Abstract
In atom probe tomography (APT), a technique that has been used to determine 3D maps of ion compositions of metals and semiconductors at sub-nanometer resolution, controlled emissions of ions can be induced from needle-shaped specimens in the vicinity of a strong electric field. Detection of these ions in the plane of a position sensitive detector provides two-dimensional compositional information while the sequence of ion arrival at the detector provides information in the third dimension. However, the applicability of APT to imaging unstained cells has not been explored. Here, we report the use of APT to obtain 3D spatial distributions of cellular ions and metabolites from unstained, freeze-dried mammalian cells. Multiple peaks were reliably obtained in the mass spectrum from tips with diameters of ~ 50 nm and heights of ~ 200 nm, with mass-to-charge ratios (m/z) ranging from 1 to 80. Peaks at m/z 12, 23, 28 and 39, corresponding to carbon, sodium, carbonyl and potassium ions respectively, showed distinct patterns of spatial distribution within the cell. Our studies establish that APT could become a powerful tool for mapping the sub-cellular distribution of atomic species, such as labeled metabolites, at 3D spatial resolutions as high as ~ 1 nm.
Keywords: Atom probe Tomography, Focused Ion Beam Scanning Electron Microscopy (FIB-SEM), Chemical Imaging
Introduction
Atom Probe Tomography (APT) is a powerful imaging technology to obtain three-dimensional (3D) compositional maps of substrates at <3 Å resolutions (Kelly and Miller, 2007; Miller, 2000; Seidman, 2007). The technique is based on a phenomenon called field evaporation, where ions are desorbed from a needle-shaped sample by application of a very intense electric field (~ several volts per nanometer) under very high vacuum and at cryogenic temperatures, and then detected by a single-ion sensitive detector (Muller, 1956). Although the first atoms were successfully imaged in a field ion microscope more than half a century ago (Müller, 1951), recent significant advances in the technology now enable the routine spatial and compositional imaging of semiconductors (Gorman et al., 2007; Perea et al., 2006; Prosa et al., 2010b), alloys (Miller et al., 2005; Mousa et al., 2010) and polymers (Prosa et al., 2010a) for visualization of features such as grain boundaries (Colijn et al., 2004), coarse precipitates (Kvist et al., 1996) and subsurface dislocations (Thompson et al., 2005). Three-dimensional compositional mapping is achieved by the use of a standing electric field in conjunction with either a pulsed electric fields or pulsed laser at the sample tip and coupled with a position-sensitive detector, allowing for time-of-flight mass spectrometric identification of each ion desorbed from a heterogeneous sample. The high spatial fidelity of APT is primarily a function of tip shape and dimensions. The electric field at the highly curved apex of the sample produces a concentration of field lines, causing an intensification of the field at the tip. The ensuing spatial divergence of evaporated ions results in a lateral magnification factor of ~106, since atoms generated from a tip measuring no more than100 nm across are detected over an area of several mm. At equilibrium evaporation conditions, atoms are field evaporated from the tip surface in an orderly and layered fashion such that the sequence of atoms arriving at the detector is directly related to the depth of those atoms within the sample tip, allowing accurate mapping along the z axis (specimen axis). A reverse point projection algorithm is then used to convert millions of detector events into a reconstruction of the entire volume of the sample tip at near-atomic resolutions (Bas et al., 1995).
Successful implementation of APT and sample reconstruction is dependent upon the geometry and robustness of the specimen. Samples with good electrical conductivity are required for voltage-pulsed analysis, but even electrical insulators (alumina, SiO2) can yield well-resolved field evaporation using laser-pulsing conditions (Kuduz et al., 2004). Specimens must be needle- or wire-shaped with sufficient sharpness and projection above the surface to produce the high electric field required for field evaporation. Optimal tips have smooth sides that taper gently (~10°) from an apex ~ 100 nm in diameter. Samples possessing inappropriate geometries can be shaped using electropolishing, micropolishing, broad ion beams (Liddle et al., 1988; Miller et al., 1996; Thompson et al., 2005) and Focused Ion Beam (FIB) based techniques (Larson et al., 1998; Miller et al., 2005; Thompson et al., 2007). In the latter, a finely focused gallium ion beam is used to first cut away a small portion of the bulk sample and then sputter away surrounding material using various milling patterns to attain the required shape. Although FIB-based protocols do impregnate the sample with gallium, recent improvements permit reliable and precise fabrication of tips from a wide range of materials with damage limited to the near-surface (Colijn et al., 2004; Kuhlman et al., 2001; Kuhlman et al., 2004; Larson et al., 2004; Miller and Russell, 2006; Thompson et al., 2007). Coupling the FIB with a Scanning Electron Microscope (SEM) enables visualization of specimen preparation while minimizing FIB-mediated damage. So far, both the sample preparation protocols and the harsh conditions of an APT run have largely restricted this method to metallurgical specimens.
The challenge to obtain three-dimensional images of biological materials with APT has proven daunting for decades. Despite limited sample preparation methods, Panitz and co-workers (Panitz, 1982; Panitz, 1983; Panitz and Giaver, 1981) attempted to visualize strands of DNA attached via a charged biopolymer to a metal substrate, with marginal success. The group also pioneered specimen preparation methods that allowed for imaging of monolayers of ferritin that were buried in a volume of benzene ice. Field-desorption of the benzene did reveal the surfaces of individual ferritin molecules but the fields required to perform large scale APT of biomolecules far exceeded the strength of the deposited material. Lack of sample stability has been a significant and lingering roadblock in APT analysis of bulk biological specimens, with the exception of a report describing the chemical analysis of a sub-volume of a tooth, which is an unusually hard biological specimen (Gordon and Joester, 2011). In addition to these difficulties, biological materials are poor electrical conductors, extremely heterogeneous in shape and local composition, and are not easily amenable to shaping. Further, they are soft, and when treated with dehydrating agents or fixatives, become brittle and lose their chemical properties.
Other methods to obtain three-dimensional maps of cellular components are also not without serious caveats. Fluorescence microscopic analysis of specific analytes (e.g. Fura-2 for calcium) do not give a complete map of all the ions within a cell and are limited to light microscopic resolutions. Simulated Raman spectroscopy enables visualization of unstained metabolites that exhibit strong Raman scattering such as certain cellular lipids (Freudiger et al., 2008; Wang et al., 2011), but the resolution is limited and molecules of interest must contain a recognizable Raman spectra. Secondary Ion Mass Spectrometry (SIMS) and Multi-isotope Imaging Mass Spectrometry (MIMS) have also been used to map specific metabolites, ions and lipid components within cells. MIMS, while amenable to sub-100 nm lateral resolutions, is restricted to ions that contain an isotopic tag (Lechene et al., 2007). SIMS can detect multiple ionic species at high resolution (Ostrowski et al., 2004; Szakal et al., 2011), but is primarily a surface phenomenon meaning that sputtering or abrasion is required to obtain any information along the z-axis. Depth profiling has been accomplished with SIMS (sometimes using a second sputtering beam), but this has come at the expense of resolution, primarily because of low ionic yields and beam-induced chemical damage (Fletcher et al., 2011). Thus, despite the many advances in chemical imaging in biology, generating a three-dimensional map of specific chemical constituents within cells at high resolution remains a formidable endeavor.
