Summary
A mutation in the LMNA gene is responsible for the most dramatic form of premature aging, Hutchinson-Gilford progeria syndrome (HGPS). Several recent studies have suggested that protein products of this gene might have a role in normal physiological cellular senescence. To explore further LMNA's possible role in normal aging, we genotyped 16 SNPs over a span of 75.4 kb of the LMNA gene on a sample of long-lived individuals (US Caucasians with age ≥95 years, N=873) and genetically matched younger controls (N=443). We tested all common non-redundant haplotypes (frequency ≥ 0.05) based on subgroups of these 16 SNPs for association with longevity. The most significant haplotype, based on 4 SNPs, remained significant after adjustment for multiple testing (OR = 1.56, P=2.5×10−5, multiple-testing-adjusted P=0.0045). To attempt to replicate these results, we genotyped 3448 subjects from four independent samples of long-lived individuals and control subjects from 1) the New England Centenarian Study (NECS) (N=738), 2) the Southern Italian Centenarian Study (SICS) (N=905), 3) France (N=1103), and 4) the Einstein Ashkenazi Longevity Study (N=702). We replicated the association with the most significant haplotype from our initial analysis in the NECS sample (OR = 1.60, P=0.0023), but not in the other three samples (P>.15). In a meta-analysis combining all five samples, the best haplotype remained significantly associated with longevity after adjustment for multiple testing in the initial and follow-up samples (OR = 1.18, P=7.5×10−4, multiple-testing-adjusted P=0.037). These results suggest that LMNA variants may play a role in human lifespan.
Keywords: longevity gene, human, longevity, genetics
Introduction
Single gene mutations have been shown to extend the lifespan of worms, yeast, and mice (Kenyon 2005). Similarly, twin studies have shown that 25–30% of the variance in human lifespan is due to genetic differences (Herskind et al. 1996; Ljungquist et al. 1998; Skytthe et al. 2003). With the completion of the International HapMap Project, power to detect common variants for common and complex diseases has increased (Manolio et al. 2008), and the search for genetic contributors to human aging and lifespan has been accelerated. The results thus far have been somewhat discouraging: only APOE and FOXO3A have been consistently associated with longevity across populations in candidate gene studies (Christensen et al. 2006; Novelli et al. 2008). Even strong candidates such as the WRN gene associated with the premature aging syndrome, Werner syndrome (Castro et al. 2000), have displayed inconsistent results (Christensen et al. 2006). Similarly, all longevity-associated SNPs reaching genome-wide significance in two recently published genome-wide association studies were associated with the APOE gene (Deelen et al. 2011; Nebel et al. 2011). However, because variants with subtler associations with longevity will not be detectable at genome-wide significance levels with conventional sample sizes, candidate gene studies remain a useful approach.
One major candidate gene for longevity that has not yet been explored is LMNA. Defects in the LMNA gene, which encodes the nuclear envelope proteins lamin A and lamin C, have been associated with at least 13 diseases, including Hutchinson-Gilford progeria syndrome (HGPS) (Gruenbaum et al. 2005; Capell & Collins 2006). HGPS is a rare but devastating condition in which symptoms resembling premature aging appear at a very young age, typically within a year of birth. Due to accelerated cardiovascular disease leading to eventual heart attack or stroke, individuals with HGPS have an average life expectancy of 13 years (Capell et al. 2007; Merideth et al. 2008).
Scientists have long questioned whether HGPS is truly a model of normal human aging. Although displaying many features typically associated with normal aging (alopecia, skin wrinkling, atherosclerosis), HGPS apparently lacks other common features such as tumors or dementia. However, recent evidence obtained since the identification of mutations in LMNA as the cause of HGPS suggests that there may in fact be a molecular relationship. For example, there is evidence that a very small amount of the mutant lamin A protein produced in HGPS, termed “progerin,” is produced in normal cells (Scaffidi & Misteli 2006; Cao et al. 2007). Levels of the progerin transcript increase with age in normal cells (Rodriguez et al. 2009), and work using a monoclonal antibody specific for progerin has demonstrated that progerin accumulates in human skin in an age-dependent manner (McClintock et al. 2007). Further work in normal human fibroblasts has demonstrated that progerin production is activated by progressive telomere damage during cellular senescence (Cao et al. 2011). Finally, it has been established that numerous other abnormalities present in HGPS are also common in cells from aged individuals. These include nuclear blebbing, particular epigenetic changes, and increased levels of DNA damage. Most remarkably, suppressing the production of the mutant progerin protein through the use of an antisense morpholino reversed all of these abnormalities in cell culture (Scaffidi & Misteli 2006).
