Ectodomain shedding of EGFR ligands by ADAM17 is a key step of transactivation of epidermal growth factor receptor (EGFR) and the downstream signaling network. In this study, we identified cell surface annexins as regulators of ectodomain shedding of amphiregulin precursor. We propose that cell surface annexins act as a shedding platform to determine the substrate selectivity of ADAM17.
Abstract
A disintegrin and metalloproteinase (ADAM) is a family of enzymes involved in ectodomain shedding of various membrane proteins. However, the molecular mechanism underlying substrate recognition by ADAMs remains unknown. In this study, we successfully captured and analyzed cell surface transient assemblies between the transmembrane amphiregulin precursor (proAREG) and ADAM17 during an early shedding phase, which enabled the identification of cell surface annexins as components of their shedding complex. Annexin family members annexin A2 (ANXA2), A8, and A9 interacted with proAREG and ADAM17 on the cell surface. Shedding of proAREG was increased when ANXA2 was knocked down but decreased with ANXA8 and A9 knockdown, because of enhanced and impaired association with ADAM17, respectively. Knockdown of ANXA2 and A8 in primary keratinocytes altered wound-induced cell migration and ultraviolet B–induced phosphorylation of epidermal growth factor receptor (EGFR), suggesting that annexins play an essential role in the ADAM-mediated ectodomain shedding of EGFR ligands. On the basis of these data, we propose that annexins on the cell surface function as “shedding platform” proteins to determine the substrate selectivity of ADAM17, with possible therapeutic potential in ADAM-related diseases.
INTRODUCTION
The epidermal growth factor receptor (EGFR) is a potent mediator of diseases such as cancer, as well as of cell growth and development (Baselga and Swain, 2009). Transactivation of EGFRs has been shown to play a crucial role in signaling by G protein–coupled receptors, cytokine receptors, receptor tyrosine kinases, and integrins in multiple cellular responses (Hackel et al., 1999; Moghal and Sternberg, 1999). Transactivation of EGFRs is mainly mediated by ectodomain shedding of EGFR ligand precursors, which include EGF, amphiregulin (AREG), heparin-binding EGF-like growth factor (HB-EGF), transforming growth factor α (TGFα), epiregulin (EREG), betacellulin, epigen, and neuregulins and are synthesized as type I transmembrane protein (proforms; Higashiyama et al., 2008). In response to a wide variety of stimulations, EGFR ligand precursors are shed by membrane-type metalloproteases of the a disintegrin and metalloproteinase (ADAM) family, on the cell surface, and subsequently release their ectodomain to activate EGFR and intracellular signaling pathways.
Aberrations in ectodomain shedding of precursors of HB-EGF (proHB-EGF) in soluble HB-EGF or uncleavable proHB-EGF knock-in mice resulted in severely abnormal skin and heart developments, because of the insufficient activation of EGFRs (Yamazaki et al., 2003). Moreover, proAREG shedding in mammary epithelial cells is required for mammary ductal morphogenesis (Sternlicht et al., 2005). These findings indicate that balanced shedding of EGFR ligand precursors is essential for development and maintenance of cellular functions. We have previously reported that ectodomain shedding of proHB-EGF evokes two independent signaling pathways: 1) EGFR signaling induced by soluble forms of HB-EGF and 2) signaling driven by a remnant peptide composed of the transmembrane and cytoplasmic fragment (CTF; Nanba et al., 2003; Hieda et al., 2008). HB-EGF-CTF–mediated signaling resulted in the reversal of promyelocytic leukemia zinc finger and B-cell lymphoma 6–mediated gene repression (Nanba et al., 2003; Kinugasa et al., 2007). Insufficiency of HB-EGF-CTF signaling evoked by proHB-EGF shedding enhanced hypoxia-induced cell death in rat cardiomyoblasts via the activation of caspase-3 and c-Jun N-terminal kinase (JNK; Uetani et al., 2009). We have also shown that proAREG and its CTF translocated into the inner nuclear membrane in response to the shedding stimulant 12-O-tetradecanoylphorbol-13-acetate (TPA). Subsequently, they interacted with lamin A/C, resulting in the induction of heterochromatinization and global transcriptional suppression (Isokane et al., 2008). Shedding of EGFR ligand precursors yields the coordinated extra- and intracellular signaling pathways, which suggests that the regulatory mechanism of ectodomain shedding is indispensable in controlling cell signaling networks (Higashiyama and Nanba, 2005; Higashiyama et al., 2008, 2011).
ADAMs are key enzymes involved in the ectodomain shedding of a large variety of membrane proteins; they are also crucial for numerous developmental and pathological events (Edwards et al., 2008). In particular, ADAM17 (tumor necrosis factor α-converting enzyme [TACE]) has emerged as a sheddase with an extremely broad substrate range, including EGFR ligand precursors, and fundamental links to EGFR signaling. Previous reports showed that adam17−/− mice (Peschon et al., 1998; Shi et al., 2003) displayed the abnormal eyelids and hair and whisker development that had also been observed in TGFα-deficient mice (Luetteke et al., 1993), and aberrant heart development, as seen in uncleavable proHB-EGF knock-in mice (Yamazaki et al., 2003). In contrast, knockdown of ADAM17 using a small interfering RNA (siRNA) or treatment with a metalloprotease inhibitor caused the malignant phenotype to revert to a normal phenotype in a breast cancer cell line. This resulted from prevention of shedding of proAREG and proTGFα (Kenny and Bissell, 2007). The activity of ADAMs is tightly controlled by gene expression, associating proteins, tissue inhibitor of metalloproteinases (TIMP), and posttranslational processes (Edwards et al., 2008). Although much has been learned about the regulatory mechanism of ADAM activities, the molecular mechanisms underlying substrate recognition and selectivity by ADAM17 have not yet been elucidated. In our study, we have successfully captured and analyzed transient assembly at the cell surface between ADAM17 and proAREG, which enabled the identification of cell surface annexins as components of the shedding complex. We characterized cell surface annexins and propose that they have a role in determining the substrate selectivity of ADAM17. These findings provide us with new insights into the process of ADAM-mediated ectodomain shedding of EGFR ligand precursors.
RESULTS
ProAREG assembles during TPA-induced shedding
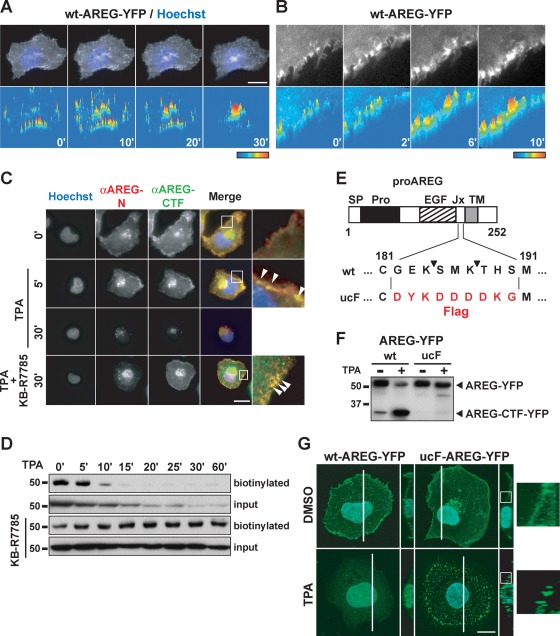
To investigate the process of proAREG shedding, we first focused on the behavior of proAREG during the early phase of proAREG shedding following treatment with the shedding stimulant, TPA. We performed a time-lapse analysis of HT1080 cells stably expressing wild-type (wt) proAREG, with yellow fluorescent protein (YFP) fused to the carboxy terminus. The proAREG-YFP and AREG-CTF-YFP translocated into and accumulated at the perinuclear region after TPA stimulation (Figure 1A; Isokane et al., 2008). We noticed the YFP signal assembled at the plasma membrane around 6 min after TPA treatment (Figure 1B). We also examined the subcellular localization of proAREG by immunofluorescence staining with antibodies recognizing the extracellular (anti–AREG-N) and intracellular (anti–AREG-CTF) domains of proAREG (Figure 1C). Five minutes after TPA treatment, both antibodies detected proAREG at the plasma membrane. Thirty minutes after TPA treatment, however, the assembly of proAREG at the plasma membrane was not observed. When cells were cultured in the presence of the metalloprotease inhibitor KB-R7785, proAREG assembly was maintained at the plasma membrane more than 30 min after TPA stimulation. These data suggest that proAREG was temporarily assembled at the plasma membrane in response to TPA and was subsequently shed by metalloproteases.
