Summary
TRiC/CCT is a highly conserved and essential chaperonin that uses ATP cycling to facilitate folding of approximately 10% of the eukaryotic proteome. This 1 MDa hetero-oligomeric complex consists of two stacked rings of eight paralogous subunits each. Previously proposed TRiC models differ substantially in their subunit arrangements and ring register. Here, we integrate chemical crosslinking, mass spectrometry and combinatorial modeling to reveal the definitive subunit arrangement of TRiC. In vivo disulfide mapping provided additional validation for the crosslinking-derived arrangement as the definitive TRiC topology. This subunit arrangement allowed the refinement of a structural model using existing X-ray diffraction data. The new structure explains all available crosslink experiments, provides a rationale for previously unexplained structural features and reveals a surprising asymmetry of charges within the chaperonin folding chamber.
Introduction
The eukaryotic chaperonin TRiC/CCT (herein, TRiC) is essential for cell survival, employing ATP hydrolysis to fold ~10 % of the proteome (Yam et al., 2008), including many essential proteins such as cytoskeletal components and cell cycle regulators (Hartl et al., 2011; Spiess et al., 2004). The folding of many of these substrates is strictly dependent on TRiC. The TRiC subunits are related to the simpler archaeal chaperonin, the thermosome (Ditzel et al., 1998; Pereira et al., 2010; Shomura et al., 2004). Most thermosomes and TRiC consist of two eight-membered rings that are stacked back-to-back. Many archaeal species have just one thermosome gene (Zhang et al., 2010). In stark contrast, the eukaryotic complex consists of eight different, but related, subunits (CCT1 to CCT8), all of which are essential in yeast. The subunit specialization occurred very early in eukaryote evolution (Archibald et al., 2001), and is conserved to such an extent that the sequence identity between orthologous mammalian and yeast subunits of the same type is nearly 60%, whereas the sequence identity between paralogous subunits in the same organism is only about 30%. Each of the eight TRiC subunits may differ in substrate specificity; as a result, non-native polypeptides engage the chaperonin through combinatorial interaction with selected subunits (Feldman et al., 2003; Llorca et al., 2001; Munoz et al., 2011; Spiess et al., 2006). This mode of recognition dictates the topology of bound substrates, thereby influencing their folding trajectory (Douglas et al., 2011).
The original proposition for the TRiC subunit arrangement came from Western blot analysis of low molecular weight subcomplexes found in very low amounts in crude mammalian cell extracts (Liou and Willison, 1997). Similar electrophoretic mobility was used to infer neighbors in the intact complex. Although these low abundance entities were never characterized further, they remain the foundation for a large body of structural work on TRiC (Llorca et al., 2000; Llorca et al., 1999; Martin-Benito et al., 2004; Martin-Benito et al., 2007; Rivenzon-Segal et al., 2005), including the recent crystal structure of the closed conformation (Dekker et al., 2011). Under the assumption that the fragmentation was always preceded by dissociation into single rings, the incomplete data (subunit θ was apparently not part of any microcomplex) were consistent with the proposed arrangement, CCT 6-5-1-7-4-8-3-2 (i.e.TCP ζ-ε-α-η-δ-θ-γ-β). Later electron microscopy (EM) studies of TRiC with bound subunit-specific antibodies seemed to confirm this arrangement (Martin-Benito et al., 2007). Because of the complexity of the problem, the data employed was sparse, and the assignment of the subunits was only possible under far-reaching assumptions. The inherent ambiguity of the antibody decoration approach is underscored by the inability to predict the correct inter-ring register even from 3D-reconstructions of such complexes (Martin-Benito et al., 2007). Unfortunately, the quality of the subsequent electron microscopy and X-ray crystallographic data was not sufficient to unequivocally establish the correct subunit arrangement (Cong et al., 2010; Dekker et al., 2011; Martin-Benito et al., 2007).
Understanding the architecture and detailed mechanism of large multi-subunit complexes is commonly limited by this inability to obtain high-resolution structural information. In the absence of atomic resolution data, orthogonal structural information is needed for accurate interpretation. An emerging structure determination technique that has the potential to obtain a highly redundant, three-dimensional map of constraints is cross-linking coupled with mass-spectrometry (XL-MS) (reviewed in (Leitner et al., 2010; Rappsilber, 2011)). In this approach, the native protein complex is incubated with a cross-linking reagent capable of forming specific covalent bonds with exposed and frequently occurring sidechains. Most commonly, amine-reactive reagents such as disuccinimidyl suberate (DSS) for cross-linking of lysine residues are used, although a variety of reagents have been introduced (Petrotchenko and Borchers, 2010). Next, the complex is proteolytically digested and subjected to MS analysis for identification of the cross-linked peptides (Figure 1A). The cross-linked anchor sites provide a comprehensive three-dimensional map as a framework for molecular modeling. Previously, the application of the XL-MS approach had been limited to individual proteins and small complexes (reviewed by (Sinz, 2006)). Recent advances in MS instrumentation and the development of more powerful analysis software have permitted the application of XL-MS to a number of increasingly complex assemblies (Bohn et al., 2010; Chen et al., 2010; Maiolica et al., 2007; Schulz et al., 2007). Multi-subunit complexes studied by XLMS include the 26S proteasome (Bohn et al., 2010; Lasker et al., 2012), eukaryotic RNA polymerases (Chen et al., 2010), and the ribosome (Lauber and Reilly, 2011).
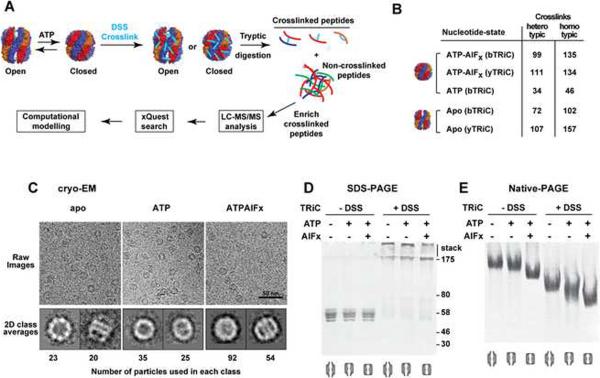
Figure 1. Mass spectrometry analysis of crosslinked TRiC yields specific intersubunit crosslinks.
(A) TRiC was incubated with or without nucleotide to generate the desired conformational state, treated with crosslinking reagent and proteolyzed to generate an ensemble of crosslinked and non-crosslinked peptides. Crosslinked peptides were chromatographically enriched and analyzed by LC-MS-MS. The identity of the peptides and anchor lysine residues was determined using xQuest (Rinner et al., 2008). Validated crosslinks were used for TRiC model building. (B) Summary of crosslinks identified using TRiC purified from two different species, bovine (bTRiC) and yeast (yTRiC). (C) Cryo-EM imaging evidence for the structural integrity of crosslinked TRiC in the apo (left), ATP (middle) and ATP+AlFx (right) states. Top and bottom panels: representative cryo-EM images and corresponding characteristic top and side views of the reference-free 2D class averages of the cross-linked TRiC; numbers of raw particle images used to derive the averages are indicated. (D–E) SDS- (D) and Native-PAGE (E) analysis of bTRiC in indicated nucleotide states without (lanes 1–3) or with (lanes 4–6) crosslinking. See also Figure S1 and Table S2.