The corollary to this observation is the possibility that variants of LMNA that produce less progerin might be protective and thus promote a longer lifespan. To investigate further the potential links of HGPS, LMNA, and progerin to the normal aging process, we sought to determine whether common variants in the LMNA gene might be associated with extreme human longevity. Towards this end, we tested variants of the LMNA gene for association with extreme longevity in five independent samples of long-lived individuals (LLI) and control subjects.
Results
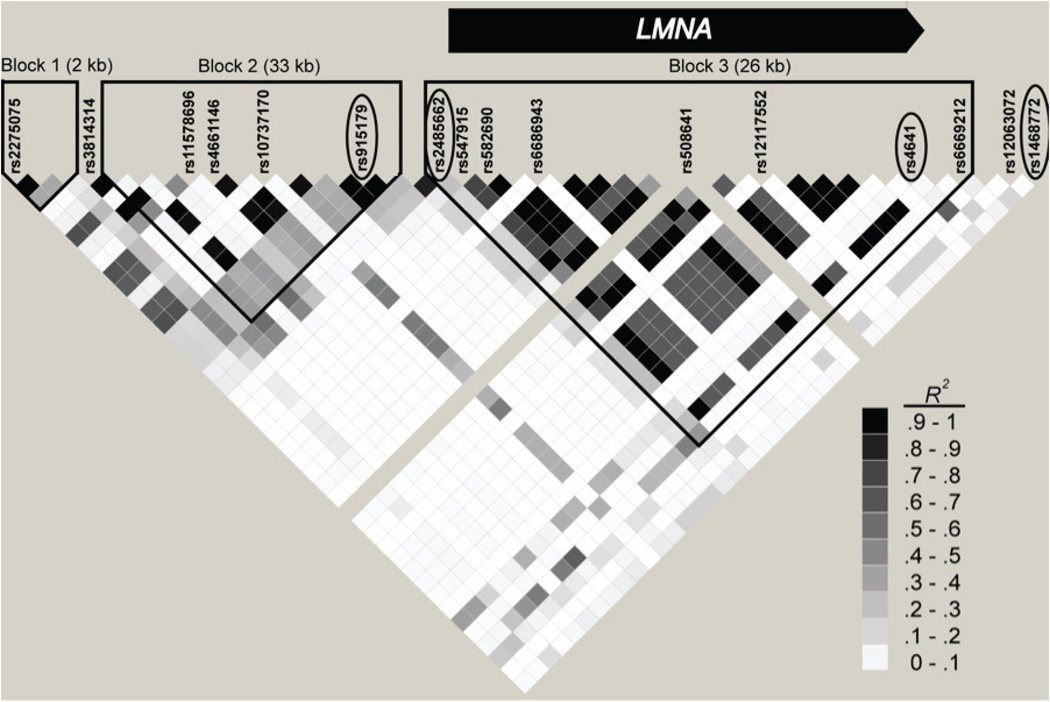
Characteristics of the five study samples are shown in Table 1. In the Stage 1 sample (US Caucasians with age ≥95 years, N=873) and genetically matched younger controls (N=443), we genotyped 16 SNPs covering two coherent linkage disequilibrium (LD) blocks, corresponding to the LMNA gene and promoter region (Figure 1). 2414 non-redundant haplotypes (i.e., haplotypes not in perfect linkage disequilibrium with another haplotype) involving two to sixteen SNPs were present with estimated frequency ≥0.05 in our Stage 1 analysis. We tested all SNPs and all 2414 haplotypes for association with longevity. SNP 6 showed the strongest single-SNP association (P=0.0137) in Stage 1, but was not significant upon correction for multiple testing via permutation tests. However, an excess of significant haplotypes over what is expected under no association was evident. We examined the proportion of haplotypes that were significant at a range of significance thresholds (α = 0.00005, 0.0001, 0.0005, 0.001, 0.005, 0.01, and 0.05) and in all cases observed significantly more than expected under no association. P-values for association tests of each of these haplotypes are plotted against their quantiles in Figure S1. The most significant haplotype, involving SNPs 6, 7, 13, and 15 (GTCT), remained significant after adjustment for multiple testing via permutation tests (P = 2.5×10−5, multiple-testing-adjusted P = 0.0045). Haplotypes involving SNPs 4 and 5 were also significant, and the most significant 6-SNP haplotype involved SNPs 4, 5, 6, 7, 13, and 15 (GAGTCT) (P = 4.0×10−5). 140 non-redundant haplotypes involving different combinations of two to six of these six SNPs were present with estimated frequency>0.05.
