Abstract
Human embryonic stem cell (hESC)-derived dopaminergic (DA) neurons hold potential for treating Parkinson’s disease (PD) through cell replacement therapy. Generation of DA neurons from hESCs has been achieved by co-culture with the stromal cell line PA6, a source of stromal cell-derived inducing activity (SDIA). However, the factor(s) produced by stromal cells that constitute SDIA are largely undefined. We previously reported that medium conditioned by PA6 cells can generate functional DA neurons from NTera2 human embryonal carcinoma stem cells. Here we show that PA6-conditioned medium can induce DA neuronal differentiation in both NTera2 cells and the hESC I6 cell line. To identify the factor(s) responsible for SDIA, we used large-scale microarray analysis of gene expression combined with mass spectrometric analysis of PA6-conditioned medium (CM). The candidate factors, hepatocyte growth factor (HGF), stromal cell-derived factor-1 α (SDF1α), secreted frizzled-related protein 1 (sFRP1), and vascular endothelial growth factor D (VEGFD) were identified and their concentrations in PA6 CM were established by immunoaffinity capillary electrophoresis. Upon addition of SDF1α, sFRP1 and VEGFD to the culture medium we observed an increase in the number of cells expressing tyrosine hydroxylase (a marker for DA neurons) and beta-III tubulin (a marker for immature neurons) in both the NTera2 and I6 cell lines. These results indicate that SDF1α, sFRP1 and VEGFD are major components of SDIA, and suggest the potential use of these defined factors to elicit DA differentiation of pluripotent human stem cells for therapeutic intervention in PD.
Keywords: dopaminergic neurons, neuronal differentiation, stromal cell derived inducing activity, embryonic stem cells
INTRODUCTION
Pluripotent stem cells (PSCs) can be propagated indefinitely, have the ability to generate cells of any germ layer origin, and provide the opportunity to study fundamental mechanisms of development or to generate cells for potential restoration of tissue damaged by disease or injury (Thomson et al., 1998). The major symptoms of Parkinson’s disease (PD) result from the loss of dopaminergic (DA) neurons in the substantia nigra. Due to their well-defined location and synaptic connections, the replacement of these neurons using cell transplantation-based therapy is a rational therapeutic strategy. DA neurons generated from PSCs therefore hold promise for cell replacement therapy for PD.
Researchers have reported moderate success in clinical trials with fetal midbrain transplants (Freed et al., 1992; Kordower et al., 1995; Piccini et al., 1999) and, although promising, transplantation of human embryonic stem cell (hESC)-derived DA neurons in animal models has proven to be challenging (Schulz et al., 2004; Cho et al., 2008). The majority of differentiation paradigms for generating human DA neurons from PSCs involve the use of glial cells (Buytaert-Hoefen et al., 2004; Roy et al., 2006), meningeal cells (Hayashi et al., 2008), or stromal cell lines such as PA6 or MS5 cells (Kawasaki et al., 2000; Schwartz et al., 2005). Co-culture protocols are straightforward and efficient methods for deriving DA neurons from hESCs; however, the factors responsible for the differentiation process remain largely unknown. Clinical translation of PSC-based therapy necessitates the use of defined factors in order to avoid the transplantation of undesired feeder cells, transmittance of pathogens, and adverse immune responses.
Initially it was hypothesized that the DA neuron-inducing potential of PA6 and MS5 stromal cell lines, termed stromal cell-derived inducing activity (SDIA), resides on the surface of cells as anchored proteins or membrane-associated secreted proteins (Kawasaki et al., 2000). However, ourselves and other groups have demonstrated that PA6 conditioned medium (CM) can induce DA differentiation, implicating secreted factors in SDIA (Schwartz et al., 2005; Hayashi et al., 2008; Vazin et al., 2008; Swistowska et al., 2010). It is likely that multiple stromal factors direct sequential stage-specific differentiation, ultimately resulting in DA neuron generation. Several factors including, Wnt5a (Hayashi et al., 2008), stromal cell-derived factor-1 α (SDF1α), pleiotrophin (PTN), insulin-like growth factor 2 (IGF2), ephrin B1 (EFNB1) (Vazin et al., 2009), and sonic hedgehog (Shh) (Swistowska et al., 2010) have been identified by gene expression analysis in PA6 cells. Perturbation (Vazin et al., 2009) or ablation (Hayashi et al., 2008; Swistowska et al., 2010) studies have resulted in the implication of these factors as DA neuron-inducing factors in SDIA; however, thus far no factor has been shown to be secreted by PA6 cells or definitively present in PA6 CM. Identification of the DA neuron-inducing components of SDIA would provide a more defined and efficient system for generating DA neurons that would likely advance pre-clinical research.
Here we used both microarray and mass spectrometry analyses to identify specific PA6 SDIA factors, and determined their concentration in PA6 CM using immunoaffinity capillary electrophoresis (ICE). The three identified PA6 factors SDF1α, sFRP1 and VEGFD, resulted in enhanced neuronal and DA differentiation in both NTera2 cells (a human embryonal carcinoma stem cell (hECSC) line) and I6 cells (a hESC line). Our results suggest that SDF1α, sFRP1 and VEGFD are the major DA neuron-inducing components of SDIA. The ability to reproducibly differentiate human PSCs in defined, serum-free culture conditions paves the way for future pre-clinical studies and clinical trials in patients with neurodegenerative disorders.
MATERIALS AND METHODS
Cell Lines and Cell Culture
The human embryonal carcinoma stem cell line NTera2 was obtained from the American Type Culture Collection (ATCC, Manassas, VA) and cultured as previously described (Schwartz et al., 2005). Cells were maintained in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA), supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT) and 50 U/mL antibiotic/antimycotic (Anti/Anti; Invitrogen). Cells were passaged at a 1:3 ratio using a cell scraper when cells were approximately 90–95% confluent.
The human ESC line I6 (NIH Registry Name TE06) was cultured on mitomycin C- treated mitotically-inactivated mouse embryonic fibroblasts (ATCC). The cells were cultured in ESC medium consisting of D-MEM/F12 (ATCC), supplemented with 5% Knockout Serum Replacement (Invitrogen), 15% FBS, 2.0 mM L-Alanyl-L-Glutamine (Invitrogen), 0.1 mM non-essential amino acids (Invitrogen), 0.1 mM beta-mercaptoethanol (Sigma, St. Louis, MO), 4 ng/mL bFGF (R&D Systems, Minneapolis, MN), penicillin (100 I.U./ml) and streptomycin (100 μg/mL) (ATCC). An additional 4 ng/mL of bFGF was added 24 hours after thawing the cells. Daily medium changes began following the initial 48 hours in culture. Cells were passaged using collagenase IV (200 Units/mL) (Invitrogen) every 6–7 days.
The PA6 cell line was obtained from Riken BioResource Center Cell Bank (Japan). Cells were grown on 0.1% gelatin (Sigma) coated flasks in medium that contained α Minimum Essential Medium with ribonucleosides and deoxyribonucleosides (MEM; Invitrogen) supplemented with 10% FBS and 50 U/mL Anti/Anti. Dr. Katsu Itoh (Kyoto University, Japan) kindly provided the MS5 cell line. Cells were cultured in high glucose DMEM medium supplemented with 20% horse serum (HyClone) and 50 U/mL of Anti/Anti. Both the PA6 cell line and MS5 cell line were sub-cultured at a ratio of 1:5 using TrpLE (Invitrogen).
Mouse embryonic fibroblasts (MEFs) were obtained from Millipore (Billerica, MA) and cultured in high glucose DMEM supplemented with 10% FBS and 50 U/mL of Anti/Anti. Cells were subcultured with TrpLE and were not used past passage 6. All cells were maintained at 37°C in a 5% CO2/95% air, humidified atmosphere.
Cell Differentiation
MEF cells were grown until confluent and then mitotically inactivated with 10 μg/mL Mitomycin C (Sigma) for 4 hours at 37°C. Following inactivation cells were washed 10 times with medium and allowed to recover overnight. PA6 cells were grown on 0.1% gelatin coated flasks until confluent. Cells were washed several times with differentiation medium prior to the addition of differentiation medium for conditioning and subsequent collection. Differentiation medium consisted of Glasgow Minimum Essential Medium (GMEM; Invitrogen) supplemented with 10% knockout serum replacement, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate (Invitrogen), 0.1 mM β-mercaptoethanol, and 50 μg/ml of Anti/Anti. For conditioned medium purposes, 100 μg/mL of Heparin (Sigma) was added following the addition of medium to the either the MEF or PA6 cells. All differentiation medium used for comparison purposes to PA6 CM also contained 100 μg/mL of heparin. Conditioned medium was collected every 24 hours for a maximum of 1 week and stored at −20°C for no longer than 1 month.
For all co-culture experiments MEF cells and PA6 cells were grown as described above and were plated on 0.1% gelatin coated dishes. Once MEF cells reached confluency they were mitotically inactivated as described above. Prior to pluripotent cell seeding, both PA6 cells and MEF cells were washed three times with differentiation medium.
NTera2 cells were lifted with a cell scrapper and dissociated into single cells or clusters of 2–3 cells by gentle trituration prior to seeding cells at a density of 1000–5000 cells/cm3. For co-culture differentiation with either MEF or PA6 cells, medium was changed 3 days following seeding and every other day thereafter. For conditioned medium experiments and testing of identified stromal factors, NTera2 cells were seeded on 0.1% gelatin coated plates and allowed to recover overnight in growth medium. The following day growth medium was removed and cells were washed once and cultured further with corresponding medium used for differentiation. Medium was changed every other day from this point forward. Identified stromal factors were added to differentiation medium in the following concentrations: HGF 25, 50, or 100 ng/mL (Peprotech, Rocky Hill, NJ); SDF1α 25, 50, or 100 ng/mL (Peprotech); VEGF-D 5, 25, or 50 ng/mL (R & D Systems); sFRP1 2.5, 5.0, or 10.0 μg/mL (R & D Systems, Peprotech and kindly provided by Dr. Jeff Rubin, National Institute of Diabetes and Digestive and Kidney Diseases).