Here, we show that HeLa cell samples can be prepared for analysis in a way that can withstand the rigors of tip-shaping and the high-field, low-vacuum, cryogenic environment of APT. We and others have previously shown (Porter and Anderson, 1982; Szakal et al., 2011) that freeze-drying protocols, where a sample is rapidly plunged frozen at liquid nitrogen temperatures in the absence of fixatives and dyes and then gradually returned to room temperature under conditions of high vacuum, can be used to effect the gentle dehydration of a eukaryotic cell without causing large-scale perturbations in the structure and chemical make-up of the sample. Appropriate applications of thin metal coats is essential to protect the more brittle freeze dried cell sample, to reduce charging effects during FIB preparation, and to provide some local electrical conductivity for the APT data collection. Using these steps, we generated suitable tips and successfully recorded cellular APT data. Reconstructions of cellular sub-volumes at high resolution reveal a surprising amount of spatial heterogeneity of specific chemical species within the cell.
Results
Specimen Preparation for APT
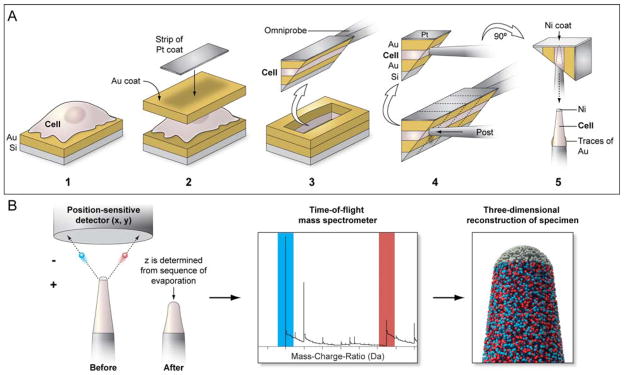
An overall schematic of the sample preparation and acquisition run is shown in Figure 1. We chose the adherent mammalian cell line HeLa for our experiments. Silicon substrates were sputter-coated with a thick layer of gold, then HeLa cells were seeded on the substrate and allowed to grow overnight in media. The substrates with the cells were then plunge frozen, freeze-dried and sputter coated again with another thick layer of gold (see Materials and Methods). Thus, the cells were sandwiched in gold, yielding a stable, conductive volume that could be imaged in a SEM. The overall morphology of the cells does not appear to be affected by the freeze-drying and coating (Szakal et al., 2011) (Fig. 2a). From here on, most of the steps were carried out in an FEI Nova Nanolab 600 or an FEI Nova 2000 Ion Abrasion Scanning Electron Microscope (IA-SEM). We deposited a ~100 nm thin layer of FIB-based platinum onto an area of interest on a HeLa cell (Fig. 2b). Following this, the chosen area was extracted using FIB milling protocols and attached with FIB-based platinum to a micromanipulator tip, revealing the profile of the extracted HeLa cell (Fig. 2c). Some flattening of the cell was observed, presumably as a result of the extraction of water and layering of a heavy gold coat on top of the cell. A thin nickel coat was applied to the back (not visible) side of the face shown in Fig. 2c, after which several sections were cut away with the FIB and attached via platinum onto posts. The dotted lines and arrow in Figure 2c show a typical FIB cut and direction of approach of the post, respectively. This resulted in an approximately 1 micron thick triangular shaped sub-volume of the sample attached to the post, where the cell was protected on two sides by the gold coating and on top by the thinner nickel coat (Fig. 2d). At this point, the sample was deemed ready for final shaping by FIB. Using annular milling patterns of decreasing radius, the sub-volume was gradually shaped into the needle-shaped tip that was tapered and ~100 nm wide at the apex, as required for APT analysis (Fig. 2d–g).
Figure 1. Schematic for the cellular sample preparation and APT data acquisition run.
(a) HeLa cells grown on gold-coated silicon substrates were coated with a thick layer of gold followed by a strip of platinum for further protection. Using FIB based protocols, a wedge containing a “sandwiched” cell volume was extracted and a sub-volume was attached to a microtip post in an orthogonal orientation. Following another protective nickel coat, the sub-volume was shaped into an APT-amenable tip by further FIB milling. (b) During an APT acquisition run, ions from the tip are extracted and are detected by a position-sensitive detector; this part of the tip is consumed as a result. Time-of-flight measurements of these detected ions allows their chemical identification in a mass spectrum, and a reverse-point algorithm can then be used to spatially place the ions in a high resolution reconstruction of the original tip.
Figure 2. Biological sample preparation for APT.
(a) HeLa cells grown on gold-coated silicon substrates, flash-frozen and sputter-coated with gold. (b) A sub-area of a selected cell protected with a layer of platinum (black arrowhead); Focused Ion Beam (FIB) milling reveals the profile of the cell sandwiched between layers of gold (white arrowhead). (c) Cellular material after extraction as a wedge and attachment to an Omniprobe tip. The cell was attached to a post from the side (arrow) and separated from the rest of the cell by FIB milling (dotted lines). (d) Sub-volume of the HeLa cell stably attached to a probe tip. (e–g) Gradual sculpting of the sample into the needle-shape required for APT by annular FIB milling with decreasing radius. Scale Bars: a, b, 10μm; c, 5 μm; d–f, 2 μm; g, 1 μm.