Table 1.
Samples tested
| Sample | N | Age range | Mean age |
|---|---|---|---|
| Stage 1 (US) | 873 LLI | ≥95 | 101.5 |
| 443 controls | <50 | 38.6 | |
| NECS (US) | 545 LLI | 96–113 | 103.2 |
| 193 controls | 55–92 | 76.8 | |
| French | 557 LLI | ≥99 | 103.1 |
| 546 controls | 18–70 | 51.2 | |
| SICS (Southern Italy) | 455 LLI | 90–109 | 97.6 |
| 450 controls | 18–46 | 32.9 | |
| Ashkenazi Jewish | 354 LLI | 95–109 | 97.9 |
| 348 controls | 43–90 | 70.7 |
Figure 1.
Linkage disequilibrium (R2 in HapMap CEU sample) between SNPs in LMNA region. Labeled SNPs indicate SNPs tested in Table 2. Encircled SNP names indicate SNPs involved in GTCT haplotype in Table 3.
To follow up on these 140 haplotypes, we selected SNPs 4, 5, 6, 7, 13, and 15 for genotyping in our four Stage 2 samples (Table 1). One of the six SNPs was not successfully genotyped in the French sample, but for purposes of haplotype inference we were able to impute genotypes for this SNP using MaCH 1.0 (Li et al. 2010) in conjunction with phased haplotype information from the HapMap CEU sample (estimated r2 between true underlying genotype and imputed genotype = 0.97). Additional LMNA SNPs beyond SNPs 4, 5, 6, 7, 13, and 15 were also genotyped based on convenience and were included in the SNP association analysis in Table 2 but not the haplotype analysis. SNP association results for all five samples are shown in Table 2. When we combined results across samples, SNP 15 showed the strongest single-SNP association in the meta-analysis (OR=1.17, P=0.0056) with odds ratios ranging from 1.09–1.22 in the four samples tested, but was not significant after correction for multiple testing via permutation tests (multiple-testing-adjusted P=0.105).
Table 2.
Trend test and meta-analysis P-values for SNPs across five samples
| SNP | RefSNP ID | Stage 1 (N=1316) |
NECS (N=738) |
French (N=1103) |
SICS (N=905) |
Ashkenazi Jewish (N=702) |
Meta-analysis |
|---|---|---|---|---|---|---|---|
| 1 | rs2275075 | 0.54 | 0.73 | 0.78 | |||
| 2 | rs3814314 | 0.017 | 0.81 | 0.039 | |||