I6 cells were dissociated into small colonies as described above prior to seeding small colonies of 50–100 cells at a density of 5–10 colonies/cm3. Cells were co-cultured with either MEF or PA6 cells and differentiated in PA6 CM, differentiation medium or differentiation medium plus identified stromal factors. The factors and corresponding concentrations are the same as listed above for the NTera2 cells with the exception of sFRP1 (0.5, 1 or 2.5 μg/mL). Medium was changed 3 days following seeding and every other day thereafter. Cells were differentiated for either 2 weeks or 3 weeks at which point cells were processed for either immunochemistry or RNA isolation.
Immunochemistry
All solutions and washing steps used were made in Dulbecco’s Phosphate Buffered Saline with calcium and magnesium (dPBS, Invitrogen) to help prevent cell lifting. Cells were washed 3 times and then fixed for 10 minutes at 4°C in 4% paraformaldehyde (Sigma). Following fixation cells were washed 3 times and stored at 4°C. Fixed cells were incubated for 30 minutes with blocking buffer containing 0.1% Triton X-100 (Sigma) and 5% goat or donkey serum (Sigma). Primary antibody was diluted in blocking buffer and incubated overnight at 4°C. Primary antibodies and dilutions used were as follows: tyrosine hydroxylase (TH; Pel Freeze, Rogers, AR) 1:500; TH (Millipore) 1:200; TuJ1 (Sigma) 1:1000; Nestin (Millipore) 1:200; DBH (Millipore) 1:500; Sox1 (Millipore) 1:500; GFAP (Millipore) 1:200. Cells were then washed 3 times and incubated for 1 hour at room temperature with the appropriate secondary antibody and fluorochrome (Molecular Probes). Cells were washed and counterstained with DAPI (Sigma). Images were obtained with an Olympus fluorescence microscope.
All cell-counting analyses were performed on at least three independent experiments. Each experiment consisted of cell counts from ten randomly chosen fields repeated a minimum three times and averaged across experiments. In total, over 6000 cells and 100 colonies were scored for each experiment. All data are represented as means with standard deviations. P values were calculated using Student’s t-test or ANOVA as indicated.
RNA extraction and RT-PCR
Medium was removed from cells and total RNA was extracted using Stat-60 (TelTest, Friendswood, TX) following the manufacturer’s recommendations. Complementary DNA (cDNA) was synthesized using 1 μg of total RNA and a reverse transcription kit (SuperScript III First-Strand Synthesis System; Invitrogen) for Reverse Transcriptase-PCR (RT-PCR) according to the manufacturer’s specifications. All primers were synthesized by Integrated DNA Technologies (Coralville, IA). Primer sequences are listed in Supporting Information Table 1. PCR was performed using the following components: 1 μl of cDNA (generated as described above from 1 μg RNA) diluted 1:10 in DEPC water, 1 μl of 10 μM forward primer, 1 μl of 10 μM reverse primer, 22 μl of DEPC water, 25 μl of RedTaq (Sigma). The thermal cycling parameters for the PCR reactions were as follows: an initial denaturation step for 3 minutes at 94°C followed by denaturation for 1 min at 94°C; annealing for 1 min at 60°C; extension for 1 min at 72°C and final extension for 7 min at 72°C. Reactions were run for 30–35 cycles. To ensure that RNA samples were not contaminated with genomic DNA during RNA extraction, all samples were tested by running the reverse transcriptase reaction without SuperScript III and PCR was then carried out with β-Actin and GAPDH primers as recommended by the manufacturer.
Microarray Analysis
Total RNA was isolated from MEF and PA6 cells as described above. RNA was reverse-transcribed, labeled and analyzed using the Illumina Mouse Expression BeadChip microarray platform (Illumina, San Diego, CA). Arrays were processed according to the manufacturer’s recommendations. Expression values were normalized and averaged between biological replicates. Detection levels above 0.95 were considered for all data analysis. Significantly up-regulated genes (ratio ≥ 5.0 and p value < 0.05) in the PA6 cells were categorized using the Ingenuity software. The top 500 genes most highly expressed genes in either PA6 or MEF cells were compared using the Venn diagram software Venny. Microarray data can be found at the GEO website (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=zhexdsseqomsule&acc=GSE20503).
Mass Spectrometry
PA6 or MEF cells were grown to confluency under conditions specified above. Cells were washed 10 times with DMEM to remove possible contamination with FBS. A minimal volume of medium was added to each flask and cultures were incubated for 24 hours. Medium was then removed and centrifuged to remove contaminating debris. Supernatant collected from 20–40, T75 flasks were used for each experiment. A total of 3 mass spectrometry experiments were performed on biological replicates.
Proteins in supernatant were precipitated using trichloroacetic acid (TCA) (Sigma), washed with cold acetone twice, and dissolved with 8 M urea (Invitrogen), 50 mM NH4HCO3 (Sigma). After reduction and carbamidomethylation of cysteine residues, protein solution was diluted to 1 M urea with 50 mM NH4HCO3. Proteins were digested overnight at 37°C with 1 μg of sequencing grade trypsin (Promega, Madison, WI). The samples were desalted using Sep-Pak Vac C18 Cartridges 1cc/50mg (Waters, Milford, MA) and further purified through strong cation exchange (SCX) chromatography that was performed using a SCX column packed with PolySULFOETHYL A™ resins (5 μm 300 Å; PolyLC, Columbia, MD). The peptide solutions were desalted with ZipTip (Millipore), lyophilized, and stored at −80°C.
Peptides were suspended with 0.1% formic acid and pressure-loaded onto 11 cm X 100-μm fused silica capillary needles packed with 5 μm C18 beads (Magic C18 5 μm 300 Å; Michrom Bioresources, Auburn, CA). The peptides were separated in a 0 50% ACN (0.1% formic acid) gradient solution for 60 min at 200 nL/min and analyzed by LXQ ion trap mass spectrometer (Thermo Fisher Scientific). Peptide ions were detected in a full scan mode from 400 to 1400 m/z followed by four data-dependent MS/MS scans in a completely automated fashion. MS raw files were directly loaded to Mascot Daemon v2.2.2 connected to the NIH Mascot server, and proteins were identified using the mouse proteome database search with the following criteria: enzyme, trypsin (KR/P); full enzymatic cleavage; missed cleavage sites, 3; peptide tolerance, 2.0 amu; fragment ions tolerance, 1.0 amu; variable modifications, carbamidomethylation (+57 Da), methionine oxidation (+16 Da); decoy database, on. For the purpose of keeping the false discovery rate (FDR) below 0.5%, the search results were further filtered with various P values.
Immunoaffinity Capillary Electrophoresis
PA6 or MEF cells were grown to confluency as described above. Cells were washed 10 times with differentiation medium prior to the addition of medium for conditioning. Differentiation medium containing 100 μg/mL heparin or DMEM medium was conditioned for 24 hours prior to removal for processing. Biological triplicates were used for all experiments. The samples were analyzed using chip-based immunoaffinity capillary electrophoresis (ICE) with laser-induced fluorescence (LIF) detection as described previously (Phillips and Wellner et al., 2007; Arumugam et al., 2010). Briefly, specific antibodies to the analyte of interest and their respective antigens (R & D Systems) were reconstituted to stock solutions of 1 μg/mL in 0.1M phosphate buffer, pH 7.4. The antibodies were reduced to F(Ab)’2 fragments using a Pierce Biotechnology F(Ab)’2 digestion kit according to the manufacturer’s instructions and further reduced to FAb fragments by incubation with 200 mM DTT solution. Equal parts of each digested FAb fragments were mixed and immobilized onto a 2-mm thiol-derivatized glass disk. The disk was placed in the immunoaffinity port of a lab-on-a-chip and 200 nL of sample added to the port. The sample was allowed to reside in the immunoaffinity port for 3 minutes, during which the immobilized FAb fragments captured their respective antigens. The sample was removed and the disk washed with 0.5 μL of 0.1M phosphate buffer. AlexaFluor 633 laser dye (Molecular Probes) was introduced into the immunoaffinity port, labeling the bound antigens. This reaction was allowed to take place for 2 minutes before again washing the port with 0.5 μL of phosphate buffer. Finally, the bound antigens were eluted from the FAbs by introducing 0.5 μl of acid buffer (0.1M phosphate buffer titrated with 1N HCl to pH 1.0) and electro-kinetically moving the released antigens into the separation channel where they were separated at 6kV and the individual peaks measured in-channel by the LIF detector. The concentration of each separated peak was calculated from calibration curves constructed from known standards of each antigen, run under identical conditions. Data are presented as means with standard deviations. P values were calculated using Student’s t-test and ANOVA.
Cell-Based Assay for Sonic Hedgehog signaling with Luciferase reporter
Conditioned medium containing Shh (for a positive assay control) was generated from NIH 293 cells over expressing Shh-N as described previously (Hyman et al., 2009). Cells were cultured in DMEM (Invitrogen) supplemented with 10% FBS and 50 U/mL of Anti/Anti, until cells were approximately 80% confluent. Growth medium was removed and replaced with DMEM supplemented with 2% FBS and 50 U/mL of Anti/Anti. Conditioned medium was collected following 24 hours and stored short-term at 4°C or for longer periods at −20°C.
NIH-3T3 cells stably containing the Gli-dependent firefly luciferase reporter and the SV40-Renilla luciferase reporters, referred to as the Light-2 cell line, were cultured as described previously (Taipale et al., 2000; Hyman et al., 2009) in DMEM supplemented with 10% FBS and 50 U/mL of Anti/Anti. Once cells reached confluency they were passaged into a 24 well plate and cultured for an additional 4 days until the cells were fully confluent. Medium was removed and replaced with medium consisting of DMEM supplemented with 0.5% FBS and 50 U/ml of Anti/Anti alone or with a 1:20 dilution of the following: Shh-conditioned medium as prepared above to serve as a positive control; Shh-conditioned medium plus the Shh inhibitor cyclopamine served as a negative control; medium conditioned by MEF cells; medium conditioned by MEF cells in the presence of 100 μg/mL of heparin; medium conditioned by PA6 cells; and medium conditioned by PA6 cells in the presence of 100 μg/mL of heparin. Cells were further cultured for 30 hours and then medium was removed and cells were promptly lysed with 25 μl/well of Passive Lysis Buffer (Promega). Cell lysates were transferred to a luminometer microplate and samples were prepared with Dual Luciferase Kit (Promega) according to the manufacturer’s instructions prior to analysis. All samples were analyzed for both Renilla-luciferase (to normalize for cell density) and Gli-luciferase signals (to measure biologically active Shh). Relative reporter activity was calculated by dividing the Gli-luciferase signal by the Renilla-luciferase signal. Data are presented as means and standard deviations. P values were calculated using Student’s t-test.
