by SDS–PAGE and immunoblotting. (G) Exponentially growing H1299 cells expressing HA-Rad18 were treated for 1 h with 1 μM SP600125 or left untreated for controls. Control and SP600125-treated cells were treated with UVC (20 J/m2) or sham irradiated. After 2 h, Rad18 was immunoprecipitated from cell extracts. Immune complexes were resolved by SDS–PAGE and probed sequentially with antibodies against Rad18 pS409 and HA. (H) H1299 cells were cotransfected with expression vectors encoding HA-Rad18 (WT), HA-Rad18 (S409A), FLAG-MKK4, and FLAG-JNK1a1 or with “empty” vector control plasmid as indicated. At 48 h posttransfection, cells were lysed and normalized for protein content. Lysates were immunoprecipitated with anti-HA, and the resulting immune complexes were resolved by SDS–PAGE, transferred to nitrocellulose, and probed sequentially with anti–Rad18 pS409 and anti-HA antibodies. Total cell lysates were also resolved and transferred to nitrocellulose and then probed with anti-FLAG to validate MKK4 and JNK expression.
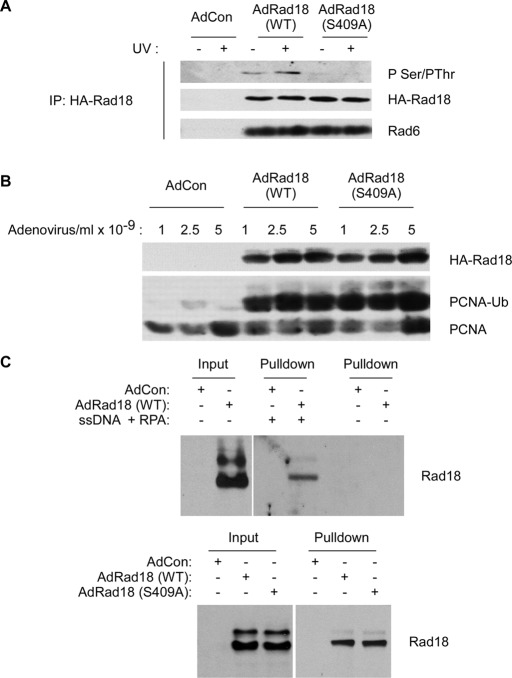
FIGURE 4:
Rad18 S409 is dispensable for Rad18–Rad6 interactions, PCNA-directed E3 ligase activity, and binding to RPA-coated ssDNA. (A) HA-Rad18–expressing H1299 cells were treated with UV (20 J/m2) or left untreated for controls. Anti-HA immune complexes were prepared from chromatin fractions and analyzed by SDS–PAGE and immunoblotting with anti-HA and anti-Rad6 antibodies. (B) H1299 cells were infected with different doses of AdCon, AdRad18WT, and AdRad18 S409A. Twenty-fours hours later, chromatin and soluble fractions were prepared and analyzed by SDS–PAGE and immunoblotting with HA and PCNA antibodies. (C) RAD18−/− cells were infected with AdCon, AdRad18 WT, or Rad18 S409A. After 24 h, soluble extracts were prepared from the resulting cells. RPA/ssDNA-interacting proteins were captured as described in Materials and Methods. Proteins bound to the ssDNA-coated beads were analyzed by SDS–PAGE and immunoblotting. Top, the RPA/ssDNA-dependency of Rad18 binding to streptavidin-coated beads. Bottom, the comparison of WT Rad18 and Rad18 S409A binding to RPA/ssDNA-beads (and ssDNA and RPA were present in all pull-down samples).
Because Rad18 S409 phosphorylation was Chk1 dependent, we considered the possibility that Chk1 phosphorylated this site directly. Therefore we performed in vitro kinase assays in which we tested recombinant baculovirus–encoded Rad18–Rad6 as well as bacterial glutathione S-transferase (GST)–Rad18 as potential Chk1 substrates. In those experiments, Rad18 was phosphorylated very inefficiently compared with Cdc25C, a bona fide Chk1 substrate. The modest Chk1-mediated in vitro phosphorylation of recombinant Rad18 was unaffected by S409A mutation (unpublished data). Furthermore, the conserved sequences flanking Rad18 S409 do not correspond to the preferred substrate motifs for Chk1 (O'Neill et al., 2002). Therefore we conclude that Rad18 S409 is not a direct Chk1 target.
Because S409 precedes a proline residue, we hypothesized that cyclin-dependent kinases (CDKs) and/or SAPKs, representing the two major families of proline-directed protein kinases (Miller et al., 2008), were likely candidate kinases responsible for Rad18 S409 phosphorylation. SAPKs (but not CDKs) are activated by DNA-damaging agents such as UV. Therefore we considered it most likely that SAPK member(s) mediate Rad18 S409 phosphorylation. Using in vitro kinase assays with recombinant CDK1 and CDK2, we were unable to detect Rad18 S409 phosphorylation, and we conclude that CDKs are most likely not relevant protein kinases for this site. We noticed that the residues surrounding Rad18 S409 were very similar to the consensus target sites for c-Jun N-terminal kinase (Figure 2C). Indeed, full-length Rad18–Rad6 complex was phosphorylated very efficiently by JNK1α1 in vitro (Figure 2D). Under our standard experimental conditions ∼1 mol of ATP was incorporated per mole of Rad18, indicating that Rad18 phosphorylation by JNK is very efficient.
To identify the Rad18 region(s) phosphorylated by JNK, we expressed a series of partially overlapping GST fusion fragments of Rad18 spanning the entire length of the Rad18 protein (Day et al., 2010). The various GST-Rad18 fragments (comprising residues 1–98, 1–121, 110–173, 165–251, 247–295, 267–402, and 398–495) were tested as possible in vitro substrates for JNK. Of the Rad18 fragments, only GST-Rad18 398–495 (comprising the C-terminal 103 amino acids of Rad18) was phosphorylated by JNK (Figure 2E). A mutant form of GST-Rad18 398–495 harboring a serine → alanine substitution at S409 (GST-Rad18 S409A) was not phosphorylated by JNK (Figure 2E). Therefore, although Rad18 harbors multiple SP sites (including S158 and S164), JNK phosphorylates Rad18 very specifically at S409. In similar experiments, p38 (another SAPK family member) did not phosphorylate Rad18 in vitro (unpublished data). We conclude that Rad18 phosphorylation by JNK is relatively specific (although we have not yet performed an exhaustive analysis of Rad18 phosphorylation by other SAPKs, and it is possible that additional JNK-related protein kinases contribute to Rad18 phosphorylation at S409).
Because the results of Figure 2 identified a JNK phosphorylation site in very close proximity to previously described Dbf4-dependent kinase (DDK) phosphorylation sites (including S421–434; Day et al., 2010), we asked whether Rad18 S409 phosphorylation by JNK influenced its phosphorylation by DDK (or vice versa). Therefore we performed kinase assays in which we measured phosphorylation of recombinant Rad18–Rad6 complex by JNK and DDK individually or in combination. In those experiments, no synergistic enhancement or inhibition of phosphorylation was observed (for either substrate) when we coincubated DDK and JNK (unpublished data). Similar results were obtained when we performed these kinase assays using a synthetic peptide substrate corresponding to Rad18 residues 393–437 (unpublished data). We conclude that there is no phosphopriming or hierarchical sequence of Rad18 phosphorylation by JNK and DDK and that S409 and S-box phosphorylations are independent events.