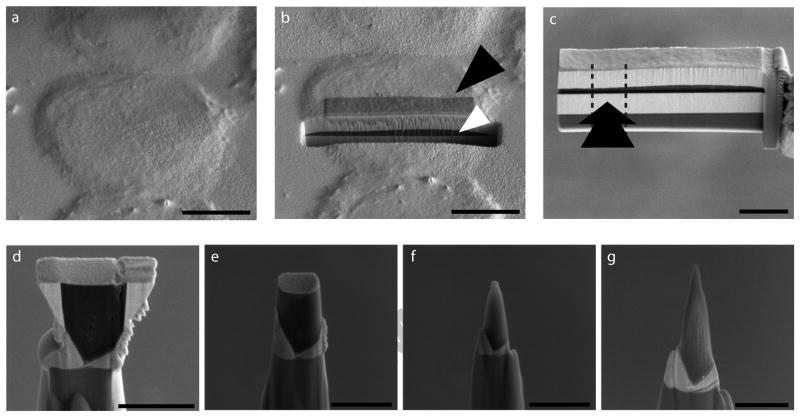
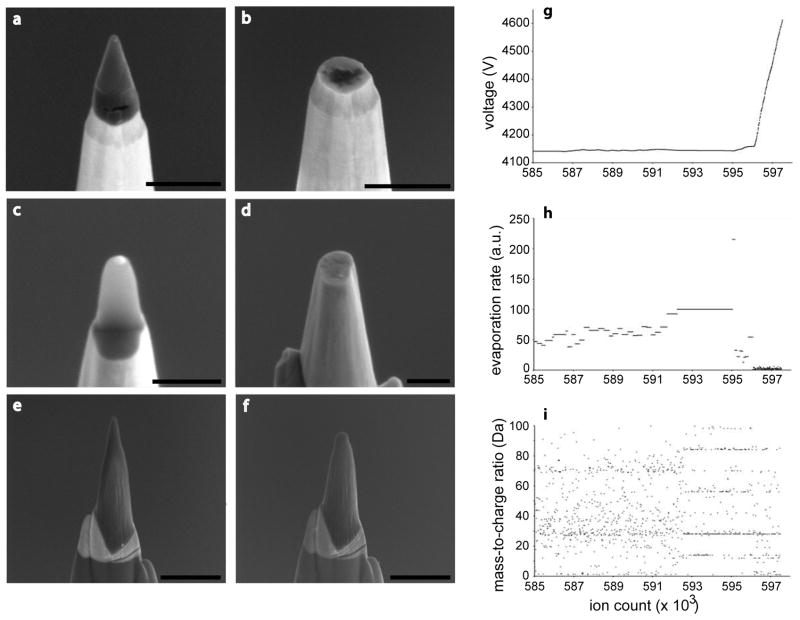
The geometry of extraction, coating and shaping was critical to the successful generation of APT-ready tips. For example, the most straightforward approach was one where the tips were milled from a “top-down” orientation, resulting in horizontal layers of metal coats and cell on the posts, with the bottom surface of the cell closer to the apex. We found that the adhesion between cell and gold coat could not withstand the extractive forces of APT, resulting in complete tip failure (Fig. 3a, b). Similarly, a “bottom-up” orientation also failed, even when extra protective metal coats were applied (Fig. 3c, d). However, when the cells were coated and deposited on the posts orthogonally, the extracted volume was stable through the experiment. When analyzed by APT, the extraction of ions from the tip resulted in the steady removal of approximately 300 nm from the top of the tip constituting a single data collection run (Fig. 3e, f). In Figures 3g–i we show a representative set of real-time data acquisition plots from a tip that failed; plots of the voltage evolution, evaporation rate and mass-over-charge ratio respectively, over an ion sequence that clearly shows dramatic changes at the point of tip failure. There is a short period of time, presumably as the tip loses structural integrity (around 593 × 103 ions into the run), where both the voltage and the evaporation rate remain static over time. This is due to a surge of ion events overwhelming the instrument response. As shown in Fig. 3i, the ions emitted during this period have a different mass spectrum than the previous ions. The interpretation is that the apex of the tip partially ruptured forming extremely sharp surface features (perhaps below the cell portion of the tip, revealing different material) that emitted ions at very high rates. Before the tip could re-equilibrate, the tip fully ruptured around (around 596 × 103 ions into the run) and the ion emission rate fell to zero rapidly causing the instrument to increase the voltage to try and compensate, at which point the experiment was manually aborted.
Figure 3. Orientation of sample and protection by metal coating is essential for stable tip generation.
(a, b) Tips generated by top down, (c, d) backside, or (e, f) cross-section methods, where the direction of attachment to the microtip post was from the top, bottom, or side of the cell, respectively, imaged by SEM before and after an APT experiment. A long and controlled APT run could consume ~300 nm of a stable tip. (g) Plots of ion sequence number versus voltage, (h) ion sequence number versus mass-to-charge ratio, and (i) ion sequence number versus evaporation rate, of a tip that failed during an APT experiment. Scale Bars: a–d, 500 nm; e, f, 1 μm.
Stable data acquisition from cellular sample
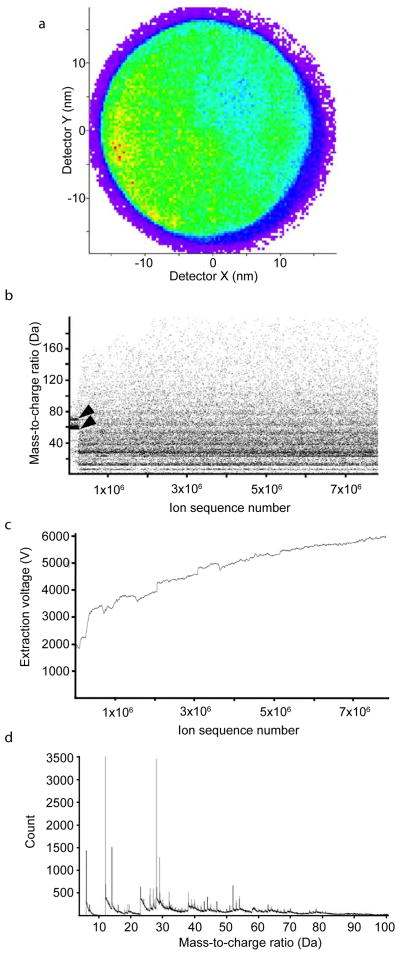
In Figure 4, we present raw data from the APT experiment performed on the tip shown in Figure 2 and 3e and f. We were able to detect approximately 7.7 × 106 ions continuously from the tip until the end of the experiment; a “heat map” of all the events recorded on the position-sensitive detector (PSD) shows that the ions field evaporated from the tip were evenly distributed over the surface of the detector, allowing for high spatial fidelity during the downstream analysis and reconstruction (Fig. 4a). In addition, the time of flight (TOF) capabilities of LEAP instrument allow for the simultaneous measurement of mass-to-charge ratios (m/z) of the evaporated ions from the tip. While data was collected to accommodate species of up to 400 Da, the plot of m/z of extracted ions over ion sequence number from the same tip (Fig. 4b) shows that ions up to ~100 Da were efficiently and reliably detected. At the start of the acquisition run, multiple events tightly clustered around 58 Da (nickel) and 69 Da (gallium) were detected, from the protective cap and FIB milling, respectively. Plotting the extraction voltage (Ve) over the ion sequence number reveals a rapid increase in Ve from 1800 V to 3200 V, followed by a steady increase to 5800 V (Fig. 4c), corresponding to the field evaporation of the nickel cap and the cellular material. The steady change in Ve is suggestive of a tip that steadily yielded ions without catastrophic failure or tip breakage. Thus, two observations could be made from the raw data: There appears to be minimal cross contamination between cellular material and the metallic cap, and there was no interrupted or abnormal data collection from the tip, even across the metal-cell interface. Thus our protocols allow the generation of a tip of cellular origin that is robust enough to withstand the harsh conditions of an APT acquisition run, yielding a rich spectrum with well defined peaks up to almost 100 Da (Fig. 4d).
Figure 4. Cellular samples are amenable to APT.
(a) Ion density observed at the detector and integrated over an entire APT acquisition run of a representative HeLa cell derived tip. The resulting heat map of the Position Sensitive Detector shows an even distribution of ions across the surface of the detector. (b) Plot of mass-to-charge ratio versus ion sequence number, showing the identity of ions emitted in sequence during the experiment. Nickel (58 Da) and gallium (69 Da) corresponding to the protective cap at the tip apex are detected at the start of the experiment (arrowheads) (c) Corresponding plot showing the evolution of extraction voltage as a function of ion sequence number (d) Mass spectrum obtained from part of the acquisition, showing well defined peaks.