FIGURE 1:
ProAREG assembles during TPA-induced shedding. (A and B) HT1080 cells were treated with 10 μg/ml cycloheximide for 4 h to block de novo protein synthesis. The time-lapse images of wt-proAREG-YFP were obtained at the indicated time points after stimulation with 20 nM TPA. The fluorescence signal was visualized as color images using LuminaVision. Scale bar: 20 μm. (C) HT1080 cells stably expressing wt-proAREG were incubated with 20 nM TPA for 30 min in the absence or presence of the metalloprotease inhibitor, KB-R7785 (10 μM). Cells were immunostained with anti–AREG-N and anti–AREG-CTF antibodies. The arrowheads indicate the proAREG complex. Scale bar: 40 μm. (D) The wt-proAREG-YFP cells were incubated with 100 nM TPA for the indicated time periods in the absence or presence of KB-R7785, and biotinylated with a membrane-impermeable biotinylation reagent. The biotinylated proteins were immunoprecipitated with an anti-GFP antibody and analyzed by Western blotting using HRP-conjugated streptavidin. Aliquots of cell lysates used for immunoprecipitation were analyzed by the anti-GFP antibody, and the expression of proAREG-YFP was indicated as the input. (E) Schematic structure of proAREG. SP, signal peptide; EGF, EGF-like domain; Jx, juxtamembrane domain; TM, transmembrane domain. The arrowheads indicate the ADAM17-mediated proAREG cleavage sites. (F) HT1080 cells were transiently transfected with YFP-fused wt- or ucF-proAREG expression vectors. Cells were incubated with 100 nM TPA for 60 min. Western blotting was performed with an anti-GFP antibody. (G) Fluorescent images of wt- and ucF-proAREG-YFP in HT1080 cells. The photographs are three-dimensional reconstructions of the Z-stack captured with a confocal microscope. The right panels show the Y- and Z-axis orthogonal view of the sections made through the stack image, as indicated by white lines. Hoechst nuclear staining is shown in blue. Scale bar: 20 μm. The higher magnification images are indicated by the white box.
A combination assay involving cell surface biotinylation and Western blotting showed that the cell surface proAREG-YFP and the whole proAREG-YFP disappeared after TPA stimulation (Figure 1D). However, disappearance of the cell surface proAREG-YFP was more rapid, because of the shedding. The proAREG-YFP still localized at the cell surface in the presence of KB-R7785, which indicated that the TPA-induced proAREG-YFP complex localized at the cell surface (Figure 1, C and D). Thus, under inhibitory conditions for shedding, intermediates of the proAREG shedding complex could be detected as a proAREG assembly at the plasma membrane and might contain regulatory molecules for shedding.
To investigate the intermediates of the TPA-induced proAREG shedding complex in greater detail, we attempted to produce an uncleavable proAREG mutant. Consistent with a previous study showing the involvement of ADAM17 in AREG shedding (Sahin et al., 2004), we observed a significant decrease in TPA-induced proAREG shedding in HT1080 cells transfected with a siRNA against ADAM17 (unpublished data). As shown in Figure 1E, proAREG contains two ADAM17-dependent cleavage sites (Hinkle et al., 2004; Sahin et al., 2004). We prepared three mutants that had alanine substitutions at these ADAM17 cleavage sites: E183A/K184A; M186A/K187A; and E183A/K184A/M186A/K187A (Supplementary Figure S1A). Western blot analysis revealed that these mutants were shed by TPA stimulation (Figure S1A). We made another proAREG mutant, in which the cleavage site was replaced with the Flag-tag sequence (ucF-proAREG). Our results indicated that ucF-proAREG was completely resistant to ectodomain shedding (Figures 1F and S1, A and B). We used this ucF-proAREG as an uncleavable mutant, then analyzed the subcellular localization of wt- and ucF-proAREG in HT1080 cells. The ucF-proAREG was diffusely localized at the plasma membrane in the steady state, while it assembled by TPA stimulation (Figures 1G and S1C). The Y- and Z-axis orthogonal-view images indicated that the TPA-induced ucF-proAREG-YFP complex localized at the plasma membrane, whereas wt-proAREG-YFP was not evident at the plasma membrane in response to TPA (Figure 1G). Time-lapse image analysis and cell surface biotinylation revealed that TPA-induced ucF-proAREG-YFP assembly was retained on the cell surface (Figure S1, D–F).
ANXA8 regulates ectodomain shedding of proAREG
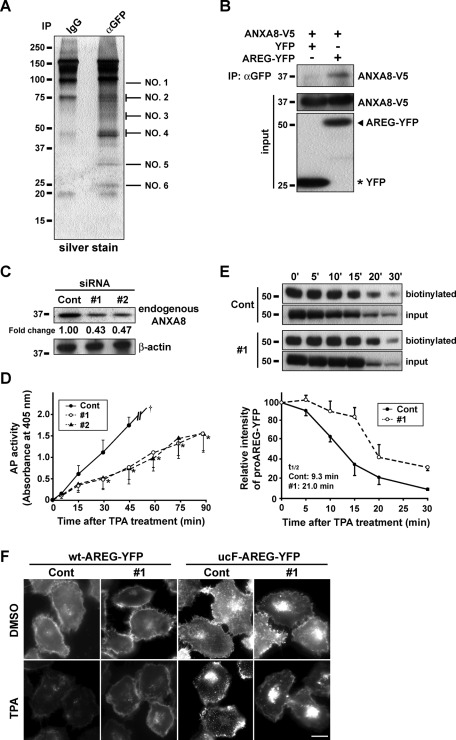
To comprehensively identify components of the TPA-induced ucF-proAREG assembly complex, we performed mass spectrometry (MS) analysis using ucF-proAREG-YFP cells. By comparing these cells with the control, we identified molecules that coimmunoprecipitated with ucF-proAREG-YFP in the presence of TPA (Figure 2A and Supplemental Table S1). Of these molecules, we focused on a member of the annexin family, ANXA8. Immunoprecipitation experiments confirmed that ANXA8 interacted with proAREG (Figure 2B). To investigate the possible role of ANXA8 in the regulation of proAREG shedding, siRNA was used to reduce endogenous protein levels of ANXA8. Following transfection of ANXA8-specific siRNA (#1, #2), endogenous ANXA8 protein expression was reduced by ∼55% in HT1080 cells (Figure 2C). To determine the effect of ANXA8 siRNA on proAREG shedding, we conducted a quantitative alkaline phosphatase (AP) assay, which estimates the efficiency of proAREG shedding (Tokumaru et al., 2000). As shown in Figure 2D, AP activity was significantly reduced during 90-min incubation after TPA stimulation in ANXA8 siRNA-treated cells (#1, #2) compared with a control. Similarly, a majority of proAREG-YFP was still retained at the plasma membrane 20 min after TPA stimulation in ANXA8 knockdown cells, because proAREG shedding was prevented (Figure 2E). We also noted that knockdown of ANXA8 prevented the assembly of ucF-proAREG-YFP by TPA stimulation (Figure 2F), suggesting that ANXA8 is involved in the formation of the proAREG assembly complex and is crucial for proAREG shedding.
FIGURE 2:
ANXA8 regulates proAREG shedding. (A) The ucF-proAREG-YFP cells were treated with 100 nM TPA for 30 min, and membrane fraction proteins were immunoprecipitated with an anti-GFP antibody. The precipitates were separated by SDS–PAGE and visualized by silver staining. Six bands were numbered and identified by MALDI-TOF MS analysis using the MASCOT software. (B) HT1080 cells were transiently transfected with ANXA8-V5 and YFP or wt-proAREG-YFP expression vectors. Cells were immunoprecipitated with an anti-GFP antibody. Western blotting was carried out using an anti-V5 antibody. (C) The effect of ANXA8 siRNA (#1, #2) on the endogenous protein levels of ANXA8 was analyzed by Western blotting. The intensity of ANXA8 bands was normalized to their respective β-actin controls and represents the fold-change relative to controls. (D) HT1080 cells stably expressing AP-tagged proAREG were transfected with control or ANXA8 siRNA (#1, #2). The conditioned media were collected at the indicated time points after TPA stimulation (20 nM), and AP activity was measured. Data represent the mean ± SEM, *p < 0.01. The dagger indicates the upper limit of the absorbance at 405 nm. (E) The wt-proAREG-YFP cells were transfected with siRNA and stimulated with 20 nM TPA, instead of 100 nM TPA as in Figure 1D, to detect the maximal effects of ANXA8 knockdown on proAREG shedding. Cells were incubated for the indicated time periods, and were biotinylated with a membrane-impermeable biotinylation reagent. The biotinylated proteins were immunoprecipitated with an anti-GFP antibody and were analyzed by Western blotting using HRP-conjugated streptavidin. Total protein was detected using anti-GFP antibody and acted as a way to monitor the expression of proAREG used for immunoprecipitation. The intensity of biotinylated proAREG-YFP bands is represented as a fold-change relative to baseline levels (0 min). Data represent the mean ± SEM from three independent experiments. The half-life of biotinylated proAREG-YFP (t1/2) was obtained from the kinetic equation of control siRNA-treated cells (y = 100e−0.033x [R2 = 0.7889]) and ANXA8 siRNA-treated cells (y = 100e−0.074x [R2 = 0.9537]). (F) The effect of siRNA on the subcellular localization of proAREG-YFP. The wt- and ucF-proAREG-YFP cells were transfected with siRNA, and stimulated with 20 nM TPA for 15 min. Scale bar: 20 μm.