We performed experiments to determine whether JNK contributes to Rad18 S409 phosphorylation in intact cells. The kinetics of JNK phosphorylation and PCNA ubiquitination in UV-treated cells were similar, indicating a temporal correlation between JNK signaling and TLS (Figure 2F). Therefore we determined the effect of JNK inhibition on Rad18 S409 phosphorylation. As shown in Figure 2G, acute (1 h) pretreatment with the pharmacological JNK inhibitor SP600125 ablated UV-induced phosphorylation of Rad18 at S409. The apparent SP600125-resistant basal S409 phosphorylation may represent unmodified Rad18 (detected due to the incomplete discrimination between unphosphorylated and phosphorylated epitopes observed for many commonly used commercial antibodies). Alternatively, there are many SAPKs, and it is already known that there is considerable overlap in substrates among different SAPK members. For example, in their recent study Cook and colleagues found that both JNK and p38 contributed to phosphorylation of Cdt1 (Chandrasekaran et al., 2011). Nevertheless, the reduced UV-dependent Rad18 S409 phosphorylation in SP600125-treated cells indicates that JNK contributes significantly to Rad18 phosphorylation at this site. We cannot exclude the possibility that other protein kinases also target S409 for phosphorylation either basally or following genotoxin treatment.
JNK may be activated in a DNA damage and stress–independent manner by ectopic expression of active forms of its upstream kinase mitogen-activated protein kinase kinase 4 (MKK4). Therefore we determined the effects of coexpressing MKK4 and JNK on Rad18 S409 phosphorylation. As shown in Figure 2H, coexpression of MKK4 and JNK led to a 1.9-fold increase in Rad18 S409 phosphorylation, even in the absence of UV irradiation. As expected, MKK4/JNK–inducible phosphorylation of a Rad18 S409A mutant was not observed. We noticed that a DDK phosphorylation–resistant Rad18 mutant (designated Rad18 S-box-A) was phosphorylated at S409 more efficiently than Rad18 wild type (WT). These data may indicate that failure to phosphorylate the Rad18 S-box is associated with compensatory phosphorylation at S409. Taken together, the results of Figure 2 indicate that JNK is a Rad18-directed kinase that contributes to UV-inducible S409 phosphorylation.
Rad18 S409 phosphorylation specifically promotes Rad18–Polη interactions
To determine the significance of Rad18 S409 phosphorylation, we compared the various activities of WT and S409A-mutant forms of Rad18. First we asked whether S409A substitution affected its association with chromatin. In cell fractionation experiments, there was no difference in association of ectopically expressed hemagglutinin (HA)-Rad18 (WT) and HA-Rad18 S409A with the detergent-insoluble chromatin (Supplemental Figure S1).
Rad18 contributes to TLS in part via its DNA damage–inducible interaction with Polη, an association that helps chaperone the TLS polymerase to sites of replication stalling (Watanabe et al., 2004; Day et al., 2010). Because S409 resides in the Polη-binding domain of Rad18, we considered the possibility that phosphorylation of this residue might influence associations with Polη (Watanabe et al., 2004).
To test this hypothesis, we expressed wild-type HA-Rad18 or HA-Rad18 S409A in RAD18−/− HCT116 cells. Control and Rad18-expressing cells were treated with UV to induce DNA damage, and then Rad18 immunoprecipitates from solubilized chromatin extracts were analyzed by immunoblotting.
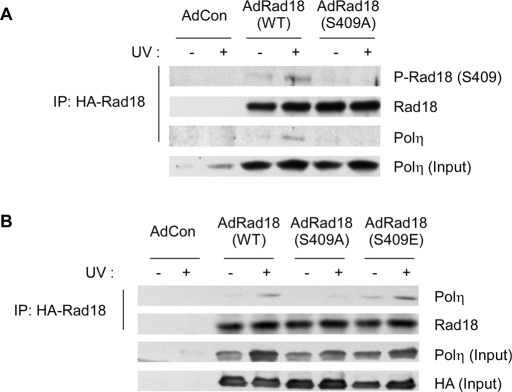
As expected, S409A substitution largely ablated the basal and DNA damage–induced phosphorylation of Rad18 and also inhibited the UV-inducible association between Rad18 and Polη (Figure 3A). Phosphomimetic substitutions are sometimes conferred when serine or threonine sites are mutated to acidic residues. Therefore we generated an adenovirus vector encoding Rad18 harboring an S409 → E substitution. However, basal and UV-inducible associations with endogenous Polη were only very modestly increased in the Rad18 S409E mutant relative to WT Rad18 (Figure 3B), most likely because the glutamate residue is an imperfect chemical mimic of a phosphate moiety (unpublished data). Nevertheless, those data indicate that the inhibitory effect of the Rad18 S409A mutation on Polη association is relatively specific. We conclude that Rad18 phosphorylation at S409 promotes formation of complexes containing Polη.
FIGURE 3:
Rad18 S409 phosphorylation promotes association with Polη. (A) RAD18−/− cells were coinfected with adenovirus vectors encoding HA-Rad18 WT or HA-Rad18 S409A or with an “empty” adenovirus vector (AdCon). Cells were treated with UV (20 J/m2) and harvested after 6 h. Chromatin fractions from the cells were solubilized and immunoprecipitated with anti-HA antibodies. The resulting immune complexes were resolved by SDS–PAGE and analyzed by immunoblotting with antibodies against Rad18 S409, HA, and Polη. Total chromatin input fractions were also analyzed using anti-Polη. (B) RAD18−/−cells were infected with adenovirus vectors encoding HA-Rad18 WT, HA-Rad18 S409A, HA-Rad18 S409E, or with an “empty” adenovirus vector (AdCon). Cells were UV irradiated, and chromatin fractions were prepared and analyzed by SDS–PAGE and immunoblotting using antibodies against Rad18 and Polη as described in A.
To test whether S409 phosphorylation specifically affected Rad18–Polη association, we determined the effect of the S409A mutation on additional Rad18 activities, including Rad6 association, PCNA ubiquitination, and ssDNA binding. Rad6 is an E2 ubiquitin–conjugating enzyme that binds and cooperates with Rad18 to ubiquitinate PCNA and other Rad18 substrates. In coimmunoprecipitation experiments, Rad18 S409A formed complexes with Rad6 as efficiently as WT Rad18 (Figure 4A). In subcellular fractionation studies, the binding of Rad18 S409A to chromatin was not impaired (Figure 4B), and ectopic expression of Rad18 S409A in RAD18−/− cells induced DNA damage–independent PCNA ubiquitylation at least as efficiently as WT Rad18 (Figure 4B). It was suggested that the recruitment of Rad18 to stalled replication forks is mediated via its associations with RPA-coated ssDNA (Davies et al., 2008; Tsuji et al., 2008). Indeed, as shown in Figure 4C, WT HA-Rad18 (derived from RAD18−/− cell extracts after complementation with Ad HA-Rad18) was recovered by streptavidin beads in an ssDNA- and RPA-dependent manner. Using in vitro pull-down assays, we detected no difference in RPA-ssDNA binding of HA-Rad18 S409A compared with WT HA-Rad18. Taken together, the results of Figure 4 indicate that S409 phosphorylation of Rad18 does not generally affect Rad18 activities. We conclude that the effect of S409 phosphorylation on associations between Rad18 and Polη is relatively specific.
