SUMMARY
RECQL4 is associated with Rothmund-Thomson Syndrome (RTS), a rare autosomal recessive disorder characterized by premature aging, genomic instability and cancer predisposition. RECQL4 is a member of the RecQ-helicase family, and has many similarities to WRN protein, which is also implicated in premature aging. There is no information about whether any of the RecQ helicases play roles in mitochondrial biogenesis, which is strongly implicated in the aging process. Here, we used microscopy to visualize RECQL4 in mitochondria. Fractionation of human and mouse cells also showed that RECQL4 was present in mitochondria. Q-PCR amplification of mitochondrial DNA demonstrated that mtDNA damage accumulated in RECQL4-deficient cells. Microarray analysis suggested that mitochondrial bioenergetic pathways might be affected in RTS. Measurements of mitochondrial bioenergetics showed a reduction in the mitochondrial reserve capacity after lentiviral knockdown of RECQL4 in two different primary cell lines. Additionally, biochemical assays with RECQL4, mitochondrial transcription factor A and mitochondrial DNA polymerase γ showed that the polymerase inhibited RECQL4’s helicase activity. RECQL4 is the first 3′ to 5′ RecQ helicase to be found in both human and mouse mitochondria and the loss of RECQL4 alters mitochondrial integrity.
Keywords: RecQ helicase, mtDNA, Rothmund-Thomson Syndrome
INTRODUCTION
RECQL1, WRN, BLM, RECQL4 and RECQL5 are five RecQ helicases present in human cells (for recent reviews see (Bohr, 2008; Capp et al., 2010). Interestingly, mutations within RECQL4 are causative in three disorders, Rothmund-Thomson (OMIM 268400), RAPADILINO (OMIM 266280) and Baller-Gerold (OMIM 218600), whereas mutations in WRN or BLM genes give rise to Werner (OMIM 277700) or Bloom syndromes (OMIM 210900), respectively. Patients afflicted with these disorders suffer from significant growth retardation and some features of accelerated aging. Common characteristics among the RECQL4-related syndromes include skeletal abnormalities, especially radial ray defects, feeding problems and sometimes diarrhea (for recent reviews see (Siitonen et al., 2009; Larizza et al., 2010)).
RecQ helicase proteins possess catalytic activities of DNA strand annealing, ATPase and 3′ to 5′ helicase function. Originally, it was reported that RECQL4 lacked helicase activity (Macris et al., 2006), but more recently it has been documented that RECQL4 has a weak inherent helicase activity (Rossi et al., 2010) which is masked by its strong strand annealing activity (Xu and Liu, 2009). It was determined that addition of excess single stranded DNA to RECQL4 helicase reactions was sufficient to visualize the helicase reaction products. Further biochemical characterization of RECQL4 by our laboratory has revealed that the helicase activity of RECQL4 can be directly assayed, without the need for excess ssDNA, by employing short fork duplex DNA oligonucleotides (Rossi et al., 2010). Additionally, we have shown that RECQL4 has a narrower substrate range than the other mammalian helicases. Thus, RECQL4 possesses the classic 3′ to 5′ RecQ helicase, DNA-stimulated ATPase, DNA strand annealing and DNA binding properties of the human RecQ helicases (Macris et al., 2006; Xu and Liu, 2009; Suzuki et al., 2009; Rossi et al., 2010).
Unlike the other RecQ helicases, RECQL4 is found in multiple compartments within the cell such as the nucleus, nucleolus and the cytoplasm (Yin et al., 2004; Petkovic et al., 2005; Woo et al., 2006; Burks et al., 2007). Using cell fractionation and immunofluorescence microscopy, Yin et al. reported that in transformed cells, including HeLa, MCF7 and Jurkat cells, endogenous RECQL4 was found in cytoplasmic extracts, whereas in WI38 cells, which are non-transformed human diploid fibroblasts, RECQL4 was mostly, but not exclusively, found in nuclear extracts (Yin et al., 2004). Other studies have reported predominantly nuclear RECQL4 staining without cytoplasmic involvement (Petkovic et al., 2005; Woo et al., 2006). Interestingly, one of the mutations found in RECQL4, which is present in the majority of RAPADILINO patients, causes RECQL4 to be preferentially localized to the cytoplasm (Burks et al., 2007). Clearly, the localization of RECQL4 can be cell type specific. Additional analysis of cytoplasmic RECQL4 is important since there is no indication of what role it might play in the cytoplasm.
RECQL4 is a DNA damage responsive protein because its localization pattern changes after cells are treated with oxidative stressors or agents that induce DNA double strand breaks (DSBs). Specifically, our laboratory has shown that RECQL4 localizes to focal laser-induced DSBs (Singh et al., 2010) and using chromatin immunoprecipitation it was observed that Xenopus RECQL4 becomes chromatin bound following DSBs (Kumata et al., 2007). Dietschy et al. also demonstrated that RECQL4 was shuttled into the cytoplasm when cells were treated with the deacetylase inhibitor, Trichostatin A (Dietschy et al., 2009). Therefore the intracellular localization of RECQL4 is dynamic and can be modulated by both deacetylation inhibitors and DNA damaging agents (Werner et al., 2006; Woo et al., 2006; Dietschy et al., 2009; Singh et al., 2010).
It has been proposed that RECQL4 participates in nuclear DNA replication because the N-terminal domain of RECQL4 resembles Sld2, a protein involved in DNA replication in yeast (Sangrithi et al., 2005). Also, Xenopus RECQL4 has been reported to recruit DNA polymerase α (Sangrithi et al., 2005). Consistent with this observation, it was recently shown that human RECQL4 interacts with proteins important for nuclear DNA replication initiation such as the MCM and GINS complexes in human cells (Im et al., 2009; Xu et al., 2009; Thangavel et al., 2010).
Our laboratory and others have shown that RTS patient samples are sensitive to a variety of DNA damaging agents (Jin et al., 2008; Schurman et al., 2009). Based on these results, we proposed and showed that RECQL4 could modulate DNA base excision repair (BER), a process that removes oxidative lesions from DNA (Schurman et al., 2009). Specifically, we showed that RECQL4 could stimulate the DNA repair activities of the BER proteins APE1, FEN1 and pol β. It is well known that multiple BER proteins exist in the nucleus and mitochondria. Since RECQL4 has been shown to localize to the cytoplasm this raises the question as to whether RECQL4 might localize to mitochondria and participate in mitochondrial DNA (mtDNA) replication or DNA repair.
RECQL4 is the least well characterized RecQ helicase that causes human disease (the others are WRN and BLM). Here, we sought to explore the extra nuclear localization of human RECQL4 and evaluate RECQL4’s potential role in mt DNA metabolism. Our results reveal that both human and mouse endogenous RECQL4 protein partially co-localizes to mitochondria which contrasts with the other human RecQ helicases. In order to further explore the mitochondrial findings, we performed microarray analysis on RECQL4 patient samples and conducted mitochondrial bioenergetics analysis on scrambled (SCR) and RECQL4 knockdown (KD) human primary cells and observed a decline in the reserve capacity of RECQL4 knockdown cells. The loss of RECQL4 also leads to an accumulation of mtDNA damage. Biochemical studies suggested that the replicative DNA polymerase in mitochondria, pol γ inhibited the helicase of RECQL4. Taken together our results show that among the RecQ helicases only RECQL4 is significantly found in mitochondria and loss of RECQL4 is associated with dysfunctional mitochondria.