RESULTS
Medium conditioned by PA6 cells generates dopaminergic neurons from hECSCs and hESCs
To determine the ability of PA6 CM to induce DA neuron differentiation, NTera2 (hECSC) and I6 (hESC) cells were differentiated for 3 weeks and assessed for DA and neuronal markers (Fig. 1). We first evaluated DA neuronal differentiation via immunoreactivity to the neuronal marker beta-III tubulin (TuJ1) and the DA marker tyrosine hydroxylase (TH) following 3 weeks of differentiation in PA6 CM in the absence of support cells (Fig. 1A). I6 cells were not evaluated in these conditions due to poor cell survival in the first few days following plating. We further examined the DA neuron differentiation efficiency of PA6 CM in both the NTera2 (Fig. 1B) and I6 (Fig. 1C) cell lines co-cultured with mouse embryonic fibroblast (MEF) cells in the presence of PA6 CM (MEF+PA6 CM). Initial examination of NTera2 and I6 cell lines following 3 weeks of differentiation demonstrated that MEF+PA6 CM cultures generated a high number of TuJ1 and TH positive colonies similar to PA6 co-cultures (PA6) These conditions generated many more positive colonies than did MEF co-cultures (MEF) (Fig. 1D). NTera2 cells grown in the absence of feeders and the presence of PA6 CM generated a similar percentage of TuJ1 and TH positive cells compared to MEF+PA6 CM cultures. NTera2 cells co-cultured with PA6 cells generated slightly more DA neurons. In contrast, very few neurons were generated in MEF, MEF CM and differentiation media (medium which was not conditioned by any cell type) cultures (Fig. 1E). As with the NTera2 cell line, the I6 cell line differentiated for 3 weeks by MEF+PA6 CM resulted in a similar number of TuJ1- and TH-positive cells compared to PA6 co-cultures, which was significantly greater than MEF the percentage of DA neuron-like cells in MEF co-cultures (Fig. 1F).
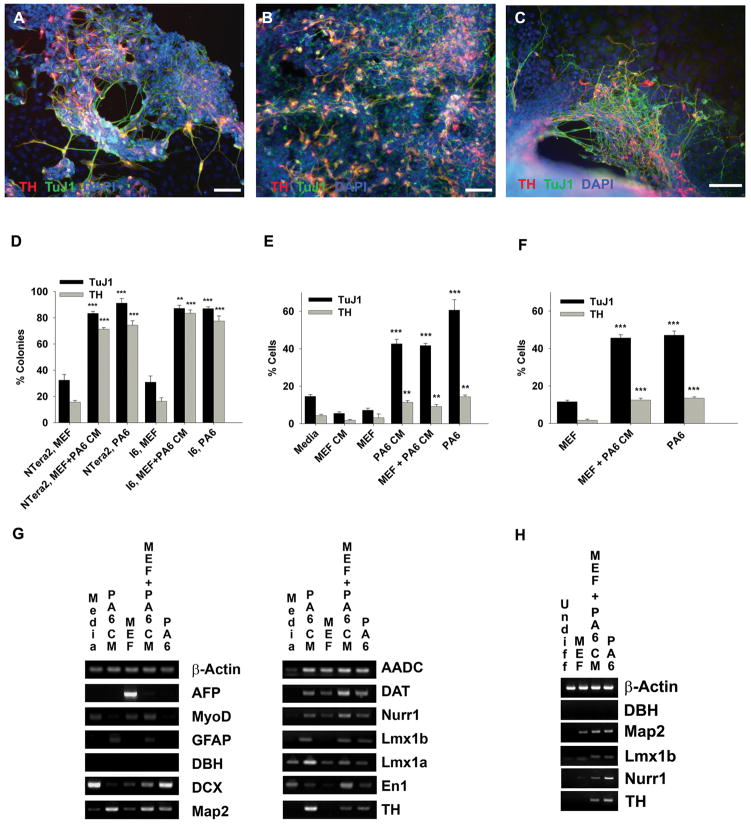
Figure 1.
PA6 conditioned medium induces dopaminergic neuron differentiation in NTera2 cells (a human embryonal carcinoma stem cell line) and I6 cells (a human embryonic stem cell line). A,B,C: Immunocytochemistry double labeling of TH (red) and TuJ1 (green) positive cells counterstained with DAPI (blue). NTera2 cells differentiated in the presence of PA6 CM (A), NTera2 cell differentiated by MEF co-culture in the presence of PA6 CM (MEF + PA6 CM) (B), and the I6 cell line differentiated by MEF+PA6 CM (C). Scale bar = 100 μm. D,E,F: The percentage of both TH and TuJ1 positive colonies or cells. The percentage of TUJ1 and TH positive colonies in MEF +PA6 CM and PA6 cultures were compared to the percentage of positive colonies in MEF cultures for both the NTera2 and I6 cell line (D). The percentage of TUJ1 and TH positive cells in PA6 CM cultures was compared to MEF CM and Media cultures in the NTera2 cell line (E). The percentage of TUJ1 and TH positive cells in MEF +PA6 CM and PA6 cultures in the I6 cell line were statistically compared to the percentage of positive colonies in MEF cultures (F). G, H: RT-PCR analysis of midbrain dopaminergic and neuronal markers for NTera2 (G) and I6 (H) cells differentiated for 3 weeks in the indicated conditions. Statistical analyses were performed using both ANOVA and Student’s t-test, both tests resulted in similar significance, Student’s t-test is shown (* = p≤0.05, ** = p≤0.01, *** = p≤0.001).
After 3 weeks of culture, NTera2 (Fig. 1G) and I6 (Fig. 1H) cells were also assessed by RT-PCR for additional DA and neuronal markers. The NTera2 cultures were monitored for alpha-fetoprotein (Afp; a marker for endodermal differentiation) and MyoD (a marker for mesodermal differentiation) to examine differentiation in the non-ectodermal lineages. Both Afp and MyoD transcripts were expressed in cells differentiated by MEF co-culture and not detected in cells differentiated by PA6 CM (Fig. 1G). The astrocytic marker glial fibrillary acidic protein (Gfap) was detected in NTera2 cells differentiated in PA6 CM and MEF+PA6 CM cultures (Fig. 1G). Elevated GFAP immunoreactivity was also observed following differentiation with PA6 CM (data not shown). Dopamine beta-hydroxylase mRNA (Dbh), which is associated with noradrenergic neurons, was not detected by RT-PCR in any of the cultures of differentiated I6 or NTera2 cells (Fig. 1G, H). We also examined DBH immunoreactivity in all NTera2 and I6 cultures and found very few positive cells, less than 0.1% of the total cells in the culture. The neuron-specific protein microtubule associated protein 2 mRNA (Map2) and doublecortin mRNA (Dcx), a marker of immature neurons, were expressed in PA6 CM and MEF+PA6 CM cultures (Fig. 1G). The midbrain DA neuron transcription factors engrailed 1 (En1), LIM homeobox transcription factors 1a and 1b (Lmx1a and Lmx1b), as well as the Nuclear receptor related 1 (Nurr1) were expressed in both PA6 CM and MEF+PA6 CM cultures and also weakly expressed in MEF cultures (Fig. 1G, H). The mRNAs encoding enzymes involved in dopamine synthesis (TH and aromatic L-amino acid decarboxylase (Aadc)) and transport (dopamine transporter (Dat)) were present in cultures differentiated by PA6 CM (Fig. 1G, H). Undifferentiated NTera2 and I6 cells were not positive for any of the above transcripts (Fig. 1H and data not shown).
Genetic and proteomic analysis of candidate stromal factors
We prospectively identified candidate PA6 cell-specific DA-inducing factors by comparing global gene expression profiles of MEF and PA6 cells (Fig. 2). Among the top 500 genes most highly expressed by either PA6 or MEF cells, 405 were shared (Fig. 2A), indicating considerable genetic similarity between these cells lines. As SDIA is likely due to one or more secreted and/or surface anchored proteins, we selected for up-regulated transcripts whose protein products were either secreted or membrane-associated (Supp. Info. Table 2). The genes encoding the candidate secreted proteins HGF, SDF1α, sFRP1 and VEGFD were up-regulated more than 5 fold, as shown by a log-based scatter plot of PA6 versus MEF cell gene expression (Fig. 2B).
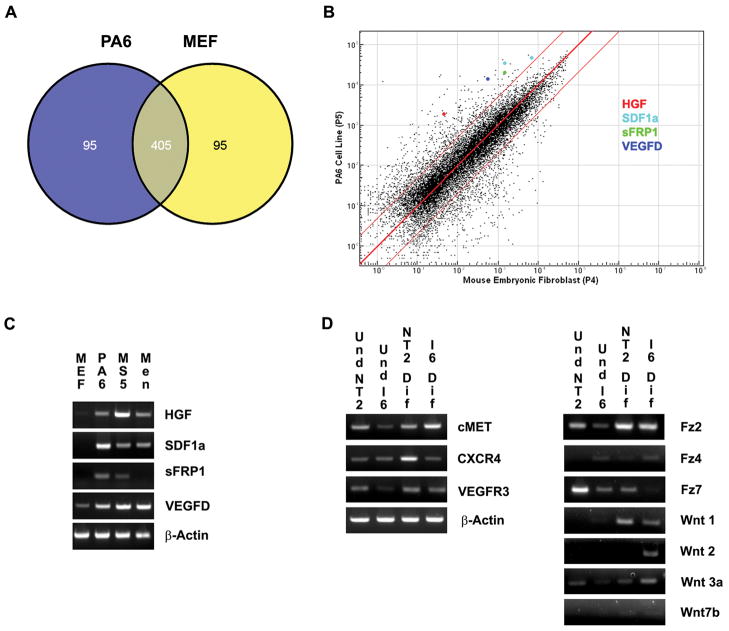
Figure 2.