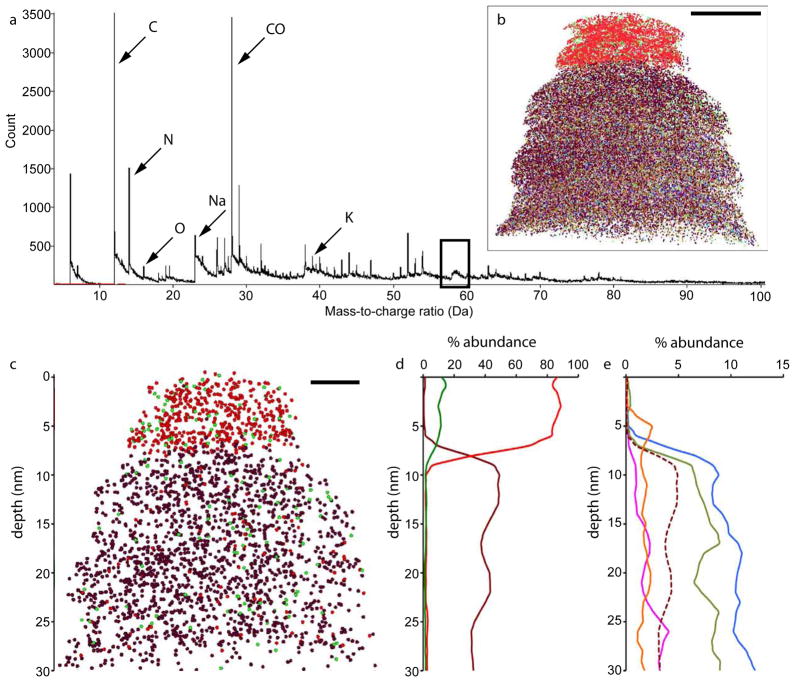
Before we attempted to reconstruct the entire cell sub-volume, we wanted to ascertain that the back-projection algorithm could work with a fraction of the tip. The nickel cap offered a suitable test – we therefore initially analyzed a fraction of the tip that contained the cap and part of the cell. Using the IVAS 3.1.1 software, we first analyzed the ions generated from the initial 1.5 × 106 ions (see Materials and Methods); the mass spectrum from this sub-volume of the tip is rich in peaks (Fig. 5a), and a broad peak consistent with the molecular nickel peak at 58 Da is clearly visible (boxed). The m/z spread of the nickel signal likely stems from a combination of TOF aberrations resulting from inadequate TOF corrections for initial evaporation events originating from a tip shape equilibrating with the application an electric field and TOF correction optimization based on peaks originating from the sub-nickel surface ions. The peaks that appear subsequently in the run are clearly defined. As would be expected from a cellular sample, peaks appeared at 12 Da (carbon), 14 Da (nitrogen), 16 Da (oxygen), 23 Da (sodium), 28 Da (silicon or carboxyl group) and 39 Da (potassium), with more modest peaks appearing at 31 Da (phosphorus) and 32 Da (sulphur). Prominent peaks from the spectrum were selected and mapped onto a 3D volume representing the chosen sub-volume (Fig. 5b), and parameters were adjusted to give a volume that was consistent with the physical measurements of that fraction of the tip e.g. the volume allotted to each ion, set at 0.1 nm3 to compensate for the volume previously made up by water (the freeze-dried sample is a dehydrated volume where ~80% of the original volume occupied by water is lost).
Figure 5. Cellular tips can be accurately reconstructed.
(a) Mass spectrum of first 1.5 × 106 ions field evaporated from the tip, plotted in 0.01 Da bins. The broad peak corresponding to nickel is boxed, and relevant ions plotted in the reconstruction are marked. (b) Corresponding 3D reconstruction of this part of the tip. (c) 10 nm slice from the central plane of the reconstruction, showing distribution of selected ions nickel (red), gallium (green) and carbon (brown). (d) Corresponding plot of % abundance of these ions versus depth along the z axis. (e) Plot of % abundance versus depth of less abundant ions, nitrogen (blue), potassium (green), oxygen (orange) and sodium (pink) of the same slice. Carbon has been plotted as a brown dotted line at 1/10th of its original abundance. Scale bars: b, 10 nm; c, 5 nm.
High resolution 3D reconstruction of cellular sub-volumes
It was apparent that APT was able to measure and successfully reconstruct the nickel-cell transition of the tip, and we directly quantified this transition. We selected and reconstructed several peaks from the mass spectrum of this sub-volume. In Figure 5c, we display the local signals from carbon (brown), nickel (red) and gallium (green) from a 10 nm thick slice around the central xy plane of the reconstruction – this removes artifacts arising from the conical shape of the reconstructed volume of the sample – and in Figure 5d we plot the relative abundance of each of these ions with tip depth. As expected, nickel (red) is the predominant species of the protective cap, while carbon (brown) is a major component of the cellular portion, and we see only a modest amount of cross-contamination of each of these ions. Nickel ions constitute 85.51% of all ions detected from the top 14 nm of the tip and 2.38% of the rest of the volume excluding a 2 nm wide region around the metal-cell interface, while only 1.16% of the ions reconstructed to the cap region corresponded to carbon (37.87% in the cellular portion of the tip). The nickel composition is consistent with other measurements of the nickel sputtering target used which is known to have 10–15% non-nickel impurities. The interface itself appears to be more like a sharp transition zone, restricted to a width of 2 nm, but this is likely an approximation caused by multiple factors: erratic evaporation events and limitations of reconstruction, as well as some non-planarity of the metal-cell interface. There is also a significant signal from gallium (green) in the topmost segment of the tip (~11.55% of all ions), which was expected since gallium is the ion source for the FIB used in the polishing and final shaping of the tip. Significantly, we observe minimal gallium implantation within the bulk of the cellular sample (~1.63%). Thus, it appears that the nickel cap largely limits gallium induced damage to the rest of the sample. That the sacrificial nickel cap was necessary is reflected in the final thickness of the cap, measured at 15 nm; we infer that approximately 35 nm of the metal cap was destroyed during milling and possibly at the very start of the APT run. We note the apparent curvature of the cellular portion of the reconstruction; this is an artifact of the reconstruction algorithm. The apparent size (and curvature) changes for the uppermost cellular region arise from the changing Ve which is consequence of the disparate evaporation fields required to field evaporation material from two material types.
We also plotted other cellular ions in this volume (Fig. 5d). A dotted line indicating carbon (plotted at 1/10th of its original abundance) is included as a reference. We found that there was no nitrogen (blue), potassium (green), or sodium (pink) in the nickel cap area of the tip; there was a small signal corresponding to oxygen. From these data, it appears that the separation between cellular and non-cellular components appears to have been measured with high fidelity. Also, the distribution of each ion appears to be unique; interestingly, carbon has a banded pattern, while sodium and potassium have similar patterns to each other at this scale, that are different from the other ions. Both ions are enriched in the trough between the bands of carbon abundance. The ratio of sodium to potassium ions (2.53% vs. 8.12% abundance, respectively) is approximately 1:3.2. This is lower than the accepted estimates of ~10:1, but confirms that we are sampling an intercellular volume, and is an intriguing hint of widely variant ionic concentrations within the cell that can only be captured at these high resolutions.