RESULTS
RECQL4 localizes to mitochondria
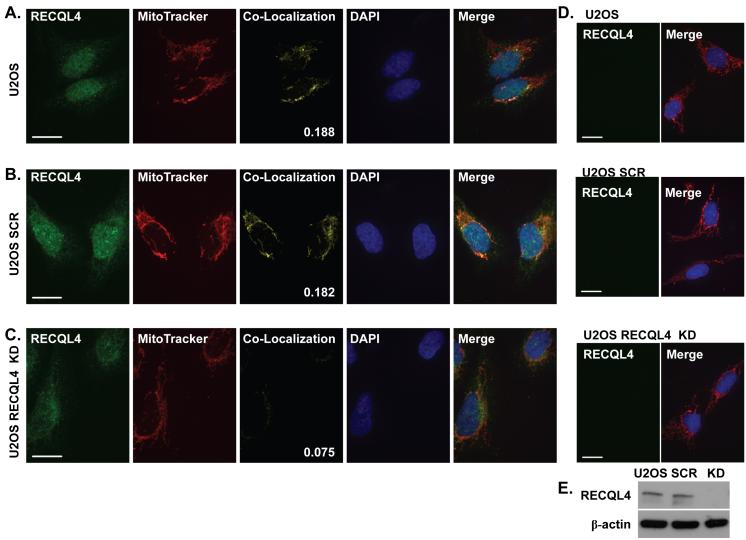
RECQL4 is found in both the nucleus and cytosolic compartments of the cell, yet we have no knowledge of what function the protein has in the cytosol. To explore the observed extra nuclear staining of RECQL4, we performed immunofluorescence staining of U2OS osteosarcoma cells. Our antibody was raised to amino acids 26-176 within the N-terminus of RECQL4 (Rossi et al., 2010). MitoTracker Red was used to visualize the mitochondria. Additionally, a co-localization channel was generated to visualize the relative RECQL4-MitoTracker Red overlap. The coefficient of co-localization of cytosolic RECQL4 with Mito Tracker Red is shown in the lower right corner of the individual co-localization panel. As seen in Fig. 1A, RECQL4 is predominantly localized to the nucleus with weak cytoplasmic staining. In order to feel confident that the staining that we observed was due to RECQL4 and not some other non-specific antibody reactive protein, a lentivirus RECQL4 knockdown was prepared. Upon RECQL4-specific lentivirus treatment (U2OS RECQL4 KD), the nuclear RECQL4 signal is significantly diminished relative to the scrambled control (U2OS SCR, compare Fig. 1 B, C). Additionally, a majority of the signal in the mitochondrial co-localization channel is lost upon RECQL4 KD, Fig. 1 (compare panels B and C for the co-localization channel signal). The average coefficient of co-localization of fifteen cells with our anti-RECQL4 antibody was 0.182, 0.188, and 0.075 for U2OS, U2OS SCR and U2OS RECQL4 KD, respectively. All of the images were captured using the same exposure settings. Additionally, to confirm that the MitoTracker Red signal was not bleeding into the green channel and artificially creating co-localization, the cells were stained with MitoTracker Red only, panel D. As can be seen, no signal was observed in the RECQL4 channel, while the Mito Tracker and DAPI are visible in the merge panels. Western blots were prepared to confirm the relative knockdown of RECQL4 and are shown in Fig. 1E.
Fig. 1. Immunofluorescence demonstrates RECQL4 is found in mitochondria.
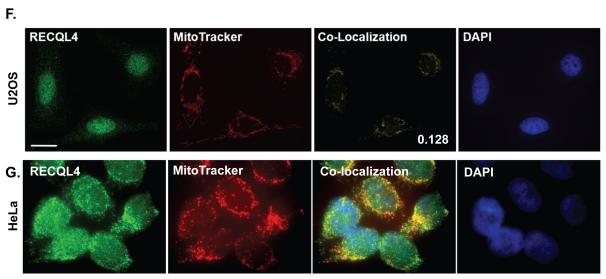
Confocal images of representative U2OS cells fixed and stained using a custom RECQL4 antibody and MitoTracker Red as described in the Experimental Procedures; (A) normal, (B) scrambled shRNA treated, (C) RECQL4 KD. (F) Representative images using Sigma anti-RECQL4 antibody on U2OS cells. (G) Representative Zeiss microscope images using a Santa Cruz anti-RECQL4 antibody on HeLa cells. The images are pseudo colored: green-RECQL4 and red-MitoTracker, with yellow areas indicating co-localization. The coefficient of co-localization is indicated on the confocal co-localization panel images. DAPI stain of the nucleus is in blue. The merge panel is the merged image of all channels. Panel D are the same cells as in A-C however the cells were only stained with MitoTracker (red) and DAPI (blue). For all images (A-D & F) the white bar denotes 20 μm. Panel E displays a western blot of RECQL4 and β-actin from 80 μg of protein from whole cell extracts documenting the level of knockdown obtained after lentivirus transduction.
Two other independent RECQL4 antibodies were also tested for their ability to detect mitochondrial RECQL4. In Fig. 1F, a Sigma anti-RECQL4 antibody was used and it detected the endogenous RECQL4 signal co-localizing with MitoTracker Red. The average coefficient of co-localization was 0.128 for the Sigma antibody and the co-localization was predominantly perinuclear. In Fig. 1G, we used HeLa cells and a Santa Cruz anti-RECQL4 antibody which detected a portion of RECQL4’s fluorescence signal co-localizing with MitoTracker Red in the cytoplasm. Thus, RECQL4 partially co-localizes to mitochondria as detected by multiple different RECQL4 primary antibodies in two different cell types. We conclude that RECQL4 is the first RecQ helicase to be found in mitochondria.
Many, but not all, mitochondrial proteins possess mitochondrial targeting sequences (Schmidt et al., 2010). To determine if RECQL4 possessed a targeting sequence, the protein sequence was analyzed by a variety of programs and scored for mitochondrial localization. PSORT v6.4 and TargetP v1.1 each predicted that RECQL4 might be targeted to mitochondria (probability 0.8 and 0.82, respectively) whereas both PSORT II and MitoProt II, did not predictthat RECQL4 would go to mitochondria, probability 0.6 and 0.16. Thus, there was no consistent prediction based on targeting for mitochondrial localization of RECQL4.
Cell fractionation supports that RECQL4 is present inside mammalian mitochondria
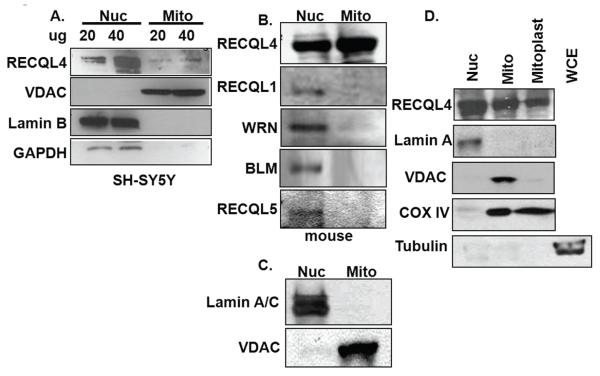
While the immunofluorescence provided supports that RECQL4 is partially associated with mitochondria, we also sought to detect RECQL4 in mitochondria from fractionated cell extracts. Differential and Percoll gradient centrifugation were used to isolate mitochondria from human SH-SY5Y cells which contain a significant amount of mitochondria. This cell fractionation method revealed that RECQL4 was present in both the nuclear and mitochondrial sub fractions (Fig. 2A). The mitochondrial preparation was free of nuclear contamination as judged by the lack of Lamin B staining. Additionally, the mitochondrial preparation was free of cytoplasmic contamination as judged by western blotting for GAPDH (Fig. 2A). As expected, VDAC immunoreactivity was only seen in the mitochondrial fraction. Additionally, we fractionated HeLa cells and observed RECQL4 in the mitochondrial compartment as well (supplementary data Fig. S1). While there is a trace of PCNA remaining in the mitochondrial preparations after proteinase K digestion, it represents an extremely small fraction of the PCNA signal in the total cell extract. Therefore, using two different human cell types we can see RECQL4 in highly purified mitochondria.