| 3 | rs11578696 | 0.21 | 0.93 | 0.39 | |||
| 4 | rs4661146 | 0.12 | 0.17 | 0.42 | 0.65 | 0.72 | 0.97 |
| 5 | rs10737170 | 0.11 | 0.016 | 0.84 | 0.54 | 0.45 | 0.075 |
| 6 | rs915179 | 0.014 | 0.0086 | 0.96 | 0.66 | 0.59 | 0.020 |
| 7 | rs2485662 | 0.054 | 0.11 | 0.81 | 0.82 | 0.35 | 0.026 |
| 8 | rs547915 | 0.11 | 0.84 | 0.84 | 0.29 | ||
| 9 | rs582690 | 0.078 | 0.42 | 0.35 | |||
| 10 | rs6686943 | 0.51 | |||||
| 11 | rs508641 | 0.014 | 0.95 | 0.044 | |||
| 12 | rs12117552 | 0.21 | 0.26 | 0.71 | |||
| 13 | rs4641 | 0.50 | 0.14 | 0.61 | 0.30 | 0.45 | 0.98 |
| 14 | rs12063072 | 0.36 | 0.060 | 0.065 | |||
| 15 | rs1468772 | 0.034 | 0.35 | 0.41 | 0.14 | 0.0056 | |
| 16 | rs6669212 | 0.041 | 0.48 | 0.36 | 0.18 |
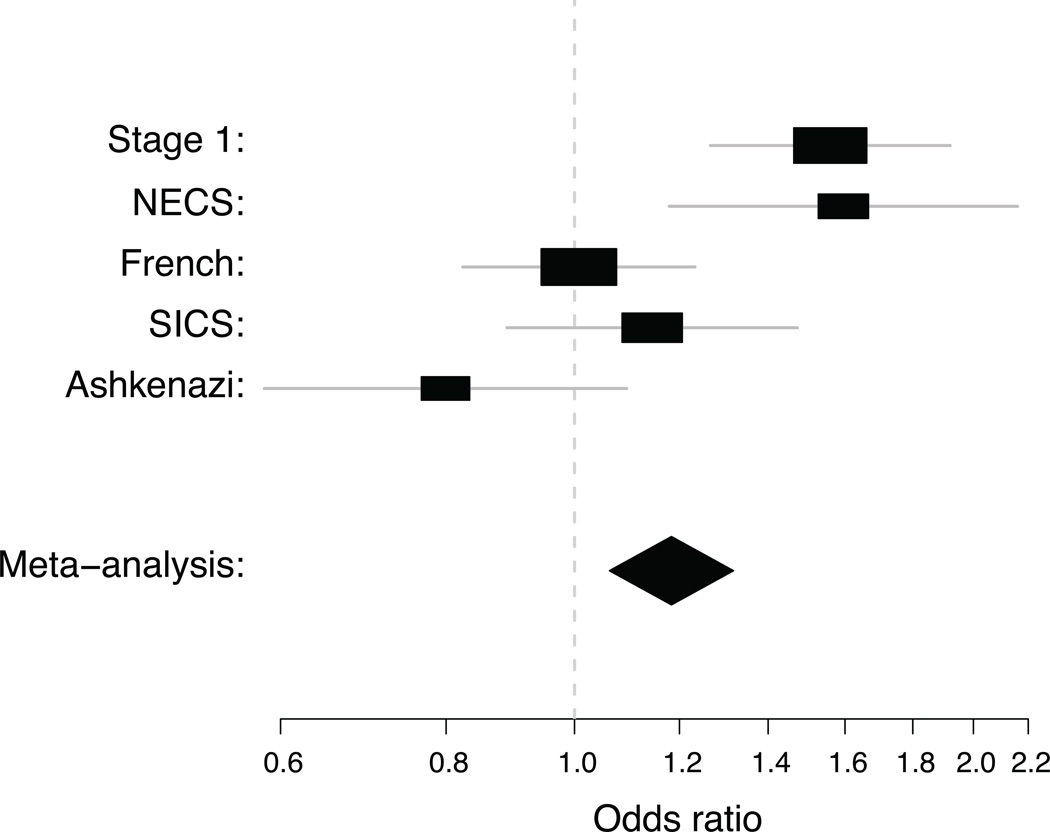
In replication analyses of the 140 haplotypes of SNPs 4, 5, 6, 7, 13, and 15 in the four Stage 2 samples, an excess of significant haplotypes was observed in the NECS sample (Figure S2; P < 0.01) for all significance thresholds examined (α= 0.00005, 0.0001, 0.0005, 0.001, 0.005, 0.01, and 0.05) but not in the other three Stage 2 samples. When the results for all 140 followed-up haplotypes were combined across all five samples in a meta-analysis (Table 3 and Figure 2), the original GTCT haplotype of SNPs 6, 7, 13, and 15 was again the most significant (P=7.5×10−4) and remained significant after permutation-based adjustment for multiple testing in both stages of the analysis (multiple-testing-adjusted P = 0.037).
Table 3.
Meta-analysis of haplotype GTCT on SNPs 6, 7, 13, and 15 across five samples
| Sample | N | case | control | OR | Z statistic | P-value | Adjusted P-value |
|---|---|---|---|---|---|---|---|
| Stage 1 | 1316 | 0.235 | 0.163 | 1.56 | 4.21 | 0.000025 | 0.0045 (single-stage) |
| NECS | 738 | 0.239 | 0.163 | 1.60 | 3.05 | 0.0023 | |
| French | 1103 | 0.208 | 0.207 | 1.01 | 0.074 | 0.94 | |
| SICS | 905 | 0.175 | 0.157 | 1.14 | 1.05 | 0.29 | |
| Ashkenazi | 702 | 0.114 | 0.139 | 0.80 | −1.40 | 0.16 | |
| Full meta-analysis | 1.18 | 3.37 | 0.00075 | 0.037 (two-stage) | |||
Figure 2.