We used the XL-MS approach to investigate the order and orientation of the 16 subunits in the 1 MDa complex TRiC/CCT. Structural data of TRiC has been obtained at near-residue resolution, 4.0 and 3.8 Å, by single-particle averaging cryo-electron microscopy (cryo-EM) and X-ray crystallography (Cong et al., 2010; Dekker et al., 2011). The derived models agree in that both rings have a specific subunit order, and that the two rings are related by 2-fold symmetry, creating two homomeric contacts across the equator. However, the proposed subunit orders completely disagree (CCT 6-5-1-7-4-8-3-2 versus CCT 8-4-5-7-1-6-2-3, for Dekker et al and Cong et al, respectively). Here, we resolve this issue by the orthogonal XL-MS approach, and present the definite model for the TRiC/CCT structure.
Results
Crosslinking tandem mass spectrometry approach
Our experimental strategy (Figure 1A) exploited recent advances in chemical crosslinking combined with mass spectrometry (Rinner et al., 2008) to identify residues in close spatial proximity in functionally competent TRiC/CCT complexes. These distance constraints then guided the selection of the most likely subunit arrangement by molecular modeling. The number of distance constraints was maximized by applying this strategy to TRiC purified from two evolutionary distant organisms, Bos taurus and Saccharomyces cerevisiae (bTRiC and yTRiC). At the peptide level, the complexes from each species are expected to yield virtually unrelated tryptic cleavage products. Furthermore, approximately 40% of the surface lysine positions available for crosslinking are scrambled between the bovine and yeast orthologs, resulting in an improved sampling of the subunit surfaces (Table S1).
Nucleotide-free TRiC is conformationally highly heterogeneous, resulting in greater structural ambiguity. ATP hydrolysis leads to a more compact state, whereby a built-in lid closes over the central TRiC folding chamber (Meyer et al., 2003). To facilitate the subsequent modeling analysis, TRiC was crosslinked following incubation with ATP or ATP+AlFx; both conditions induce the closed conformation for which highly reliable structural models derived from archaeal chaperonins exist (Ditzel et al., 1998; Pereira et al., 2010; Shomura et al., 2004). Native protein complexes were incubated with two different isotopically labeled forms of DSS (Muller et al., 2001), which crosslinks exposed primary amino groups found in lysine side chains and polypeptide N-termini. The complex was then digested with trypsin and samples enriched for crosslinked peptides (Leitner, 2012) were analyzed by capillary liquid chromatography tandem mass spectrometry (LC-MS-MS) and the resulting complex fragment ion spectra were assigned to the corresponding peptide sequences using xQuest (Rinner et al., 2008) (Figure 1A). Under our experimental conditions the extent of lysine modification approached saturation. For example, yTRiC has a total of 334 lysines and of these, 151 were involved in crosslinks in the corresponding ATP−AlFx dataset. Furthermore, many crosslinks were identified by multiple peptide pairs. Overall, we identified 997 peptide pairs across all experiments with an estimated false discovery rate (FDR) of less than 5% (Figure 1B and Table S2). They consisted of 423 heterotypic crosslinks i.e. crosslinks between different subunits in the TRiC complex, and 574 homotypic crosslinks, i.e. crosslinks within the same subunit or between two identical subunits. Of the 423 heterotypic crosslinks, 302 mapped to likely ordered parts of the subunit homology models; these were used for determining the overall topology of the complex (see below and Figure S2A). The remainder mapped primarily to the unstructured N- and C-terminal tails (Figure S2A).
Integrity of the complex during crosslinking
To verify that the complex integrity was not affected by crosslinking, we assessed the conformation of crosslinked and native TRiC by EM and gel electrophoresis (Figures 1C–E and S1). bTRiC samples incubated with or without ATP or ATP+AlFx were analyzed before and after DSS treatment. Two-dimensional class averages of cryo-EM single particles of TRiC indicated that the conformations before (Cong et al., 2010) and after crosslinking were virtually indistinguishable at low resolution (Figure 1C, bottom panel). Thus, TRiC integrity was not detectably compromised by crosslinking. SDS-PAGE of DSS-crosslinked TRiC yielded high molecular weight species consistent with full crosslinking of all TRiC subunits (Figure 1D). DSS-treated TRiC migrated as a single band in native gels, indicating the stabilization of a coherent complex population (Figure 1E). The faster migration of DSS-treated TRiC is expected due to the overall reduction in charge by the crosslinker. Of note, the ATP and ATP+AlFx - induced closed states exhibited a characteristic mobility shift, consistent with the cryo-EM analysis. Similar results were obtained for yTRiC (Figure S1). We conclude that that the crosslinks identified in this study are derived from structurally intact chaperonin complexes.
Mapping of the crosslinks onto a structural model
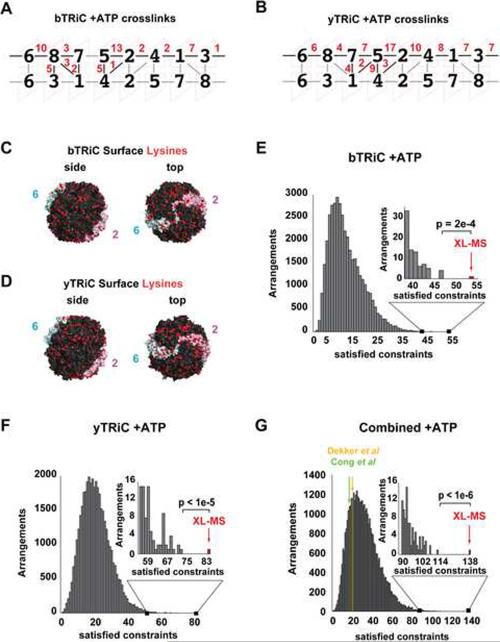
The identified intermolecular crosslinks were next employed as spatial constraints to derive the most likely TRiC/CCT subunit arrangement (Figure 2 and S2). Homology models were first generated for each of the eight subunits using the crystal structure of the related archaeal chaperonin from Methanococcus maripaludis in the nucleotide bound state (Pereira et al., 2010). The crosslinked lysine positions obtained in the ATP and ATP+AlFx-induced states were then mapped onto the homology models. Of note, only heterotypic crosslinks that mapped to ordered parts of the structure were used in the subsequent calculations to evaluate the compatibility of different geometries between two different subunits (i.e. crosslinks involving residues in loops of unclear conformation and flexible tails were discarded, see Experimental Procedures and Figure S2A). Importantly, identical results were obtained using other archaeal group II chaperonin structures as templates (see below, Figure S3). For each pair of crosslinked subunits, the fifteen possible pairwise orientations in the hexadecamer were generated (Figure S2B) and the respective lysine distances calculated (Figure S2C,D). The contour length between two Cα atoms of DSS-crosslinked lysines is approximately 24 Å (Muller et al., 2001). We applied a slightly longer Cα-Cα distance cut-off of 30 Å to account for protein dynamics and potential model inaccuracies (see also below, Figure S3). We also checked whether these crosslinks were physically possible, eliminating any crosslinks that would traverse the protein core. For the complexes of both species, the same unique TRiC/CCT subunit order, namely CCT 6-8-7-5-2-4-1-3 (Fig. 2a; i.e. TCP ζ-θ-η-σ-β-δ-α-γ), was obtained. Both rings are related by 2-fold symmetry, as predicted by previous structural analysis, with CCT6/ζ and CCT2/β engaging in homotypic inter-ring contacts. This subunit arrangement, determined by XL-MS, was thus independently determined from two unrelated datasets for TRiC from two evolutionarily distant species (Figure 2A,B). Of note, the heterotypic crosslinked peptides were different in yTRiC and bTRiC; this likely reflects the variability of surface exposed lysines in the two TRiC complexes (Figure 2C, D). The set of unambiguous crosslinks was complete for the closed conformation of yTRiC: Every directional intra-ring neighbor pair relationship and the inter-ring register was established by individual crosslinks (Figure 2B). For bTRiC, only one intra-ring neighbor pair (CCT5-CCT7) relationship was not directly established by crosslinks (Figure 2A). In case of the closed conformation dataset of yTRiC, each intra-ring subunit contact was established by at least four different crosslinked peptide pairs. Thus a wrong assignment of any individual neighbor pair relationship at an FDR of 5 % is highly unlikely (probability 6.25 10−6 or less). This shows that the assignment must be correct beyond reasonable doubt.