Rad18 S409 phosphorylation facilitates recruitment of Polη to PCNA
We and others have shown that Rad18–Polη interactions help guide the TLS polymerase to sites of replication fork stalling (Watanabe et al., 2004; Day et al., 2010). Because the results of Figure 3 showed that Rad18 S409A associates with Polη inefficiently (compared with WT Rad18), we asked whether Rad18-induced formation of nuclear Polη foci was affected by the S409A mutation. Therefore RAD18−/− cells were complemented with WT Rad18 or Rad18 S409A.
We examined formation of yellow fluorescent protein (YFP)–Polη foci in the Rad18-complemented cells using fluorescence microscopy. As expected, YFP-Polη was distributed broadly and diffusely in the nuclei of Rad18-depleted H1299 cells, and this pattern was insensitive to UV treatment in the absence of Rad18 (Figure 5A). Complementation of Rad18-depleted cells with small interfering RNA (siRNA)–resistant WT Rad18 resulted in redistribution of YFP-Polη and conferred a basal level of YFP–Polη focus formation (Figure 5A).
FIGURE 5:
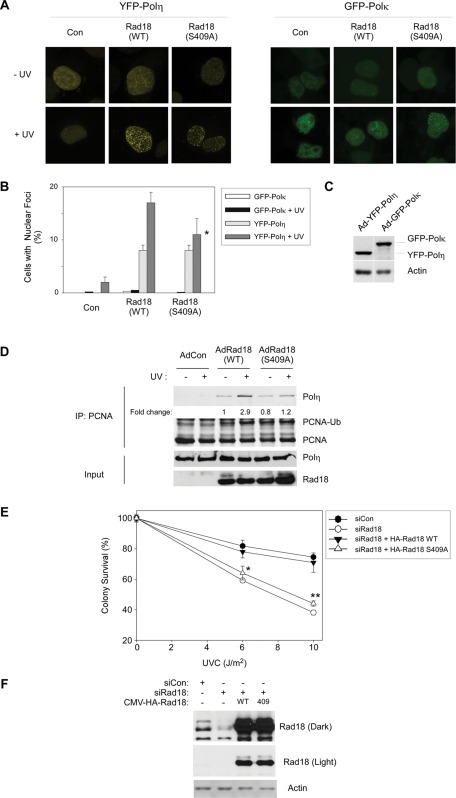
Rad18 S409A does not promote efficient recruitment of Polη to PCNA and fails to complement the UV sensitivity of Rad18-depleted cells. (A) Triplicate cultures of H1299 cells were depleted of endogenous Rad18 and complemented with siRNA-resistant HA-Rad18 (WT) and HA-Rad18 S409A. The Rad18 dependence of UV-induced YFP-Polη and GFP-Polκ redistribution to nuclear foci was determined as described in Materials and Methods. For each experimental condition (performed in triplicate) 60 cells were scored as nuclei containing >20 TLS polymerase foci. Representative images of cells displaying YFP-Polη foci are shown in A. For each treatment, number of cells positive for nuclear foci was expressed as a percentage of YFP/GFP-polymerase–positive cells (B). For GFP-Polκ– and YFP-Polη–expressing cells we performed analysis of variation (ANOVA) followed by Tukey's test to correct for experiment-wise error rates between multiple comparisons. For UV-irradiated cells coexpressing YFP-Polη and Rad18 WT or YFP-Polη and Rad18 S409A, the difference in number of foci was significant (p < 0.05). For GFP-Polκ–expressing cells under these experimental conditions there were no statistically significant differences between groups (p > 0.05). (C) We performed immunoblotting (with anti-GFP antibodies that recognize both GFP and YFP) to confirm that expression levels of GFP-Polκ and YFP-Polη were similar under these experimental conditions. (D) H1299 cells were infected with AdCon, AdRad18 WT, or AdRad18 S409A and treated with UV (20 J/m2) or left untreated for controls. PCNA was immunoprecipitated from the resulting cells, and immunoprecipitates (as well as appropriate input fractions) were analyzed by SDS–PAGE and immunoblotting. Relative levels of PCNA-associated Polη in the adjacent lanes were calculated after densitometric analysis of the Polη immunoblot. (E) H1299 cells were transfected with siRNA directed against the 3′ untranslated region of endogenous Rad18 mRNA or with a nontargeting control siRNA, siCon. The resulting cells were transfected with CMV-driven mammalian expression plasmids encoding HA-Rad18 (WT), HA-Rad18 S409A, or “empty” pAC.CMV vector for control. Control and HA-Rad18–complemented cells were trypsinized and replated in replicate (five replicates per experimental condition) and then treated with varying doses of UV and analyzed for clonogenic survival as described in Materials and Methods. For each siRNA/complementation, the number of surviving colonies from UV-treated cultures was expressed as a percentage of colony number from unirradiated cells. On the survival curves, each data point represents the mean of five replicate determinations, and error bars represent the range. For each dose of UV, we performed ANOVA between groups followed by Tukey's multiple comparison of means test. For cells that received 6 or 10 J/m2 of UV, in ANOVA the p value was <0.0001, which is significant. Results of the Tukey test were as follows: siCon vs. siRad18, p < 0.001 (indicating reduced UV tolerance); siCon vs. siRad18 + WT Rad18, p > 0.05 (indicating no significant difference and therefore full complementation by WT Rad18); siRad18 + Rad18 WT vs. siRad18 + Rad18 S409A, p < 0.001 (indicating significant difference in phenotype between Rad18 WT and Rad18 S409A, and indicated by asterisk and double asterisk for 6 and 10 J/m2, respectively).
In WT Rad18–expressing cells, the numbers of Polη foci increased approximately twofold in response to DNA damage, as we and others previously reported (Kannouche et al., 2004; Bi et al., 2006). Of interest, however, Rad18 S409A failed to fully correct the UV-induced formation of Polη foci in cells depleted of endogenous Rad18 (Figure 5B). In the experiment shown in Figure 5, A and B, the UV-inducible component of YFP-Polη focus formation was reduced by ∼66% (p < 0.05, n = 3) in Rad18 S409A–expressing cells compared with Rad18 WT–expressing cultures. For the purpose of comparison, in the same experiment we also determined the effects of Rad18 on subcellular distribution of another Y-family TLS polymerase, namely Polκ. In contrast with YFP-Polη, the subcellular distribution of GFP-Polκ (expressed at similar levels to that of YFP-Polη as determined by immunoblotting experiments shown in Figure 5C) was largely unaffected by UV treatment, regardless of Rad18 expression. The relative insensitivity of GFP-Polκ to UV irradiation is consistent with previous studies indicating that Polκ does not participate significantly in bypass of CPD (Zhang et al., 2000). We conclude that Rad18 phosphorylation at S409 specifically facilitates efficient recruitment of Polη to sites of UV-induced replication fork stalling.