Forest plot of odds ratios for meta-analysis of haplotype GTCT on SNPs 6, 7, 13, and 15 across five samples. For each sub-sample, the black box is centered at the estimated odds ratio, and the gray line represents a 95% confidence interval. Area of the black box varies in proportion to the precision of each estimate. For the meta-analysis, the black diamond is centered at the estimated odds ratio and spans the 95% confidence interval for the meta-analysis odds ratio.
If the genetic load is greater for older individuals, restricting the definition of LLI to include only centenarians should lead to a stronger genetic effect (Tan et al. 2008). For the four samples where age and sex information was available (Stage 1, NECS, SICS, and the Ashkenazi sample), we re-performed the association test for the GTCT haplotype within each sample after restricting the definition of LLI to include individuals with age ≥ 100. However, the odds ratio for GTCT did not change substantially in any of the four samples. We also performed a sex-stratified version of the haplotype analysis. Similar results were observed in the sex-stratified analysis for each sample, and results were similar for males and females within each sample.
The Stage 1 sample had previously been tested for the presence of population stratification between LLI and control subjects (see Supporting Information) (Geesaman et al. 2003). However, to ensure that the association with the GTCT haplotype was not due to subtle population stratification, we re-fit the model including the principal components of genotypes from 73 ancestry-informative markers (AIMs) from a published panel of European substructure AIMs (Tian et al. 2008). Inclusion of the first four principal components from the AIM genotypes as covariates in the analyses did not substantially diminish the strength of the association. For the 1242 individuals for whom AIMs were available, a similar association between the GTCT haplotype and longevity was observed before (OR=1.47, P=0.00039) and after (OR=1.46, P=0.00060) inclusion of the principal components. Although a Tracy-Widom test (Patterson et al. 2006) suggested that four principal components was sufficient to account for potential population stratification, the observed result was robust to the number of principal components included in the model (P ranged from 0.00057 to 0.00062 when one to ten principal components were included). Similarly, the top two principal components from genome-wide data were available for 671 of the NECS individuals; inclusion of these components as covariates in the analysis slightly strengthened the result. In both cases, the inclusion of principal components in the analyses affected the results only minimally, suggesting that the observed haplotype association in the Stage 1 and NECS samples is not driven by population stratification. Similarly, analysis of the first two principal components did not reveal evidence of population stratification in a genome-wide study of SICS cases and controls (Malovini et al. 2011). The French sample had been previously tested for population stratification on 57 randomly selected microsatellite markers. As described in Supporting Information, χ2≤1.00 in all cases, indicating that the French LLI and controls were well-matched.
Discussion
We observed evidence for association with longevity involving a 4-SNP haplotype, and an excess of significant associations among the same set of correlated haplotypes, in an initial sample of centenarians and younger controls. This result was replicated in one of four follow-up samples, and the association was sufficiently strong in those two samples to remain significant when combined in a meta-analysis of all five.
Had our initial haplotype association been unreplicated in all four follow-up samples, it would have been difficult to argue against the possibility of a false positive. However, the probability of observing a second false positive of similar strength is low (<0.005): in the same 10,000 permutations used to adjust the meta-analysis P-value for multiple testing, the best haplotype observed in the Stage 1 sample was followed up with an association in the same direction with |Z| > 3.05 in one or more of the four follow-up samples only 46 times.