Figure 2. Mass spectrometry derived constraints reveal the TRiC subunit arrangement.
(A–B) Subunit arrangement for (A) bTRiC and (B) yTRiC derived from datasets for the closed state. CCTx subunits are shown as black numbers. The total number of heterotypic crosslinks supporting this arrangement is denoted in red. (C–D) Surface representation of the bTRiC and yTRiC complexes, showing the surface distribution of lysines (shown in red) (see also Supplementary Table 1); CCT2 (cyan) and CCT6 (pink) are highlighted for orientation. (E–G) Combinatorial analysis of the heterotypic crosslinking constraints. A histogram showing the distribution of numbers of constraints satisfying the 30 Å cutoff in each conceivable arrangement for closed bTRiC (E), closed yTRiC (F) and the combined datasets (G). Inset: right tail of the distribution. The XL-MS arrangement satisfies the largest number of constraints (indicated by red arrow), which are 54 of 64, 84 of 102 crosslinks for the bTRiC, yTRiC closed state datasets, respectively; i.e. 138 of total 166 for the combined closed state datasets. XL-MS p-value indicates statistical significance over the second best arrangement. The previously proposed arrangements (Cong et al; Dekker et al) are consistent with only 17 (green) and 23 (yellow) of the 166 crosslinks in the combined bTRiC and yTRiC closed state datasets. See also Figure S2 and Tables S1 and S3.
Combinatorial analysis of distance constraints
The statistical significance of the arrangement determined by XL-MS as the unique solution to the experimental distance constraints was further investigated by an unbiased combinatorial approach that determined the number of constraints satisfied for each of the 40,320 possible subunit arrangements (see Experimental Procedures for details). This approach explicitly evaluated the ambiguity of several plausible pairs of subunit orientations satisfying a given distance cut-off (see Experimental Procedures section for details) (Figure 2 and Table S3). The distribution of arrangements satisfying these constraints is shown for both the individual (Figures 2E, F) and the combined closed TRiC datasets (Figure 2G) and demonstrates that the arrangement determined by XL-MS is the only subunit ordering that can explain the majority of the heterotypic crosslinks, satisfying 85% (Figure 2E) and 82% (Figure 2F) of the crosslinks for the individual datasets and 83% for the combined dataset (Figure 2G). The secondary solutions (see Table S3 and Experimental Procedures section for details) are significantly worse than the XL-MS determined arrangement; indeed the correctness of the XL-MS determined arrangement is statistically significant, relative to the second best arrangement, with p-values of 2×10−4 and <10−5, respectively, for the bovine and yeast datasets. Combining the yTRiC and bTRiC data increased the statistical significance of the XL-MS determined arrangement (p-value <10−6; Figure 2G) with respect to the second best arrangement. Importantly, the previously proposed TRiC subunit arrangements (Cong et al., 2010; Dekker et al., 2011) explain only a minor fraction (10% and 13%, respectively) of the observed crosslinks (Figure 2G), and thus are essentially incompatible with our extensive crosslink dataset.
Application of XL-MS analysis to the dynamic open state of TRiC
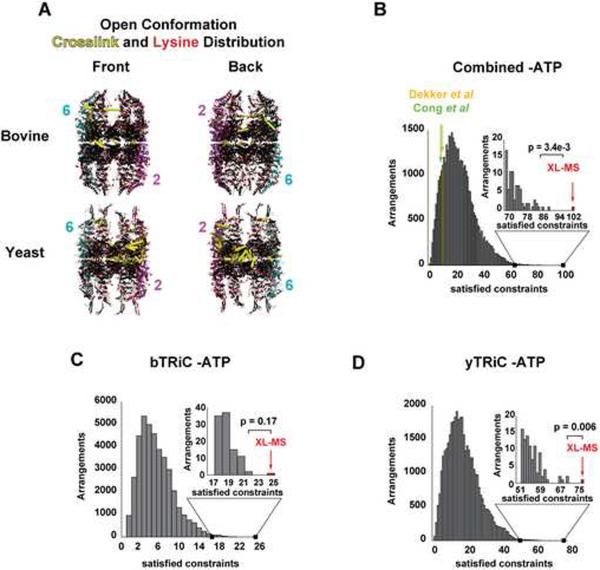
To assess whether the XL-MS and modeling strategy can be applied to structurally less well-defined complexes, we next analyzed crosslinks obtained for the more flexible open state of TRiC without nucleotide using the coordinates of the open state of Mm-Cpn as a model (Pereira et al., 2010) (Figure 3). For both bTRiC and yTRiC a similar number of identified peptide pairs was obtained as in the closed state (Figures 1E and S2A), but fewer constraints passed the 30 Å distance cut-off, particularly for the highly dynamic apical domains (Figure 3A, yellow lines). To account for the increased flexibility of the open state, and the lower confidence level of available structural models, the distribution of matching crosslinks over the considered models was computed using a 36 Å distance cutoff (Figure 3B–D). This analysis also yielded the XL-MS determined arrangement as the best solution, satisfying 75% of the crosslinks (p value 3.4×10−3, 6.1×10−3and 0.17 for the combined yTRiC and bTRiC datasets, respectively; see Figure 3B–D, Table S3 and Experimental Procedures for details), highlighting the power of our cross species strategy to model the subunit topology even for structurally flexible, less well-characterized complexes. As shown below (Figure S8), these larger distances likely reflect inadequacies of our initial homology model.
Figure 3. Global analysis of mass spectrometry derived constraints for TRiC in the open conformation.
(A) Mapping the crosslinked lysines (yellow lines) onto open state models of bTRiC or yTRiC (colored as in Figure 2C,D). The crosslinks preferentially map to the equatorial domains, consistent with increased flexibility of the apical domains in the open state. (B–D) Combinatorial analysis of heterotypic crosslinking constraints from open conformation data. The number of constraints satisfying the 36 Å cutoff in each conceivable arrangement is shown as a histogram for (B) combined open bTRiC and yTRiC, (C) open bTRiC and (D) open yTRiC dataset. Inset: right tail of the distribution. The XL-MS arrangement satisfies the largest number of constraints (indicated by red arrow), for the three respective datasets these are 102 of 136 (combined), 25 of 36 (bTRiC) and 77 of a total of 100 (yTRiC). The p-value indicates statistical significance of XL-MS over the second best arrangement. The previously proposed arrangements (Cong et al; Dekker et al) are consistent with only 10 (green) and 11 (yellow) of the 136 crosslinks in the combined bTRiC and yTRiC closed state datasets. See also Figure S3 and Table S3.
To systematically explore how the choice of template and distance cutoff influence our analysis, we next computed the number of satisfied constraints as a function of distance using the different available group II chaperonin structures as templates (Figure S3) (Ditzel et al., 1998; Pereira et al., 2010; Shomura et al., 2004). For the closed datasets, this analysis indicated a clear convergence between 24 Å and 30 Å (Ditzel et al., 1998; Pereira et al., 2010; Shomura et al., 2004) (Figure S3B–F). Notably, the quality of the optimal arrangement was not sensitive to the exact structural group II chaperonin template employed to build the models (Figure S3). For longer distance cut-offs the number of satisfied constraints approached the total number of constraints but decreased the discrimination between the optimal arrangement and the median of random solutions (data not shown), supporting our choice of distance cut-off (Figure S3BF).