It was of interest to determine whether recruitment of endogenous Polη to replication and repair foci was similarly dependent on Rad18 S409 phosphorylation. Unfortunately, however, we have been unable to detect endogenous levels of Polη by immunofluorescence microscopy using available antibodies against Polη. Therefore, in an alternative approach for studying recruitment of endogenous Polη to replication forks, we investigated the Rad18-dependent association of endogenous Polη with PCNA. In the experiment presented in Figure 5D, PCNA was immunoprecipitated from Ad HA-Rad18– or Ad HA-Rad18 S409A–infected cells. As shown in Figure 5B, UV treatment induced a 2.9-fold increase in the association of Polη with PCNA in HA-Rad18 WT–expressing cells. In cells expressing HA-Rad18 S409A, UV only induced a 1.5-fold increase in the association of Polη with PCNA. Taken together, the results of Figure 5, A and B, suggest that Rad18 phosphorylation at S409 promotes efficient Rad18-dependent recruitment of Polη to sites of stalled replication.
Polη recruitment to sites of DNA damage is considered important for DNA damage tolerance. Therefore we performed clonogenic survival assays to test whether impaired (Rad18 S409-dependent) recruitment of Polη to replication forks is associated with reduced viability after UV treatment. For reasons that are not understood, the HCT116 RAD18−/− cells used elsewhere in this study do not show UV sensitivity when compared with parental HCT116 RAD18+/+ cells (Shiomi et al., 2007). Therefore we analyzed the role of Rad18 in UV tolerance in an alternative cell line. H1299 cells were selected for these studies because we found that Polη is necessary for UV tolerance in this line. Moreover, Rad18 depletion fully phenocopies the UV sensitivity of Polη-depleted H1299 cells (Supplemental Figure S2). We depleted H1299 cells of endogenous Rad18 (using siRNA) and then complemented with siRNA-resistant WT or mutant forms of Rad18. As expected, Rad18-depleted cells were moderately sensitive to UV, and UV tolerance was fully corrected by ectopically- expressed HA-Rad18 WT (Figure 5E). Of interest, HA-Rad18 S409A failed to correct the UV sensitivity of Rad18-depleted cells, thereby indicating a role for JNK-mediated Rad18 S409 phosphorylation in DNA damage tolerance via TLS (Figure 5E).
The UV sensitivity of the Rad18 S409A–complemented cells resembles the phenotype we previously reported for the Rad18 S-box-A mutant (which harbors DDK phosphorylation-site mutations at S421–423 and S434; Day et al., 2010). It was of interest therefore to determine whether the DNA damage tolerance defects conferred by Rad18 S409 and S-box → A mutations were additive or nonadditive. Therefore we performed UV survival assays in cells expressing Rad18 mutants harboring individual or combinatorial S → A substitutions in the S-box serines and S409. As shown in Supplemental Figure S3, the survival curves for S409A, S-box → A and S409, S-box → A mutants were not significantly different. Therefore we conclude that independent phosphorylation events at S409 and S-box serine residues (mediated by JNK and DDK, respectively) are both necessary for Rad18 TLS functions and UV tolerance.
DISCUSSION
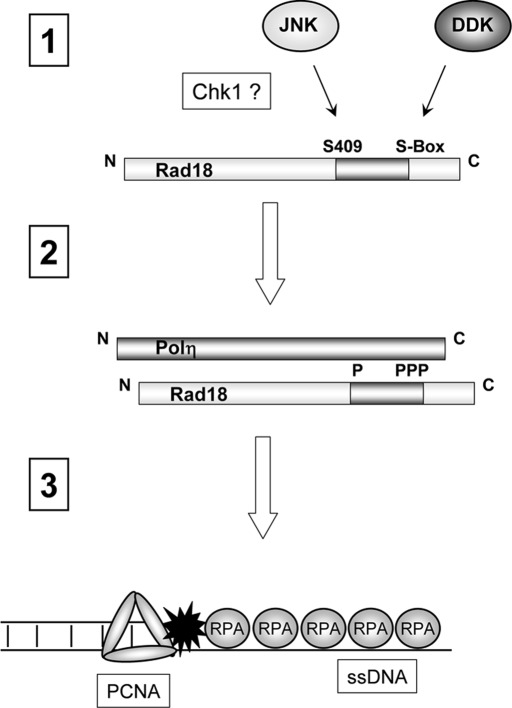
The interaction of Rad18 with Polη represents one of several mechanisms that facilitate recruitment of Polη to the replication machinery. It is generally believed that Rad18 guides Polη to sites of stalled replication (Watanabe et al., 2004) and that Rad18-mediated Polη chaperone activity promotes stable engagement of Polη with PCNA via PCNA-interacting protein (PIP) box and UBZ-mediated interactions (Bienko et al., 2005). In this study we identified a novel DNA damage–inducible Rad18 phosphorylation site at the conserved residue Ser-409 that resides in the Polη-binding domain of Rad18. We demonstrate that S409 phosphorylation is important for Rad18–Polη association and confers more efficient chaperoning of Polη to PCNA at sites of replication stalling. We previously identified a distinct cluster of DDK-dependent phosphorylation sites (serines 421–423 and 434 in the Polη-binding domain) that similarly promote Polη association and DNA damage tolerance (Day et al., 2010). However, S409 is not phosphorylated by DDK (Day et al., 2010), thereby distinguishing the new JNK-mediated pathway from the DDK-dependent mechanism that we previously described. Our finding that S409 and S-box phosphorylation-site mutations in Rad18 confer similar and nonadditive phenotypes shows that efficient Rad18-mediated DNA damage tolerance requires independent JNK- and Cdc7-mediated phosphorylation of Rad18 (Figure 6).
FIGURE 6:
Hypothetical model showing roles of DDK and JNK in UV-inducible Rad18 phosphorylation and DNA damage tolerance. UV induces JNK-mediated phosphorylation of S409 and DDK-mediated phosphorylation of the S-box serine cluster. UV-induced phosphorylation of both S409 and the S-box is Chk1 dependent (step 1). S409 and the S-box reside in the Polη-binding region of Rad18 (residues 401–445). S409 and S-box phosphorylations promote Rad18–Polη interactions (step 2), thereby facilitating Rad18-dependent chaperoning of Polη to sites of replication stalling, where the Rad18–Polη complex associates with RPA-coated ssDNA (step 3).
To our knowledge, this study represents the first demonstration of a relationship between JNK signaling and TLS. The activation of JNK in response to replication stress–inducing agents such as UV radiation (Weston and Davis, 2007) and BPDE (Li et al., 2004) has been studied extensively, yet few studies have explored the integration of SAPK and checkpoint signaling in the DNA damage response (Wang et al., 2009). Our finding that JNK regulates TLS in a manner that is Chk1 dependent further demonstrates the intricate coordination of the various effector branches of the DNA damage response, including checkpoint signaling, SAPK cascades, and TLS.