Multiple members of the annexin family interact with proAREG and ADAM17 in the regulation of proAREG shedding
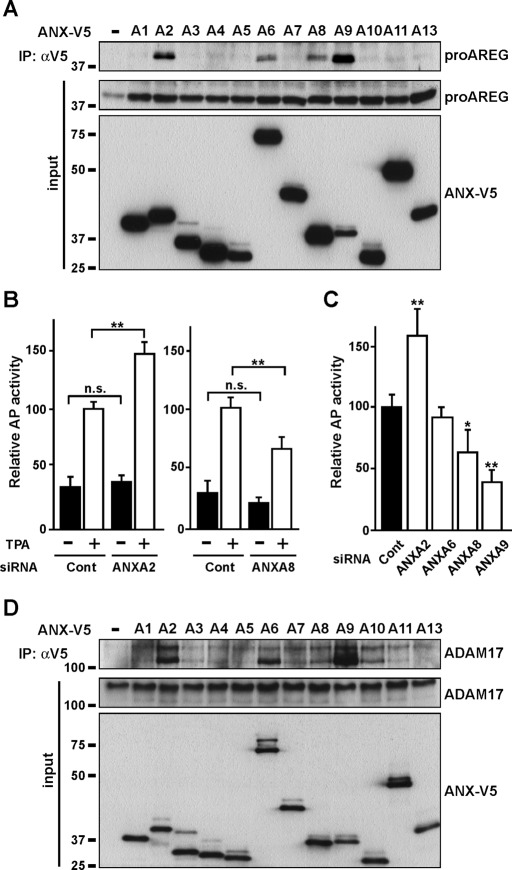
The annexin family contains a conserved structural element, an annexin repeat of some 70 amino acid residues required for binding to calcium ions. The similarity of its molecular structure at the three-dimensional level has been well established (Moss and Morgan, 2004; Gerke et al., 2005). We analyzed the interaction between all 12 members of the annexin family and proAREG, and found that ANXA2, A6, A8, and A9 interacted with proAREG (Figure 3A). We tested the effect of annexin knockdown on proAREG shedding using an AP assay. Basal shedding, which is observed in culture without addition of shedding inducers, such as TPA, accounted for ∼30% of total (basal plus TPA-induced) shedding, and there was no statistical significance of the extent of the basal shedding in control and ANXA2 and A8 knockdown cells (Figure 3B). On the other hand, TPA-induced proAREG shedding was significantly suppressed by knockdown of ANXA8 and A9, while treatment with ANXA2 siRNA significantly increased proAREG shedding (Figure 3, B and C). ANXA6 siRNA had no effect on shedding (Figure 3C). Although some annexin family members contribute to endocytic transport (Mayran et al., 2003; Goebeler et al., 2008; Grewal and Enrich, 2009), annexin knockdown did not affect the levels of cell surface proAREG (Figure S2A).
FIGURE 3:
Multiple members of the annexin family interact with proAREG and ADAM17. (A) HT1080 cells were transiently transfected with ANX-V5 and proAREG expression vectors. Cells were immunoprecipitated with an anti-V5 antibody. Western blotting was carried out using anti–AREG-N antibody. (B) AP activity was measured in the conditioned media from HT1080 cells stably expressing AP-tagged proAREG. Cells treated with siRNA were stimulated with 20 nM TPA for 30 min. Data represent the mean ± SEM; **p < 0.01; n.s., not significant. (C) AP activity was measured as in (B). Data represent the mean ± SEM; *p < 0.05; **p < 0.01. (D) HT1080 cells were transiently transfected with ANX-V5 and ADAM17 expression vectors. Cells were immunoprecipitated with an anti-V5 antibody. Western blotting was carried out using an anti-ADAM17 antibody.
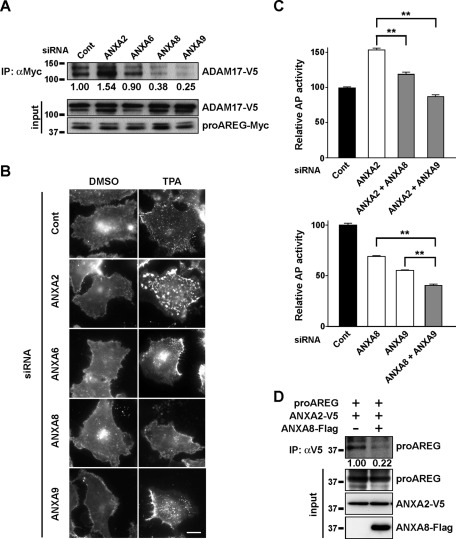
Because ADAM17 is required for proAREG shedding (Sahin et al., 2004), we investigated whether annexins would interact with ADAM17. As shown in Figure 3D, ANXA2, A6, and A9 efficiently interacted with ADAM17, whereas ANXA8 and A10 interacted to a moderate degree. It is intriguing to describe that the lower band of ADAM17 largely coprecipitated with these annexins. ANXA2, A6, A8, and A9 interacted with both proAREG and ADAM17, suggesting that these annexins can regulate the association between proAREG and ADAM17. As expected, knockdown of these annexins, except ANXA6, altered the extent of interaction between proAREG and ADAM17 (Figure 4A). Furthermore, in annexin knockdown cells, we found that the extent of TPA-induced ucF-proAREG assembly correlated with that of proAREG shedding (Figures 3, B and C, and 4B). With respect to ANXA9 knockdown, ucF-proAREG assembled to the same extent as in the controls, which could be explained by the compensation of other annexin levels and/or functions. To explore the coordinated regulation of proAREG shedding by multiple annexins, we performed the AP assay using cells cotransfected with combinations of annexin siRNAs. The inhibitory effect of ANXA8 and A9 siRNAs on proAREG shedding was dominant over ANXA2. This inhibitory effect was additive in ANXA8 and A9 knockdown cells (Figure 4C). Overexpression of ANXA8 interfered with the interaction between proAREG and ANXA2 (Figure 4D), suggesting their mutual exclusivity for interacting with proAREG, indicating a multiplicity and complexity of annexins in the regulation of proAREG shedding.
FIGURE 4:
Multiple annexin family members regulate proAREG shedding. (A) HT1080 cells stably expressing AP-AREG were incubated with siRNA and transiently transfected with the proAREG-Myc and ADAM17-V5 expression vectors. Cells were treated with the membrane-impermeable cross-linking agent DTSSP; this was followed by immunoprecipitation with an anti-Myc antibody. Western blotting was carried out using an anti-V5 antibody. The intensity of the ADAM17-V5 bands is represented as the fold-change relative to control siRNA-transfected cells. (B) The effect of siRNA on the subcellular localization of proAREG-YFP. The ucF-proAREG-YFP cells were transfected with siRNA and stimulated with 20 nM TPA for 30 min. Scale bar: 20 μm. (C) AP activity was measured in the conditioned medium of HT1080 cells stably expressing AP-AREG. Cells were cotransfected with combinations of siRNA and were stimulated with 20 nM TPA for 30 min. Data represent the mean ± SEM; **p < 0.01. (D) Prevention of the interaction between proAREG and ANXA2 by ANXA8. HT1080 cells were transfected with expression vectors encoding proAREG and ANXA2-V5 together with empty or ANXA8-Flag expression vectors. Cell lysates were immunoprecipitated with an anti-V5 antibody, and the coprecipitated proAREG was detected using an anti–AREG-N antibody.