Gene expression analysis of potential dopaminergic neuron inducing stromal cell factors. A: A Venn-diagram shows the number of genes shared (405 genes) and the number of genes uniquely expressed between MEF (95 genes) and PA6 (95 genes) cells when the top 500 genes most highly expressed were examined. B: A logarithmic scatter plot of microarray gene analysis comparing global gene expression patterns of PA6 and MEF cells, HGF (red), SDF1α (light blue), sFRP1 (green) and VEGFD (dark blue) are highlighted. The red lines indicate a 5 fold or greater change. C: Relative RT-PCR analysis of the transcripts for HGF, SDF1α, SFRP1 and VEGFD in MEF cells, PA6 stromal cells, MS5 stromal cells and E18 meningeal cells (Men). D: RT-PCR analysis of receptors for HGF (c-Met), SDF1α, (CXCR4), VEGF-D (VEGFR3) and sFRP1 (Fz2, Fz4, Fz7, Wnt1, Wnt2, Wnt3a and Wnt7b) are examined in both undifferentiated and differentiated conditions.
RT-PCR was used to confirm the relative levels of selected candidate mRNAs from the microarray results (Fig. 2C). Other cell types including the MS5 mouse stromal cell line and primary mouse E18 meningeal cells have also been shown to possess SDIA (Kawasaki et al., 2000; Hayashi et al., 2008). Therefore, in addition to confirming expression levels of Hgf, Sdf1α, sFrp1 and Vegfd in PA6 cells, we also examined their expression in the MS5 cell line and primary mouse E18 meningeal cells (Fig. 2C). Hgf, Sdf1α, and Vegfd were more highly expressed in PA6, MS5, and primary E18 meningeal cells compared to MEF cells (Fig. 2C). sFrp1 was also expressed in PA6 and MS5 cells, but was faintly present in meningeal cells and was not detected in MEF cells (Fig. 2C). Western blot analysis also indicated that sFRP1 was present in higher amounts in PA6 cells compared to MEF cells (data not shown).
To determine if undifferentiated PSCs or partially differentiated progenitor cells express the corresponding receptors for the candidate factors, we performed RT-PCR for both undifferentiated and differentiated (2 weeks) I6 and NTera2 cells (Fig. 2D). cMet (the receptor for HGF), Vegfr3 (vascular endothelial growth factor receptor 3, the receptor for Vegfd), and CXCR4 (C-X-C chemokine receptor type 4, the receptor for Sdf1α) were expressed in all cultures; however, expression was much weaker in undifferentiated PSCs, particularly in the I6 cell line (Fig. 2D). sFRP1 has several possible receptors/binding partners, including Frizzled (FZ) 2, FZ4, FZ7, WNT1, WNT2, WNT3a and WNT7b (Dennis et al., 1999; Rodriguez et al., 2005; Rosso et al., 2005; Wawrzak et al., 2007; Klein et al., 2008). We found that Fz2, Wnt3a and Fz7 transcripts were expressed in undifferentiated and differentiated cell populations. Fz2 and Wnt3a mRNAs were weakly expressed in undifferentiated I6 cells and Fz7 was weakly expressed in differentiated I6 cells. Fz4 was weakly expressed in undifferentiated and differentiated I6 cells and was not detected in either undifferentiated or differentiated NTera2 cells. Wnt1 mRNA was expressed in both differentiated NTera2 and I6 cells but was not detected in undifferentiated cells. Wnt2 mRNA was expressed in differentiated I6 cells, but was not detected in undifferentiated I6, undifferentiated NTera2 or differentiated NTera2 cells. Wnt7b was very faintly expressed in all cell cultures (Fig. 2D).
We next performed mass spectrometric analysis to determine the protein ‘secretome’ of PA6. We identified 158 different proteins present in PA6 CM; relevant proteins uniquely expressed in PA6 CM (compared to MEF CM) with known developmental and cell signaling functions are presented in Supporting Information Table 3. The relative amount of a given protein in the medium was calculated using spectral counts relative to total counts. Many of the proteins detected by LC-MS/MS analysis of PA6 CM were also highly expressed in our PA6 microarray gene expression profile. The candidate SDIA factor sFRP1 was detected in PA6 CM and not detected in MEF CM; an exemplar MS/MS spectrum for a peptide ion from sFRP1 is shown (Fig. 3A). Notably, we also detected IGF2, neural precursor cell expressed developmentally down-regulated 4 (NEDD4), nidogen 1 (NID1), and talin 1 (TLN1) in PA6 CM, all of which have been implicated in DA differentiation and/or neural development.
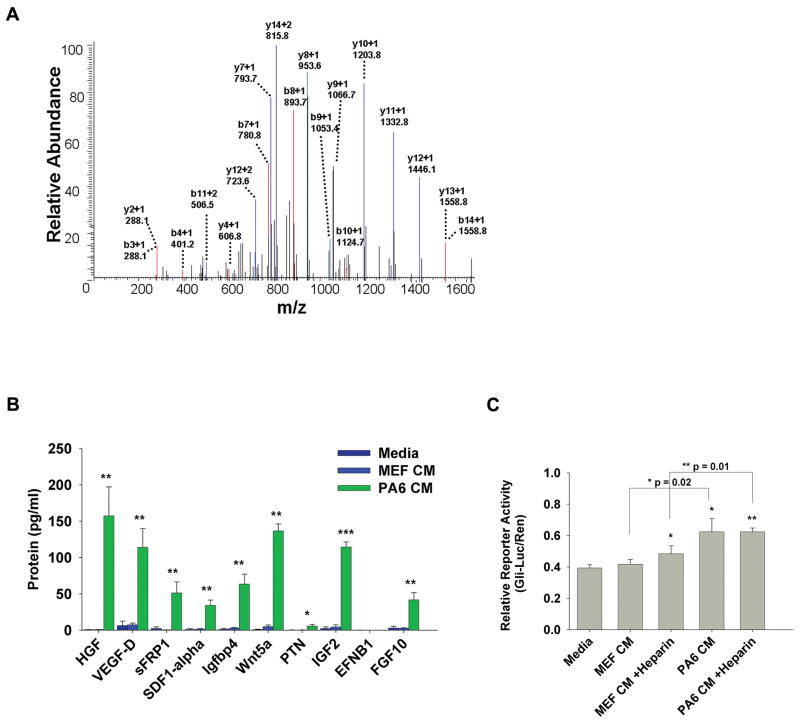
Figure 3.
Protein secretome analysis of potential dopaminergic neuron inducing stromal cell factors. A: MS/MS spectrum of a peptide with amino acid sequence, SEAIIEHLCASEFALR, from sFRP1. sFRP1 was detected in PA6 CM and not MEF CM by mass spectrometry analysis. Peptide fragments are indicated by b-ions and y-ions and their mass to charge values. B: Protein concentrations (pg/mL) using immunoaffinity capillary electrophoresis (ICE) were measured for candidate secreted stromal factors, HGF, VEGFD, SDF1α, IGFBP4, Wnt5a, PTN, IGF2, ENFB1 and FGF10 in Media, MEF CM and PA6 CM. C: A cell-based assay for Shh signaling using the Gli-luciferase reporter system detected the presence of biologically active Shh in media, MEF CM and PA6 CM. * = p≤0.05, ** = p≤0.01, *** = p≤0.001.
To confirm the presence and determine the concentration of relevant proteins in medium conditioned by either PA6 or MEF cells, we also employed immunoaffinity capillary electrophoresis (ICE) (Phillips, 2010). HGF, SDF1α, sFRP1 and VEGFD were present in significantly higher concentrations in PA6 CM compared to either MEF CM or differentiation medium (which was not conditioned by any cell type) (Fig. 3B).
We also examined other candidate factors that have been either identified or alluded to as potential DA or neuron differentiation-inducing components of SDIA including insulin-like growth factor binding protein 4 (IGFBP4) (Vazin et al., 2009; Swistowska et al., 2010), WNT5a (Roussa and Krieglstein, 2004; Hayashi et al., 2008; Chan et al., 2009; Swistowska et al., 2010), PTN (Vazin et al., 2009), IGF2 (Vazin et al., 2009; Swistowska et al., 2010), EFNB1 (Vazin et al., 2009) and fibroblast growth factor 10 (FGF10) (Vazin et al., 2009; Swistowska et al., 2010). IGFBP4, WNT5a, IGF2 and FGF10 were present in substantially higher amounts in PA6 CM compared to either MEF CM or differentiation medium. We were unable to detect EFNB1 in either PA6 CM (0.2 pg/mL), MEF CM (0.2 pg/mL) or differentiation medium (0.2 pg/mL). We also detected a small amount of PTN in PA6 CM (5 pg/mL). The above measurements were performed on PA6 CM, MEF CM and differentiation media all containing heparin. We also measured concentrations of all the above factors in the absence of heparin and the same proteins were detected although concentration levels were slightly lower (data not shown).
Sonic hedgehog (Shh) is known to play a pivotal role in both neural and DA differentiation (Roussa and Krieglstein, 2004) and its role in SDIA was investigated recently (Swistowska et al., 2010). We employed the widely accepted Gli-Luciferase reporter assay to measure biologically active Shh-N and found that PA6 CM contained 3-fold more biologically active Shh-N than either MEF CM or differentiation medium (Fig. 3C). We also examined the effects of heparin on Shh-N levels because of recently discovered interactions between Shh-N and heparan proteoglycans (Chan et al., 2009), and evidence for differing actions of PA6 CM with and without heparin (Vanzin et al., 2008; Swistowska et al., 2010). There was slightly more Shh-N (1.25 fold) in MEF CM containing heparin, but heparin did not alter the relative levels of Shh-N in PA6 CM.