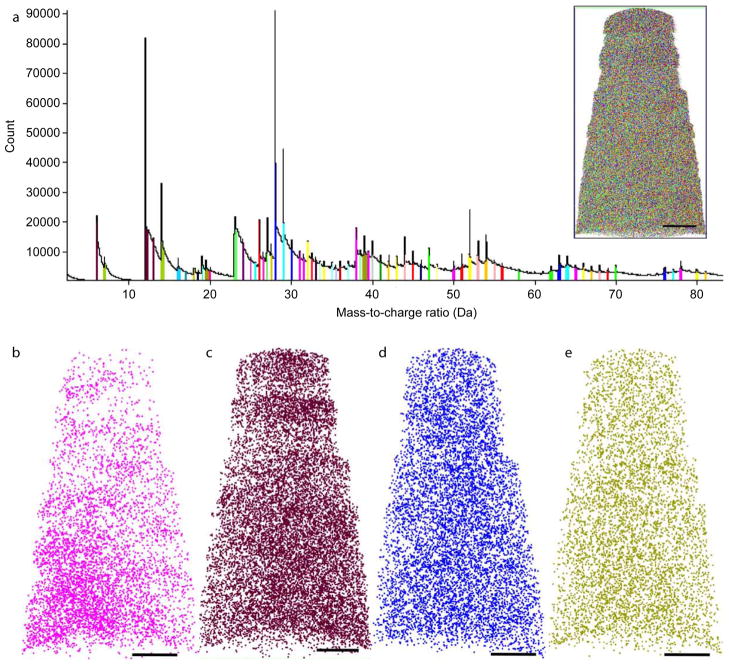
Next, we reconstructed the purely cellular portion of the tip, which was essentially free of nickel. Once again, we were able to detect multiple well-defined peaks appearing at almost every integer Da in the mass spectrum (Fig. 6a). There were sporadic peaks detected up to 200 Da, but for the purposes of this analysis, only prominent peaks < 100 Da were used for reconstruction. Some of these peaks were assigned molecular ions (marked with arbitrary colors in Fig. 6a), but it was impossible to confidently ascribe the true identity of many of the other peaks. There are multiple reasons for this: the mass resolution was set at 0.001 Da, but the width of many peaks far exceeded this number, and it was common to assign peak widths of 0.02 – 0.05 Da to various ions. At this mass resolution, it is impossible to tell apart with any certainty near-coincident peaks such as Si (28.08 Da) and CO (28.11 Da), even though some clues could be gleaned from natural isotopic abundances and expected peak intensity ratios. There are also multiple candidate ions of cellular origin for each of the peaks, especially at higher molecular weights.
Figure 6. A subset of cellular ions shows spatial heterogeneity at the nano-scale.
(a) Mass spectrum of all ions from the entire tip excepting the metal cap, in 0.01 Da bins. Prominent peaks were chosen and assigned arbitrary colors. (a, inset) Reconstruction of the entire volume, mapping all the prominent ions in the mass spectrum. Several of ions within the cellular sub-volume were mapped, including peaks at m/z (b) 12 Da, labeled C, (c) 23 Da, labeled Na (d) 28 Da, labeled CO, and (e) 39 Da, labeled K. Scale bars 20 nm.
Nevertheless, there were certain peaks that could be assigned with some confidence, as was done for the first analysis. Some of the relative abundances of these molecular ions were as follows: Carbon (6 Da and 12 Da combined) constituted 13.8% of the ions in this sub-volume of the cell; CO/silicon 8.0%; nitrogen (7 Da and 14 Da) 6.0%; potassium 5.7%; sodium 5.6%; sulphur 2.0% oxygen 1.7% (other elements such as phosphorus and calcium constituted <1.5% of the volume). The peak corresponding to water was negligible at 0.6%. With the exception of oxygen, the order of relative abundances are broadly consistent with the rough atomic composition of a dried cell, considering that we are only looking at molecular ions representing an unknown fraction of the actual abundance of the element. We hypothesize that the majority of oxygen is extracted bound to other elements, resulting in a lower peak at 16 Da. However, a reconstruction could still be carried out, assigning arbitrary assignments for each major peak (Fig. 6a, inset), and using parameters so that the dimensions of the reconstructed tip matched the physical measurements of the tip.
Spatial heterogeneity of a subset of cellular ions
An analysis of the reconstructed tip revealed that some ions, but not all, showed spatial heterogeneity in their localization within the cellular sub-volume. Figure 6d shows a panel of ions that were chosen to illustrate this phenomenon, where ions were chosen from a 10 nm thick slice around the central xy plane of the entire reconstruction as in Figure 5. A striking example of this heterogeneity at the nano-scale was observed with the ion at m/z 23 Da, assigned to sodium (Fig. 6c). Sodium appears to be enriched in a “pocket” approximately 50 nm across at the bottom left of the reconstruction, whereas it is markedly diminished nearer the top of the tip, which corresponds to the far edge of the cell. Similarly, the peaks at 6 Da and 12 Da were combined and assigned to carbon (the two peaks plotted individually show identical spatial patterns). Carbon also shows an interesting pattern of distribution across the cell sub-volume (Fig. 6b), with a slight enrichment in the bottom-right area of the reconstruction, away from and less pronounced as that seen for sodium. However, the layered pattern was seen again at the top of the reconstruction, with two denser bands of carbon approximately 10 nm wide separated by a smaller band approximately half that width. Further, the small but noticeable negative correlation between sodium and carbon in this region of the tip was also confirmed. On the other hand, not all ions show any significant heterogeneity within the volume. Two abundant ions at m/z 28 Da, designated CO, representing the carbonyl group (Fig. 6d) and at 39 Da, designated as potassium (Fig. 6e) showed very little heterogeneity within the sample, although it is impossible to rule out a contribution from silicon at m/z 28, given that substrate derived contamination is can be observed during lift-out procedures. Thus, using APT, we can detect verifiable boundaries (here, the nickel–cell layer) with high precision, and we can also observe unique patterns of spatial heterogeneity of cellular ions at the nanoscale.
Discussion
In this work, we have used a combination of techniques to generate a cellular sample that can withstand the rigors of shaping into an APT-amenable tip as well as the high-field, low-vacuum, cryogenic environment of APT itself. In order to render a eukaryotic cell sample amenable to APT, we devised a set of protocols with the aim of achieving the following: (i) processing the cell sample in a manner that would minimally perturb the order and location of chemical moieties within it, (ii) protecting and manipulating a chosen sub-volume of the cell appropriately to allow shaping of the sample into a needle shaped tip, and (iii) generating a stable tip that would allow a robust and meaningful APT readout. Eukaryotic cells are generally composed of ~ 80–90% water by weight, and it was important to leach out the water from a cellular sample for several reasons: the absence of this predominant signal would allow sensitive detection of other molecular species, and since the APT experiment is carried out at cryogenic temperatures, retention of water in such a bulk sample would have resulted in ice crystal formation and cell destruction. On the other hand, simply drying out a cell would also cause have lysis or membrane rupture due to osmotic imbalances, resulting in considerable spatial disruption of cellular constituents. We and others have previously shown(Porter and Anderson, 1982; Szakal et al., 2011) that freeze-drying protocols, where a sample is rapidly plunged frozen at liquid nitrogen temperatures and then gradually returned to room temperature under conditions of high vacuum, can be used to effect the gentle dehydration of a eukaryotic cell without causing large-scale perturbations in the structure and chemical make-up of the sample. Importantly, in the absence of fixatives and other chemical reagents the fidelity of the chemical composition of the cell is likely retained in a near-native state. However, freeze dried biological samples suffer from two important limitations, both primarily due to the leaching of water: they are brittle and uneven, and they are also poor electrical conductors. These properties mitigate the ability of the sample to withstand the high vacuum and extraction voltages of APT; thus further steps such as the application of gold or nickel coats were required at various stages of specimen preparation to protect the cell sample, to provide some local electrical conductivity for the APT data collection and to reduce charging effects during FIB preparation. Using a combination of these protocols, we were able to generate suitable tips and successfully record APT data from HeLa cells.