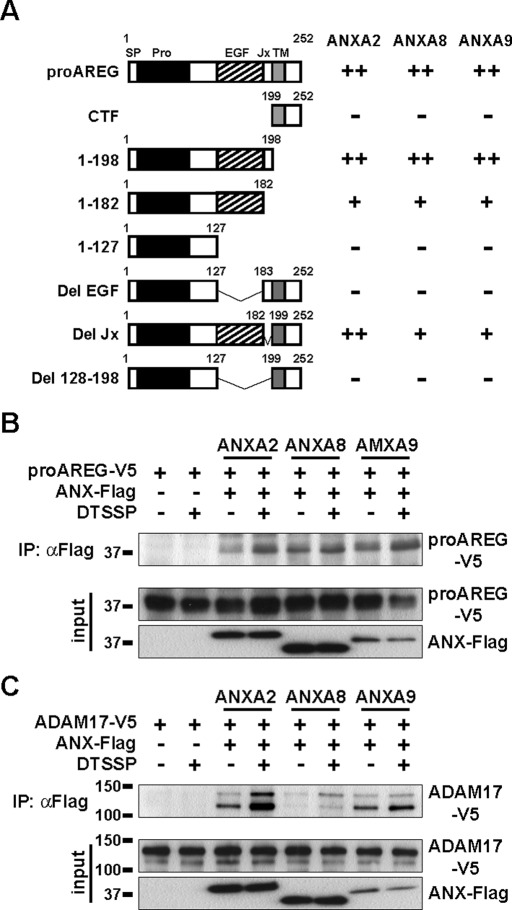
Annexins interact with proAREG and ADAM17 on the cell surface
To investigate the molecular interaction between annexins and proAREG in detail, we performed a GST pulldown assay using AREG derivatives. The EGF-like domain of proAREG (amino acids 128–182) was responsible for the interaction of ANXA2, A8, and A9, indicating that annexins interact with proAREG at the outer plasma membrane (Figures 5A and S3). Some members of the annexin family have been shown to localize and function inside and outside of the cell (Gerke et al., 2005). We confirmed that annexins were expressed on the cell surface, as assessed by a biotinylation study of the annexin family (Figure S2B). Moreover, a membrane-impermeable cross-linking agent, 3, 3′-dithiobis(sulfosuccinimidylpropionate) (DTSSP), slightly enhanced the interactions among proAREG and ANXA2, A8, and A9, indicating that they interact at the outer plasma membrane (Figure 5B). Notably, the interactions among ADAM17 and ANXA2, A8, and A9 were also enhanced by DTSSP (Figure 5C). These results, together with those from the GST pulldown assay, suggest that ANXA2, A8, and A9 interact with proAREG and ADAM17 at the cell surface. Therefore we can suggest that annexins are able to control proAREG shedding by regulating the association between proAREG and ADAM17 on the cell surface.
FIGURE 5:
Annexins interact with proAREG and ADAM17 on the cell surface. (A) Schematic representation of a result of the pulldown assay shown in Figure S3. V5-tagged AREG derivatives were pulled down by GST-annexins. The binding properties were estimated based on the relative intensity of the bands as compared with the proAREG band, and are indicated as ++ (>50%), + (10–50%), and − (<10%). (B and C) Effects of the membrane-impermeable cross-linking agent on the association between proAREG, annexins, and ADAM17. HT1080 cells were transiently transfected with proAREG-V5 or ADAM17-V5, with or without an ANX-Flag expression vector. Cells were incubated in the absence or presence of DTSSP; this was followed by immunoprecipitation with an anti-Flag antibody. Western blotting was carried out using an anti-V5 antibody. The upper and lower bands in the ADAM17 immunoblot indicate the proform and mature ADAM17, respectively (Sahin et al., 2004).
Annexins are required for EGFR transactivation induced by various stimuli in keratinocytes
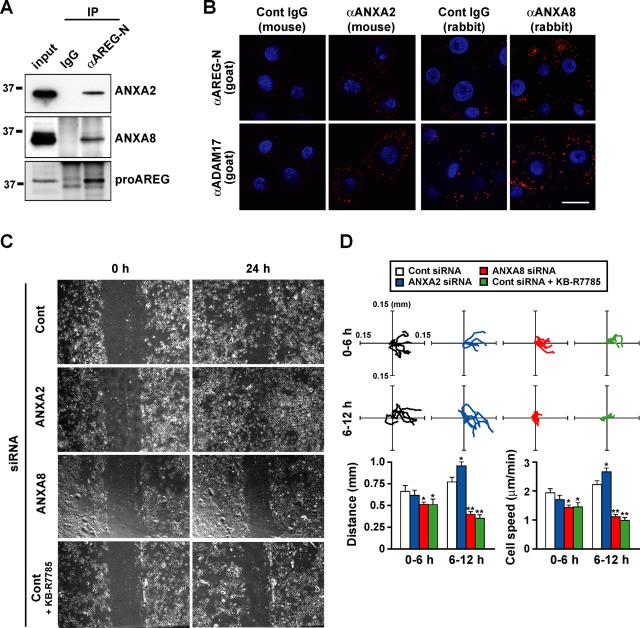
To study the role of the annexin-regulated shedding machinery under physiological conditions, we used a functional transactivation assay for EGFR with various stimulations in primary human keratinocytes. We examined the expression levels of annexins and confirmed that ANXA2 and A8 were normally expressed in keratinocytes, whereas the expression of A6 and A9 was low (Figure S4). We confirmed the physical interaction between endogenous proAREG and ANXA2 or A8 in keratinocytes by coimmunoprecipitation (Figure 6A). We also performed an in situ proximity assay (Fredriksson et al., 2002), demonstrating that both endogenous proAREG and ADAM17 interact with ANXA2 and A8 in keratinocytes (Figure 6B).
FIGURE 6:
Annexins are indispensable to keratinocyte migration in response to wounding. (A) Keratinocytes were immunoprecipitated with normal goat IgG or anti–AREG-N antibodies, and samples were analyzed by Western blotting. Aliquots of whole-cell lysates used for immunoprecipitation were loaded in the left lanes as the input to confirm the molecular weight of immunoprecipitated proteins (0.1, 1.0, and 1.5% of lysates were loaded for the detection of ANXA2, ANXA8, and AREG, respectively). (B) Visualization of proAREG/ANXA2, proAREG/ANXA8, ADAM17/ANXA2, and ADAM17/ANXA8 interaction at endogenous levels in keratinocytes. Keratinocytes were fixed and incubated with primary antibodies as indicated. The next day, primary antibodies were detected by an in situ proximity ligation assay. Red fluorescence signals indicate PLA probes bound to the primary antibodies that are in close proximity. Nuclei were stained with 4′,6-diamidino-2-phenylindole. Note that the higher background signals in the four right panels were due to the lower specificity of anti-ANXA8 antibodies compared with anti-ANXA2 antibody. Scale bar: 50 μm. (C and D) A cell motility assay. Keratinocytes were transfected with siRNA. The day before the wound assay, cells were incubated with BHE-free medium. Cells were wounded by scraping with the tip of a micropipette, washed, and given fresh medium. The remaining cells were incubated for 24 h in the absence or presence of 10 μM KB-R7785 (C). Trajectories of five cells from 0–6 h and from 6–12 h after wound stimulation are shown in the top panels. The bottom panels represent the total distance and average motility of cells (D). Data represent the mean ± SEM; *p < 0.01; **p < 0.001.
It has been reported that EGFR activation and signaling mediated by shedding of EGFR ligand precursors are essential for keratinocyte migration (Tokumaru et al., 2000; Shirakata et al., 2005). To examine the involvement of annexins in wound-induced cell migration, the motility of keratinocytes treated with ANXA2 and A8 siRNA was analyzed using wound assays (Figure 6, C and D). The ANXA2 knockdown cells migrated faster than controls. In contrast, cell motility was suppressed by knockdown of ANXA8, as well as by addition of KB-R7785. This indicated the role of annexins in the keratinocyte migration. The cell migration analysis showed that cell motility was markedly regulated 6–12 h after wounding. On the basis of these results, we speculate that the altered shedding of EGFR ligand precursors in annexin knockdown cells would gradually affect the transmission of EGFR activation to the cells surrounding the wound edge by up- or down-regulating the autocrine loop of AREG.