Candidate SDIA factors SDF1α, sFRP1 and VEGFD increase dopaminergic neuronal differentiation
We first tested candidate DA neuron-inducing PA6 factors using the NTera2 cell line to determine appropriate factors and concentrations (Fig. 4 and Supp. Info. Fig. 1), followed by further testing in the hESC line I6 (Fig. 5, Supp. Info. Fig. 1, and Supp. Info. Fig. 2). NTera2 cells were differentiated for 3 weeks without support cells in differentiation medium (medium that is not conditioned by any cell type) alone or supplemented with identified stromal factors HGF, SDF1α, sFRP1 and/or VEGFD (Fig. 4 and Supp. Info. Fig. 1). Cells were assessed for immunoreactivity to TuJ1 and TH antibodies counterstained with DAPI to label all cells (Fig. 4A–F); the percentages of total positive cells were determined for each antigen (Fig. 4G). We observed no detectable change in the number of TuJ1 or TH positive cells following the addition of HGF (100 ng/mL) compared to differentiation medium (Fig. 4B and Fig. 4G). Larger or smaller doses of HGF failed to have an effect on differentiation (Supp. Info. Fig. 1D). The addition of stromal factors SDF1α (100 ng/mL), sFRP1 (2.5 μg/mL), VEGFD (50 ng/ml) or a combination of all factors (at the above concentrations) generated a greater percentage of TuJ1 and TH positive cells than differentiation media (Fig. 4C–G and Supp. Info. Fig. 1). The total number of cells positive for TuJ1 and TH tended to increase with increasing amounts of SDF1α and VEGFD (Supp. Info. Fig. 1). Interestingly, using several sources of recombinant sFRP1 protein, we observed that concentrations over 2.5 μg/mL of sFRP1 resulted in massive cell death (Supp. Info. Fig. 1).
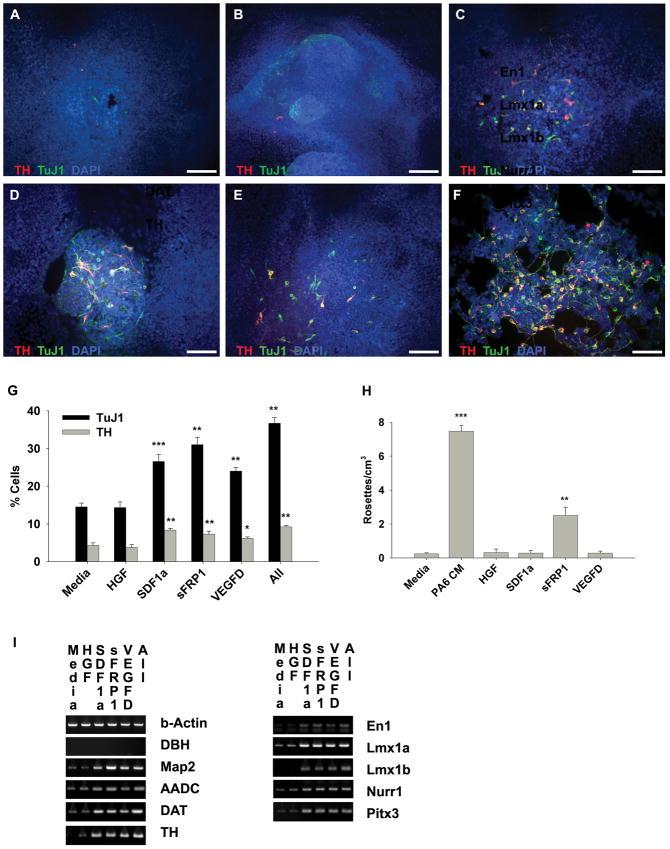
Figure 4.
Recombinant stromal factors SDF1α, sFRP1 and VEGF-D induce dopaminergic neuron differentiation in NTera2 cells, a human embryonal carcinoma stem cell line. A–F: Immunoctyochemistry double labeling of TH (red) and TuJ1(green) positive cells counterstained with DAPI (blue) differentiated in the presence of media (A), or media supplemented with HGF (100 ng/mL) (B), SDF1α (100 ng/mL) (C), sFRP1 (2.5 μg/mL) (D), VEGFD (50ng/mL) (E), or the combination of all factors (F). G: The percentage of both TH and TuJ1 positive cells was increased compared to media alone following 3 weeks of differentiation in the presence of SDF1α, sFRP1, VEGF-D or the combination of all factors. H: The relative number of neural rosettes observed following 2 weeks of differentiation in the indicated conditions. I: Relative RT-PCR analysis for dopaminergic and neuronal markers is shown for differentiated NTera2 cells grown for 3 weeks in the indicated conditions. * = p≤0.05, ** = p≤0.01, *** = p≤0.001. Scale bar = 100 μm.
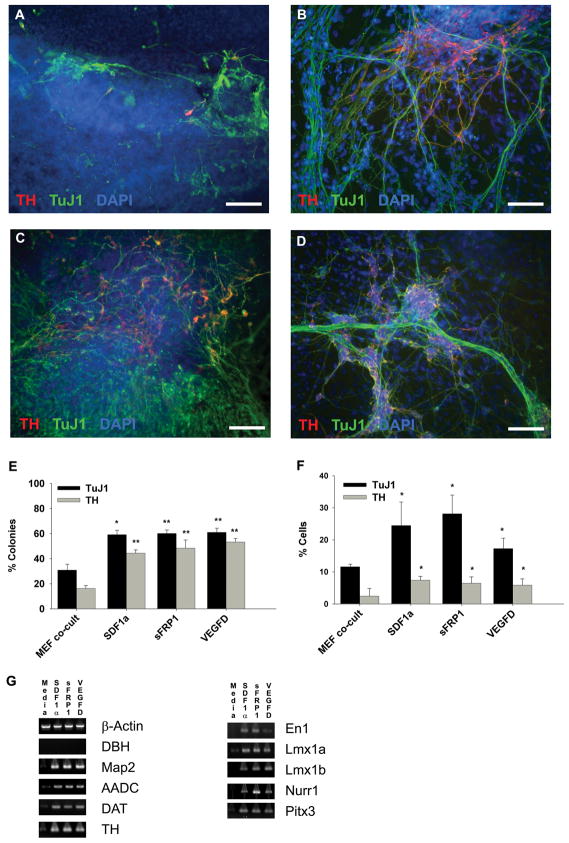
Figure 5.
Stromal factors SDF1α, sFRP1 and VEGF-D induce dopaminergic neuron differentiation in I6 cells, a human embryonic stem cell line. A–D: Immunoctyochemistry double labeling of TH (red) and TuJ1(green) positive cells counterstained with DAPI (blue) differentiated in the presence of MEF co-culture (A), SDF1α (100ng/mL) (B), sFRP1 (2.5 μg/mL) (C), and VEGFD (50ng/mL) (D). E,F: The percentage of TH- and TuJ1-doubly positive colonies (E) and cells (F) following 3 weeks of differentiation in the indicated conditions are presented as a bar graph. G: RT-PCR analysis for dopaminergic and neuronal markers is shown for differentiated I6 cells grown for 3 weeks in the indicated condition. * = p≤0.05, ** = p≤0.01, *** = p≤0.001. Scale bar = 100 μm.
Previously, we observed that NTera2 cells cultured in the presence of PA6 CM form neural rosette-like structures (Schwartz et al., 2005) and so quantified the number of neural rosettes formed following 12 days of differentiation in differentiation medium alone or supplemented with HGF, SDF1α, sFRP1 or VEGFD (Fig. 4H). sFRP1 generated a 10-fold increase in the number of rosette structures, compared to differentiation medium, or medium plus either HGF, SDF1α, sFRP1 or VEGFD (Fig. 4H).
To further evaluate DA and neural markers present in cultures differentiated for 3 weeks we performed RT-PCR analysis on NTera2 cells cultured in differentiation medium or supplemented with either HGF, SDF1α, sFRP1, VEGFD or the combination of all factors (Fig. 4I). Dbh mRNA was not expressed in any of the cultures; similarly, few cells were immunoreactive for DBH (data not shown). Map2, Aadc, Dat, Th, En1, Lmx1a, Lmx1b, Nurr1 and Pitx3 were all more highly expressed in cultures differentiated with SDF1α, sFRP1, VEGFD, or a combination of all factors compared to cultures differentiated with differentiation media alone or differentiation media supplemented HGF.
As the NTera2 cells and hESCs have similar abilities to generate DA neurons and respond to differentiating stimuli (Schwartz et al., 2005), we performed comparable perturbation experiments with the hESC line I6. I6 cells were differentiated for 3 weeks by MEF co-culture, as described previously described in Figure 1, and medium was supplemented with SDF1α, sFRP1 or VEGFD (Fig. 5). Experiments with HGF were not performed because there was no evidence of an effect on NTera2 cell differentiation. Cells were assessed for immunoreactivity to TuJ1 and TH and counterstained with DAPI (Fig. 5A–D) and initially examined for positive colonies (Fig. 5E) followed by total positive cells (Fig. 5F,G). Numbers of TuJ1 and TH positive colonies at 2 weeks of differentiation were similar to those at 3 weeks of differentiation, albeit the effects of the factors were less pronounced (Supp. Info. Fig. 2). Following 3 weeks of differentiation we observed an increase in the number of the colonies containing TuJ1 and TH positive cells following exposure to SDF1α, sFRP1, or VEGFD compared to MEF co-culture (Fig. 5E). To further quantify differentiation we counted TuJ1 and TH immunoreactive cells as a percentage of total cells and found results that paralleled the colony count data, and our previous results with the NTera2 cell line (Fig. 5F). The addition of SDF1α, sFRP1, or VEGFD resulted in an increase in the number of TuJ1 and TH positive cells compared to differentiation medium alone (Fig. 5G). Numbers of cells positive for TuJ1 and TH tended to increase with increasing amounts of SDF1α, sFRP1, and VEGFD (Supp. Info. Fig. 1E–G). We also quantified the number of nestin-positive colonies following 2 weeks of differentiation (Supp. Info. Fig. 2B,C); none of the factors significantly altered the number of nestin-positive colonies.
RT-PCR analysis demonstrated that Dbh mRNA was not expressed in any of the cultures (Fig. 5G); similarly, few cells were immunoreactive for DBH (less than 0.1% of total cells in culture). Map2, Aadc, Dat, Th, En1, Lmx1a, Lmx1b, Nurr1 and Pitx3 were all more highly expressed in cultures differentiated with SDF1α, sFRP1, or VEGFD than in cultures grown in differentiation medium (Fig. 5G). Interestingly, we noticed a much higher level of Nurr1 mRNA in cultures differentiated with sFRP1 compared to those treated with either SDF1α or VEGFD.