The refined XL-MS structural model
Prior attempts to generate an accurate structural model for TRiC/CCT were confounded by the low resolution of available cryo-EM and X-ray data. The previous cryo-EM model was based on the visual analysis of density features in the apical domains (Cong et al., 2010). Reanalysis of these cryo-EM data (Cong et al., 2010) with more quantitative and statistical procedures (see Table S4) suggests that the quality of the map suffices for rough backbone tracing but lacks the resolvability to distinguish the highly similar TRiC subunits, so this previous interpretation has to be revoked. The X-ray diffraction data from the closed conformation suffer from model bias since no experimental phases are available. We refined a structural model representing the XL-MS determined subunit arrangement against these X-ray diffraction data, carefully avoiding overt model bias (see Supplemental Experimental Procedures for details) (Dekker et al., 2011). Our final XL-MS structural model has clearly improved refinement statistics and model geometry compared to the published model based on the original subunit topology (Table 1). Strikingly, unanticipated features of the refined XL-MS-based structure provide a rationale for several crosslinks mapping to regions not included in the homology model. Indeed, the refined XL-MS-derived structure could explain approximately 94% of heterotypic (Figure 4A, B) and 97% of homotypic (data not shown) crosslinks, according to the 30 Å criterion. This is much better than the thermosome-based homology models. Thus, the new XL-MS-based structure explains virtually all experimentally obtained crosslinks; the fraction of outliers corresponds to the 5% FDR for the MS assignment.
Table 1.
Crystallographic refinement statistics and model geometry.
| Model | 3P9D + 3P9E (Dekker et al.) | XL-MS (Refmac, NCS, no TLS) | XL-MS (Refmac, NCS, TLS) |
|---|---|---|---|
| resolution limits | 30 – 3.8 | 30 – 3.8 | 30 – 3.8 |
| Rwork / Rfree | 0.3178 / 0.3513 | 0.2696 / 0.3279 | 0.2568 / 0.3046 |
| Figure of merit | 0.672 | 0.715 | 0.751 |
| number of atoms | |||
| Protein | 110444 | 119056 | 119056 |
| Ligand/ion | 784 | 1024 | 1024 |
| Water | 7 | 0 | 0 |
| average B factors | |||
| Protein (Å2) | 141 | 125 | 139 |
| Ligand / ion (Å2) | 130 | 103 | 123 |
| Water (Å2) | 43 | - | - |
| r.m.s. deviations | |||
| bonds (Å) | 0.012 | 0.007 | 0.007 |
| angles (°) | 0.986 | 1.052 | 1.068 |
| Ramachandran plot | |||
| % preferred (Coot) | 85.8 % | 89.5 % | 90.1 % |
| % outliers (Coot) | 4.68 % | 3.16% | 2.89 % |
| number non-Proline cis peptides | 184 | 0 | 0 |
In order to allow a fair comparison with the original model (Dekker; PDB codes 3P9E/3P9D), the XL-MS model was also refined without TLS B-factor parameterization (middle column). The statistics for the Dekker model were determined using Refmac using the default values from CCP4i.
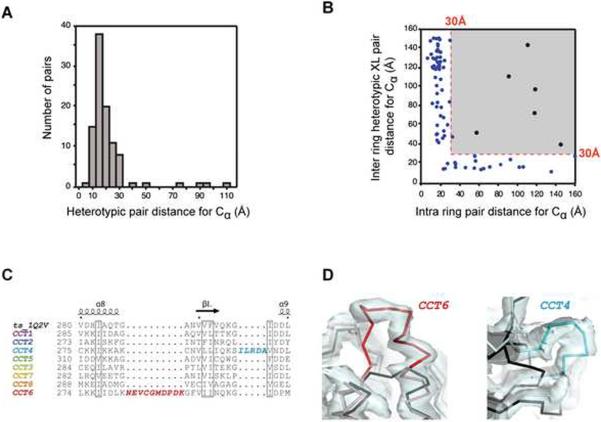
Figure 4. Cross-validation of crystal structure and crosslink data for yTRiC.
(A) Distance distribution for the closed state yTRiC heterotypic crosslink dataset. The median heterotypic Cα-Cα crosslink distance in the model is 16.4 Å. (B) Heterotypic crosslink Cα pair distances for inter ring and intra ring subunit pairings observed in the refined XL-MS-based crystal structure. The crosslinks compatible with the XL-MS arrangement are highlighted in blue; crosslinks mapping to the grey box exceed the cutoff. (C) Alignment showing unique insertions in yTRiC subunits CCT6 and CCT4. (D) Unbiased 2Fo-Fc electron density for these insertions at 1 σ. The thermosome structure is shown in black for comparison. See also Figure S4 and Table S4.
The XL-MS-derived structure is also more plausible with regard to TRiC sequence features. The new model accounts for several large insertions unique to individual TRiC subunits, which are well-defined in the electron density. For instance, CCT6 has a unique 10-residue insertion after helix α8 (residues 282–291), which elongates this helix by two turns (Figure 4C, D). This feature is clearly discernible in unbiased difference maps (Figures 4D and S4A). The XL-MS model furthermore explains structurally defined distinctive insertions in CCT4 (residues 291–295 and 371–374), CCT1 (341–345 and 484–495) and CCT6 (481–485) (Figures 4C, 4D, S4B and not shown). In the construction of the Dekker model, these aberrant density features, which are clearly present in the map, had been mostly ignored (Figure S4).
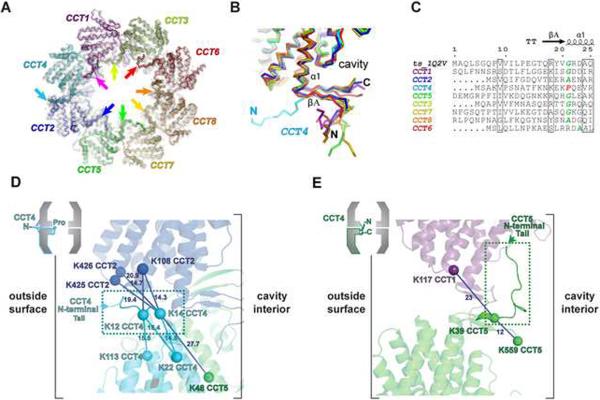
Another striking finding of our model is that most of the N-termini preceding strand β1 are resolved in the density. This revealed two unexpected structural features validated by crosslinking data. First, we find in our model that CCT4 is the single subunit that has an outward pointing N-terminal density in the map (Figure 5A). In contrast, CCT5 was the corresponding subunit with an outward pointing N-terminus in the original model (Dekker et al., 2011). Strikingly, CCT4 is the only CCT subunit that has a conserved proline at the N-terminal junction to helix α1 (Figures 5B, C and S5A). This provides an evolutionary and structural rationale of why CCT4 is the only CCT subunit with an outward pointing N-terminus, explaining the aberrant density (Figure 5A). In contrast, CCT5 has a glycine at this position, as do most other TRiC subunits and archaeal subunits (Figure 5C), all of which have inward pointing N-termini (Figure 5A). Of note, the outward conformation of the CCT4 N-terminus is strongly corroborated by a series of crosslinks within our dataset, establishing contacts of K12 and K14 to residues on the complex exterior (Figure 5D). These crosslinks are incompatible with an inward facing N-terminus, but are entirely consistent with the subunit docking and the CCT4 sequence data. Similarly, crosslinks between the N-terminus of CCT5 and residues on the cavity walls support the location of the CCT5 N-terminus inside the complex (Figure 5E). Altogether, these observations ascertain the validity of the XL-MS model.