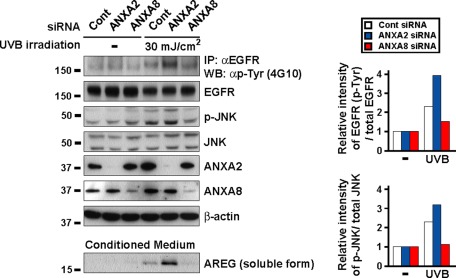
We investigated the role of annexins in ultraviolet B (UVB)-induced EGFR transactivation in keratinocytes, because skin cancer is a common human cancer, and UVB radiation in sunlight is a major etiological factor. Several reports have shown that shedding of proAREG and proHB-EGF is required for UV-induced EGFR transactivation in keratinocytes and have suggested the broad relevance of the UV-ADAM-proligand-EGFR pathway and its significance in skin cancer (Seo et al., 2007; Singh et al., 2009). Consistent with a previous report (Singh et al., 2009), treatment with KB-R7785 or AREG siRNAs inhibited the UVB-induced phosphorylation of EGFR and JNK (Figure S5), indicating that UVB-induced EGFR transactivation is dependent on shedding of EGFR ligand precursors in keratinocytes. Knockdown of ANXA2 increased UVB-induced phosphorylation of EGFR and JNK, while knockdown of ANXA8 inhibited UVB-induced phosphorylation of EGFR and JNK (Figure 7). These data correlated with the amount of soluble AREG in the conditioned medium (Figure 7, bottom panel). Our results indicate that annexins are indispensable for shedding of an adequate amount of EGFR ligand precursors, such as proAREG, and EGFR signal transduction in response to natural stimulation in keratinocytes.
FIGURE 7:
Annexins are involved in UVB-induced EGFR transactivation via ADAM-mediated proAREG shedding in keratinocytes. Keratinocytes were transfected with siRNA. The day before the UVB experiment, cells were incubated in BHE-free medium. After 30 min of UVB irradiation, cells were immunoprecipitated with an anti-EGFR antibody. Cell lysates and precipitates were analyzed by Western blotting. The intensity of UVB-induced phosphorylated EGFR and JNK was normalized to the respective total protein content and represents fold-change relative to UVB-untreated cells. Soluble peptides present in the conditioned medium were extracted with a 20% TCA solution. Released AREG in the conditioned medium was detected by anti–AREG-N antibodies.
DISCUSSION
It has been reported that impaired shedding of EGFR ligand precursors causes abnormal development (Yamazaki et al., 2003; Sternlicht et al., 2005) and weakness of cells against stressors (Uetani et al., 2009). We also showed that ectodomain shedding of EGFR ligand precursors is a key event in receptor cross-talk and in intercellular signaling by the CTF pathway to directly regulate gene expression (Bao et al., 2003; Nanba et al., 2003; Hieda et al., 2008; Higashiyama et al., 2008; Isokane et al., 2008; Stoeck et al., 2010). These findings indicate that regulatory mechanisms of ectodomain shedding are crucial for the maintenance of cellular functions and are possible therapeutic targets for certain diseases, such as cancers. However, the molecular mechanisms underlying substrate selectivity by ADAMs remains unknown. To resolve this issue, we needed to determine how EGFR ligand precursors associate with ADAMs and to identify the key molecule(s) controlling ADAMs on the cell surface. Because ADAM-mediated shedding is a rapid and transient event, precise analysis of the ADAM-substrate complex formation in the early phase has been difficult. In this study, we visualized the intermediates of the proAREG complex on the cell surface during shedding and found that annexins are key molecules for the regulation of proAREG shedding.
Annexins are calcium-dependent, phospholipid-binding proteins involved in several biological events (Moss and Morgan, 2004; Gerke et al., 2005). In our study, we found that ANXA2, A8, and A9 interacted with both the EGF-like domain of proAREG (Figure 5A) and mature ADAM17 (Figure 3D). TPA treatment had no effect on proAREG–annexins or ADAM17–annexins interaction (unpublished data), suggesting their constitutive interaction. However, knockdown of these annexins altered the extent of interaction between proAREG and ADAM17 (Figure 4A), and the administration of a cell surface cross-linking agent enhanced the association between proAREG and annexins (Figure 5B), and ADAM17 and annexins (Figure 5C). Both sets of evidence strongly suggest that the proAREG–annexins–ADAM17 complex needs to localize at the cell surface for shedding of proAREG to occur.
We clearly showed that these annexins interacted with proAREG and ADAM17 outside the cell, although annexins mainly localize at the cytosol and peripheral inner cell membrane. It is a characteristic of certain annexins that they have functionally distinct roles inside and outside cells. Extracellular ANXA1 can bind to chemoattractant receptors of the formyl peptide receptor family in neutrophils and monocytes in response to glucocorticoids. ANXA1 is implicated in dexamethasone-induced l-selectin shedding, which may contribute to the anti-inflammatory system (Walther et al., 2000; de Coupade et al., 2003). ANXA2 on the cell surface acts as a coreceptor for tissue plasminogen activator, which is important for the degradation of fibrin, and maintenance of fibrinolytic homeostasis (Ling et al., 2004). Extracellular ANXA5 works as an antithrombotic shield and is involved in thrombosis and pregnancy loss in antiphospholipid syndrome (Rand, 2000). In contrast, whether ANXA8 and A9 localize and function at the outer cell membrane is poorly understood. We have detected cell surface ANXA8 and A9 using a biotinylation study (Figure S2B), and our data have provided new insight regarding the role of annexins and their regulation in proAREG shedding on the cell surface. Annexins have a crucial role in controlling EGFR signal transduction in response to physiological stimulation in keratinocytes. It is interesting to note that although annexin family members lack a signal peptide sequence, they are exported from the cell; therefore their export mechanism(s) requires further study.
Previous reports have documented a number of adaptors implicated in ectodomain shedding. Tetraspanin CD9 and N-arginine dibasic convertase bind to proHB-EGF and ADAMs in the regulation of proHB-EGF shedding (Yan et al., 2002; Nishi et al., 2006). It has also been previously reported that calmodulin regulates ADAM17-mediated cleavage of L-selectin by interactions with the L-selectin cytoplasmic domain (Kahn et al., 1998). However, it is still unclear how ADAMs recognize a large variety of substrates, and how their activity is controlled. We have demonstrated that proAREG shedding was positively or negatively regulated by multiple annexins. Considering that annexins organize membrane domains and platforms for protein interaction (Gerke et al., 2005), it is plausible that multiple annexins interact with proAREG and ADAM17 directly or indirectly, thereby contributing to the substrate selectivity of ADAMs. Previous reports have shown that the hypervariable region (HVR) of ADAMs may have a significant effect on substrate recognition and may adjust the spatial alignment of the catalytic and adhesion sites (Takeda et al., 2006; Igarashi et al., 2007). Furthermore, the HVR segment is an exosite for capturing substrates directly or via binding to an associated protein (Takeda, 2009). It is possible that annexins modulate the affinity of ADAM17 to substrates by binding to the HVR and regulating ectodomain shedding. Because none of the constructed ADAM17 deletion mutants interacted with annexins, the whole structure of ADAM17 is required to determine the presence of the relevant binding domains to annexins (unpublished data). Therefore x-ray crystallography analysis of the proAREG-annexin-ADAM17 complex should be conducted.
We propose a novel role for cell surface annexins as “shedding platform” proteins for proAREG to modulate the substrate selectivity of ADAM17. The composition of a shedding platform might determine the efficiency of proAREG shedding by changing the annexins that interact with proAREG and ADAM17 in response to various stimuli. We also tested whether annexins were involved in the shedding of other EGFR ligand precursors, including proHB-EGF, proTGFα, and proEREG (Figure S6). Knockdown of ANXA2 increased the efficiency of proHB-EGF and proTGFα shedding, whereas ANXA8 knockdown reduced that efficiency. Shedding of proTGFα and proEREG decreased in ANXA9 knockdown cells. We also detected interactions between these EGFR ligand precursors and annexins, with the unique binding partnership of annexins to EGFR ligand precursors (unpublished data). We speculate that annexins have a broad effect on shedding of EGFR ligand precursors, and further elucidation of the interaction between other ADAMs and annexins would expand our knowledge of the regulation of shedding of EGFR ligand precursors.
The deregulation of annexins is implicated in human diseases, such as cancer (Gerke and Moss, 2002). We showed that ANXA2 negatively regulated cell migration and UVB-induced EGFR phosphorylation in keratinocytes (Figures 6, C and D, and 7). The expression of ANXA2 negatively correlates with the progression of metastasis in prostate cancer, which is consistent with our study (Chetcuti et al., 2001; Liu et al., 2003). In some cases, however, the overexpression of ANXA2 has been reported in breast, lung, and gastric and colonic malignant tumors (Gerke and Moss, 2002). We found that ANXA8 positively regulated the cell migration and UVB-induced EGFR phosphorylation; proAREG shedding was prevented by knockdown of ANXA8 (Figures 6, C and D, and 7). A previous report showed that ANXA8 inhibited the cell migratory and metastatic characteristics in cholangiocarcinoma cells (Lee et al., 2009). These contradictory results may arise from the fact that the expression level and/or function of annexins depends on tissue distribution, tumor development and progression stage, and various factors, such as hormones and glucose (Mussunoor and Murray, 2008; Fatimathas and Moss, 2010), which eventually may affect the components of the shedding platform. Pharmaceutical compounds that directly inhibit metalloproteases have been evaluated for therapy of several diseases (Higashiyama et al., 2008); it is believed that cell surface annexins and components of the shedding platform could also have therapeutic potential for ADAM-related diseases, such as cancer, rheumatoid arthritis, and Alzheimer's disease.