Other possible explanations for incomplete replication include undetected population structure, use of under-powered replication samples, and heterogeneity between samples. However, adjustment for potential population stratification via principal components only modestly diminished the associations reported here. Similarly, differential power based on sample size differences does not appear to explain the incomplete replication since the NECS sample was arguably the least well-powered due to small sample size and skewed case-control ratio. Heterogeneity between samples can occur for a variety of reasons. For example, the GTCT haplotype could tag a functional variant in some samples but not others due to differential LD patterns. Alternatively, a functional variant tagged by GTCT could correlate with the longevity phenotype only in the NECS and Stage 1 samples; this can result from differential ascertainment of cases or from between-sample differences in environmental exposures (McCarthy et al. 2008). One potential source of heterogeneity is differential ascertainment of LLI, since the minimum age for inclusion varied by study (Table 1). Notably, the association with GTCT was observed in two of the three samples that had ascertained older sets of LLI (mean age > 100); Tan et al. (2008) has argued that the power of association studies investigating exceptional longevity markedly increases with the use of centenarian subjects compared to nonagenarians. However, in the three samples where individual ages were available, we observed similar results when the definition for cases was narrowed to include only centenarians. Another potential source of heterogeneity is environmental differences across samples due to region of origin. The association between GTCT and longevity was observed in two of the three US samples, but not in the European samples. One region-specific environmental exposure which could interact with aging-related genes to affect longevity is diet. Dietary differences between Europe and the US have been proposed as explanations for regional differences in rates of coronary mortality (Renaud & de Lorgeril 1992; de Lorgeril et al. 1994), and caloric restriction and several dietary antioxidants, including resveratrol, found in red wine, can alter expression of aging-related genes in the hearts and brains of mice (Park et al. 2009).
Replicating genetic association results across diverse populations has generally been difficult in the longevity literature (Christensen et al. 2006). For example, a strong association between longevity and a FOXO3A SNP (Willcox et al. 2008) replicated in some samples (Flachsbart et al. 2009; Li et al. 2009), including males from the SICS (Anselmi et al. 2009), but was not present in others (Kuningas et al. 2007) (Pawlikowska et al. 2009; Soerensen et al. 2010), including females from the SICS (Anselmi et al. 2009), although two of these studies reported associations with other FOXO3A SNPs Reasons proposed for the lack of replication in some samples included those discussed above, as well as lack of gender stratification in some analyses (Anselmi et al. 2009), although as described in Results, this does not appear to be an issue in the present study.
The meta-analysis results presented here complement a growing body of experimental evidence (Scaffidi & Misteli 2006; McClintock et al. 2007; Cao et al. 2011) in suggesting a potential role for LMNA and progerin in aspects of normal aging. However, given the incomplete replication we observed, further work is needed to validate and interpret the results reported here. One of the four SNPs in our reported haplotype, rs4641, is a coding SNP for which allele-specific expression has been demonstrated (Rodriguez & Eriksson, 2011), with 2.3-fold greater expression for transcripts with the C allele, which is part of the GTCT haplotype associated with greater longevity in our meta-analysis. Another SNP in our reported haplotype, rs915179, associated with longevity in a recent genome-wide association study (Sebastiani et al. 2012), though this study included most of our NECS cases and controls along with other subjects. Additional association studies of LMNA variants in independent samples and additional experiments focusing on functional consequences of the associated variants would be particularly valuable in establishing and elucidating the role of LMNA in the normal aging process.
Experimental Procedures
Samples
We analyzed one initial (Stage 1) and four follow-up (Stage 2) samples of LLI and younger controls. Samples collected for the Stage 1 study by Elixir Pharmaceuticals were Caucasians primarily of Northern European ancestry. Samples were also obtained from the New England (NECS), French, Southern Italian (SICS), and Ashkenazi Jewish Centenarian Studies. Details of sample collection, as well as steps taken to ensure that the study samples did not overlap, can be found in Supporting Information.
SNP selection
We downloaded all SNP genotype data for the CEU population from −40 kb to +10 kb of LMNA from the website of the International HapMap (International HapMap Consortium 2005): http://www.hapmap.org. These genotype data in raw HapMap format were uploaded to Tagger (http://www.broad.mit.edu/mpg/tagger/), and our multiplex assay design (see Supporting Information for details) was created from the list of tag SNPs to capture all variants of interest in our LMNA region of interest. We used r2 >0.8 as the threshold for accepting tag SNPs. 10 SNPs were selected in HapMap Phase I, release #16 along with the synonymous coding SNP, rs4641, which was not in HapMap Phase I. Upon release of HapMap Phase II, release #19 we selected SNPs in the LMNA region again and added 5 additional SNPs not tagged (r2<0.8) by the previously selected SNPs. Based on Phase II of the HapMap, these 16 SNPs offered comprehensive coverage and representation of common variants in the LMNA region and were genotyped in Stage 1.