Fig. 2. Cell fractionation supports that RECQL4 is found in mitochondria in both human and mouse cells.
(A) Nuclear (nuc) and mitochondrial (mito) extracts were prepared from undifferentiated SHSY-5Y neuroblastoma cells. Mitochondria were prepared as described in the Experimental Procedures. The extracts were probed by western blotting for RECQL4; VDAC, a mitochondrial antigen; Lamin B, a nuclear antigen; and GAPDH, a cytoplasmic antigen. (B) Nuclear and mitochondrial extracts were prepared from mouse liver extracts. The western blots were probed for each of the mammalian RecQ helicases. (C) The quality of the mouse liver mitochondrial preparation was assessed by western blotting for both Lamin A/C and VDAC. (D) Nuclear (nuc), mitochondrial (mito), mitoplast and whole cell extracts (WCE) were immunoblotted for RECQL4, Lamin A, VDAC, COX IV and tubulin. Isolated mitochondria used in panels B and C were treated with digitonin to strip off the mitochondrial outer membrane. Extracts were western blotted for RECQL4, Lamin A, VDAC, COX IV and Tubulin to determine the relative distribution of RECQL4 and the purity of the mitoplast preparation. For B, C and D, 70 μg of protein were loaded in each lane. The western blots have been repeated at least in triplicate and representative blots are shown.
Since isolation of mitochondria from mouse tissues is easier and often results in greater purity and yield, mitochondria were isolated from mouse liver using the same method. The cell fractionation of mouse liver revealed a significant proportion of RECQL4 in the mitochondrial compartment (Fig. 2B). Additionally, we probed these extracts for all of the other mammalian RecQ helicases: RECQL1, WRN, BLM and RECQL5. There was no detectable RECQL1, BLM and RECQL5 in the mitochondrial fraction; however, there was a very slight band when the blot was probed for WRN (Fig. 2B). Purity of the extracts was determined by lack of Lamin A/C immunoreactivity in the mitochondrial fraction (Fig. 2C).
To demonstrate that the RECQL4 was present inside the mitochondria and not merely attached to the outer membrane, mitoplasts were prepared by treatment with digitonin. VDAC, an outer mitochondrial protein marker, was shed upon treatment with digitonin whereas RECQL4 was retained inside the mitoplast (Fig. 2D). Cox IV was used as a marker for the inner membrane proteins and tubulin was used to demonstrate that the mitochondrial preparation was free of cytosolic contamination. Thus, among the human RecQ helicases, RECQL4 is by far the most abundant RecQ helicase found inside the mitochondria and the first 3′-5′ RecQ helicase to be described in this compartment.
Characterization of RTS patient samples and RECQL4 knockdown cells
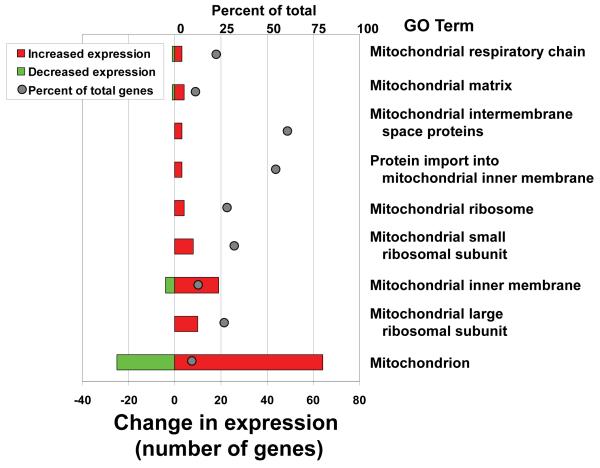
We have previously reported some results from a microarray analysis on three confirmed RTS patient samples that do not express RECQL4 due to inactivating mutations (Schurman et al., 2009). Further microarray analysis indicated that among the top 100 gene ontology term changes, several were mitochondrial-related groups and all were among the most highly up-regulated groups (Fig. 3). In addition to the gene ontology analysis, we also examinedthe significantly changed genes within these gene ontology terms. A list of these genes, along with their corresponding z-ratio, p-values and fold changes, is shown in supplementary data, Table S2. Several were mitochondrial ribosomal proteins, cytochrome oxidase subunits and components of the ATP synthase complex. It is likely that dysregulation of these components impact mitochondrial function in RTS patients. It should be noted that the list does not report all the genes changed from the Gene Set Enrichment analysis but only the statistically significantly changed genes. Up-regulation of mitochondrial proteins and pathways is a common feature among mitochondrial myopathies and may be a general indicator of mitochondrial stress. If RTS patients have an up-regulation of genes for mitochondria they may also have more mtDNA. To test this hypothesis, relative mtDNA copy number was determined for each cell line (Table 1). Relative mitochondrial DNA amplification was measured by real-time PCR as described in the Methods. Our analysis showed that two out of the three RTS patient samples have more mtDNA relative to their corresponding age and sex matched controls. Furthermore, we were interested in determining whether the deficiency of RECQL4 also translated into changes in mitochondrial function in the RTS cells. Accordingly, experiments were designed to test if the loss of RECQL4 might alter the mitochondrial bioenergetics of these patient primary cells. The mitochondrial bioenergetics XF-24 (Seahorse Biosciences) analyzer was employed to measure cellular oxygen consumption rates of cells in a micro plate format, in real-time. The assay was conducted as described in the Materials and Methods to determine basal (supplementary data Fig. S2), ATP-linked (supplementary data Fig. S2-B) and FCCP-stimulated (supplementary data Fig. S2-C) oxygen consumption rates (OCR) from the three normal and the RTS cells. The patient samples showed no statistically significant changes. Even though these were early passage primary cell lines, the RECQL4-deficient cell lines are genetically heterogeneous and could have altered their gene expression profiles to compensate for the loss of RECQL4.
Fig. 3. Microarray analysis of RTS patient fibroblasts.
Multiple mitochondrial-related GO terms are up-regulated in RTS patient samples. Gene set enrichment analysis was used to parse genes into Gene Ontology terms. The set of significantly changed mitochondrial-related GO terms are shown on the right. The percent of total genes in the group, top X-axis legend, represents the number of significant genes within each pathway, relative to all the genes in that pathway, that were altered in RTS cells relative to control cells and is shown by the gray balls. The number of genes that were up- or down-regulated, lower X-axis legend, is depicted by the red and green bar graphs, respectively.
Table 1. Relative mtDNA copy number analysis.
| SN | Age/sex | Sample # | Group | β-actin Cta | mtDNA Cta | ΔCtb | Rcc |
|---|---|---|---|---|---|---|---|
| 1 | 11/M | GM00323 | Normal | 27.59 | 23.42 | 4.17 | 18 |
| 2 | 11/M | GM01864 | Normal | 28.33 | 23.84 | 4.48 | 22.35 |
| 3 | 2/F | GM00969 | Normal | 30.81 | 23.31 | 7.50 | 180.81 |
| 4 | 10/M | AG05013 | RTS | 29.83 | 23.18 | 6.66 | 100.92 |
| 5 | 12/M | AG18371 | RTS | 28.91 | 22.07 | 6.85 | 115.13 |
| 6 | 4/F | AG17524 | RTS | 28.17 | 22.50 | 5.67 | 50.98 |
Sex and age matched samples are similarly colored.
Ct is average cycle number from three independent experiments.
ΔCt = Ctβ-actin − CtmtDNA.