Figure 5. Features of the TRiC crystal structure model based on the XL-MS subunit order.
(A) Electron density for XL-MS crystal structure model. The view from the equator shows the cavity of one ring. The final 2Fo-Fc density at 1.5 σ is shown as meshwork. The N-terminal β-strands of TRiC subunits 1, 2, 3, 5, 6, 7 and 8 are highlighted by arrows. The N-terminus of CCT4 (cyan) is inserted between CCT4 and CCT2. Please note that side chain density is hardly visible at all, and thus cannot be used for sequence docking. (B) Superposition of the yeast TRiC subunits, highlighting the aberrant CCT4 geometry at the N-terminus (cyan). (C) Alignment of the N-terminal sequences of the thermosome and the yTRiC subunits. The junction residue between βA and α1 is shown in italics, highlighting residues compatible (green) or incompatible (red) with the thermosome geometry. The sharp transition is also facilitated by small helix residues facing the β-strands, as observed in CCT6. Numbering and secondary structure elements refer to the thermosome structure (PDB code 1Q2V (Shomura et al., 2004)). (D) Validation of the CCT4 N-terminus geometry by crosslinking. The location of the CCT4 N-terminal tail (dashed box) is corroborated by specific crosslinks to residues on the outside surface. The backbones of CCT2, CCT4 and CCT5 are shown in blue, cyan and green, respectively. The Cα atoms of lysines are indicated by spheres, and crosslinks in between represented by dashed lines. The distance between lysine Cα's is denoted in Å. (D) Localization of the CCT5 N-terminus in the cavity by crosslinking. The location of the CCT5 N-terminal tail (dashed box) is corroborated by specific crosslinks to residues on the cavity surface. CCT1 and CCT5 are indicated in magenta and green, respectively. See also Figure S5.
The new TRiC structure also provides unanticipated insights into inter-ring interactions between the N-termini of CCT1 and CCT8. In the crystal structure there is an extensive direct interaction between the N-termini of the CCT8 subunits across the equator (Figure S5B). Perhaps these unique structural features help to correctly establish the subunit topology in TRiC by stabilizing the ring-ring interface. They might also contribute to allosteric rearrangements during the functional cycle. The extensive interactions between the CCT8 N-termini are consistent with previous crosslinking and 2D-gel data (Cong et al., 2010), which had suggested direct contacts between CCT8 subunits (Figure S5C, D). Indeed, all the crosslinks observed in Cong et al, by themselves insufficient to unambiguously determine the correct arrangement, are fully consistent and explained by the XL-MS architecture.
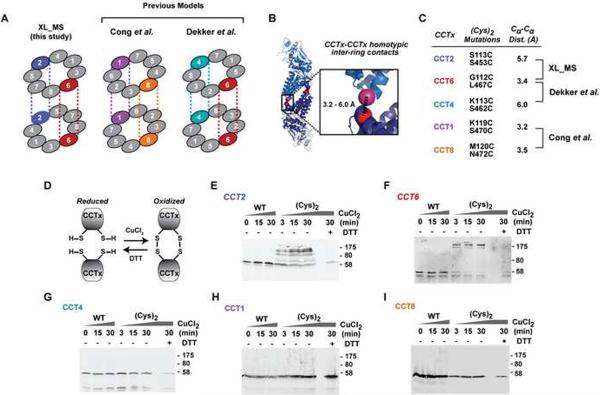
In vivo validation of XL-MS architecture using disulfide mapping
To independently validate the intra-ring subunit order and inter-ring subunit register determined by XL-MS we next employed in vivo near-neighbor disulfide engineering (Figures 6 and S6). The XL-MS determined arrangement predicts that subunits CCT2 and CCT6 form inter-ring homotypic contacts (Figures 2AB and 6A). Previous models predict homotypic contacts for either CCT4 and CCT6 (Dekker et al., 2011) or CCT1 and CCT8 (Cong et al., 2010) (Figure 6A). We engineered cysteine pairs at residues predicted to be proximal (Cα-Cα <6 Å) in a homotypic inter-ring interface, and thus permitting disulfide bond formation (Figure 6B, C). Importantly, the yTRiC inter-ring interface is otherwise free of cysteines. The CCTx-(Cys)2 genes supported normal growth of yeast lacking the corresponding wild type gene (Figure S6A). Disulfide crosslinking of TRiC obtained from CCTx-(Cys)2 cells was induced by oxidation with CuCl2 (Figures 6D and S6D). As predicted by the XL-MS-based model, disulfide-crosslinked dimers occurred in a time- and oxidant-dependent manner only in TRiC from CCT2-(Cys)2 and CCT6-(Cys)2 cells (Figure 6E, F). No such dimers were observed for CCT1-(Cys)2, CCT4-(Cys)2 and CCT8-(Cys)2 (Figure 6F, H, I), indicating that these subunits do not form homotypic contacts in TRiC. In conjunction with the wealth of evidence from the crosslinking distance constraints and crystallographical analysis, this orthogonal in vivo approach definitively validates the XL-MS-derived arrangement as the correct topology of TRiC across eukaryotes.
Figure 6. In vivo validation of the inter-ring register using disulfide crosslinking.
(A) Different TRiC models predict distinct pairs of homotypic contacts: XL-MS (this study) proposes CCT2 and CCT6 inter-ring pairs; previous studies proposed CCT1 and CCT8 (Cong et al., 2010) or CCT4 and CCT6 pairs (Dekker et al., 2011). (B) Model of the inter-ring interface highlighting residues substituted by cysteines for disulfide bond formation. (C) Summary of relevant cysteine replacements and inter-Cys distances. All CCTx-(Cys)2 subunits support wild type growth (Supplementary Fig. S5). (D) Near-neighbor disulfide mapping: symmetrically related cysteine pairs will form disulfide bonds under oxidizing conditions (CuCl2), which are reversed with the reducing agent DTT. (E–I) Incubation under oxidizing conditions reveals subunits CCT2-(Cys)2 and CCT6-(Cys)2 form DTT-sensitive disulfide dimers, while WT subunits, and the (Cys)2 variants of subunits CCT4, CCT1 and CCT8 do not. See also Figure S6.
Discussion
Previous attempts to define the TRiC topology have been mired in controversy due to the pseudo-symmetry of the complex and confounded by methodological limitations. To resolve this long-standing problem we developed and applied a crosslinking tandem mass spectrometry approach to generate two complete and self-consistent sets of constraints to model the topology of the eukaryotic chaperonin TRiC. These data unambiguously assign the intra-ring subunit order in the TRiC complex and invalidate the previously proposed arrangements. Importantly, the XL-MS-derived model is also consistent with previous crosslinking data (Figure S5) and likely compatible with the subunit spacing derived from 3D cryo-EM reconstructions of TRiC decorated with antibodies (Martin-Benito et al., 2007).