JNK is widely viewed as a mediator of apoptotic responses, and it is perhaps counterintuitive that JNK may play a protective role in Rad18–Polη recruitment and fork recovery. However, not all stimuli that activate JNK lead to apoptosis, perhaps because of active survival signaling pathways that prevent the apoptotic response (Xia et al., 1995). In addition, the kinetics of JNK activation may determine whether or not there is an apoptotic outcome. For example, transient JNK activation (e.g., in response to cytokines) may mediate survival, whereas prolonged activation can mediate apoptosis (Ventura et al., 2006). Although SAPK signaling has not previously been implicated in TLS or postreplication repair, recent studies by other labs have implicated JNK in control of DNA replication. Miotto and Struhl (2011) reported that JNK phosphorylates Cdt1 (a replication licensing factor) on T29, leading to dissociation of HBO1 from replication origins and blocking DNA replication licensing. In an independent study, Cook and colleagues identified multiple JNK and p38 target sites on Cdt1 the phosphorylation of which serves to rapidly inactivate Cdt1 and inhibit origin licensing (Chandrasekaran et al., 2011). Therefore JNK helps to ensure appropriate DNA replication and genome maintenance in genotoxin-treated cells via at least two mechanisms: inhibition of origin licensing (Chandrasekaran et al., 2011; Miotto and Struhl, 2011) and stimulation of TLS at sites of ongoing DNA replication (the present study). It will be interesting to determine whether additional proteins involved in other stages of DNA synthesis (e.g., initiation) or other modes of DNA repair are similarly subject to regulation by SAPKs.
Potentially consistent with additional roles for SAPKs in DNA replication and repair, Lannigan and colleagues demonstrated that extracellular signal-regulated kinase 8 (ERK8; a SAPK family member) contains a conserved PIP box that mediates PCNA binding (and subsequent PCNA turnover via Mdm2; Groehler and Lannigan, 2010). PCNA represents a central hub for numerous DNA repair and replication processes. Therefore recruitment of ERK8 and perhaps additional PIP box–containing mitogen-activated protein kinase (MAPK) family members to PCNA may provide the basis for cross-talk between MAPK signaling and DNA replication and repair events. By analogy, JNK2 was recently detected at RPA–coated ssDNA (a replication intermediate generated by DNA damage that acts as a proximal trigger of both the TLS and ATR/Chk1 pathways; Chen et al., 2010. Therefore, similar to ERK8, JNK2 may be poised to regulate DNA replication and repair events in the vicinity of stalled replication forks. Further work is needed to investigate potential regulators and targets of the SAPKs at sites of DNA replication and repair.
Increasingly it is apparent that TLS is coordinated with other elements of the DNA damage response. JNK- and DDK-dependent Rad18 phosphorylation (of S409 and the S-box serines, respectively) is clearly Chk1 dependent (Day et al., 2010), demonstrating further links between S-phase checkpoint signaling and TLS. The Chk1 dependence of these Rad18 phosphorylation events may indicate that both JNK and DDK activities are influenced by Chk1 or that Chk1 inhibits phosphatase(s) that target the S409- and S-box–phosphorylated forms of Rad18. Alternatively, the effect of Chk1 status on Rad18 phosphorylation might be indirect, possibly resulting from Chk1-dependent stabilization of stalled forks, which promotes a state that is permissive for regulated changes in Rad18 phosphorylation. In preliminary experiments, Chk1 inhibition did not influence phosphorylation of other JNK or DDK substrates. Therefore we favor the hypothesis that Chk1 influences putative Rad18-directed protein phosphatases or that Chk1 promotes formation or stabilization of replication fork structures that facilitate Rad18 phosphorylation by its upstream kinases, including JNK and DDK. Further studies are needed to distinguish between these (non–mutually exclusive) mechanisms of Chk1 action.
There is a strong precedent for phosphorylation-dependent regulation of E3 ubiquitin ligases. For example, sustained (but not transient) JNK signaling mediates tumor necrosis factor α–induced cell death via the E3 ligase Itch (which degrades the caspase inhibitor cFLIPL; Chang et al., 2006b). Another E3 ligase, Siah2, is regulated by p38-dependent phosphorylation (Khurana et al., 2006). Rad18 plays a central role in TLS, DSB repair (Huang et al., 2009; Palle and Vaziri, 2011), and perhaps additional DNA repair pathways. Therefore Rad18 phosphorylation has the potential to regulate multiple effector pathways, perhaps by influencing associations with its substrates (e.g., PCNA, RFC2, 53BP) or regulatory binding partners (e.g., Polη, Rad6, Rad5). Rad18 phosphorylation may serve as a molecular switch for determining various physiological outcomes including but not limited to TLS. Unpublished mass spectrometry data from our lab and global proteomic profiling data from other labs suggest that Rad18 is phosphorylated on multiple sites (Nousiainen et al., 2006). Further work is underway to validate additional Rad18 phosphorylation sites, test the physiological significance of these putative phosphorylation events, and identify the relevant kinases.
We also consider it likely that many other TLS proteins will be subject to regulation by phosphorylation. Rad6 is also phosphorylated by CDKs (Sarcevic et al., 2002), and this modification may provide a mechanism for coordinating Rad6-dependent ubiquitination events with specific stages of the cell cycle. A study by Cleaver and colleagues suggested that Polη recruitment to stalled replication forks is regulated via its direct phosphorylation, most likely by protein kinase C family members (Chen et al., 2008). Lehmann and colleagues also demonstrated that Polη is an ATR substrate whose phosphorylation modulates S-phase checkpoint signaling (Gohler et al., 2011). Taken together, our results in this report and a previous study (Day et al., 2010), together with studies from other labs (Chen et al., 2008; Gohler et al., 2011), indicate that multiple phosphorylation-based mechanisms facilitate the efficient recruitment of Polη to sites of replication fork stalling. Further studies are underway to determine how protein kinases regulate Rad18 and integrate TLS (and perhaps other Rad18-dependent DNA repair processes) with the cell cycle.
MATERIALS AND METHODS
Adenovirus construction and infection
Adenovirus construction and infections were performed as described previously (Guo et al., 2002; Bi et al., 2006). In brief, cDNAs encoding HA-Rad18 WT, HA-Rad18 S409A, and HA-Rad18 S409E were subcloned into pAC-CMV. The resulting shuttle vectors were cotransfected into 293T cells with the pJM17 plasmid to generate recombinant adenovirus as described previously (Bi et al., 2005). H1299 and HCT 116 cells were routinely infected with adenovirus at a multiplicity of infection of 20 and 50, respectively. To control for adenoviral infection, cells received AdCon (“empty” adenovirus vector) or AdGFP.
Cell culture
Human lung carcinoma H1299 cells and RAD18−/− HCT116 cells (Shiomi et al., 2007) were cultured in DMEM supplemented with 10% fetal bovine serum, streptomycin sulfate (100 μg/ml), and penicillin (100 U/ml).
In vitro kinase assays
Recombinant Rad18–Rad6 complex from insect cells and GST-Rad18 from bacteria were expressed, purified, and tested as DDK and JNK substrates using in vitro phosphorylation reactions as described previously (Day et al., 2010). Recombinant DDK was purified from insect cells as described previously (Day et al., 2010), and recombinant JNK1α1 was purchased from Upstate (Millipore, Billerica, MA).