MATERIALS AND METHODS
Antibodies
The following antibodies were used in this study: affinity-purified rabbit polyclonal antibody against the cytoplasmic region of proAREG (anti–AREG-CTF; IBL, Fujioka, Gunma, Japan; Isokane et al., 2008); goat polyclonal antibody against the extracellular region of proAREG (anti–AREG-N, clone AF262; R&D Systems, Minneapolis, MN); mouse monoclonal anti-ANXA2 (clone 5/Annexin II; BD Biosciences, San Jose, CA); mouse monoclonal anti-ANXA9 (H00008416-B01; Novus Biologicals, Littleton, CO); rabbit polyclonal anti-ANXA6 (HPA009650; Sigma-Aldrich, St. Louis, MO); rabbit polyclonal anti-ANXA8 (JM-3637R-3; MBL, Nagoya, Aichi, Japan); rabbit polyclonal anti-ADAM17 (PC491; Calbiochem, San Diego, CA); and mouse monoclonal anti-phosphotyrosine antibody (4G10; Millipore, Bedford, MA). The rabbit monoclonal anti-EGFR (C74B9), polyclonal anti-JNK (#9252), and polyclonal anti–phospho-JNK (Thr183/Tyr185; #9251) were purchased from Cell Signaling Technology (Danvers, MA). The goat polyclonal anti-ADAM17 (C-15), normal mouse immunoglobulin G (IgG), and rabbit IgG were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell culture and plasmid transfection
Human fibrosarcoma HT1080 cells and their transfectants were grown in Eagle's minimal essential medium (EMEM) containing 10% fetal bovine serum (FBS) and nonessential amino acids (Invitrogen, Carlsbad, CA). Human keratinocytes were cultured in optimized nutrient medium MCDB153 (Nissui, Tokyo, Japan) supplemented with 5 μg/ml insulin, 0.5 μM hydrocortisone, 0.1 mM ethanolamine, 0.1 mM phosphoethanolamine, and 150 μg/ml bovine hypothalamic extract (BHE), as described previously (Hashimoto et al., 1994). All cells were cultured in a humidified incubator at 37°C/5% CO2. Cells were transfected with plasmids using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol.
RNA interference
Control and ANXA8 (#1: Hs_ANXA8_8, #2: Hs_ANXA8_10) siRNA were purchased from Qiagen (Chatsworth, CA). ON-TARGETplus SMARTpool siRNA for ANXA2, A6, and A9 were purchased from Dharmacon (Lafayette, CO). The AREG siRNA used in this study was the same as in a previous report (Gschwind et al., 2003) and was obtained from B-Bridge International (San Jose, CA). The transfection of siRNA (20 nM) was performed with Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's protocol.
Imaging of YFP fusion proteins and immunofluorescence staining
For time-lapse analysis, wt-proAREG-YFP or ucF-proAREG-YFP cells were cultured in glass-bottom dishes (Matsunami, Osaka, Japan). Cells were treated with 10 μg/ml cycloheximide for 4 h and were given serum-free EMEM 30 min before TPA stimulation. Time-lapse observations were made with a BIOREVO fluorescence microscope (Keyence, Osaka, Japan). The fluorescence signals were visualized as color images using LuminaVision (Mitani, Tokyo, Japan). For immunofluorescence staining, cells were fixed with 4% paraformaldehyde and incubated with primary antibodies at 4°C overnight. Cells were viewed by fluorescence (Olympus, Tokyo, Japan) or confocal (Nikon, Tokyo, Japan) microscopy.
Cell surface biotinylation
Cell surface biotinylation was carried out as described in a previous report (Goishi et al., 1995). Briefly, cells were biotinylated with 0.01% sulfo-NHS-biotin (Pierce Rockford, IL) in 0.1 M HEPES and 0.15 M NaCl (pH 8.0) for 15 min at 4°C at the indicated each time point (Figures 1D and 2E). Excess reagent was quenched and removed with ice-cold EMEM/10% FBS. Cells were lysed and immunoprecipitated with appropriate antibodies.
Cross-linking
For cross-linking of cell surface proteins, cells were incubated with a thiol-cleavable cross-linking agent, DTSSP (Pierce), on ice for 2 h, which was followed by quenching with 20 mM Tris-HCl (pH 7.4).
Immunoprecipitation
Cell lysates were incubated with the indicated antibodies for 2 h at 4°C, which was followed by incubation with protein G-Sepharose 4 Fast Flow beads (GE Healthcare, Milwaukee, WI) for 1 h. The samples were dissolved in SDS sample buffer containing 5% β-mercaptoethanol and boiled for 5 min.
Western blotting
Samples were separated by SDS–PAGE and transferred to a nitrocellulose membrane. The membranes were blocked with 4% skim milk in PBS-T (0.05% Tween-20 in phosphate-buffered saline [PBS]) for 30 min, which was followed by incubation with primary antibodies. After being washed with PBS-T, the membranes were incubated with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies. For biotinylated protein detection, HRP-conjugated streptavidin was used. Immunoreactivity was detected by using enhanced chemiluminescence detection reagents.
MS
The ucF-AREG-YFP cells were treated with eight volumes of hypotonic buffer (5 mM Tris-HCl, pH 7.4) and homogenized until more than 90% of the cells were broken. Then 0.25 volumes of compensation buffer (20 mM Tris-HCl, 0.95 M sucrose, 0.15 M NaCl, pH 7.4) were added to restore isotonicity. Nucleus fractions were separated from the rest of the cell extract by centrifugation at 3900 rpm for 7 min. The supernatant was further centrifuged at 100,000 × g for 30 min at 4°C (Himac; Hitachi Koki, Tokyo, Japan). The pellet was dissolved in RIPA buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP40, 0.1% SDS, and 1% sodium deoxycholate) containing Protease Inhibitor Cocktail Tablets (Roche, Basel, Switzerland), and used as a membrane fraction. ProAREG-YFP in the membrane fraction was immunoprecipitated with a mouse anti–green fluorescent protein (GFP) monoclonal antibody. The immunoprecipitates were separated by SDS–PAGE, and silver staining was carried out using a Silver Stain MS kit (Wako, Osaka, Japan). After in-gel digestion, the analysis was performed by matrix-assisted laser desorption/ionization time-of-flight MS (MALDI-TOF MS). The MALDI-TOF MS system used was an AXIMA-TOF2 (Shimadzu, Kyoto, Japan). MASCOT (Matrix Sciences, Boston, MA) was used as the search engine for MS analysis.
AP assay
HT1080 cells stably expressing AP-tagged proAREG were incubated with serum-free EMEM 30 min before TPA stimulation. The conditioned media were collected at indicated time points after TPA stimulation (20 nM) and heated for 15 min at 65°C to inactivate endogenous APs. An equal volume of a 2× AP mixture (2 M diethanolamine, pH 9.8, 1 mM MgCl2, 20 mM l-homoarginine, and 24 mM p-nitrophenylphosphate) was added. AP activity was determined by measuring the absorbance at 405 nm (Tokumaru et al., 2000).
Pulldown assay
GST or GST-annexin were produced from Escherichia coli strain BL21, induced by treatment with 0.1 mM isopropyl-1-thio-β-d-galactopyranoside at 20°C for 20 h. Cells were suspended in PBS containing 1% NP40, 1 mM EDTA, 5 mM dithiothreitol, 0.2 mM p-amidinophenyl methanesulfonyl fluoride hydrochloride, and 2 μg/ml aprotinin. The mixture was sonicated for 20 s, and the supernatants were obtained by centrifugation at 9300 × g for 10 min at 4°C. The supernatants were incubated with glutathione-Sepharose 4B beads (GE Healthcare). Extracts from HT1080 cells expressing various V5-tagged AREG mutants were mixed with GST or GST-annexin immobilized on glutathione-Sepharose beads for 2 h at 4°C. The bound proteins were analyzed by SDS–PAGE, which was followed by Western blotting using an anti-V5 antibody.
In situ proximity assay
Keratinocytes were cultured in type I collagen-coated glass-bottom dishes (Matsunami). After fixation, cells were incubated with anti–AREG-N (AF262), anti-ANXA2 (Clone: 5/Annexin II), anti-ANXA8 (JM-3637R-3), anti-ADAM17 (C-15), or normal mouse and rabbit IgGs at 4°C overnight. In situ proximity assays were performed using a Duolink II Kit including PLA probes for anti-Mouse PLUS, anti-Rabbit PLUS, and anti-Goat MINUS according to the manufacturer's instructions. Images were acquired using confocal microscopy (Nikon).