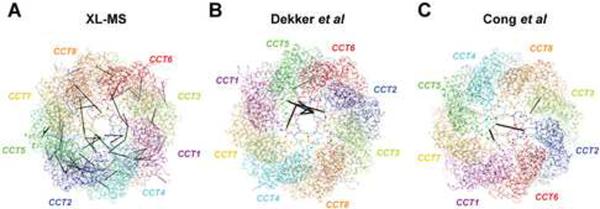
Importantly, the prior models of TRiC are entirely incompatible with our data, because their subunit orders diverge significantly from ours (Cong et al., 2010; Dekker et al., 2011). Figure 7 shows the crosslinks obtained from the closed conformation of yTRiC or bTRiC mapped onto the three respective final structure models. It is evident that while the XL-MS model explains ~95% of the obtained crosslinks, only a small fraction of the crosslinks fit to the previous models. The few consistent inter-subunit crosslinks locate close to the apical pore where all eight subunits meet, i.e. these ambiguous crosslinks fit to the majority of conceivable subunit topologies. In contrast, XL-MS data is consistent with the previously reported crosslinking data from Cong et al, which alone cannot discriminate between the Cong et al and XL-MS-derived models (Figure S5).
Figure 7. Consistency of TRiC structural models with crosslinking data.
Heterotypic crosslinks obeying the 30 Å criterion were mapped onto ribbon representations of the XL-MS (A), Dekker et al. (B) and Cong et al. (C) structural models of TRiC. See also Figure S7 and Supplementary Movie 1.
The subunit docking into the density of the original crystallographic yTRiC model seemed to be corroborated by antibody binding to a FLAG epitope fused to the exposed N-terminus of CCT5 in the presence of ATP (Dekker et al., 2011). However, yeast has an anomalously long CCT5 N-terminal peptide that could easily reach out from the cavity through the apical opening (Figure S7). Because pore closure in TRiC is not stringently induced by the addition of only ATP, it allows transient exposure to the antibody, which would explain the reported experimental result. Our crosslinking data on the closed conformation of yTRiC unambiguously show that the N-terminus of CCT4 is located on the exterior surface of the complex, close to the equator of the complex (Figure 5D), whereas the N-terminal segment of CCT5 was involved in crosslinks to the interior (Figure 5E). Taken together with the conserved proline in the CCT4 N-terminus, this provides strong evidence for the XL-MS model and against the Dekker subunit docking.
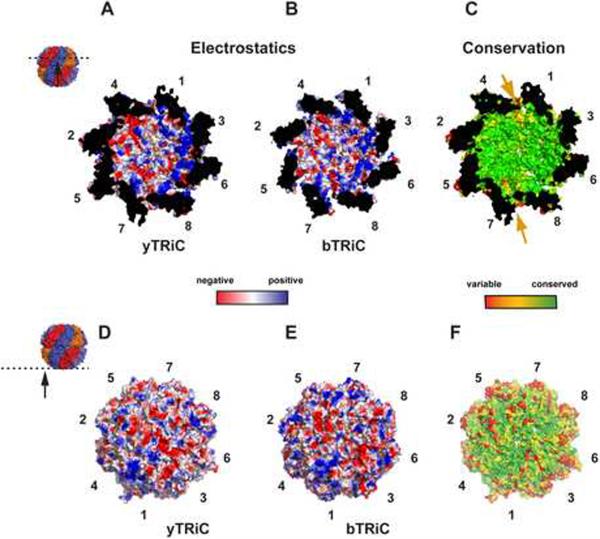
The XL-MS-derived model of the eukaryotic chaperonin uncovers unexpected structural features instrumental to understand its function. Strikingly, it shows that the conserved and highly charged surface of the closed chamber of TRiC has a conspicuous segregation of positive and negative charges contributed by subunits CCT5-2-4 and CCT3-6-8, respectively, and results in a bipolar distribution within the folding chamber (Figure 8A, B). The high conservation of the inner surface suggests functional importance in the folding of encapsulated substrate proteins (Figure 8C). Indeed, the bacterial chaperonin GroEL has a negatively charged chamber that is critical for folding (Tang et al., 2008). In comparison, the charge patterning on the outside surface of TRiC is less conserved (Figure 8D–F). The least conservation within the chamber occurs at the interface between the positive and negative hemispheres, likely reflecting interspecies variation in the charge asymmetry boundaries (see arrow in Figure 8C).
Figure 8. Chemical properties of the XL-MS derived TRiC chaperonin structure.
Analysis of the electrostatic charge distribution of yeast (A, D) and bovine (B, E) TRiC complexes. (A,B) The folding chamber for yTRiC and bTRiC reveals a striking asymmetry of charged residues on the inside of the cavity, where subunits CCT1-CCT3-CCT6-CCT8 are positively charged (blue) and subunits CCT7-CCT5-CCT2-CCT4 are neutral (white) or acidic (red). (D,E) In contrast, the outside surface of yTRiC and bTRiC show moderate conservation of charged residues. (C,F) Surface conservation of TRiC. The similarity scores from aligning each 100 orthologous sequences were mapped onto the yTRiC structure. A color gradient from green to red indicates decreasing conservation. The internal cavity surface is strikingly conserved. Interestingly, interfacial regions between pairs of subunits (CCT4/CCT1 and CCT7/CCT8) are less conserved as indicated by arrows. Consistent with the charge variability between bTRiC and yTRiC, the outside surface of the TRiC complex is not highly conserved across orthologs. See also Figure S8.
An interesting feature that is shared between the EM and X-ray structures of the open TRiC conformations is pairwise association of the apical domains, yielding a four-fold pseudo-symmetry (Cong et al., 2011; Munoz et al., 2011) (Figure S8). This is also apparent in our open conformation datasets: In the yTRiC dataset, we find multiple crosslinks between the apical domains of CCT1–3 (4 crosslinks), CCT6–8 (2), CCT7–5 (3), and CCT2–4 (6), but only one or no crosslinks for the other apical intra-ring pairs. The pattern is less pronounced in the bTRiC open state dataset. These open-state apical domain contacts may help propagate allosteric rearrangements throughout the ring (Reissmann et al., 2007; Rivenzon-Segal et al., 2005).
In the light of the new XL-MS-derived topology earlier data on CCT-substrate and CCT-cofactor complexes will have to be reinterpreted (Dekker et al., 2011; Llorca et al., 2000; Llorca et al., 1999; Munoz et al., 2011) (Cuellar et al., 2008; Martin-Benito et al., 2004). Here we examine only the crystallographic information on tubulin binding (Dekker et al., 2011; Munoz et al., 2011). The position of the two-fold inter-ring axis cannot be directly derived from the crystal structure of the TRiC-tubulin complex because of extensive disorder in one ring (Munoz et al., 2011). However, comparison with the EM structure of TRiC in the open conformation (Cong et al., 2011) suggests that the subunit with the most retracted apical domain orients perpendicular to the axis (subunit 3 in (Cong et al., 2011), chain G in (Munoz et al., 2011)), i.e. should be assigned either CCT1 or CCT7, and consequently the tubulin density sits on top of the axis. The reported crosslink between tubulin and the C-terminus of CCT2 (Munoz et al., 2011) suggests that tubulin interacts with the equatorial domains of TRiC subunits CCT5-2-4 and the aberrant apical domain belongs to CCT7 (Figure S8B). Interestingly, tubulin appears to bind near the negatively charged region of the cavity. In contrast, we could not detect meaningful density for actin in the cavity of the closed state crystal structure, unlike previously reported (Dekker et al., 2011). This suggests that TRiC associated actin present in the crystal may be poorly ordered.