DISCUSSION
Co-culture with the mouse stromal cell line PA6 can efficiently generate DA neurons from various PSCs including mouse ESC (Kawasaki et al., 2000; Cajánek et al., 2009), non-human primate ESC (Morizane et al., 2002), hESCs (Buytaert-Hoefen et al., 2004; Roy et al., 2006; Hayashi et al., 2008), and hECSCs (Schwartz et al., 2005). The spectrum of cell types possibly affected by SDIA, in combination with potentially numerous unknown SDIA factors has complicated efforts to elucidate the factor(s) responsible for generating DA neurons. Several groups have implicated PA6 cell factors including SDF1α, IGF2, PTN, EFNB1 (Vazin et al., 2009), Wnt5a (Hayashi et al., 2008), and Shh (Swistowska et al., 2010); however, none of these factors have previously been shown to either be secreted by PA6 cells or present in PA6 CM. We identified SDF1α, sFRP1 and VEGFD as novel factors present in PA6 CM that contribute to DA differentiation in both hECSCs and hESCs. Our findings offer a reproducible and simplified protocol for DA derivation of PSCs, and provide novel insight into possible mechanisms controlling DA differentiation that may aid in advancement towards clinically-relevant PSC-derived DA neurons for cell replacement therapy.
It was originally proposed that SDIA is associated with the cell surface and exists as a membrane-anchored protein(s) (Kawasaki et al., 2000). However, similar to previous findings (Schwartz et al., 2005; Yamazoe et al., 2005; Hayashi et al., 2008; Vazin et al., 2008) we found that PA6 CM can generate DA neurons with midbrain characteristics from PSCs, suggesting that secreted factors contribute to SDIA. Conflicting results have been reported with respect to the DA differentiation capability of PA6 CM on PSCs. We believe these differences could be due to the presence or absence of heparin during the preparation of PA6 CM. Heparin or endogenous heparin sulfate proteoglycans exist at the cell surface and also in the extracellular matrix where they interact with growth factors to potentiate their biological activity (Yanagishita et al., 1992); therefore, heparin is commonly added in the preparation of CM or in instances of growth factor supplementation. PA6 CM generated in the absence of heparin was unable to generate DA neurons from ESCs (Kawasaki et al., 2000; Swistowska et al., 2010). Vazin and colleagues (Vazin et al., 2008) examined the effects of heparin addition to PA6 CM on differentiation of hESCs or neural progenitors and found that PA6 CM generated with heparin, but not without heparin, resulted in DA neuron differentiation of hESCs. In our experiments heparin was added during the preparation of PA6 CM and resulted in effective DA differentiation of both hECSCs and hESCs. We have previously observed DA differentiation of hECSC-derived neural progenitors in PA6 CM prepared in the absence of heparin (Schwartz et al., 2005). Together this suggests that heparin bound soluble factor(s) likely contribute to neural and DA neuron induction from PSCs, while soluble non-heparin bound factor(s) could affect DA differentiation at a progenitor stage. It is also possible that neural and/or DA neuron inducing factors from PA6 cells are autocrine soluble factors and the addition of heparin further releases these factors in sufficient concentrations for DA neuron induction. Endogenous expression of heparin sulfate proteoglycans in PA6 cells has not yet been examined. However, we did detect high expression of perlecan and other enzymes involved in proteoglycan biosynthesis, transport and metabolism in our PA6 microarray data set, consistent with autocrine growth factor release.
We comparatively examined large-scale gene expression profiles of PA6 and MEF cells. Consistent with previous findings (Hayashi et al., 2008; Vazin et al., 2009; Swistowska et al., 2010), we observed an up-regulation of delta-like 1 homolog (Dlk1), growth arrest-specific 6 (Gas6), Igfbp4 and Wnt5a transcripts in our PA6 microarray results. However, other factors previously suggested as candidate SDIA factors and/or up-regulated in PA6 cells including Ptn, Igf2, Fgf10, Efnb1 and transforming growth factor β3 (Tgfβ3) (Hayashi et al., 2008; Vazin et al., 2009; Swistowska et al., 2010), were not consistently up-regulated in our PA6 microarray results. Of additional interest, we observed high expression of the BMP inhibitors Gremlin and Chordin in PA6 cells, which are known neural inducers and may play a role in observed SDIA neural induction. Moreover, we observed an up-regulation of several novel factors including Vegfd, SPARC related modular calcium binding 1 (Smoc1), chemokine (C-C motif) ligand 9 (Ccl9), spondin 2 (Spon2) and neuritin 1(Nrn1), which have roles in embryonic development, neuronal survival and/or dendritic outgrowth (Burstyn-Naeve et al., 1997; Cohen et al., 1999). We choose to focus on genes up-regulated in PA6 cells encoding for potentially secreted proteins, and so selected candidate factors HGF, SDF1α, sFRP1 and VEGFD which are each heparin-binding proteins (Matsumoto et al., 1993; Orlandini and Oliviero, 2001) and available as recombinant proteins.
Importantly, we established that HGF, SDF1α, sFRP1 and VEGFD are present in PA6 CM. Several groups have implicated PA6 cell factors including SDF1α, IGFBP4, IGF2, PTN, EFNB1, FGF10, Wnt5a and Shh (Hayashi et al., 2008; Vazin et al., 2009; Swistowska et al., 2010) based on gene expression studies similar to ours; however, thus far none of these factors have been shown to either be secreted by PA6 cells or present in PA6 CM. Our mass spectrometry and ICE studies presented here are the first studies to examine the proteins present in PA6 CM. Of particular importance, our mass spectrometry analysis prospectively identified our candidate factor sFRP1 as present in PA6 CM. We also detected IGF2 in PA6 CM by mass spectrometry, a factor which has been demonstrated to have roles in neuronal differentiation and survival (Pera et al., 2003; Silva et al., 2009). Vazin and colleagues recently identified IGF2 as a SDIA component that increased DA neuron differentiation of hESC-derived progenitors (Vazin et al., 2009). Our ICE analysis confirmed the presence of the SDIF candidates HGF, SDF1α, sFRP1 and VEGFD in PA6 CM and also determined that their protein concentrations were significantly higher in PA6 CM compared to MEF CM or differentiation medium not conditioned by any cell type. As several recent studies have either alluded to or examined the roles of IGFBP4, IGF2, Wnt5a, PTN, EFNB1 and FGF10 in SDIA DA induction (Hayashi et al., 2008; Vazin et al., 2009; Swistowska et al., 2010), we also determined their presence and concentration in PA6 CM by ICE. We found that PA6 CM contained much higher concentrations of Wnt5a, IGF2, IGBP4, and FGF10 proteins than MEF CM and differentiation medium. Therefore, based on previous studies, these factors likely affect DA differentiation (Catelo-Branco; Hayashi et al., 2008; Parish et al., 2008; Vazin et al., 2009). Although, PTN and EFNB1 were also highly expressed in the present and previously reported microarray analysis of PA6 cells (Vazin et al., 2009), our ICE experiments revealed that levels of these two proteins were very low in PA6 CM, indicating PA6 cells likely do not secrete these factors efficiently.
Recent efforts in studies of ESC-derived DA neuron-like cells have focused on identifying the diffusible factors involved in SDIA (Hayashi et al., 2008; Vazin et al., 2009; Swistowska et al., 2010; Yamazoe et al., 2005) and elucidating the mechanisms operating during development (Simeone, 2005; Abeliovich and Hammond, 2007) in order to generate DA neurons efficiently. Based on the presence of our candidate factors HGF, VEGFD, SDF1α and sFRP1 in both PA6 cells and PA6 CM, we pursued perturbation studies, first in the NTera2 hECSCs and later in the I6 hESCs, to determine whether these proteins promote DA neuron differentiation. Although the HGF receptor c-Met mRNA was expressed in both undifferentiated and differentiated NTera2 and I6 cells, and despite the high concentration of HGF protein in PA6 CM determined by ICE studies, we were unable to detect an effect of recombinant HGF on DA or neuronal differentiation. However, it is possible that HGF may play a role in a process other than differentiation, such as neuronal survival (Hamanoue et al., 2006; Lan et al., 2008), or have a stage-specific effect. CXCR4, the receptor for our candidate factor SDF1α, is known to be expressed in developing midbrain DA neurons while SDF1α is expressed in the meningeal tissue surrounding the ventral mesencephalon where it affects DA neuron migration and process orientation during development (Edman, 2009). Similar to the results of Edman and colleagues (Edman, 2009), we found that CXCR4 with TH immunoreactivities were co-localized in the developing murine midbrain and in our NTera2-derived DA neurons (C.M.S., unpublished findings). In the present study we found that Cxcr4 mRNA is weakly expressed in undifferentiated NTera2 and I6 cells and more strongly expressed in their differentiated progeny suggesting these cells possess the necessary signaling components to respond to SDF1α. Treatment with SDF1α increased DA neuronal differentiation in both the NTera2 and I6 cells. Similar to our findings, Vazin et al. showed that addition of SDF1α to hESC-derived progenitors resulted in increased DA neuron differentiation (Vazin et al., 2009).
We found that recombinant sFRP1 increased the number of neural rosettes observed in the NTera2 cell line and the DA neuronal differentiation in both the NTera2 and I6 cell line, indicating that sFRP1 may have both neuronal- and DA-inducing effect. The sFRP family of proteins shares a cysteine-rich domain that permits interactions with Fz receptors as well as Wnts. sFRP1 is known to bind to Wnt1 (Dennis et al., 1999), Wnt2 (Klein et al., 2008), Wnt 3a (Wawrzak et al., 2007) and Wnt 7b (Rosso et al., 2005), therefore reducing its ability to activate downstream signaling cascades. Recently, Čajánek et al. found that loss of Wnt1 signaling or treatment of mESCs with a Wnt/ β-catenin pathway inhibitor resulted in increased neuroectoderm and DA differentiation (Canjánek et al., 2009). We observed that Wnt1 mRNA was expressed in very low levels in undifferentiated cells and up-regulated in progenitors. Therefore, possible blockade of Wnt1 signaling through sFRP1 may explain why we see increased DA differentiation in hECSCs and hESCs and neural rosettes in hECSCs. Uren and colleagues showed sFRP1 acts as a biphasic modulator of Wnt signaling; low concentrations enhance Wnt signaling, whereas high concentrations inhibit Wnt’s actions. Consistent with these results, Kele and colleagues demonstrated that sFRP1 is expressed in the developing ventral midbrain and acts as a competitive antagonist of Wnt signaling at low concentrations, whereas high concentrations of sFRP1 resulted in cell death (Kele-Olovsson, 2009). Similarly we observed massive cell death of undifferentiated NTera2 cells and derived neuronal progenitors following high doses of sFRP1. Although it is unclear how sFRP1 signals in PSCs or their derived progenitors, we found that sFRP1 is a DA neuron-inducing component of SDIA.