Wound and migration assays
For wound and migration experiments, keratinocytes were seeded on type I collagen-coated dishes or glass-bottom dishes. Cells were treated with siRNA, and on the day before the wound assay, cells were incubated with BHE-free medium. Cells were wounded by the tip of a micropipette, washed once with fresh medium to remove floating cells, and refed with fresh medium with or without KB-R7785 (10 μM). Cell movement was observed after 24 h. For the migration assay, time-lapse observations were performed with a BioStation IM (Nikon) every 20 min for 12 h after wound stimulation. Distance and average motility speed of keratinocytes (at least 10 cells) were determined by tracking single cells using the Volocity software (Perkin Elmer-Cetus, Foster City, CA).
UVB treatment
Keratinocytes were exposed to UVB with FL20SE30 fluorescence sunlamps (Toshiba Medical Supply, Tokyo, Japan). A Kodacel filter was mounted in front of the tubes to filter any wavelength below 290 nm. Irradiation intensity was monitored using a photodetector. Cells were seeded on type I collagen-coated dishes. The day before the UVB experiment, cells were incubated with BHE-free medium. Thirty minutes before UVB exposure, the BHE-free medium was refreshed. Cells were irradiated with UVB light (30 mJ/cm2) and incubated for 30 min. The UVB-irradiated keratinocyte-conditioned media were collected at the indicated time points. Cells were immunoprecipitated with an anti-EGFR antibody and Western blotting was carried out using an anti-phosphotyrosine antibody (4G10). Soluble peptides present in the conditioned medium were extracted with a 20% trichloroacetic acid (TCA) solution.
Statistical analysis
All assays were performed independently three times. The results are represented as the mean ± SEM. The two groups were compared using Student's t test. Analysis of variance using Scheffe's post hoc test was conducted for multiple comparisons. A p value of less than 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank the members of the Higashiyama lab for helpful comments and discussion and the members of the Hashimoto lab for advice in the UV experiment, especially S. Tokumaru, T. Tsuda, and E. Tan for technical support. We also thank S. Matsuda (Division of Anatomy and Embryology, Ehime University Graduate School of Medicine) for the technical support. We acknowledge the help of the Shimadzu Co. (mass spectrometry) and the Keyence Co. (time-lapse imaging). This work was supported by a Grant-in-Aid for Scientific Research to H.N. from Ehime University, a Grant-in-Aid for Scientific Research (no. 20390082) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and a Grant-in-Aid for Scientific Research (no. S2207) from the Strategic Young Researcher Overseas Visiting Program for Accelerating Brain Circulation, Japan Society for the Promotion of Science, Japan, to S.H.
Abbreviations used:
- ADAM
a disintegrin and metalloproteinase
- ANXA2/A8/A9
annexin A2/A8/A9
- AP
alkaline phosphatase
- AREG
amphiregulin
- BHE
bovine hypothalamic extract
- CTF
cytoplasmic fragment
- EGFR
epidermal growth factor receptor
- EMEM
Eagle's minimal essential medium
- EREG
epiregulin
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- HB-EGF
heparin-binding EGF-like growth factor
- HRP
horseradish peroxidase
- HVR
hypervariable region
- IgG
immunoglobulin G
- JNK
c-Jun N-terminal kinase
- MALDI-TOF MS
matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- MS
mass spectrometry
- PBS
phosphate-buffered saline
- proAREG
precursor of amphiregulin
- proHB-EGF
precursor of HB-EGF
- siRNA
small interfering RNA
- TCA
trichloroacetic acid
- TGFα
transforming growth factor α
- TIMP
tissue inhibitor of metalloproteinases
- TPA
12-O-tetradecanoylphorbol-13-acetate
- UVB
ultraviolet B
- wt
wild type
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-08-0683) on March 21, 2012.
REFERENCES
- Bao J, Wolpowitz D, Role LW, Talmage DA. Back signaling by the Nrg-1 intracellular domain. J Cell Biol. 2003;161:1133–1141. doi: 10.1083/jcb.200212085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9:463–475. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- Chetcuti A, Margan SH, Russell P, Mann S, Millar DS, Clark SJ, Rogers J, Handelsman DJ, Dong Q. Loss of annexin II heavy and light chains in prostate cancer and its precursors. Cancer Res. 2001;61:6331–6334. [PubMed] [Google Scholar]
- de Coupade C, Solito E, Levine JD. Dexamethasone enhances interaction of endogenous annexin 1 with L-selectin and triggers shedding of L-selectin in the monocytic cell line U-937. Br J Pharmacol. 2003;140:133–145. doi: 10.1038/sj.bjp.0705413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med. 2008;29:258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatimathas L, Moss SE. Annexins as disease modifiers. Histol Histopathol. 2010;25:527–532. doi: 10.14670/HH-25.527. [DOI] [PubMed] [Google Scholar]
- Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, Ostman A, Landegren U. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- Gerke V, Creutz CE, Moss SE. Annexins: linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol. 2005;6:449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- Gerke V, Moss SE. Annexins: from structure to function. Physiol Rev. 2002;82:331–371. doi: 10.1152/physrev.00030.2001. [DOI] [PubMed] [Google Scholar]
- Goebeler V, Poeter M, Zeuschner D, Gerke V, Rescher U. Annexin A8 regulates late endosome organization and function. Mol Biol Cell. 2008;19:5267–5278. doi: 10.1091/mbc.E08-04-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goishi K, Higashiyama S, Klagsbrun M, Nakano N, Umata T, Ishikawa M, Mekada E, Taniguchi N. Phorbol ester induces the rapid processing of cell surface heparin-binding EGF-like growth factor: conversion from juxtacrine to paracrine growth factor activity. Mol Biol Cell. 1995;6:967–980. doi: 10.1091/mbc.6.8.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal T, Enrich C. Annexins—modulators of EGF receptor signalling and trafficking. Cell Signal. 2009;21:847–858. doi: 10.1016/j.cellsig.2009.01.031. [DOI] [PubMed] [Google Scholar]
- Gschwind A, Hart S, Fischer OM, Ullrich A. TACE cleavage of proamphiregulin regulates GPCR-induced proliferation and motility of cancer cells. EMBO J. 2003;22:2411–2421. doi: 10.1093/emboj/cdg231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackel PO, Zwick E, Prenzel N, Ullrich A. Epidermal growth factor receptors: critical mediators of multiple receptor pathways. Curr Opin Cell Biol. 1999;11:184–189. doi: 10.1016/s0955-0674(99)80024-6. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Higashiyama S, Asada H, Hashimura E, Kobayashi T, Sudo K, Nakagawa T, Damm D, Yoshikawa K, Taniguchi N. Heparin-binding epidermal growth factor-like growth factor is an autocrine growth factor for human keratinocytes. J Biol Chem. 1994;269:20060–20066. [PubMed] [Google Scholar]
- Hieda M, Isokane M, Koizumi M, Higashi C, Tachibana T, Shudou M, Taguchi T, Hieda Y, Higashiyama S. Membrane-anchored growth factor, HB-EGF, on the cell surface targeted to the inner nuclear membrane. J Cell Biol. 2008;180:763–769. doi: 10.1083/jcb.200710022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama S, Iwabuki H, Morimoto C, Hieda M, Inoue H, Matsushita N. Membrane-anchored growth factors, the epidermal growth factor family: beyond receptor ligands. Cancer Sci. 2008;99:214–220. doi: 10.1111/j.1349-7006.2007.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama S, Nanba D. ADAM-mediated ectodomain shedding of HB-EGF in receptor cross-talk. Biochim Biophys Acta. 2005;1751:110–117. doi: 10.1016/j.bbapap.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Higashiyama S, Nanba D, Nakayama H, Inoue H, Fukuda S. Ectodomain shedding and remnant peptide signalling of EGFRs and their ligands. J Biochem (Tokyo) 2011;150:15–22. doi: 10.1093/jb/mvr068. [DOI] [PubMed] [Google Scholar]