The unequivocal solution to the TRiC/CCT topology will prove critical to understand its assembly, mechanism and allosteric regulation. The XL-MS-derived model reveals a surprising degree of asymmetry in this ring-shaped chaperonin, for the surface properties of the chamber and probably also for allosteric transitions and substrate binding. The conserved hetero-oligomeric structure of TRiC provides the structural basis for these asymmetric features. This study highlights the power of mass spectrometry-guided approaches to facilitate structural modeling of hetero-oligomeric complexes. Accurate model building of many large dynamic macromolecular complexes using data from X-ray crystallography and cryo-EM alone is often extremely difficult. The successful application to the challenging case of the pseudo-symmetrical TRiC/CCT suggests that XL-MS, in combination with low-resolution structural data and computational modeling can reveal the topology of other complexes, even if they consist of highly homologous subunits.
Experimental Procedures Summary
bTRiC was purified as described previously (Feldman et al., 2003); yTRiC was affinity-purified using His6- and Strep-tagged Plp2p, followed by Heparin affinity and Superose-6 size exclusion chromatography. DSS-treated TRiC complexes were characterized by SDS-PAGE, native-PAGE and cryo-EM 2D class averages to confirm the structural integrity of the crosslinked complex. DSS-crosslinked TRiC samples were treated with trypsin, enriched for crosslinked peptides by size exclusion chromatography and analyzed by tandem mass spectrometry. Crosslinked peptides were identified by xQuest (Rinner et al., 2008). The anchor lysine residues were mapped onto homology models of bTRiC and yTRiC subunits arranged in all pairwise subunit combinations (representing 15 possible spatial orientations) and Cα-Cα distances were computed. The distance matrix was used to evaluate all possible arrangements of the hexadecameric complex and deduce the best arrangement. A parametric bootstrap test was used to evaluate the significance of the best with respect to the second best arrangements as simulated according to a binomial distribution function. Plasmids of the indicated yTRiC subunits containing introduced cysteine pairs (Cys)2 at putative homotypic interface contacts were inserted in the respective cctxΔ by plasmid shuffling; the corresponding TRiC complexes were tested for the formation of specific disulfide bonds using SDS-PAGE and western blot. The XL-MS topology model was refined against the deposited crystal structure factors (Dekker et al., 2011) using Refmac (Murshudov et al., 1997). For manual model editing, Coot was employed (Emsley and Cowtan, 2004). Ortholog CCT sequences were retrieved from NCBI (Sayers et al., 2009), aligned using ClustalW (Thompson et al., 1994) and the conservation scores were calculated using Rate4site (Pupko et al., 2002), mapped onto the XL-MS structure using Consurf (Ashkenazy et al., 2010) and visualized using Pymol.
Supplementary Material
Highlights
Model explaining structural and functional asymmetry of hetero-oligomeric TRiC/CCT.
Chemical crosslinking, mass spectrometry and modeling reveal the architecture of TRiC.
In vivo disulfide mapping confirms the crosslinking based TRiC subunit topology.
The new structural model explains all available data and uncovers unexpected features.
Acknowledgements
This work was supported by NIH grants to JF, SJL, WC, SH; an NIH fellowship to LAJ; the European Union Seventh Framework Program PROSPECTS (Proteomics Specification in Space and Time Grant HEALTH-F4-2008-201648) to RA and FUH, and the Swiss Initiative for Systems Biology and by funds from the ERC advanced grant “Proteomics v3.0” (grant# 233226) to RA. We thank Rachel Bond for help in CCTx-Cys2 experiments and Ramya Kumar for help in bTRiC purification. Stephan Nickell and Marius Boicu helped us in the EM analysis of yTRiC. Expert assistance by Stefan Pinkert in yTRi XL-MS data analysis is gratefully acknowledged.
Footnotes
AL and TW performed crosslinking experiments and analyzed mass spectrometry data, LAJ purified and biochemically characterized bTRiC for XL-MS, modeled the crosslinking data for the bTRiC and yTRiC datasets, designed the CCTx-Cys2 mutations and analyzed the structural data, AB carried out the crystallographic refinement of the XL-MS model and analyzed the structural data, BC generated and analyzed the CCTx-Cys2 mutations, LM purified and biochemically characterized yTRiC for XL-MS, LAJ and SP wrote software for the combinatorial analysis, SH computed the statistical significance of the results; BM and WC performed cryo-EM analysis of bTRiC, YC, SJL and WC carried out analyses of previous cryo-EM map; FUH, RA and JF designed and discussed experiments. All authors contributed to writing the MS.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers Atomic coordinates for the refined XL-MS-derived structure of the yeast TRiC complex have been deposited in the Protein Data Bank, http://www.pdb.org (PDB code 4D8Q and 4D8R, respectively). The structure factor amplitudes were previously deposited under accession code 3P9D (Dekker et al., 2011).
Author information The authors declare no competing financial interest.
Supplemental Information Supplemental Information includes four tables, eight figures, one movie and can be found online at XXXXX.
References
- Archibald JM, Blouin C, Doolittle WF. Gene duplication and the evolution of group II chaperonins: implications for structure and function. J Struct Biol. 2001;135:157–169. doi: 10.1006/jsbi.2001.4353. [DOI] [PubMed] [Google Scholar]
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn S, Beck F, Sakata E, Walzthoeni T, Beck M, Aebersold R, Forster F, Baumeister W, Nickell S. Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proc Natl Acad Sci U S A. 2010;107:20992–20997. doi: 10.1073/pnas.1015530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZA, Jawhari A, Fischer L, Buchen C, Tahir S, Kamenski T, Rasmussen M, Lariviere L, Bukowski-Wills JC, Nilges M, et al. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. The EMBO journal. 2010;29:717–726. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Baker ML, Jakana J, Woolford D, Miller EJ, Reissmann S, Kumar RN, Redding-Johanson AM, Batth TS, Mukhopadhyay A, et al. 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement. Proc Natl Acad Sci U S A. 2010;107:4967–4972. doi: 10.1073/pnas.0913774107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Schroder GF, Meyer AS, Jakana J, Ma B, Dougherty MT, Schmid MF, Reissmann S, Levitt M, Ludtke SL, et al. Symmetry-free cryo-EM structures of the chaperonin TRiC along its ATPase-driven conformational cycle. The EMBO journal. 2011 [Google Scholar]
- Cuellar J, Martin-Benito J, Scheres SH, Sousa R, Moro F, Lopez-Vinas E, Gomez-Puertas P, Muga A, Carrascosa JL, Valpuesta JM. The structure of CCT-Hsc70 NBD suggests a mechanism for Hsp70 delivery of substrates to the chaperonin. Nat Struct Mol Biol. 2008;15:858–864. doi: 10.1038/nsmb.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker C, Roe SM, McCormack EA, Beuron F, Pearl LH, Willison KR. The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins. The EMBO journal. 2011;30:3078–3090. doi: 10.1038/emboj.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzel L, Lowe J, Stock D, Stetter KO, Huber H, Huber R, Steinbacher S. Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT. Cell. 1998;93:125–138. doi: 10.1016/s0092-8674(00)81152-6. [DOI] [PubMed] [Google Scholar]