We observed that VEGFD induced DA differentiation in hECSC and hESC. VEGFD is known to activate the VEGFR3 receptor. In a recent study, Englund-Johansson et al. found that VEGFD was among several factors possibly regulating survival of human neural progenitors in a retinal explant model (Englund-Johansson et al., 2010). A detailed study by Choi et al. examined distribution of VEGFR3 in the developing forebrain and eye and found expression was mainly distributed in proliferating progenitors and some mature post-mitotic neurons (Choi et al., 2010). We found the mRNA expression of Vegfr3 to be higher in hECSC- and hESC-derived progenitors compared to undifferentiated cells. Although the functions of VEGFD in the CNS have not been established, VEGFB is neuroprotective in rat midbrain cultures exposed to DA neurotoxins (Falk et al., 2009) and VEGF has numerous functions in the CNS including neuroprotection and neurogenesis (Yasuhara et al., 2004).
Supplementary Material
. Differentiation dose response of NTera2 cells and I6 cells to identified factors. A–D: The percentage of TH and TuJ1 positive cells following 3 weeks of differentiation of NTera2 cells in SDF1α (A), sFRP1 (B), VEGFD (C) and HGF (D). All statistical comparisons were performed using the Student’s t-test comparing Media versus each condition (A–D). E–G: The percentage of TH and TuJ1 positive cells following 3 weeks of differentiation of I6 cells in SDF1α (E), sFRP1 (F), VEGFD (G). All statistical comparisons were performed using the Student’s t-test comparing MEF versus each condition. * = p≤0.05, ** = p≤0.01, *** = p≤0.001.
Addition of identified stromal factors influences the differentiation of I6 cells. A: Following 2 weeks of differentiation with the addition of either SDF1α (100 ng/mL), sFRP1 (2.5 μg/mL) or VEGFD (50 ng/mL) colonies were assessed TuJ1 and TH immunoreactivity. B: The percentage of Nestin positive colonies in the indicated condition was assessed following 2 weeks of differentiation. C: A representative image Nestin (green) counterstained with DAPI (blue) of I6 cells differentiated for 2 weeks in the presence of sFRP1 (2.5 μg/mL). * = p≤0.05, ** = p≤0.01, *** = p≤0.001.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institute on Aging and by the Intramural Research Program of the National Institute for Biomedical Imaging and Bioengineering. We thank Dr. Jeff Rubin, National Institutes of Health, National Cancer Institute for kindly providing recombinant sFRP1. We would also like to thank Dr. Kevin Becker and William Wood at the National Institute for Aging, Microarray Core Facility for running the microarray experiments and their assistance with analysis. We thank Jessica DeLeon, Dr. Michael O’Connell and Dr. Ashani Weeraratna at the National Institute on Aging for their time, advice and initial work with western blot experiments. This research was included as part of Catherine Schwartz’s Doctoral dissertation titled “Derivation, Enrichment and Characterization of Dopaminergic Neurons from Pluripotent Stem Cells”, http://diss.kib.ki.se/2010/978-91-7409-867-9/thesis.pdf, Karolinska Institutet Press, 2010.
Funding acknowledgements: This research was supported by the Intramural Research Program of the National Institute on Aging and by the Intramural Research Program of the National Institute for Biomedical Imaging and Bioengineering.
References
- Arumugam TV, Phillips TM, Cheng A, Morrell CH, Mattson MP, Wan R. Age and energy intake interact to modify cell stress pathways and stroke outcome. Ann Neurol. 2010;67:41–52. doi: 10.1002/ana.21798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeliovich A, Hammond R. Midbrain dopamine neuron differentiation: factors and fates. Dev Biol. 2007;304:447–454. doi: 10.1016/j.ydbio.2007.01.032. [DOI] [PubMed] [Google Scholar]
- Burstyn-Cohen T, Tzarfaty V, Frumkin A, Feinstein Y, Stoeckli E, Klar A. F-Spondin is required for accurate pathfinding of commissural axons at the floor plate. Neuron. 1999;23:233–246. doi: 10.1016/s0896-6273(00)80776-x. [DOI] [PubMed] [Google Scholar]
- Buytaert-Hoefen KA, Alvarez E, Freed CR. Generation of tyrosine hydroxylase positive neurons from human embryonic stem cells after coculture with cellular substrates and exposure to GDNF. Stem Cells. 2004;22:669–674. doi: 10.1634/stemcells.22-5-669. [DOI] [PubMed] [Google Scholar]
- Cajánek L, Ribeiro D, Liste I, Parish CL, Bryja V, Arenas E. Wnt/beta-catenin signaling blockade promotes neuronal induction and dopaminergic differentiation in embryonic stem cells. Stem Cells. 2009;27:2917–2927. doi: 10.1002/stem.210. [DOI] [PubMed] [Google Scholar]
- Castelo-Branco G, Wagner J, Rodriguez FJ, Kele J, Sousa K, Rawal N, Pasolli HA, Fuchs E, Kitajewski J, Arenas E. Differential regulation of midbrain dopaminergic neuron development by Wnt-1, Wnt-3a, and Wnt-5a. Proc Natl Acad Sci USA. 2003;100:12747–12755. doi: 10.1073/pnas.1534900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JA, Balasubramanian S, Witt RM, Nazemi KJ, Choi Y, Pazyra-Murphy MF, Walsh CO, Thompson M, Segal RA. Proteoglycan interactions with Sonic Hedgehog specify mitogenic responses. Nat Neurosci. 2009;12:409–417. doi: 10.1038/nn.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MS, Lee YE, Kim JY, Chung S, Cho YH, Kim DS, Kang SM, Lee H, Kim MH, Kim JH, Leem JW, Oh SK, Choi YM, Hwang DY, Chang JW, Kim DW. Highly efficient and large-scale generation of functional dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci USA. 2008;105:3392–3397. doi: 10.1073/pnas.0712359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JS, Shin YJ, Lee JY, Yun H, Cha JA, Choi JY, Chun MH, Lee MY. Expression of vascular endothelial growth factor receptor-3 mRNA in the rat developing forebrain and retina. J Comp Neurol. 2010;518:1064–1081. doi: 10.1002/cne.22263. [DOI] [PubMed] [Google Scholar]
- Dennis S, Aikawa M, Szeto W, d’Amore PA, Papkoff J. A secreted frizzled related protein, FrzA, selectively associates with Wnt-1 protein and regulates wnt-1 signaling. J Cell Sci. 1999;112:3815–3820. doi: 10.1242/jcs.112.21.3815. [DOI] [PubMed] [Google Scholar]
- Edman LC. Chemokines and their role in dopaminergic development. Larserics digital print. Karolinska Institutet Press; Stockholm, Sweden: 2009. [Google Scholar]
- Englund-Johansson U, Mohlin C, Liljekvist-Soltic I, Ekström P, Johansson K. Human neural progenitor cells promote photoreceptor survival in retinal explants. Exp Eye Res. 2010;90:292–299. doi: 10.1016/j.exer.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Falk T, Zhang S, Sherman SJ. Vascular endothelial growth factor B (VEGF-B) is up-regulated and exogenous VEGF-B is neuroprotective in a culture model of Parkinson’s disease. Mol Neurodegener. 2009;4:49. doi: 10.1186/1750-1326-4-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed CR, Breeze RE, Rosenberg NL, Schneck SA, Kriek E, Qi J, Lone T, Zhang Y, Snyder JA, Wells TH, Ramig LO, Thompson L, Mazziotta JC, Huang SC, Grafton ST, Brooks D, Sawle G, Schroter G, Ansari AA. Survival of implanted fetal dopamine cells and neurologic improvement 12 to 46 months after transplantation for Parkinson’s disease. N Engl J Med. 1992;327:1549–1555. doi: 10.1056/NEJM199211263272202. [DOI] [PubMed] [Google Scholar]
- Hamanoue M, Takemoto N, Matsumoto K, Nakamura T, Nakajima K, Kohsaka S. Neurotrophic effect of hepatocyte growth factor on central nervous system neurons in vitro. J Neurosci Res. 1996;43:554–564. doi: 10.1002/(SICI)1097-4547(19960301)43:5<554::AID-JNR5>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Morizane A, Koyanagi M, Ono Y, Sasai Y, Hashimoto N, Takahashi J. Meningeal cells induce dopaminergic neurons from embryonic stem cells. Eur J Neurosci. 2008;27:261–268. doi: 10.1111/j.1460-9568.2008.06027.x. [DOI] [PubMed] [Google Scholar]
- Hyman JM, Firestone AJ, Heine VM, Zhao Y, Ocasio CA, Han K, Sun M, Rack PG, Sinha S, Wu JJ, Solow-Cordero DE, Jiang J, Rowitch DH, Chen JK. Small-molecule inhibitors reveal multiple strategies for Hedgehog pathway blockade. Proc Natl Acad Sci USA. 2009;106:14132–137. doi: 10.1073/pnas.0907134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki H, Mizuseki K, Nishikawa S, Kaneko S, Kuwana Y, Nakanishi S, Nishikawa SI, Sasai Y. Induction of midbrain dopaminergic neurons from ES cells by stromal cell-derived inducing activity. Neuron. 2000;28:31–40. doi: 10.1016/s0896-6273(00)00083-0. [DOI] [PubMed] [Google Scholar]
- Kele-Olovsson JM. Regulation Of Midbrain Dopaminergic Neuron Development By Wnts, Sfrps And bHLH Proteins. Karolinska Institutet Press; Stockholm, Sweden: 2009. Larserics digital print. [Google Scholar]
- Klein D, Demory A, Peyre F, Kroll J, Augustin HG, Helfrich W, Kzhyshkowska J, Schledzewski K, Arnold B, Goerdt S. Wnt2 acts as a cell type-specific, autocrine growth factor in rat hepatic sinusoidal endothelial cells cross-stimulating the VEGF pathway. Hepatology. 2008;47:1018–1031. doi: 10.1002/hep.22084. [DOI] [PubMed] [Google Scholar]