Rc = relative mtDNA copy number given by 2ΔCt
Loss of mitochondrial reserve capacity following RECQL4 knockdown
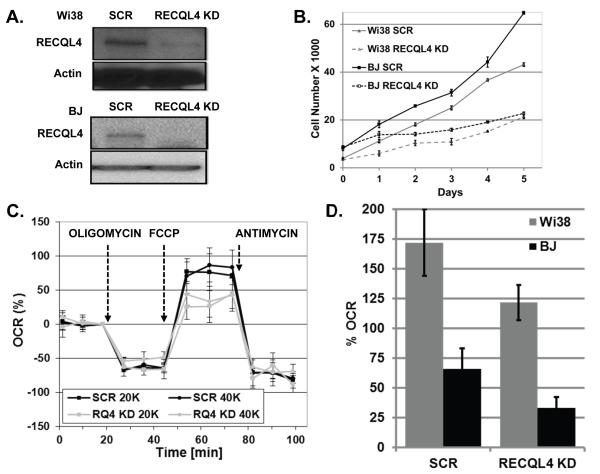
To search for a functional role of RECQL4 in mitochondria, we sought to determine if loss of RECQL4 induced any mitochondrial functional defects in an isogenic background. We generated lentiviral particles containing a shRNA against a scrambled sequence or RECQL4 gene. Normal primary Wi38 and BJ cells were transduced with packaged virus particles and after four days, cells were seeded into XF-24 microplates. First, western blotting and Q-PCR was performed to confirm the RECQL4 knockdown, and these assays estimated the depletion of RECQL4 to about 85-90% (Fig. 4A). Secondly, RECQL4 is known to be important for DNA replication, therefore growth curves were generated to determine if growth rates were altered after RECQL4 KD. Knockdown cells were generated and plated after four days for growth assays. Cell numbers were counted in triplicate each day for six days. As can be seen, loss of RECQL4 caused significant growth retardation relative to the SCR treated cells, Fig. 4B.
Fig. 4. RECQL4 knockdown cells show lower reserve capacity.
(A) Typical knockdown of RECQL4 that we obtained after lentivirus treatment in Wi38 and BJ cells. (B) Growth curves of scrambled or RECQL4 KD treated Wi-38 or BJ cells upon lentivirus treatment. (C) Graph showing the percent oxygen consumption (OCR) profile of BJ cells treated with scrambled (SCR) or RECQL4 (RQ4 KD) lentivirus. Cells were seeded at two densities, 20,000 (20K) and 40,000 (40K) cells. Cells were treated with the indicated oligomycin, FCCP and antimycin at the indications times. For more details see the Materials and Methods section. (D) Graph showing relative loss of FCCP-stimulated OCR between the SCR and RECQL4 KD Wi-38 and BJ cells. The average percent OCR between two biological experiments is shown with the standard deviation from at least two or more wells measured at three different time points, BJ p-value 0.002 and Wi-38 p-value 0.003.
To determine if there were any bioenergetic consequences to the cells with a knockdown of RECQL4, scramble or RECQL4 KD treated Wi-38 and BJ cells were evaluated for cellular respiration on the XF-24 analyzer. This instrument uses a fluorescent probe to monitor the change in oxygen concentration in a small volume of media over adherent cells (see Methods and supplementary information). After optimizing the experiment for these cell types, we tested the cells at two different cell densities, and representative BJ curves, at two different cell densities, are shown in Fig. 4C. This data are baseline corrected for ease of analysis. Oligomycin was used to inhibit the mitochondrial ATP synthase, complex V, of the electron transport system. The difference in oxygen consumption between pre-and post oligomycin addition is considered ATP-linked respiration. Next, p-trifluoromethoxy carbonyl cyanide phenyl hydrazone, FCCP, was added to the cells to dissipate the electrochemical gradient. The difference between basal and the FCCP-stimulated OCR is the reserve capacity of the cells. After FCCP treatment, the cells were treated with antimycin A. This compound inhibits complex III and eliminates oxygen consumption from mitochondria. By this analysis, we found that the loss of RECQL4 in BJ cells contributes to the specific loss of reserve capacity. We repeated this analysis in another primary cell type, Wi-38 and had a similar finding. In the BJ cells, we observed a 50% loss of reserve capacity whereas in Wi-38 cells a 29% loss of reserve capacity was seen following depletion of RECQL4 (Fig. 4D). Since reserve capacity is thought to represent spare potential energy that a cell could rely upon when energy demands increase (Hill et al., 2009), our finding that RECQL4-depleted cells have reduced reserve capacity may render them more susceptible to cell death due to an inability to meet energy demands. Our data indicate a role for RECQ4 in mitochondrial bioenergetics but does not demonstrate any direct role of the protein on electron transport proteins.
RECQL4-deficient cells have increased levels of mtDNA damage
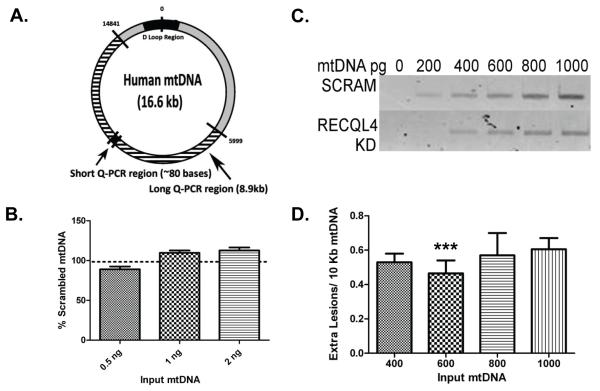
RTS cells have higher endogenous levels of 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG), and we have previously reported that both RTS and RECQL4-deficient cells have defective repair of H2O2-induced lesions (Schurman et al., 2009). Hence, RECQL4 has a direct role in the modulation of BER, a repair pathway also integral to DNA maintenance in the mitochondria. We employed a quantitative PCR strategy to determine whether a reduction of RECQL4 would lead to increased amounts of endogenous mtDNA damage. This technique is based upon the notion that many DNA lesions block the progression of a polymerase, hence by measuring the amount of amplification using DNA from RECQL4-deficient cells compared to DNA from scrambled cells we can estimate the relative amount of lesions present in mtDNA (Santos et al., 2002). A diagram of our amplified mtDNA fragments, in relation to the entire mitochondrial genome, is shown Fig. 5A. Since mtDNA amplification is proportional to the amount of DNA used as input and RECQL4 loss might alter mtDNA copy number, we first determined relative mtDNA copy number for three different biological knockdown cell experiments. As can be seen in Fig. 5B, loss of RECQL4 did not significantly alter copy number between the scramble or RECQL4 knockdown cells. Although in two out of three experiments there was a trend toward increased DNA copy number between the samples.
Fig. 5. RECQL4-deficient cells have higher amounts of endogenous mtDNA damage.
Quantitative PCR was used to establish the relative amount of mtDNA damage between scrambled and RECQL4 lentivirus treated HeLa cells. (A) Depicts the mitochondrial genome showing where our PCR primers reside and the location of the amplified mtDNA fragments. (B) Q-PCR of a short fragment of mtDNA shows that there is no change in mtDNA copy number between SCR and RECQL4 KD cells. (C) Relative amplification was assayed across a range of DNA concentrations that gave a linear response according to the amount of input mtDNA. A representative agarose gel image is shown of the relative amplification products. (D) Graphic representation of the lesion frequencies at each concentration of DNA tested. Two or more independent knockdown experiments were conducted for each mtDNA concentration. (*** denotes p< 0.01, n=3).
Following optimization of the experiment for HeLa cells, amplification measurements were taken across a range of mtDNA concentrations, using only those verified to be within the linear range of the assay. The specificity of the mtDNA PCR products was verified on an agarose gel (Fig. 5C). The amplification data was then analyzed using the Poisson distribution and it revealed that untreated RECQL4-deficient cells have at least 0.45 extra lesions for every 10 kb of mtDNA than scrambled control cells, when measured in the linear range (see Figure legend for additional details, Fig. 5D). The variability on this measurement is ±0.14 lesions and thus it is unlikely that the extra lesions reported are due simply to experimental error. Also, to put this level of DNA damage into context, similar lesion levels were generated when cells were treated with 200 μM hydrogen peroxide for 15 min. (Santos et al., 2004). Therefore, the loss of RECQL4 causes elevated mtDNA damage. This result confirms that even after only a relatively short period of RECQL4 deficiency, mtDNA maintenance is compromised leading to the accumulation of toxic lesions.