Although data was successfully collected from the cellular tips, certain caveats must be noted with these APT reconstructions: first, freeze-drying results in some collapse of the cell onto its more structured components such as the nucleus and cytoskeletal network. We do not know the extent of molecular displacements at the resolutions afforded by APT. Additionally, the roughness of the surface at this scale translates to multiple fine-scale emitting features rather than a single atomically smooth surface, which may contribute to erratic evaporation events and a complicated projection from the specimen surface to the detector, for which the reconstruction algorithms may not fully account. For example, although the transition from protective nickel cap to the cell appears sharp and obvious to the eye (Fig. 4b), we observed some sporadic evaporation of material during field evaporation through the nickel-cell interface. This suggests that some uncontrolled evaporation was occurring through the interface, which complicates the reconstruction of ion positions with very high precision for that region of the analysis. Similarly, reconstruction of the tip in its entirety was unsuccessful, on account of the sudden change in characteristics during the transition from the metallic cap to the cellular portion of the tip. Evaporation field differences required for evaporation of nickel versus non-nickel ions would be expected to result in a generic reconstruction aberration that occurs when analyzing between materials with disparate evaporation fields(Vurpillot et al., 2004), leading to reconstructions that did not match well with the calculated tip dimensions. Finally, contamination from extracellular sources during cell processing can be limited with careful experimentation but are difficult to rule out entirely. In our case, the major contaminant is likely substrate-derived silicon, whose m/z peak overlaps sufficiently with another ubiquitous cellular signal, the carbonyl ion so as to be indistinguishable at these mass resolutions. The corresponding NH ion from peptide bonds is not prominent and has no discernable pattern, hence we refrain from interpreting this peak.
The use of APT for routine biological applications is clearly in a very early phase. Rearrangements on the scale of 1–10 nm may occur with freeze-drying that will limit the spatial resolution that can be achieved. The necessity of using small sample sizes is yet another limitation. Further, many more methodological improvements will need to take place before the use of APT, possibly in conjunction with other mass spectrometric and imaging methods leads to new mechanistic insights. Notwithstanding these limitations, we present these results as a significant advance in the potential of mapping the chemical composition of cells at near-atomic resolution in three dimensions. There are multiple lines of research that we envisage in the future based on this technique. Currently, the volume sampled constitutes < 0.01% of the volume of a typical eukaryotic cell, but the use of APT on bacterial cells (especially rod-shaped bacteria, which could be well suited for APT, a technique dependent on tight radii of curvature) could provide insights into the steady state chemical composition of bacteria. Well-defined biological materials in the sub-micron size range such as viruses may also be amenable to APT, however, technical challenges in positioning and shaping these samples in order to carry out a successful experiment need to be solved. Another important application is likely to be in localizing drugs and metabolites at high spatial resolution in mammalian cells, especially in conjunction with the use of isotope tags, and with correlative fluorescence microscopy. We thus expect that the further standardization of sample preparation and reconstruction protocols will enable APT to become a more useful tool for targeted high-resolution chemical imaging studies of small biological samples in the future.
Materials and Methods
Preparation of cell sample
An overall schematic of the sample preparation and acquisition run is shown in Figure 1. Pre-cut and sterilized silicon substrates were sputter coated with gold to a thickness of 1–2 microns. Cells from the adherent mammalian cell line HeLa were allowed to grow overnight in RPMI medium containing 10% Fetal Bovine Serum on gold quantifoil grids. The cells were >90% viable and sub-confluent at the time of the experiment. The gold coated substrates were then washed in 5 mL of phosphate-buffered saline (PBS) and 1 % bovine serum albumin (BSA) at 37 °C for 5 minutes, blotted for 2 seconds, and immediately plunge-frozen in liquid ethane. The cell samples were transferred at liquid nitrogen temperatures to a liquid nitrogen-cooled SEM prep chamber for freeze drying. Freeze-drying was performed slowly such that the cells were maintained at liquid nitrogen temperatures for 5 hours, before warming to room temperature over a period of 10 hours. The resulting freeze-dried cells on silicon pieces were gently rinsed in a small volume of HPLC-grade water for 5 seconds, and the water quickly wicked away with filter paper. The cells were then sputter coated again with gold to a thickness of approximately 1 micron; thus the freeze-dried cell layer was stably “sandwiched” between layers of gold and transferred to the FIB-SEM for tip shaping.
“Cross Section” Tip preparation
Stable cellular tips were successfully generated using a cross-section lift-out prep approach (Lawrence et al., 2008; Miller et al., 2005; Thompson et al., 2007). All specimen preparation and imaging were performed on an FEI Nova Nanolab 600 or an FEI Nova 2000 (Hillsboro, OR).
First, a 100 nm thick layer of focused-ion-beam (FIB) deposited platinum (~4 × 15 μm) was applied to a chosen HeLa cell to mark and protect the region of interest. Next, two 3 μm wide trenches were cut into the substrate using a 6 nA ion beam current to undercut a region for removal at 52° and 17° stage tilt respectively. A cantilever of material was formed by cutting one end of the undercut region. In order to remove the cantilevered wedge of material from the bulk, an Omniprobe micromanipulator (Dallas, TX) was first attached to the free end of the wedge via FIB-platinum applied at 28 pA beam current, then the remaining attached end of the wedge was cut free enabling transfer of the extracted material to an Axial Rotational Manipulator (ARM) (Lawrence et al., 2008). Once transferred to the ARM, the micromanipulator was cut from the wedge and the chamber was vented. Next, the ARM was rotated by 90° so that the “profile” of the cell was facing upward. The lifted-out cell sample was then coated with 50 nm of nickel to additionally protect the cell during the remaining FIB shaping steps. Finally, the ARM was returned to the FIB, where the wedge was made to have the cross-section resembling an equilateral triangle by further milling each long side of the wedge with the stage tilted to 22°.