Genotoxin treatments
BPDE (National Cancer Institute Carcinogen Repository) was dissolved in anhydrous Me2SO and added directly to the growth medium as a 1000× stock to give various final concentrations, as indicated in the figure legends. For UVC treatment, the growth medium was removed from the cells, reserved, and replaced with phosphate-buffered saline (PBS). The plates were transferred to a UV cross-linker (Stratagene, Santa Clara, CA) and then irradiated. The UVC dose delivered to the cells was confirmed with a UV radiometer (UVP BioImaging Systems, Upland, CA). The reserved medium from the cells was replaced, and cells were returned to the incubator.
RNA interference
Cells were plated into 60- or 100-mm culture dishes. Twenty-four hours later, when cells were 50–75% confluent, the cultures were placed in antibiotic-free medium (2 or 5 ml per plate). siRNA transfections were performed using Lipofectamine, using protocols recommended by the manufacturer (Invitrogen, Carlsbad, CA). After overnight transfections, the Lipofectamine-containing medium was removed and replaced with standard culture medium. All tubes, tips, and solutions used for RNA interference experiments were certified RNase free.
Clonogenic survival assays
H1299 cells were grown to ∼30–50% confluence in 60-mm plates and transfected with 4 μg of pAC.CMV GFP vector (for controls) or with pAC.CMV expression vectors encoding wild-type or mutant Rad18 using Lipofectamine. Transfected cells were treated with UVC (as described) and then split into replicate 10-cm plates at a density of 1000 cells/plate. After 10–14 d, colonies on the plates were fixed in methanol, stained with crystal violet, and counted as described previously (Day et al., 2010).
Fluorescence microscopy
H1299 cells at near-confluency were transfected with siRNA targeting the 3′ untranslated region of Rad18. Ten hours later, the cells were split into glass-bottomed dishes (MatTek Corporation, Ashland, MA) to give a density of ∼50% after 24 h. The cells were then infected with adenovirus encoding Rad18, YFP-Polη, or GFP-Polκ (or “empty” vector control). Six hours later, the virus-infected cells were transfected one more time with siRNA against Rad18. Twenty hours after the second siRNA transfection, the cells were sham or UV irradiated (10 J/m2). Two hours after UV treatment, the cells were rinsed with cold PBS, immersed briefly (<5 s) in PBS containing 0.1% Triton X-100, rinsed again in ice-cold PBS, and then fixed in PBS containing 4% formaldehyde for 5 min. Fixed nuclei were 4′,6-diamidino-2-phenylindole stained and mounted with Vectashield solution (Vector Laboratories, Burlingame, CA). Slides were imaged and analyzed using a Zeiss 710 Laser Scanning Microscope at 63× magnification with digital zoom (Zeiss, Jena, Germany). We collected 0.5 μM z-sections of representative cells from different experimental conditions and deconvolved them to generate the images presented.
Immunoblotting
Cells were separated into cytosolic and nucleosolic or chromatin fractions using CSK buffer as described previously (Bi et al., 2005; Liu et al., 2006). The resulting samples were normalized for protein content, then separated by SDS–PAGE, transferred to nitrocellulose, and analyzed by immunoblotting with the following antibodies: rabbit anti-Chk1 (Santa Cruz Biotechnology, Santa Cruz, CA), mouse monoclonal anti-PCNA (Santa Cruz Biotechnology), rabbit anti–phospho-Chk1 Ser-317 (Cell Signaling, Beverly, MA), polyclonal HA tag antibody (Santa Cruz Biotechnology), polyclonal PCNA antibody (Santa Cruz Biotechnology), polyclonal Polη (Bethyl Laboratories, Montgomery, TX), GFP (Invitrogen Molecular Probes, Eugene, OR), and Rad18 (Bethyl Laboratories). Phospho–serine/threonine antibodies were from BD Biosciences (San Diego, CA). Rad18 S409 phosphospecific antibodies were generated by immunizing rabbits with the peptide FSQSKLD[pS]PEELEPC. To ensure phosphospecificity, the resulting sera were depleted using unmodified immunogen and purified using the phosphopeptide. Peptide synthesis, immunizations, and antibody purification steps were performed by 21st Century Biochemicals (Marlboro, MA).
Coimmunoprecipitation of Polη with PCNA and Rad18
Cells were plated in 10-cm culture dishes and infected with adenovirus as described. Genotoxin treatments were performed at ∼70% confluence. In some experiments, 2% formaldehyde was added for 5 min to cross-link chromatin-associated proteins before cell lysis. Cross-linking reactions were quenched by the addition of 1 M glycine for 5 min before PBS washes and CSK lysis. Samples containing chromatin fractions were generated as described and were incubated for 30 min at room temperature with 1 unit/μl RQ DNAse (Promega, Madison, WI) and then sonicated (for three 15-s pulses at 30% maximum output). Pulses were separated by a 10-s interval on ice to prevent excessive heating. The sonicated samples were clarified by centrifugation at 10,000 × g for 5 min. Supernatants were removed and normalized for protein concentration (∼600 μg of protein in 1 ml was used for each immunoprecipitation). PCNA or HA-Rad18 was immunoprecipitated overnight at 4°C using anti-PCNA or anti-HA monoclonal antibodies. Replicate immunoprecipitations were performed using immunoglobulin G to control for specificity of protein–protein associations. After antibody incubations, 25 μl pf protein A/G beads were added to each sample for 4 h. The beads were recovered by brief centrifugation and washed five times with 1 ml CSK (5–10 min per wash). The washed immune complexes were boiled in protein loading buffer for 10 min to release and denature immunoprecipitated proteins before separation on SDS–PAGE.
ssDNA-binding assays
The binding of Rad18 to ssDNA was detected as described by Tateishi and colleagues (Tsuji et al., 2008). To prepare ssDNA-coated beads, 0.1 ml of TetraLink Tetrameric Avidin Resin (Promega) was washed twice with 1 ml of buffer A (50 mM Tris-HCl, pH 7.6, 1 mM dithiothreitol). Washed beads were suspended in 50 μl of buffer A. We mixed 10 μl of 10 μM 5′-biotinylated oligonucleotides, whose nucleotide sequences are described by Tsuji et al. (2008), with the beads and incubated them for 15 min at room temperature. In parallel reactions beads were prepared without addition of biotinylated oligonucleotides, and these were subsequently used to control for the ssDNA dependence of binding reactions. We added 1 ml of a solution containing 3% skim milk, 20 mM Tris-HCl (pH 7.5), and 150 mM NaCl to the beads and incubated the mixture for 20 min at room temperature. The beads were then collected by centrifugation, washed once with 1 ml of buffer A, and resuspended in 0.3 ml of buffer A (DNA–beads solution). Bovine serum albumin was added to the DNA–beads solution to give a final concentration of 0.1 mg/ml. We removed 20-μl aliquots of the resulting mixture and added them to fresh microfuge tubes. RPA, 0.5 μM, was added to each 20-μl aliquot of ssDNA-coated beads. We mixed 100-μl aliquots of Rad18-containing CSK extracts (normalized to a protein concentration of 1 μg/μl) with the RPA-coated beads and incubated them for 10 min at 37°C. The binding reactions were then diluted by addition of 1 ml of buffer A. The beads containing bound Rad18 were collected by centrifugation (1000 × g, 5 min) and washed three times in buffer A. After the final wash the packed beads were resuspended in 50 μl of buffer A containing 0.4 M NaCl and incubated for 5 min at room temperature. The samples were centrifuged for 5 min at 10,000 × g. A 20-μl amount of the supernatant was analyzed by SDS–PAGE and immunoblotting with ant-Rad18 antibodies.