- Hinkle CL, Sunnarborg SW, Loiselle D, Parker CE, Stevenson M, Russell WE, Lee DC. Selective roles for tumor necrosis factor α-converting enzyme/ADAM17 in the shedding of the epidermal growth factor receptor ligand family: the juxtamembrane stalk determines cleavage efficiency. J Biol Chem. 2004;279:24179–24188. doi: 10.1074/jbc.M312141200. [DOI] [PubMed] [Google Scholar]
- Igarashi T, Araki S, Mori H, Takeda S. Crystal structures of catrocollastatin/VAP2B reveal a dynamic, modular architecture of ADAM/adamalysin/reprolysin family proteins. FEBS Lett. 2007;581:2416–2422. doi: 10.1016/j.febslet.2007.04.057. [DOI] [PubMed] [Google Scholar]
- Isokane M, Hieda M, Hirakawa S, Shudou M, Nakashiro K, Hashimoto K, Hamakawa H, Higashiyama S. Plasma-membrane-anchored growth factor pro-amphiregulin binds A-type lamin and regulates global transcription. J Cell Sci. 2008;121:3608–3618. doi: 10.1242/jcs.031443. [DOI] [PubMed] [Google Scholar]
- Kahn J, Walcheck B, Migaki GI, Jutila MA, Kishimoto TK. Calmodulin regulates L-selectin adhesion molecule expression and function through a protease-dependent mechanism. Cell. 1998;92:809–818. doi: 10.1016/s0092-8674(00)81408-7. [DOI] [PubMed] [Google Scholar]
- Kenny PA, Bissell MJ. Targeting TACE-dependent EGFR ligand shedding in breast cancer. J Clin Invest. 2007;117:337–345. doi: 10.1172/JCI29518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinugasa Y, Hieda M, Hori M, Higashiyama S. The carboxyl-terminal fragment of pro-HB-EGF reverses Bcl6-mediated gene repression. J Biol Chem. 2007;282:14797–14806. doi: 10.1074/jbc.M611036200. [DOI] [PubMed] [Google Scholar]
- Lee MJ, Yu GR, Yoo HJ, Kim JH, Yoon BI, Choi YK, Kim DG. ANXA8 down-regulation by EGF-FOXO4 signaling is involved in cell scattering and tumor metastasis of cholangiocarcinoma. Gastroenterology. 2009;137:1138–1150.. doi: 10.1053/j.gastro.2009.04.015. [DOI] [PubMed] [Google Scholar]
- Ling Q, Jacovina AT, Deora A, Febbraio M, Simantov R, Silverstein RL, Hempstead B, Mark WH, Hajjar KA. Annexin II regulates fibrin homeostasis and neoangiogenesis in vivo. J Clin Invest. 2004;113:38–48. doi: 10.1172/JCI200419684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JW, Shen JJ, Tanzillo-Swarts A, Bhatia B, Maldonado CM, Person MD, Lau SS, Tang DG. Annexin II expression is reduced or lost in prostate cancer cells and its re-expression inhibits prostate cancer cell migration. Oncogene. 2003;22:1475–1485. doi: 10.1038/sj.onc.1206196. [DOI] [PubMed] [Google Scholar]
- Luetteke NC, Qiu TH, Peiffer RL, Oliver P, Smithies O, Lee DC. TGFα deficiency results in hair follicle and eye abnormalities in targeted and waved-1 mice. Cell. 1993;73:263–278. doi: 10.1016/0092-8674(93)90228-i. [DOI] [PubMed] [Google Scholar]
- Mayran N, Parton RG, Gruenberg J. Annexin II regulates multivesicular endosome biogenesis in the degradation pathway of animal cells. EMBO J. 2003;22:3242–3253. doi: 10.1093/emboj/cdg321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghal N, Sternberg PW. Multiple positive and negative regulators of signaling by the EGF-receptor. Curr Opin Cell Biol. 1999;11:190–196. doi: 10.1016/s0955-0674(99)80025-8. [DOI] [PubMed] [Google Scholar]
- Moss SE, Morgan RO. The annexins. Genome Biol. 2004;5:219. doi: 10.1186/gb-2004-5-4-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussunoor S, Murray GI. The role of annexins in tumour development and progression. J Pathol. 2008;216:131–140. doi: 10.1002/path.2400. [DOI] [PubMed] [Google Scholar]
- Nanba D, Mammoto A, Hashimoto K, Higashiyama S. Proteolytic release of the carboxy-terminal fragment of proHB-EGF causes nuclear export of PLZF. J Cell Biol. 2003;163:489–502. doi: 10.1083/jcb.200303017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi E, Hiraoka Y, Yoshida K, Okawa K, Kita T. Nardilysin enhances ectodomain shedding of heparin-binding epidermal growth factor-like growth factor through activation of tumor necrosis factor-α-converting enzyme. J Biol Chem. 2006;281:31164–31172. doi: 10.1074/jbc.M601316200. [DOI] [PubMed] [Google Scholar]
- Peschon JJ, et al. An essential role for ectodomain shedding in mammalian development. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- Rand JH. Antiphospholipid antibody-mediated disruption of the annexin-V antithrombotic shield: a thrombogenic mechanism for the antiphospholipid syndrome. J Autoimmun. 2000;15:107–111. doi: 10.1006/jaut.2000.0410. [DOI] [PubMed] [Google Scholar]
- Sahin U, Weskamp G, Kelly K, Zhou HM, Higashiyama S, Peschon J, Hartmann D, Saftig P, Blobel CP. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J Cell Biol. 2004;164:769–779. doi: 10.1083/jcb.200307137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Lee MJ, Heo JH, Lee YI, Kim Y, Kim SY, Lee ES, Juhnn YS. G protein βγ subunits augment UVB-induced apoptosis by stimulating the release of soluble heparin-binding epidermal growth factor from human keratinocytes. J Biol Chem. 2007;282:24720–24730. doi: 10.1074/jbc.M702343200. [DOI] [PubMed] [Google Scholar]
- Shi W, Chen H, Sun J, Buckley S, Zhao J, Anderson KD, Williams RG, Warburton D. TACE is required for fetal murine cardiac development and modeling. Dev Biol. 2003;261:371–380. doi: 10.1016/s0012-1606(03)00315-4. [DOI] [PubMed] [Google Scholar]
- Shirakata Y, et al. Heparin-binding EGF-like growth factor accelerates keratinocyte migration and skin wound healing. J Cell Sci. 2005;118:2363–2370. doi: 10.1242/jcs.02346. [DOI] [PubMed] [Google Scholar]
- Singh B, Schneider M, Knyazev P, Ullrich A. UV-induced EGFR signal transactivation is dependent on proligand shedding by activated metalloproteases in skin cancer cell lines. Int J Cancer. 2009;124:531–539. doi: 10.1002/ijc.23974. [DOI] [PubMed] [Google Scholar]
- Sternlicht MD, Sunnarborg SW, Kouros-Mehr H, Yu Y, Lee DC, Werb Z. Mammary ductal morphogenesis requires paracrine activation of stromal EGFR via ADAM17-dependent shedding of epithelial amphiregulin. Development. 2005;132:3923–3933. doi: 10.1242/dev.01966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeck A, Shang L, Dempsey PJ. Sequential and γ-secretase-dependent processing of the betacellulin precursor generates a palmitoylated intracellular-domain fragment that inhibits cell growth. J Cell Sci. 2010;123:2319–2331. doi: 10.1242/jcs.060830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S. Three-dimensional domain architecture of the ADAM family proteinases. Semin Cell Dev Biol. 2009;20:146–152. doi: 10.1016/j.semcdb.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Takeda S, Igarashi T, Mori H, Araki S. Crystal structures of VAP1 reveal ADAMs' MDC domain architecture and its unique C-shaped scaffold. EMBO J. 2006;25:2388–2396. doi: 10.1038/sj.emboj.7601131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumaru S, et al. Ectodomain shedding of epidermal growth factor receptor ligands is required for keratinocyte migration in cutaneous wound healing. J Cell Biol. 2000;151:209–220. doi: 10.1083/jcb.151.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetani T, Nakayama H, Okayama H, Okura T, Higaki J, Inoue H, Higashiyama S. Insufficiency of pro-heparin-binding epidermal growth factor-like growth factor shedding enhances hypoxic cell death in H9c2 cardiomyoblasts via the activation of caspase-3 and c-Jun N-terminal kinase. J Biol Chem. 2009;284:12399–12409. doi: 10.1074/jbc.M900463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther A, Riehemann K, Gerke V. A novel ligand of the formyl peptide receptor: annexin I regulates neutrophil extravasation by interacting with the FPR. Mol Cell. 2000;5:831–840. doi: 10.1016/s1097-2765(00)80323-8. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, et al. Mice with defects in HB-EGF ectodomain shedding show severe developmental abnormalities. J Cell Biol. 2003;163:469–475. doi: 10.1083/jcb.200307035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Shirakabe K, Werb Z. The metalloprotease Kuzbanian (ADAM10) mediates the transactivation of EGF receptor by G protein-coupled receptors. J Cell Biol. 2002;158:221–226. doi: 10.1083/jcb.200112026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.