Probing the function of RECQL4 in mitochondria
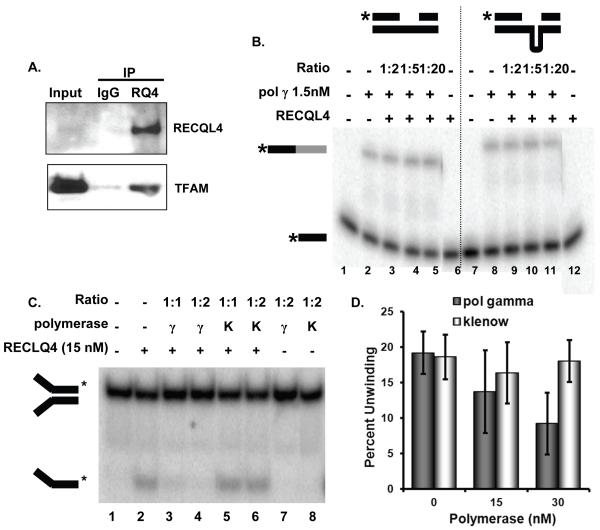
Since RECQL4 is likely involved with DNA replication in the nucleus, we wanted to determine whether RECQL4 might participate in mitochondrial DNA maintenance. In this regard, Transcription Factor A Mitochondrial (TFAM) is an abundant mitochondrial protein essential for mtDNA maintenance. It packages the mtDNA into nucleoid structures and participates in mtDNA transcription and replication (Kang and Hamasaki, 2005). Recently, we showed that TFAM inhibits base excision repair likely through competition for DNA binding with mitochondrial BER proteins (Canugovi et al., 2010). Therefore, we sought to determine if RECQL4 could physically and/or functionally interact with TFAM. Co-immunoprecipitation experiments were performed, in the presence of ethidium bromide to eliminate DNA, to determine if the proteins could physically interact. As shown in Fig. 6A, RECQL4 immunoprecipitated TFAM while the IgG sample did not.
Fig. 6. Interactions between RECQL4 and TFAM or pol γ.
(A) Immunoprecipitation reactions were performed on HeLa whole cell lysates with Santa Cruz normal rabbit IgG (lane 2) or rabbit anti-RECQL4 antibody in the presence of ethidium bromide (lane 3). Western blots were probed for RECQL4 with rabbit RECQL4 and True blot reagent and AbCam mouse TFAM antibody. (B) Synthesis by pol γ was assayed on a radiolabeled gapped heteroduplex (GH1*:GH2:GH3, 0.5nM) and a gapped hairpin heteroduplex (GH1*:GH4:GH3, 0.5 nM) in the presence of increasing concentrations of RECQL4 at the indicated molar ratios. Substrates are depicted above the figure with asterisk at the position of the radiolabel. Starred black bar shows starting substrate, and starred black/gray bar represents full length extension of the template. Representative gel is shown. (C) Helicase activity of RECQL4 was assayed on a radiolabeled 22-base pair forked duplex (T1*:B1, 0.5 nM). Either pol γ (γ) or Klenow (K) were added to the reaction mixture at the indicated molar ratios. Representative gel is shown. (D) Quantification of the relative helicase activity observed without or with the indicated polymerase present at the indicated concentration. n=3, standard deviation of the mean is shown.
To further search for the specific function of RECQL4 in mitochondria we used biochemical in vitro assays. Since RECQL4 has two catalytic activities, helicase and strand annealing activities (Rossi et al., 2010), we tested these functions in the presence of increasing concentrations of TFAM, but no functional interactions were observed (supplementary data Fig. S3-A & B). The biochemical details of these experiments are described in the Figure legends. Additionally, we tested if RECQL4 could modulate DNA binding by TFAM in our BER assay (Canugovi et al., 2010), however RECQL4 could not displace TFAM from DNA (data not shown).
RECQL4 and DNA polymerase γ
An important protein for mtDNA maintenance is pol γ, the mitochondrial DNA polymerase, which functions during both replication and repair (Graziewicz et al., 2006). Thus, to ascertain if RECQL4 could facilitate replication by pol γ, DNA polymerization and strand displacement were assayed on a variety of substrates. However, RECQL4 did not affect pol γ activity (Fig. 6B). We then tested the effect of pol γ on RECQL4 helicase activity on a 22-bp partial duplex (0.5 nM) with 15-mer splayed arms (Fig. 6C and 6D). Interestingly, RECQL4’s helicase activity was inhibited in the presence of increasing concentrations of pol γ but not in the presence of the Klenow fragment of E. coli DNA polymerase I (Fig. 6C and 6D). Thus, this inhibition appeared to be specific for DNA pol γIt is possible that the high DNA binding affinity of pol γ could preclude RECQL4 from the substrate. However, this inhibition could potentially indicate a specific functional interaction between pol γ and RECQL4.
DISCUSSION
A major theory of aging is based on the notion that mitochondrial dysfunction is central to the aging process. A number of human premature aging conditions involve dysfunction of the RecQ helicase family of proteins, and so far there has been no connection made to mitochondrial deficiencies amongst this important family of proteins.
Here we have used both immunofluorescence and cellular fractionation studies to document the localization of RECQL4 to mitochondria. This is the first study to show that RECQL4, unlike the other RecQ helicases, partially co-localizes to mitochondria in both human cells and mouse liver tissue. We next evaluated if there were any bioenergetic consequences following RECQL4 loss. We generated a knockdown of RECQL4 in primary Wi-38 and BJ cells in order to more directly evaluate the loss of RECQL4 on mitochondrial bioenergetics. When these cells were characterized on the XF-24 analyzer, the RECQL4 knockdown cells consistently displayed a lower reserve capacity. Since RECQL4 is present in mitochondria it may directly affect bioenergetics, but it is also possible that loss of RECQL4 stresses the cells in some way such that mitochondrial function is altered. For example, we have shown that RECQL4 participates in telomere maintenance (Ghosh et al., 2011) and there has been a report linking telomere dysfunction with metabolic changes via the telomere-p53-PGC1 axis (Sahin et al., 2011). Also, the altered growth properties of the knockdown cells, elevated persistent DNA damage or mtDNA mutations could all potentially contribute to the observed lower reserve capacity in these cells.
Additional support that mitochondria are altered upon loss of RECQL4 comes from the observation that there is evaluated mtDNA damage in RECQL4 KD cells. We consistently observed less amplification of mtDNA from the RECQL4 KD cells implying that these cells have more polymerase blocking mtDNA damage. We have previously shown that RTS patient cells have persistent DNA damage (Schurman et al., 2009). This DNA damage may activate PARP and other DNA repair proteins that have the potential to utilize high amounts of energy and thus RTS and RECQL4-depleted cells may have lower reserve capacities due to elevated levels of persistent DNA damage. Thus, the loss of RECQL4 has biological consequences for mitochondrial bioenergetics and mtDNA integrity which could contribute to genomic instability and segmental premature aging phenotypes.
We have documented that RECQL4 is in complex with TFAM but no functional catalytic interaction was observed. Since the RECQL4 association with TFAM was observed in cell extracts, it is possible that our in vitro reconstituted system was missing an additional protein component which mediates the interaction between the two proteins. In addition, we evaluated RECQL4’s ability to modulate DNA synthesis by the major replicative mitochondrial polymerase, pol γ. We observed that RECQL4 did not alter polymerization by pol γ on gapped heteroduplexes. When we evaluated the impact of pol γ on RECQL4 function, interestingly, we found that pol γ inhibited RECQL4 helicase activity. While the inhibition could be due to competition for substrate binding, it was specific for pol γ since another polymerase, the Klenow fragment of E. coli DNA polymerase I, did not have the same effect on RECQL4.