- Kordower JH, Freeman TB, Snow BJ, Vingerhoets FJ, Mufson EJ, Sanberg PR, Hauser RA, Smith DA, Nauert GM, Perl DP, Olanow CW. Neuropathological evidence of graft survival and striatal reinnervation after the transplantation of fetal mesencephalic tissue in a patient with Parkinson’s disease. N Engl J Med. 1995;332:1118–1124. doi: 10.1056/NEJM199504273321702. [DOI] [PubMed] [Google Scholar]
- Lan F, Xu J, Zhang X, Wong VW, Li X, Lu A, Lu W, Shen L, Li L. Hepatocyte growth factor promotes proliferation and migration in immortalized progenitor cells. Neuroreport. 2008;19:765–769. doi: 10.1097/WNR.0b013e3282fdf69e. [DOI] [PubMed] [Google Scholar]
- Maina F, Hilton MC, Ponzetto C, Davies AM, Klein R. Met receptor signaling is required for sensory nerve development and HGF promotes axonal growth and survival of sensory neurons. Genes Dev. 1997;11:3341–3350. doi: 10.1101/gad.11.24.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Tajima H, Okazaki H, Nakamura T. Heparin as an inducer of hepatocyte growth factor. J Biochem. 1993;114:820–826. doi: 10.1093/oxfordjournals.jbchem.a124262. [DOI] [PubMed] [Google Scholar]
- Morizane A, Takahashi J, Takagi Y, Sasai Y, Hashimoto N. Optimal conditions for in vivo induction of dopaminergic neurons from embryonic stem cells through stromal cell-derived inducing activity. J Neurosci Res. 2002;69:934–939. doi: 10.1002/jnr.10363. [DOI] [PubMed] [Google Scholar]
- Naeve GS, Ramakrishnan M, Kramer R, Hevroni D, Citri Y, Theill LE. Neuritin: a gene induced by neural activity and neurotrophins that promotes neuritogenesis. Proc Natl Acad Sci USA. 1997;94:2648–2653. doi: 10.1073/pnas.94.6.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandini M, Oliviero S. In fibroblasts Vegf-D expression is induced by cell-cell contact mediated by cadherin-11. J Biol Chem. 2001;276:6576–6681. doi: 10.1074/jbc.M009573200. [DOI] [PubMed] [Google Scholar]
- Parish CL, Castelo-Branco G, Rawal N, Tonnesen J, Sorensen AT, Salto C, Kokaia M, Lindvall O, Arenas E. Wnt5a-treated midbrain neural stem cells improve dopamine cell replacement therapy in parkinsonian mice. J Clin Invest. 2008;118:149–160. doi: 10.1172/JCI32273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pera EM, Ikeda A, Eivers E, De Robertis EM. Integration of IGF, FGF, and anti-BMP signals via Smad1 phosphorylation in neural induction. Genes Dev. 2003;17:3023–3028. doi: 10.1101/gad.1153603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips TM, Wellner EF. Analysis of inflammatory biomarkers from tissue biopsies by chip-based immunoaffinity CE. Electrophoresis. 2007;28:3041–3048. doi: 10.1002/elps.200700193. [DOI] [PubMed] [Google Scholar]
- Phillips TM. Immunoaffinity techniques in analysis. J Chromatogr B Analyt Technol Biomed Life Sci. 2010;878:113–114. doi: 10.1016/j.jchromb.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccini P, Brooks DJ, Björklund A, Gunn RN, Grasby PM, Rimoldi O, Brundin P, Hagell P, Rehncrona S, Widner H, Lindvall O. Dopamine release from nigral transplants visualized in vivo in a Parkinson’s patient. Nat Neurosci. 1999;2:1137–1140. doi: 10.1038/16060. [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Esteve P, Weinl C, Ruiz JM, Fermin Y, Trousse F, Dwivedy A, Holt C, Bovolenta P. SFRP1 regulates the growth of retinal ganglion cell axons through the Fz2 receptor. Nat Neurosci. 2005;8:1301–1309. doi: 10.1038/nn1547. [DOI] [PubMed] [Google Scholar]
- Rosso SB, Sussman D, Wynshaw-Boris A, Salinas PC. Wnt signaling through Dishevelled, Rac and JNK regulates dendritic development. Nat Neurosci. 2005;8:34–42. doi: 10.1038/nn1374. [DOI] [PubMed] [Google Scholar]
- Roussa E, Krieglstein K. Induction and specification of midbrain dopaminergic cells: focus on SHH, FGF8, and TGF-beta. Cell Tissue Res. 2004;318:23–33. doi: 10.1007/s00441-004-0916-4. [DOI] [PubMed] [Google Scholar]
- Roy NS, Cleren C, Singh SK, Yang L, Beal MF, Goldman SA. Functional engraftment of human ES cell-derived dopaminergic neurons enriched by coculture with telomerase-immortalized midbrain astrocytes. Nat Med. 2006;12:1259–1268. doi: 10.1038/nm1495. [DOI] [PubMed] [Google Scholar]
- Schulz TC, Noggle SA, Palmarini GM, Weiler DA, Lyons IG, Pensa KA, Meedeniya AC, Davidson BP, Lambert NA, Condie BG. Differentiation of human embryonic stem cells to dopaminergic neurons in serum-free suspension culture. Stem Cells. 2004;22:1218–1238. doi: 10.1634/stemcells.2004-0114. [DOI] [PubMed] [Google Scholar]
- Schwartz CM, Spivak CE, Baker SC, McDaniel TK, Loring JF, Nguyen C, Chrest FJ, Wersto R, Arenas E, Zeng X, Freed WJ, Rao MS. NTera2: a model system to study dopaminergic differentiation of human embryonic stem cells. Stem Cells Dev. 2005;14:517–534. doi: 10.1089/scd.2005.14.517. [DOI] [PubMed] [Google Scholar]
- Silva D, Dikkes P, Barnes M, Lopez MF. Decreased motoneuron survival in Igf2 null mice after sciatic nerve transection. Neuroreport. 2009;20:1414–1418. doi: 10.1097/WNR.0b013e328330b735. [DOI] [PubMed] [Google Scholar]
- Simeone A. Genetic control of dopaminergic neuron differentiation. Trends Neurosci. 2005;28:62. doi: 10.1016/j.tins.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Swistowska AM, da Cruz AB, Han Y, Swistowski A, Liu Y, Shin S, Zhan M, Rao MS, Zeng X. Stage-specific role for shh in dopaminergic differentiation of human embryonic stem cells induced by stromal cells. Stem Cells Dev. 2010;19:71–82. doi: 10.1089/scd.2009.0107. [DOI] [PubMed] [Google Scholar]
- Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Uren A, Reichsman F, Anest V, Taylor WG, Muraiso K, Bottaro DP, Cumberledge S, Rubin JS. Secreted frizzled-related protein-1 binds directly to Wingless and is a biphasic modulator of Wnt signaling. J Biol Chem. 2000;275:4374–4382. doi: 10.1074/jbc.275.6.4374. [DOI] [PubMed] [Google Scholar]
- Vazin T, Chen J, Lee CT, Amable R, Freed WJ. Assessment of stromal-derived inducing activity in the generation of dopaminergic neurons from human embryonic stem cells. Stem Cells. 2008;26:1517–1525. doi: 10.1634/stemcells.2008-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazin T, Becker KG, Chen J, Spivak CE, Lupica CR, Zhang Y, Worden L, Freed WJ. A novel combination of factors, termed SPIE, which promotes dopaminergic neuron differentiation from human embryonic stem cells. PLoS One. 2009;4:e6606. doi: 10.1371/journal.pone.0006606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrzak D, Métioui M, Willems E, Hendrickx M, de Genst E, Leyns L. Wnt3a binds to several sFRPs in the nanomolar range. Biochem Biophys Res Commun. 2007;357:1119–1123. doi: 10.1016/j.bbrc.2007.04.069. [DOI] [PubMed] [Google Scholar]
- Yamazoe H, Murakami Y, Mizuseki K, Sasai Y, Iwata H. Collection of neural inducing factors from PA6 cells using heparin solution and their immobilization on plastic culture dishes for the induction of neurons from embryonic stem cells. Biomaterials. 2005;26:5746–5754. doi: 10.1016/j.biomaterials.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Yanagishita M, Hascall VC. Cell surface heparan sulfate proteoglycans. J Biol Chem. 1992;267:9451–9454. [PubMed] [Google Scholar]
- Yasuhara T, Shing T, Date I. The potential role of vascular endothelial growth factor in the central nervous system. Rev Neurosci. 2004;15:293–307. doi: 10.1515/revneuro.2004.15.4.293. [DOI] [PubMed] [Google Scholar]
- Zhang F, McLellan JS, Ayala AM, Leahy DJ, Linhardt RJ. Kinetic and structural studies on interactions between heparin or heparan sulfate and proteins of the hedgehog signaling pathway. Biochemistry. 2007;46:3933–3941. doi: 10.1021/bi6025424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
. Differentiation dose response of NTera2 cells and I6 cells to identified factors. A–D: The percentage of TH and TuJ1 positive cells following 3 weeks of differentiation of NTera2 cells in SDF1α (A), sFRP1 (B), VEGFD (C) and HGF (D). All statistical comparisons were performed using the Student’s t-test comparing Media versus each condition (A–D). E–G: The percentage of TH and TuJ1 positive cells following 3 weeks of differentiation of I6 cells in SDF1α (E), sFRP1 (F), VEGFD (G). All statistical comparisons were performed using the Student’s t-test comparing MEF versus each condition. * = p≤0.05, ** = p≤0.01, *** = p≤0.001.
Addition of identified stromal factors influences the differentiation of I6 cells. A: Following 2 weeks of differentiation with the addition of either SDF1α (100 ng/mL), sFRP1 (2.5 μg/mL) or VEGFD (50 ng/mL) colonies were assessed TuJ1 and TH immunoreactivity. B: The percentage of Nestin positive colonies in the indicated condition was assessed following 2 weeks of differentiation. C: A representative image Nestin (green) counterstained with DAPI (blue) of I6 cells differentiated for 2 weeks in the presence of sFRP1 (2.5 μg/mL). * = p≤0.05, ** = p≤0.01, *** = p≤0.001.