Reproducibility
All data shown are representative of experiments that were repeated at least three times, with similar results on each occasion.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Environmental Health Sciences Grants ES09558 and ES016280 to C.V. We thank Satoshi Tateishi and Jean Cook for numerous helpful discussions during the course of this work. We thank Bonnie Gunn for advice with statistical analysis.
Abbreviations used:
- ATR
ATM- and Rad3-related
- B[a]P
benzo[a]pyrene
- BPDE
(+)-r-7,t-8-dihydroxy-t-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene
- Cdc7
cell division cycle 7
- Chk1
checkpoint kinase 1
- CPD
cyclo-butane pyrimidine dimer
- Dbf4
Dumbell former 4
- DDK
Dbf4-dependent kinase
- DDR
DNA damage response
- DSB
double-stranded break
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- JNK
c-Jun N-terminal kinase
- PCNA
proliferating cellular nuclear antigen
- PIP
PCNA-interacting peptide
- Polη
DNA polymerase eta
- Polκ
DNA polymerase kappa
- Rad6
radiation sensitive 6
- Rad18
radiation sensitive 18
- RPA
replication protein A
- ssDNA
single-stranded DNA
- TLS
trans-lesion synthesis
- UV
UV radiation
- XPV
xeroderma pigmentosum variant
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-10-0829) on March 28, 2012.
REFERENCES
- Barkley LR, Ohmori H, Vaziri C. Integrating S-phase checkpoint signaling with trans-lesion synthesis of bulky DNA adducts. Cell Biochem Biophys. 2007;47:392–408. doi: 10.1007/s12013-007-0032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Barkley LR, Slater DM, Tateishi S, Yamaizumi M, Ohmori H, Vaziri C. Rad18 regulates DNA polymerase {kappa} and is required for recovery from S-phase checkpoint-mediated arrest. Mol Cell Biol. 2006;26:3527–3540. doi: 10.1128/MCB.26.9.3527-3540.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Slater DM, Ohmori H, Vaziri C. DNA polymerase kappa is specifically required for recovery from the benzo[a]pyrene-dihydrodiol epoxide (BPDE)-induced S-phase checkpoint. J Biol Chem. 2005;280:22343–22355. doi: 10.1074/jbc.M501562200. [DOI] [PubMed] [Google Scholar]
- Bienko M, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- Bomgarden RD, Lupardus PJ, Soni DV, Yee MC, Ford JM, Cimprich KA. Opposing effects of the UV lesion repair protein XPA and UV bypass polymerase eta on ATR checkpoint signaling. EMBO J. 2006;25:2605–2614. doi: 10.1038/sj.emboj.7601123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows AE, Elledge SJ. How ATR turns on: TopBP1 goes on ATRIP with ATR. Genes Dev. 2008;22:1416–1421. doi: 10.1101/gad.1685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran S, Tan TX, Hall JR, Cook JG. Stress-stimulated mitogen-activated protein kinases control the stability and activity of the Cdt1 DNA replication licensing factor. Mol Cell Biol. 2011;31:4405–4416. doi: 10.1128/MCB.06163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DJ, Lupardus PJ, Cimprich KA. Monoubiquitination of proliferating cell nuclear antigen induced by stalled replication requires uncoupling of DNA polymerase and mini-chromosome maintenance helicase activities. J Biol Chem. 2006a;281:32081–32088. doi: 10.1074/jbc.M606799200. [DOI] [PubMed] [Google Scholar]
- Chang L, Kamata H, Solinas G, Luo JL, Maeda S, Venuprasad K, Liu YC, Karin M. The E3 ubiquitin ligase itch couples JNK activation to TNFalpha-induced cell death by inducing c-FLIP(L) turnover. Cell. 2006b;124:601–613. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Chen P, O'Neal JF, Ebelt ND, Cantrell MA, Mitra S, Nasrazadani A, Vandenbroek TL, Heasley LE, Van Den Berg CL. Jnk2 effects on tumor development, genetic instability and replicative stress in an oncogene-driven mouse mammary tumor model. PloS One. 2010;5:e10443. doi: 10.1371/journal.pone.0010443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YW, Cleaver JE, Hatahet Z, Honkanen RE, Chang JY, Yen Y, Chou KM. Human DNA polymerase eta activity and translocation is regulated by phosphorylation. Proc Natl Acad Sci USA. 2008;105:16578–16583. doi: 10.1073/pnas.0808589105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein a. Mol Cell. 2008;29:625–636. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- Day TA, Palle K, Barkley LR, Kakusho N, Zou Y, Tateishi S, Verreault A, Masai H, Vaziri C. Phosphorylated Rad 18 directs DNA polymerase η to sites of stalled replication. J Cell Biol. 2010;191:953–966. doi: 10.1083/jcb.201006043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dipple A. DNA adducts of chemical carcinogens. Carcinogenesis. 1995;16:437–441. doi: 10.1093/carcin/16.3.437. [DOI] [PubMed] [Google Scholar]
- Gohler T, Sabbioneda S, Green CM, Lehmann AR. ATR-mediated phosphorylation of DNA polymerase eta is needed for efficient recovery from UV damage. J Cell Biol. 2011;192:219–227. doi: 10.1083/jcb.201008076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groehler AL, Lannigan DA. A chromatin-bound kinase, ERK8, protects genomic integrity by inhibiting HDM2-mediated degradation of the DNA clamp PCNA. J Cell Biol. 2010;190:575–586. doi: 10.1083/jcb.201002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo N, Faller DV, Vaziri C. Carcinogen-induced S-phase arrest is Chk1 mediated and caffeine sensitive. Cell Growth Differ. 2002;13:77–86. [PubMed] [Google Scholar]
- Heffernan TP, Simpson DA, Frank AR, Heinloth AN, Paules RS, Cordeiro-Stone M, Kaufmann WK. An ATR- and Chk1-dependent S checkpoint inhibits replicon initiation following UVC-induced DNA damage. Mol Cell Biol. 2002;22:8552–8561. doi: 10.1128/MCB.22.24.8552-8561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffernan TP, Unsal-Kacmaz K, Heinloth AN, Simpson DA, Paules RS, Sancar A, Cordeiro-Stone M, Kaufmann WK. Cdc7/Dbf4 and the human S checkpoint response to UVC. J Biol Chem. 2007;282:9458–9468. doi: 10.1074/jbc.M611292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, Chen J. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol. 2009;11:592–603. doi: 10.1038/ncb1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D'Andrea AD. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- Huttner D, Ulrich HD. Cooperation of replication protein A with the ubiquitin ligase Rad18 in DNA damage bypass. Cell Cycle. 2008;7:3629–3633. doi: 10.4161/cc.7.23.7166. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- Kannouche PL, Lehmann AR. Ubiquitination of PCNA and the polymerase switch in human cells. Cell Cycle. 2004;3:1011–1013. [PubMed] [Google Scholar]
- Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- Khurana A, Nakayama K, Williams S, Davis RJ, Mustelin T, Ronai Z. Regulation of the ring finger E3 ligase Siah2 by p38 MAPK. J Biol Chem. 2006;281:35316–35326. doi: 10.1074/jbc.M606568200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Tang MS, Liu B, Shi X, Huang C. A critical role of PI-3K/Akt/JNKs pathway in benzo[a]pyrene diol-epoxide (B[a]PDE)-induced AP-1 transactivation in mouse epidermal Cl41 cells. Oncogene. 2004;23:3932–3944. doi: 10.1038/sj.onc.1207501. [DOI] [PubMed] [Google Scholar]
- Liu P, Barkley LR, Day T, Bi X, Slater DM, Alexandrow MG, Nasheuer HP, Vaziri C. The Chk1-mediated S-phase checkpoint targets initiation factor Cdc45 via a Cdc25a/Cdk2-independent mechanism. J Biol Chem. 2006;281:30631–30644. doi: 10.1074/jbc.M602982200. [DOI] [PubMed] [Google Scholar]
- Maher VM, Ouellette LM, Curren RD, McCormick JJ. Frequency of ultraviolet light-induced mutations is higher in xeroderma pigmentosum variant cells than in normal human cells. Nature. 1976;261:593–595. doi: 10.1038/261593a0. [DOI] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- Miller ML, et al. Linear motif atlas for phosphorylation-dependent signaling. Sci Signal. 2008;1:ra2. doi: 10.1126/scisignal.1159433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto B, Struhl K. JNK1 phosphorylation of Cdt1 inhibits recruitment of HBO1 histone acetylase and blocks replication licensing in response to stress. Mol Cell. 2011;44:62–71. doi: 10.1016/j.molcel.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes DA, Cortez D. Activation of ATR and related PIKKs. Cell Cycle. 2008;7:2809–2812. doi: 10.4161/cc.7.18.6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci USA. 2006;103:5391–5396. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill T, Giarratani L, Chen P, Iyer L, Lee CH, Bobiak M, Kanai F, Zhou BB, Chung JH, Rathbun GA. Determination of substrate motifs for human Chk1 and hCds1/Chk2 by the oriented peptide library approach. J Biol Chem. 2002;277:16102–16115. doi: 10.1074/jbc.M111705200. [DOI] [PubMed] [Google Scholar]
- Ohmori H, et al. The Y-family of DNA polymerases. Mol Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- Palle K, Vaziri C. Rad18 E3 ubiquitin ligase activity mediates fanconi anemia pathway activation and cell survival following DNA Topoisomerase 1 Inhibition. Cell Cycle. 2011;10:1625–1638. doi: 10.4161/cc.10.10.15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB. p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38 MAPK/MK2 pathway for survival after DNA damage. Cancer Cell. 2007;11:175–189. doi: 10.1016/j.ccr.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt HC, Yaffe MB. Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2, and MK2. Curr Opin Cell Biol. 2009;21:245–255. doi: 10.1016/j.ceb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Sarcevic B, Mawson A, Baker RT, Sutherland RL. Regulation of the ubiquitin-conjugating enzyme hHR6A by CDK-mediated phosphorylation. EMBO J. 2002;21:2009–2018. doi: 10.1093/emboj/21.8.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi N, Mori M, Tsuji H, Imai T, Inoue H, Tateishi S, Yamaizumi M, Shiomi T. Human RAD18 is involved in S phase-specific single-strand break repair without PCNA monoubiquitination. Nucleic Acids Res. 2007;35:e9. doi: 10.1093/nar/gkl979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen CS, Syljuasen RG, Lukas J, Bartek J. ATR, Claspin and the Rad9-Rad1-Hus1 complex regulate Chk1 and Cdc25A in the absence of DNA damage. Cell Cycle. 2004;3:941–945. [PubMed] [Google Scholar]
- Thakker DR, Yagi H, Lu AY, Levin W, Conney AH. Metabolism of benzo[a]pyrene: conversion of (+/-)-trans-7,8-dihydroxy-7,8-dihydrobenzo[a]pyrene to highly mutagenic 7,8-diol-9,10-epoxides. Proc Natl Acad Sci USA. 1976;73:3381–3385. doi: 10.1073/pnas.73.10.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji Y, Watanabe K, Araki K, Shinohara M, Yamagata Y, Tsurimoto T, Hanaoka F, Yamamura K, Yamaizumi M, Tateishi S. Recognition of forked and single-stranded DNA structures by human RAD18 complexed with RAD6B protein triggers its recruitment to stalled replication forks. Genes Cells. 2008;13:343–354. doi: 10.1111/j.1365-2443.2008.01176.x. [DOI] [PubMed] [Google Scholar]
- Ulrich HD. How to activate a damage-tolerant polymerase: consequences of PCNA modifications by ubiquitin and SUMO. Cell Cycle. 2004;3:15–18. [PubMed] [Google Scholar]
- Ventura JJ, Hubner A, Zhang C, Flavell RA, Shokat KM, Davis RJ. Chemical genetic analysis of the time course of signal transduction by JNK. Mol Cell. 2006;21:701–710. doi: 10.1016/j.molcel.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang M, Kar S, Carr BI. Involvement of ATM-mediated Chk1/2 and JNK kinase signaling activation in HKH40A-induced cell growth inhibition. J Cell Physiol. 2009;221:213–220. doi: 10.1002/jcp.21844. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Iwabuchi K, Sun J, Tsuji Y, Tani T, Tokunaga K, Date T, Hashimoto M, Yamaizumi M, Tateishi S. RAD18 promotes DNA double-strand break repair during G1 phase through chromatin retention of 53BP1. Nucleic Acids Res. 2009;37:2176–2193. doi: 10.1093/nar/gkp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston CR, Davis RJ. The JNK signal transduction pathway. Curr Opin Cell Biol. 2007;19:142–149. doi: 10.1016/j.ceb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Xia Z, Dickens M, Raingeaud J, Davis RJ, Greenberg ME. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- Yanagihara H, et al. NBS1 recruits RAD18 via a RAD6-like domain and Regulates Pol eta-dependent translesion DNA synthesis. Mol Cell. 2011;43:788–797. doi: 10.1016/j.molcel.2011.07.026. [DOI] [PubMed] [Google Scholar]
- Yang XH, Shiotani B, Classon M, Zou L. Chk1 and Claspin potentiate PCNA ubiquitination. Genes Dev. 2008;22:1147–1152. doi: 10.1101/gad.1632808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XH, Zou L. Dual functions of DNA replication forks in checkpoint signaling and PCNA ubiquitination. Cell Cycle. 2009;8:191–194. doi: 10.4161/cc.8.2.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Wu X, Wang M, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- Ziv O, Geacintov N, Nakajima S, Yasui A, Livneh Z. DNA polymerase zeta cooperates with polymerases kappa and iota in translesion DNA synthesis across pyrimidine photodimers in cells from XPV patients. Proc Natl Acad Sci USA. 2009;106:11552–11557. doi: 10.1073/pnas.0812548106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci USA. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.