It is interesting that among all RecQ helicases only RECQL4 displays significant mitochondrial localization. At this time we do not know why there are so many RecQ homologues in higher eukaryotes but it may involve altered cellular localization. In unperturbed cells, most of the RecQ helicases display a predominantly nucleolar staining pattern, with the exception of RECQL4. It is predominantly found in the nucleoplasm with a fraction found in the cytoplasm and mitochondrial compartments. Furthermore, it is intriguing that RECQL4 shows such significant mitochondrial localization in mouse liver tissue because liver is one of the few regenerative tissues.
This is the first report to describe a role for RECQL4, a 3′-5′ helicase, in mitochondria. Previously, a proteomics study identified RECQL4 in isolated human mitochondria, but no further analysis was conducted (Jiang et al., 2009). A major helicase in mitochondria is Twinkle, a 5′-3′ helicase, which is essential for mtDNA maintenance (Copeland, 2008). By comparing the severe symptoms associated with the dysfunction of Twinkle, we can gain insight into the potential role of RECQL4 in this organelle. Infantile onset spinocerebellar ataxia is perhaps the most severe manifestation caused by Twinkle deficiency, and characterized by very early onset ataxia and reduced mental capacity both proposed to be the result of the cerebellar atrophy in these patients (Palau and Espinos, 2006). Similarly, a subset of RTS patients also present with mental retardation associated with atrophy of the brain (Gelaw et al., 2004; Larizza et al., 2010). Why only a subset of RTS patient show these symptoms may be related to peripheral factors in these patients such as elevated mitochondrial stress. Unlike Twinkle, which plays a prominent role in many facets of mitochondrial DNA maintenance and replication, RECQL4 is likely to play a more specialized role. The necessity for a 3′-5′ helicase may not be as frequent in the mitochondria under normal conditions and this organelle may only require the part time presence of RECQL4. Additionally, mitochondrial disease is complex due the fact that altered mtDNA must rise above some threshold level before a phenotype is observed. Therefore, given that RECQL4 is not essential for mtDNA metabolism and that phenotypic expression of mitochondrial diseases is complex, a mild mitochondrial phenotype would be expected for RECQL4 deficient patients. Thus, the presence of RECQL4 in mitochondria may well explain components of the clinical phenotype. It is of great interest to further understand the biochemical function of RECQL4 in mitochondria and how it differs from its nuclear role.
EXPERIMENTAL PROCEDURES
Cells and Tissues
A summary of the source of and growth conditions used for the various cell lines used in this paper are described in the supporting information, Data S1. Generation of lentiviral mediated RECQL4 knockdown cells is described in the supporting information, Data S1.
Immunofluorescence
U2OS and HeLa cells were used for immunofluorescence. A detailed protocol for staining can be found in supplementary information, Data S1. The coefficient of co-localization (Mander’s Mx) between RECQL4, in the cytosol, and MitoTracker Red, was determined by using Volocity 6.0.1 software. First, the nuclei in the image were determined by DAPI staining. Then all RECQL4 signal in the GFP channel was selected. Minimum thresholds in both channels were determined using automatic thresholding, as implemented in Volocity. Maximum thresholds were determined from the image using Voxel Spy in order to remove spurious high intensity spots on some images. Next, GFP signals associated with the nuclei were subtracted out. Finally, the software determined the coefficient of co-localization between the remaining GFP signal and that of the mitochondria using the RFP channel. The coefficient of co-localization represents the fraction of RECQL4 co-localized with MitoTracker Red (values range from 0-1, 1 being 100% co-localization). Fifteen cells per antibody were analyzed and the average coefficient of co-localization determined.
Western Blotting
Standard differential and Percoll gradient centrifugation were used to prepare mitochondrial extracts (Maynard et al., 2010). Additional Western blotting details can be found in the supplementary information Data S1 and a list of antibodies used can be found in supplementary Table S1.
Microarray
Briefly, RNA was isolated from three normal (GM00323, GM00969, GM01864) and three RTS patient cell lines (AG18371, AG05013, AG17524). Pools of RNA, either normal or RTS, were hybridized to the 23k gene Illumina Sentrix HumanRef-8 v2 Expression BeadChips (Illumina Systems Inc., Redwood, CA, USA). The Illumina BeadStation 500GX Genetic Analysis System scanner and Illumina BeadStudio ver. 15 was used for image capture, image processing and data extraction. For further details, please see the original publication (Schurman et al., 2009).
Patient fibroblast mitochondrial DNA relative amplification
Total DNA was isolated from three sets of RTS and normal human fibroblasts. The purity and concentration of the DNA was measured using the Nanodrop spectrophotometer. The relative mitochondrial DNA amplification from 10 ng of genomic was measured by real time-PCR (Applied Biosystems 7900 HT and software Sequence Detection Systems, version SDS 2.3) where TaqMan probes (Applied Biosystems) were used to detect the amplification changes. These probes are listed in supplementary information Data S1 and were derived from (Mambo et al., 2005). The amount of mitochondrial COX1 gene amplification was normalized to nuclear β-actin gene amplification termed as the ΔCt and was calculated for all samples. The relative mitochondrial amplification is represented by the number 2ΔCt in each case (Szuhai et al., 2001).
Mitochondrial bioenergetics
The relative oxygen consumption rate (OCR) of cells, with or without RECQL4 KD, was determined using the XF-24 analyzer (Seahorse Bioscience Inc., Billerica, MA). This instrument uses a fluorescence based assay to measures oxygen consumption rates in real-time. The preparation of cells and drugs were done essentially as described by Qian and Van Houten (Qian and Van Houten, 2010) except 2-deoxyglucose was omitted. Additional details regarding the mitochondrial bioenergetics assay can be found in the supporting information Data S1.
Cell Survival Assay
Scrambled or RECQL4 knockdowns cells were counted and plated 96 h after transduction and 48 h post selection into 18, 6 cm dishes with 10,000 cells seeded per dish. Three dishes were harvested every 24 h post plating and counted using a Coulter Counter (Beckman Coulter Inc., Brea, CA, USA). The first set of three were used to normalize for plating efficiency.
Measurement of DNA damage
The assay was performed as described by (Santos et al., 2002) with minor changes see supplementary information Data S1.
Immunoprecipitation
HeLa cells were grown to near confluence in 150 mm dishes. The cells were then washed with ice cold PBS and scraped off the dishes. The cells were centrifuged briefly then lysed in lysis buffer (25 mM Tris-HCl pH-7.4, 150 mM NaCl, 0.1% Triton X-100 and EDTA-free protease inhibitor cocktail (Roche, Indianapolis, IN, USA) for 45 min at 4°C. The lysate was clarified by centrifugation at 10000 xg for 20 min at 4°C. The whole cell lysate was pre-cleared with protein G beads for 2 hr at 4°C. Pre-cleared lysate was incubated with primary antibody against the protein of interest or normal IgG as a control overnight at 4°C in the presence of ethidium bromide (10 μg/ml). Protein G beads were added for 2 hr to pull down the antibody-protein complex. Beads were washed in wash buffer (25 mM Tris-HCl pH-7.4, 150 mM NaCl, 0.01% Triton X-100) for at least four times and boiled with SDS loading buffer containing β-mercaptoethanol for 15 min. The samples were blotted as described above.