The last specimen preparation stage involved transferring sections of this wedge to free-standing microtip posts and annularly milling those sections to their final tip dimensions(Larson et al., 1999; Thompson et al., 2007). After transfer of the wedge from the ARM to the micromanipulator, the wedge was placed immediately above an available silicon microtip post (Thompson et al., 2005). A small-area (~1 μm2) of FIB-platinum was applied to the location where the bottom of the wedge intersected the top of the microtip using a short application of 28 pA current. The attached wedge was then cut along the direction parallel to the post using at 0.28 nA, leaving behind a 0.5–1.0 μm wide slice of the wedge mounted on top of the post. The wedge was then translated to the next post and the cutting process was repeated until the entire wedge was consumed and slices corresponding to adjacent areas of the cell were attached to individual microtips. Finally, the FIB stage was rotated 180° and a second platinum weld was applied to the reverse side of the wedge/microtip intersection to firmly attach the mounted cellular sample to the microtip post.
To achieve the final needle specimen shape necessary for APT analysis, a series of annular milling patterns were applied to the top of the mounted posts. Standard circle milling patterns were used with outer diameters of ~4 μm and inner diameters of decreasing size at beam currents of 0.28 nA (milling patterns proceeded from the outer to inner circumference of the circle). Typical inner diameters might proceed from 1.6 to 0.8 to 0.4 μm to produce a conically shaped tip with a gentle cone angle. Milling time and specific inner diameters were adjusted based on experience with the shape evolution of previously shaped tips. A final 5 kV low-energy clean-up mill was applied to minimize the depth of surface ion-damage produced by the milling process (Thompson et al., 2006). At the end of this process, approximately 10 tips measuring ~50–100 nm in end form radius, comprising subsections of single HeLa cells were generated.
APT analysis
Data from the cellular samples were collected on a LEAP 3000X HR (Cameca Instruments, Madison WI) operated in a pulsed laser mode and equipped with a reflectron (total flight path 382 mm) (Kelly and Miller, 2007). The base temperature and pressure were maintained at 30 K and ~1 × 10−9 Pa respectively throughout the duration of the experiment. The laser wavelength of 532 nm was operated at a pulse frequency of 200 kHz. A variety of pulse energies and evaporation rates were attempted to optimize data quality and yield, however the choice and variation of parameters were not exhaustive. Best results were obtained using pulse energies between 0.2 – 0.5 nJ, and average target evaporation rates of 0.5–1 %. Changes in pulse energy did not result in a noticeable changes in the total number of ions detected before specimen failure, and did not affect data quality. In all cases, data collection was continued until tip failure. In the case of cross-section tip preparation, up to ~10 million ions could be collected from each tip in some runs; in the representative tip shown here (Fig. 3e, f) ~7.7 million ions were collected by the LEAP instrument in a single run between scanning electron microscope (SEM) images. We note that multiple ion events were common during cell analysis, typically approaching 75% of the detected ion events.
Reconstruction
3D reconstruction of detected ions was performed using the IVAS 3.1.1 software (Cameca Instruments, Madison WI) (Bas et al., 1995; Miller, 2000). SEM imaging of a tip before and after analysis, (Figure 1d and e, respectively) indicated a final tip radius of ~110 nm and an analysis depth of > 300 nm. Using default values for the field-factor, k (3.3), and detector efficiency (0.37), an evaporation field of 17 V/nm and average ionic volume of 0.1 nm3 allowed the reconstructed volume to match the observed volume change of the specimen. Typically, a tip was reconstructed in two phases: the sharp end of the tip with the protective nickel cap was analyzed along with a representative portion of the cell (see analysis 1). The cellular portion of the tip, without the nickel cap, was analyzed separately (see analysis 2). In both analyses, the time-of-flight (TOF) histogram was collected up to 10000 ns, in 0.5 ns bins, and the corresponding mass spectrum was collected up to a maximum of 400 Da, divided into 0.001 Da bins. The spectrum was calibrated based on the prominent peaks of carbon (at m/z 6 Da and 12 Da), nitrogen (m/z 14 Da) and sodium (m/z 23 Da). In the case of analysis 1, the peaks corresponding to the atomic weight of the major cellular elements excluding hydrogen (C m/z 11.99–12.01 Da; N m/z 13.95–14.04 Da; O m/z 15.97–16.02 Da; as well as Na m/z 22.97–23.03 Da; K m/z 38.92–39.04 Da) along with the single isotope of gallium used during ion milling (m/z 68.91–68.94 Da) and various isotopes of nickel (primarily the two isotopic peaks chosen at 59.90–60.01 and 57.91–57.95 Da) were chosen and assigned a volume of 0.1 nm3 for the reconstruction. The prominent peak at m/z 28 Da was assigned to Si based on isotopic distribution, but at these mass resolutions, the contribution of the CO ion could not be ruled out. Based on the shape and size of the tip measured from SEM images before and after data collection in the LEAP instrument, a nickel compatible evaporation field of 35 V/nm (Miller, 2000) was chosen, corresponding to an initial tip of 15.5 nm. The initial voltage for the reconstruction was 1791.5 V. For analysis 2, corresponding to the cellular portion of the tip, all major peaks in the spectrum were chosen and assigned arbitrary colors, typically with 0.02–0.06 Da widths. Here, this initial radius was chosen to be 39.07 nm and shank angle 11.7°. This resulted in a reconstructed tip that was approximately 250 nm in height and 100 nm width, which corresponded well with the physically imaged measurements of the tip.