- Douglas NR, Reissmann S, Zhang J, Chen B, Jakana J, Kumar R, Chiu W, Frydman J. Dual action of ATP hydrolysis couples lid closure to substrate release into the group II chaperonin chamber. Cell. 2011;144:240–252. doi: 10.1016/j.cell.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Feldman DE, Spiess C, Howard DE, Frydman J. Tumorigenic mutations in VHL disrupt folding in vivo by interfering with chaperonin binding. Mol Cell. 2003;12:1213–1224. doi: 10.1016/s1097-2765(03)00423-4. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Lasker K, Forster F, Bohn S, Walzthoeni T, Villa E, Unverdorben P, Beck F, Aebersold R, Sali A, Baumeister W. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci U S A. 2012;109:1380–1387. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber MA, Reilly JP. Structural analysis of a prokaryotic ribosome using a novel amidinating cross-linker and mass spectrometry. J Proteome Res. 2011;10:3604–3616. doi: 10.1021/pr200260n. [DOI] [PubMed] [Google Scholar]
- Leitner A. Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol Cell Proteomics. 2012 doi: 10.1074/mcp.M111.014126. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol Cell Proteomics. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou AK, Willison KR. Elucidation of the subunit orientation in CCT (chaperonin containing TCP1) from the subunit composition of CCT micro-complexes. The EMBO journal. 1997;16:4311–4316. doi: 10.1093/emboj/16.14.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca O, Martin-Benito J, Gomez-Puertas P, Ritco-Vonsovici M, Willison KR, Carrascosa JL, Valpuesta JM. Analysis of the interaction between the eukaryotic chaperonin CCT and its substrates actin and tubulin. J Struct Biol. 2001;135:205–218. doi: 10.1006/jsbi.2001.4359. [DOI] [PubMed] [Google Scholar]
- Llorca O, Martin-Benito J, Ritco-Vonsovici M, Grantham J, Hynes GM, Willison KR, Carrascosa JL, Valpuesta JM. Eukaryotic chaperonin CCT stabilizes actin and tubulin folding intermediates in open quasi-native conformations. The EMBO journal. 2000;19:5971–5979. doi: 10.1093/emboj/19.22.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca O, McCormack EA, Hynes G, Grantham J, Cordell J, Carrascosa JL, Willison KR, Fernandez JJ, Valpuesta JM. Eukaryotic type II chaperonin CCT interacts with actin through specific subunits. Nature. 1999;402:693–696. doi: 10.1038/45294. [DOI] [PubMed] [Google Scholar]
- Maiolica A, Cittaro D, Borsotti D, Sennels L, Ciferri C, Tarricone C, Musacchio A, Rappsilber J. Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol Cell Proteomics. 2007;6:2200–2211. doi: 10.1074/mcp.M700274-MCP200. [DOI] [PubMed] [Google Scholar]
- Martin-Benito J, Bertrand S, Hu T, Ludtke PJ, McLaughlin JN, Willardson BM, Carrascosa JL, Valpuesta JM. Structure of the complex between the cytosolic chaperonin CCT and phosducin-like protein. Proc Natl Acad Sci U S A. 2004;101:17410–17415. doi: 10.1073/pnas.0405070101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Benito J, Grantham J, Boskovic J, Brackley KI, Carrascosa JL, Willison KR, Valpuesta JM. The inter-ring arrangement of the cytosolic chaperonin CCT. EMBO Rep. 2007;8:252–257. doi: 10.1038/sj.embor.7400894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AS, Gillespie JR, Walther D, Millet IS, Doniach S, Frydman J. Closing the folding chamber of the eukaryotic chaperonin requires the transition state of ATP hydrolysis. Cell. 2003;113:369–381. doi: 10.1016/s0092-8674(03)00307-6. [DOI] [PubMed] [Google Scholar]
- Muller DR, Schindler P, Towbin H, Wirth U, Voshol H, Hoving S, Steinmetz MO. Isotope-tagged cross-linking reagents. A new tool in mass spectrometric protein interaction analysis. Anal Chem. 2001;73:1927–1934. doi: 10.1021/ac001379a. [DOI] [PubMed] [Google Scholar]
- Munoz IG, Yebenes H, Zhou M, Mesa P, Serna M, Park AY, Bragado-Nilsson E, Beloso A, de Carcer G, Malumbres M, et al. Crystal structure of the open conformation of the mammalian chaperonin CCT in complex with tubulin. Nat Struct Mol Biol. 2011;18:14–19. doi: 10.1038/nsmb.1971. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Pereira JH, Ralston CY, Douglas NR, Meyer D, Knee KM, Goulet DR, King JA, Frydman J, Adams PD. Crystal structures of a group II chaperonin reveal the open and closed states associated with the protein folding cycle. J Biol Chem. 2010;285:27958–27966. doi: 10.1074/jbc.M110.125344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrotchenko EV, Borchers CH. Crosslinking combined with mass spectrometry for structural proteomics. Mass Spectrom Rev. 2010;29:862–876. doi: 10.1002/mas.20293. [DOI] [PubMed] [Google Scholar]
- Pupko T, Bell RE, Mayrose I, Glaser F, Ben-Tal N. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics. 2002;18(Suppl 1):S71–77. doi: 10.1093/bioinformatics/18.suppl_1.s71. [DOI] [PubMed] [Google Scholar]
- Rappsilber J. The beginning of a beautiful friendship: cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J Struct Biol. 2011;173:530–540. doi: 10.1016/j.jsb.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissmann S, Parnot C, Booth CR, Chiu W, Frydman J. Essential function of the built-in lid in the allosteric regulation of eukaryotic and archaeal chaperonins. Nat Struct Mol Biol. 2007;14:432–440. doi: 10.1038/nsmb1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinner O, Seebacher J, Walzthoeni T, Mueller LN, Beck M, Schmidt A, Mueller M, Aebersold R. Identification of cross-linked peptides from large sequence databases. Nat Methods. 2008;5:315–318. doi: 10.1038/nmeth.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivenzon-Segal D, Wolf SG, Shimon L, Willison KR, Horovitz A. Sequential ATP-induced allosteric transitions of the cytoplasmic chaperonin containing TCP-1 revealed by EM analysis. Nat Struct Mol Biol. 2005;12:233–237. doi: 10.1038/nsmb901. [DOI] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz DM, Kalkhof S, Schmidt A, Ihling C, Stingl C, Mechtler K, Zschornig O, Sinz A. Annexin A2/P11 interaction: new insights into annexin A2 tetramer structure by chemical crosslinking, high-resolution mass spectrometry, and computational modeling. Proteins. 2007;69:254–269. doi: 10.1002/prot.21445. [DOI] [PubMed] [Google Scholar]
- Shomura Y, Yoshida T, Iizuka R, Maruyama T, Yohda M, Miki K. Crystal structures of the group II chaperonin from Thermococcus strain KS-1: steric hindrance by the substituted amino acid, and inter-subunit rearrangement between two crystal forms. J Mol Biol. 2004;335:1265–1278. doi: 10.1016/j.jmb.2003.11.028. [DOI] [PubMed] [Google Scholar]
- Sinz A. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom Rev. 2006;25:663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- Spiess C, Meyer AS, Reissmann S, Frydman J. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 2004;14:598–604. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess C, Miller EJ, McClellan AJ, Frydman J. Identification of the TRiC/CCT substrate binding sites uncovers the function of subunit diversity in eukaryotic chaperonins. Mol Cell. 2006;24:25–37. doi: 10.1016/j.molcel.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YC, Chang HC, Chakraborty K, Hartl FU, Hayer-Hartl M. Essential role of the chaperonin folding compartment in vivo. The EMBO journal. 2008;27:1458–1468. doi: 10.1038/emboj.2008.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam AY, Xia Y, Lin HT, Burlingame A, Gerstein M, Frydman J. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat Struct Mol Biol. 2008;15:1255–1262. doi: 10.1038/nsmb.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Baker ML, Schroder GF, Douglas NR, Reissmann S, Jakana J, Dougherty M, Fu CJ, Levitt M, Ludtke SJ, et al. Mechanism of folding chamber closure in a group II chaperonin. Nature. 2010;463:379–383. doi: 10.1038/nature08701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.