Protein Expression
RECQL4 was expressed and purified from E. coli Rosetta2 (DE3) (Novagen, Gibbstown, NJ, USA) as described previously (Rossi et al., 2010). Wild type TFAM (WT) and the DNA binding mutant of TFAM (LA), were purified according to Canugovi et al. (Canugovi et al., 2010). The purity and protein concentrations of the proteins were determined by Coomassie blue staining of polyacrylamide gels. Polymerase γ p140 and p55 subunits were purified from baculoviral infected insect cells and E. coli, respectively, as described (Longley et al., 1998; Lim et al., 1999). The proteins were aliquoted, and stored at −80°C until use.
Oligonucleotide Substrates
All oligonucleotides used in this study are listed in Table 2. A detailed protocol for labeling the oligonucleotides is included in supplementary information Data S1.
Table 2. Oligonucleotides used in this study.
| Name | Sequence (5′-3′) |
|---|---|
| T1 | GTAGTGCATGTACACCACACTCTTTTTTTTTTTTTTT |
| B1 | TTTTTTTTTTTTTTTGAGTGTGGTGTACATGCACTAC |
| GH1 | GGGTGAACCTGCAGGTGGGC |
| GH2 | GACGCTGCCGAATTCTACCAGTGCCTTGCTAGGACATC TTTGCCCACCTGCAGGTTCACCC |
| GH3 | ACTGGTAGAATTCGGCAGCGTC |
| GH4* | GACGCTGCCGAATTCTACCAGTGCGTGCGAAGCACCTT GCT AGGACATCTTTGCCCACCTGCAGGTTCACCC |
Underline indicates hairpin fold.
Helicase and Strand Annealing Assays
For the helicase assays, RECQL4 protein, at concentrations shown in the fig. legends, was incubated with radiolabeled forked duplex substrate, T1:B1 (0.5 nM, Table 2) for 30 minutes at 37 °C in a reaction volume of 10 μl reaction buffer containing 30 mM Tris pH 7.4, 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 100 μg/ml BSA, 10% glycerol, and 5 mM ATP. For strand annealing assays, RECQL4 protein, at concentrations shown in the fig. legends, was incubated with radiolabeled single stranded primer T1 (0.5 nM) and its unlabeled complimentary primer B1 (1 nM) for 10 minutes at 37°C in a reaction volume of 10 μl reaction buffer, the same as above without ATP. Where indicated, TFAM or polymerase γ (pol γ) was added at the concentrations mentioned in the fig. legends. When present pol γ p140 and p55 subunits were incubated together (1:2 ratio p140:p55) on ice for 15 minutes before adding to the reaction. All reactions were stopped by addition of 5 μl of 3X native stop dye (50 mM EDTA, 40% glycerol, 0.9% SDS, 0.05% bromophenol blue, and 0.05% xylene cyanol). The reaction products were separated by electrophoresis on 12% native polyacrylamide gel, exposed to a PhosphorImager screen (GE Healthcare, Piscataway, NJ), then imaged with a Typhoon scanner (GE Healthcare, Piscataway, NJ, USA). ImageQuant version 5.2 (GE Healthcare, Piscataway, NJ, USA) was employed to analyze the phosphor images. The experiments were done at least in triplicate, and a representative gel is shown.
Polymerase γ Gap Filling Assay
Pol γ activity was measured on gapped heteroduplexes (GH1:GH2:GH3 or GH1:GH4:GH3, Table 2, 0.5 nM) in the presence of increasing amounts of RECQL4 (3, 7.5, and 30 nM). Pol γ p140 and p55 subunits were incubated together (1:2 ratio of p140:p55) on ice for 10 minutes before adding RECLQ4. Reactions were initiated by addition of substrate to pol γ and RECQL4 in 10 μl reaction buffer containing 30 mM Tris pH 7.4, 5 mM MgCl2, 5 mM ATP, 1mM DTT, 100 μg/ml BSA, 50 mM KCl, and 100μM total dNTPs for 30 min at 37°C. Klenow was used as a positive control for synthesis. The reaction products were separated by electrophoresis on 7M urea/15% polyacrylamide gels and visualized as above. The experiments were done at least in triplicate, and a representative gel is shown.
Supplementary Material
Acknowledgements
We would like to thank Dr. Avik Ghosh and Dr. Ivana Rybanska for critically reading the manuscript. We would like to thank Avik Ghosh for assistance with the purification of WT RECQL4. We would like to thank Al May (NIA) for assistance with the microscopy. We would like to thank Yongqing Zhang and Kevin Becker (Research Resources Branch, NIA) for assistance with the microarray. This work was supported by the intramural research program of the National Institute on Aging, NIH, Z01 AG000726.
Footnotes
Supporting Information
Additional supporting information may be found in the online version of this article
Reference List
- Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem. Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burks LM, Yin J, Plon SE. Nuclear import and retention domains in the amino terminus of RECQL4. Gene. 2007;391:26–38. doi: 10.1016/j.gene.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Canugovi C, Maynard S, Bayne AC, Sykora P, Tian J, de Souza-Pinto NC, Croteau DL, Bohr VA. The mitochondrial transcription factor A functions in mitochondrial base excision repair. DNA Repair (Amst) 2010;9:1080–1089. doi: 10.1016/j.dnarep.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capp C, Wu J, Hsieh TS. RecQ4: the second replicative helicase? Crit Rev. Biochem. Mol Biol. 2010;45:233–242. doi: 10.3109/10409231003786086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland WC. Inherited mitochondrial diseases of DNA replication. Annu. Rev. Med. 2008;59:131–146. doi: 10.1146/annurev.med.59.053006.104646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietschy T, Shevelev I, Pena-Diaz J, Huhn D, Kuenzle S, Mak R, Miah MF, Hess D, Fey M, Hottiger MO, Janscak P, Stagljar I. p300-mediated acetylation of the Rothmund-Thomson-syndrome gene product RECQL4 regulates its subcellular localization. J. Cell Sci. 2009;122:1258–1267. doi: 10.1242/jcs.037747. [DOI] [PubMed] [Google Scholar]
- Gelaw B, Ali S, Becker J. Rothmund-Thomson syndrome, Klippel-Feil syndrome, and osteosarcoma. Skeletal Radiol. 2004;33:613–615. doi: 10.1007/s00256-004-0798-2. [DOI] [PubMed] [Google Scholar]
- Ghosh AK, Rossi ML, Singh DK, Dunn C, Ramamoorthy M, Croteau DL, Liu Y, Bohr VA. RECQL4, the protein mutated in Rothmund-Thomson syndrome, functions in telomere maintenance. J. Biol. Chem. 2011 doi: 10.1074/jbc.M111.295063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziewicz MA, Longley MJ, Copeland WC. DNA polymerase gamma in mitochondrial DNA replication and repair. Chem. Rev. 2006;106:383–405. doi: 10.1021/cr040463d. [DOI] [PubMed] [Google Scholar]
- Hill BG, Dranka BP, Zou L, Chatham JC, rley-Usmar VM. Importance of the bioenergetic reserve capacity in response to cardiomyocyte stress induced by 4-hydroxynonenal. Biochem. J. 2009;424:99–107. doi: 10.1042/BJ20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im JS, Ki SH, Farina A, Jung DS, Hurwitz J, Lee JK. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc. Natl. Acad. Sci. U. S. A. 2009;106:15628–15632. doi: 10.1073/pnas.0908039106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Liu X, Fang X, Wang X. Proteomic analysis of mitochondria in Raji cells following exposure to radiation: implications for radiotherapy response. Protein Pept. Lett. 2009;16:1350–1359. doi: 10.2174/092986609789353646. [DOI] [PubMed] [Google Scholar]