Footnotes
Competing Interests:
Two of the authors (Ty Prosa and Thomas Kelly) are employees of Cameca Instruments, a commercial manufacturer of instruments used to perform atom probe tomography.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bas P, Bostel A, Deconihout B, Blavette D. A General Protocol for the Reconstruction of 3D Atom Probe Data. Appl Surf Sci. 1995;87/88:298–304. [Google Scholar]
- Colijn HO, Kelly TF, Ulfig RM, Buchheit RG. Site-Specific FIB Preparation of Atom Probe Samples. Microsc Microanal. 2004;10:1150–1151. [Google Scholar]
- Fletcher JS, Lockyer NP, Vickerman JC. Developments in molecular SIMS depth profiling and 3D imaging of biological systems using polyatomic primary ions. Mass Spectrom Rev. 2011;30:142–74. doi: 10.1002/mas.20275. [DOI] [PubMed] [Google Scholar]
- Freudiger CW, Min W, Saar BG, Lu S, Holtom GR, et al. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science. 2008;322:1857–61. doi: 10.1126/science.1165758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon LM, Joester D. Nanoscale chemical tomography of buried organic-inorganic interfaces in the chiton tooth. Nature. 2011;469:194–7. doi: 10.1038/nature09686. [DOI] [PubMed] [Google Scholar]
- Gorman BP, Norman AG, Yan Y. Atom Probe Analysis of III–V and Si-Based Semiconductor Photovoltaic Structures. Microsc Microanal. 2007;13:493–502. doi: 10.1017/S1431927607070894. [DOI] [PubMed] [Google Scholar]
- Kelly TF, Miller MK. Invited review article: Atom probe tomography. Rev Sci Instrum. 2007;78:031101. doi: 10.1063/1.2709758. [DOI] [PubMed] [Google Scholar]
- Kuduz M, Schmitz G, Kirchheim R. Investigation of oxide tunnel barriers by atom probe tomography (TAP) Ultramicroscopy. 2004;101:197–205. doi: 10.1016/j.ultramic.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Kuhlman K, Martens R, Kelly T, Evans ND, Miller MK. Fabrication of Specimens of Metamorphic Magnetite Crystals for Field Ion Microscopy and Atom Probe Microanalysis. Ultramicroscopy. 2001;89:169–176. doi: 10.1016/s0304-3991(01)00116-4. [DOI] [PubMed] [Google Scholar]
- Kuhlman KH, Kelly TF, Miller MK. Atomic-Scale Analysis of Metamorphic Magnetite using Field Ion Microscopy and Atom Probe Tomography. Microsc Microanal. 2004;10:512–513. [Google Scholar]
- Kvist A, Andren HO, Lundin L. A specimen preparation technique for atom probe analysis of the near-surface region of cemented carbides. Appl Surf Sci. 1996;94/95:356–361. [Google Scholar]
- Larson DJ, Petford-Long AK, Ma YQ, Cerezo A. Information Storage Materials: nanoscale Characterization By Three-Dimensional Atom Probe Analysis. Acta Materialia. 2004;52:2847–2862. [Google Scholar]
- Larson DJ, Petford-Long AK, Cerezo A, Smith GDW, Foord DT, et al. Three-Dimensional Atom Probe Field-Ion Microscopy Observation of Cu/Co Multilayer Film Structures. Appl Phys Lett. 1998;73:1125–1127. [Google Scholar]
- Larson DJ, Foord DT, Petford-Long AK, Liew H, Blamire MG, et al. Field-Ion Specimen Preparation Using Focused Ion-Beam Milling. Ultramicroscopy. 1999;79:287–293. [Google Scholar]
- Lawrence D, Alvis R, Olson D. Specimen Preparation for Cross-Section Atom Probe Analysis. Microsc Microanal. 2008;14:1004–1005. [Google Scholar]
- Lechene CP, Luyten Y, McMahon G, Distel DL. Quantitative imaging of nitrogen fixation by individual bacteria within animal cells. Science. 2007;317:1563–6. doi: 10.1126/science.1145557. [DOI] [PubMed] [Google Scholar]
- Liddle JA, Norman A, Cerezo A, Grovenor CRM. Pulsed laser atom probe analysis of Ternary and Quaternary III–V Epitaxial Layers. J Phys (Paris) Colloq. 1988;49:509–514. [Google Scholar]
- Miller MK. Atom Probe Tomography: Analysis at the Atomic Level. Kluwer Academic/Plenum Publishers; New York: 2000. [Google Scholar]
- Miller MK, Russell KF. FIB-based Atom Probe Specimen Preparation of Powders. Microsc Microanal. 2006;12:1294–1295. doi: 10.1017/S1431927607070845. [DOI] [PubMed] [Google Scholar]
- Miller MK, Russell KF, Thompson GB. Strategies for fabricating atom probe specimens with a dual beam FIB. Ultramicroscopy. 2005;102:287–298. doi: 10.1016/j.ultramic.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Miller MK, Cerezo A, Hetherington MG, Smith GDW. Atom Probe Field Ion Microscopy. Oxford University Press; Oxford: 1996. [Google Scholar]
- Mousa M, Wanderka N, Timpel M, Singh S, Kruger M, et al. Modification of Mo-Si alloy microstructure by small additions of Zr. Ultramicroscopy. 2010;111:706–710. doi: 10.1016/j.ultramic.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Muller EW. Resolution of the Atomic Structure of a Metal Surface by the Field Ion Microscope. J Appl Phys. 1956;27:474–476. [Google Scholar]
- Müller EW. Z Phys. 1951;131:136–142. [Google Scholar]
- Ostrowski SG, Van Bell CT, Winograd N, Ewing AG. Mass spectrometric imaging of highly curved membranes during Tetrahymena mating. Science. 2004;305:71–3. doi: 10.1126/science.1099791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panitz JA. Point-Projection Imaging of Macromolecular Contours. J Microsc. 1982;125:3–23. [Google Scholar]
- Panitz JA. Direct visualization of unstained nucleic acids on a metal substrate. Ultramicroscopy. 1983;11:161–6. doi: 10.1016/0304-3991(83)90232-2. [DOI] [PubMed] [Google Scholar]
- Panitz JA, Giaver I. Ferritin Deposition on Field-Emitter Tips. Ultramicroscopy. 1981;6:3–6. [Google Scholar]
- Perea DE, Allen JE, May SJ, Wessels BW, Seidman DN, et al. Three-Dimensional Nanoscale Composition Mapping of Semiconductor Nanowires. Nano Lett. 2006;6:181–185. doi: 10.1021/nl051602p. [DOI] [PubMed] [Google Scholar]
- Porter KR, Anderson KL. The structure of the cytoplasmic matrix preserved by freeze-drying and freeze-substitution. Eur J Cell Biol. 1982;29:83–96. [PubMed] [Google Scholar]
- Prosa TJ, Keeney SK, Kelly TF. Atom probe tomography analysis of poly(3-alkylthiophene)s. J Microsc. 2010a;237:155–67. doi: 10.1111/j.1365-2818.2009.03320.x. [DOI] [PubMed] [Google Scholar]
- Prosa TJ, Alvis R, Tsakalakos L, Smentkowski VS. Characterization of dilute species within CVD-grown silicon nanowires doped using trimethylboron: protected lift-out specimen preparation for atom probe tomography. J Microsc. 2010b;239:92–98. doi: 10.1111/j.1365-2818.2010.03375.x. [DOI] [PubMed] [Google Scholar]
- Seidman DN. Three-Dimensional Atom-Probe Tomography: Advances and Applications. Annu Rev Mat Res. 2007;37:127–58. [Google Scholar]
- Szakal C, Narayan K, Fu J, Lefman J, Subramaniam S. Compositional mapping of the surface and interior of mammalian cells at submicrometer resolution. Anal Chem. 2011;83:1207–13. doi: 10.1021/ac1030607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson K, Larson DJ, Ulfig R. Pre-sharpened and Flat-top Microtip Coupons: a Quantitative Comparison for Atom-Probe Analysis Studies. Microsc Microanal. 2005;11:882. [Google Scholar]
- Thompson K, Gorman BP, Larson DJ, vanLeer B, Hong L. Minimization of Ga Induced FIB Damage Using Low Energy Clean-up. Microsc Microanal. 2006;12:1736CD. [Google Scholar]
- Thompson K, Lawrence DJ, Larson DJ, Olson JD, Kelly TF, et al. In-Situ Site-Specific Specimen Preparation for Atom Probe Tomography. Ultramicroscopy. 2007;107:131–139. doi: 10.1016/j.ultramic.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Vurpillot F, De Geuser F, Da Costa G, Blavette D. Application of Fourier transform and autocorrelation to cluster identification in the three-dimensional atom probe. J Microsc. 2004;216:234–40. doi: 10.1111/j.0022-2720.2004.01413.x. [DOI] [PubMed] [Google Scholar]
- Wang MC, Min W, Freudiger CW, Ruvkun G, Xie XS. RNAi screening for fat regulatory genes with SRS microscopy. Nat Methods. 2011;8:135–8. doi: 10.1038/nmeth.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]