- Jin W, Liu H, Zhang Y, Otta SK, Plon SE, Wang LL. Sensitivity of RECQL4-deficient fibroblasts from Rothmund-Thomson syndrome patients to genotoxic agents. Hum. Genet. 2008;123:643–653. doi: 10.1007/s00439-008-0518-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D, Hamasaki N. Mitochondrial transcription factor A in the maintenance of mitochondrial DNA: overview of its multiple roles. Ann. N. Y. Acad. Sci. 2005;1042:101–108. doi: 10.1196/annals.1338.010. [DOI] [PubMed] [Google Scholar]
- Kumata Y, Tada S, Yamanada Y, Tsuyama T, Kobayashi T, Dong YP, Ikegami K, Murofushi H, Seki M, Enomoto T. Possible involvement of RecQL4 in the repair of double-strand DNA breaks in Xenopus egg extracts. Biochim. Biophys. Acta. 2007;1773:556–564. doi: 10.1016/j.bbamcr.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Larizza L, Roversi G, Volpi L. Rothmund-Thomson syndrome. Orphanet. J. Rare. Dis. 2010;5:2. doi: 10.1186/1750-1172-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SE, Longley MJ, Copeland WC. The mitochondrial p55 accessory subunit of human DNA polymerase gamma enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance. J. Biol. Chem. 1999;274:38197–38203. doi: 10.1074/jbc.274.53.38197. [DOI] [PubMed] [Google Scholar]
- Longley MJ, Prasad R, Srivastava DK, Wilson SH, Copeland WC. Identification of 5′-deoxyribose phosphate lyase activity in human DNA polymerase gamma and its role in mitochondrial base excision repair in vitro. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12244–12248. doi: 10.1073/pnas.95.21.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macris MA, Krejci L, Bussen W, Shimamoto A, Sung P. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair (Amst) 2006;5:172–180. doi: 10.1016/j.dnarep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Mambo E, Chatterjee A, Xing M, Tallini G, Haugen BR, Yeung SC, Sukumar S, Sidransky D. Tumor-specific changes in mtDNA content in human cancer. Int. J. Cancer. 2005;116:920–924. doi: 10.1002/ijc.21110. [DOI] [PubMed] [Google Scholar]
- Maynard S, de Souza-Pinto NC, Scheibye-Knudsen M, Bohr VA. Mitochondrial base excision repair assays. Methods. 2010;51:416–425. doi: 10.1016/j.ymeth.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palau F, Espinos C. Autosomal recessive cerebellar ataxias. Orphanet. J. Rare. Dis. 2006;1:47. doi: 10.1186/1750-1172-1-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkovic M, Dietschy T, Freire R, Jiao R, Stagljar I. The human Rothmund-Thomson syndrome gene product, RECQL4, localizes to distinct nuclear foci that coincide with proteins involved in the maintenance of genome stability. J. Cell Sci. 2005;118:4261–4269. doi: 10.1242/jcs.02556. [DOI] [PubMed] [Google Scholar]
- Qian W, Van Houten B. Alterations in bioenergetics due to changes in mitochondrial DNA copy number. Methods. 2010;51:452–457. doi: 10.1016/j.ymeth.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Rossi ML, Ghosh AK, Kulikowicz T, Croteau DL, Bohr VA. Conserved helicase domain of human RecQ4 is required for strand annealing-independent DNA unwinding. DNA Repair (Amst) 2010;9:796–804. doi: 10.1016/j.dnarep.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin E, Colla S, Liesa M, Moslehi J, Muller FL, Guo M, Cooper M, Kotton D, Fabian AJ, Walkey C, Maser RS, Tonon G, Foerster F, Xiong R, Wang YA, Shukla SA, Jaskelioff M, Martin ES, Heffernan TP, Protopopov A, Ivanova E, Mahoney JE, Kost-Alimova M, Perry SR, Bronson R, Liao R, Mulligan R, Shirihai OS, Chin L, DePinho RA. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470:359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, Venkitaraman AR. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 2005;121:887–898. doi: 10.1016/j.cell.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Santos JH, Mandavilli BS, Van Houten B. Measuring oxidative mtDNA damage and repair using quantitative PCR. Methods Mol Biol. 2002;197:159–176. doi: 10.1385/1-59259-284-8:159. [DOI] [PubMed] [Google Scholar]
- Santos JH, Meyer JN, Skorvaga M, Annab LA, Van Houten B. Mitochondrial hTERT exacerbates free-radical-mediated mtDNA damage. Aging Cell. 2004;3:399–411. doi: 10.1111/j.1474-9728.2004.00124.x. [DOI] [PubMed] [Google Scholar]
- Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat. Rev. Mol Cell Biol. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- Schurman SH, Hedayati M, Wang Z, Singh DK, Speina E, Zhang Y, Becker K, Macris M, Sung P, Wilson DM, III, Croteau DL, Bohr VA. Direct and indirect roles of RECQL4 in modulating base excision repair capacity. Hum. Mol. Genet. 2009;18:3470–3483. doi: 10.1093/hmg/ddp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siitonen HA, Sotkasiira J, Biervliet M, Benmansour A, Capri Y, Cormier-Daire V, Crandall B, Hannula-Jouppi K, Hennekam R, Herzog D, Keymolen K, Lipsanen-Nyman M, Miny P, Plon SE, Riedl S, Sarkar A, Vargas FR, Verloes A, Wang LL, Kaariainen H, Kestila M. The mutation spectrum in RECQL4 diseases. Eur. J. Hum. Genet. 2009;17:151–158. doi: 10.1038/ejhg.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DK, Karmakar P, Aamann M, Schurman SH, May A, Croteau DL, Burks L, Plon SE, Bohr VA. The involvement of human RECQL4 in DNA double-strand break repair. Aging Cell. 2010;9:358–371. doi: 10.1111/j.1474-9726.2010.00562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Kohno T, Ishimi Y. DNA helicase activity in purified human RECQL4 protein. J. Biochem. 2009;146:327–335. doi: 10.1093/jb/mvp074. [DOI] [PubMed] [Google Scholar]
- Szuhai K, Ouweland J, Dirks R, Lemaitre M, Truffert J, Janssen G, Tanke H, Holme E, Maassen J, Raap A. Simultaneous A8344G heteroplasmy and mitochondrial DNA copy number quantification in myoclonus epilepsy and ragged-red fibers (MERRF) syndrome by a multiplex molecular beacon based real-time fluorescence PCR. Nucleic Acids Res. 2001;29:E13. doi: 10.1093/nar/29.3.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangavel S, Mendoza-Maldonado R, Tissino E, Sidorova JM, Yin J, Wang W, Monnat RJ, Jr., Falaschi A, Vindigni A. Human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol Cell Biol. 2010;30:1382–1396. doi: 10.1128/MCB.01290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner SR, Prahalad AK, Yang J, Hock JM. RECQL4-deficient cells are hypersensitive to oxidative stress/damage: Insights for osteosarcoma prevalence and heterogeneity in Rothmund-Thomson syndrome. Biochem. Biophys. Res. Commun. 2006;345:403–409. doi: 10.1016/j.bbrc.2006.04.093. [DOI] [PubMed] [Google Scholar]
- Woo LL, Futami K, Shimamoto A, Furuichi Y, Frank KM. The Rothmund-Thomson gene product RECQL4 localizes to the nucleolus in response to oxidative stress. Exp. Cell Res. 2006;312:3443–3457. doi: 10.1016/j.yexcr.2006.07.023. [DOI] [PubMed] [Google Scholar]
- Xu X, Liu Y. Dual DNA unwinding activities of the Rothmund-Thomson syndrome protein, RECQ4. EMBO J. 2009;28:568–577. doi: 10.1038/emboj.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Rochette PJ, Feyissa EA, Su TV, Liu Y. MCM10 mediates RECQ4 association with MCM2-7 helicase complex during DNA replication. EMBO J. 2009;28:3005–3014. doi: 10.1038/emboj.2009.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Kwon YT, Varshavsky A, Wang W. RECQL4, mutated in the Rothmund-Thomson and RAPADILINO syndromes, interacts with ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Hum. Mol. Genet. 2004;13:2421–2430. doi: 10.1093/hmg/ddh269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.