For Stage 2 samples, we genotyped the 6 SNPs from the most significant 6-SNP haplotype in Stage 1. Additional SNPs from the original 16 were genotyped in some Stage 2 samples based on convenience; these SNPs were tested for association (Table 2) but were not included in the haplotype analysis. Details on SNP genotyping are provided in Supporting Information.
Statistical analysis
We used the MaCH 1.0 haplotype inference and imputation software (Li et al. 2010) to infer 16-SNP haplotypes for all individuals in Stage 1, and 6-SNP haplotypes for all individuals in Stage 2. For individuals with missing genotypes, the most likely genotype was inferred based on haplotype frequencies within the sample. We imputed genotypes for SNP rs1468772 that was not genotyped in the French sample, using MaCH (Li et al. 2010) in conjunction with phased haplotype information from the HapMap CEU sample.
We performed association tests in all five samples and combined the samples in a meta-analysis using R (R Development Core Team 2011). We performed all SNP and haplotype association tests as Cochran-Armitage tests (Cochran 1954; Armitage 1955) for a linear trend between the number of copies of a reference allele (0, 1, or 2) and the likelihood that an individual belongs to the case or control group. We first tested each SNP for association with centenarian status. We computed meta-analysis statistics for all followed-up SNPs and haplotypes as the weighted sum of z-statistics from the trend tests. The weights for sample i were defined as , where ni= sample size for samples i = 1,…,5 and .
To account for the testing of multiple correlated SNPs and haplotypes in Stage 1, we performed permutation tests by 1) randomly permuting case-control status and 2) re-computing the test statistics for all SNPs and haplotypes. Based on 10,000 permutations, we then computed adjusted P-values that reflected the probability of observing each result by chance alone. To establish whether our most extreme P-value was significantly different than what would be expected if no true association existed, we computed an empirical P-value as the proportion of permutations in which a more extreme result was observed in any of the SNPs or haplotypes analyzed. To establish whether a significant excess of haplotypes with P<α was observed for a given α-level, we computed empirical P-values as the proportion of permutations in which we observed as many haplotypes with P<α as in our original data.
To account for multiple testing in the two-stage analysis, we performed permutation tests in R to obtain an empirical P-value for the best observed meta-analysis test statistic. Permutation tests for two-stage studies are complicated by the fact that only partial genotyping is performed in the second stage of the study. Our strategy for dealing with this was, for permutation purposes only, to use MACH 1.0, along with inferred haplotypes from the HapMap CEU sample, to impute the full 16-SNP haplotypes for all Stage 2 samples. We were then able to perform a full two-stage analysis in each permutation as follows:
Randomly re-assign case-control status within each of the five samples.
Compute trend test statistics for all haplotypes (or SNPs) in the permuted Stage 1 sample.
To mimic the selection of 6 SNPs for genotyping in the Stage 2 samples, as was done in the original study, select the most significant 6-SNP haplotype from the (permuted) Stage 1 analysis for follow-up. Perform trend tests of association for all 6 SNPs and for all haplotypes composed of 2–6 SNP combinations of these 6 SNPs in the four permuted Stage 2 samples.
Compute meta-analysis statistics for all haplotypes (or SNPs) tested on all five permuted samples; record the value of the most significant meta-analysis test statistic.
We performed 10,000 permutations, and recorded the best meta-analysis test statistic for each permutation. We computed the empirical P-value as the proportion of permutations in which the best statistic from the permutation was greater in magnitude than the best statistic from the original data analysis.
Population stratification
As described in Supporting Information, the Stage 1 and French samples had previously been tested for the presence of population stratification between LLI and control subjects and the Stage 1 sample had been genetically matched. There was no evidence of population stratification in the French sample (mean χ2=1.00 across 57 random microsatellite markers) or in the genetically matched Stage 1 sample (mean χ2=0.95 across 80 random SNPs) (Geesaman et al. 2003).
For the Stage 1 sample, we also genotyped AIMs from a published panel specific to European genetic substructure (Tian et al. 2008); genotyping and quality control described in Supporting Information. We used R to compute principal components and to identify the most important principal components via a Tracy-Widom test (Patterson et al. 2006). To perform an analysis adjusted for population stratification, we included the top principal-components as covariates in the original analysis.
Principal components based on genotypes from the 370K Illumina SNP chip (collected as part of a separate study – Sebastiani et al. 2012) were available for a subset of the NECS sample. We used EIGENSTRAT (Price et al. 2006) to compute the principal components and identified the principal components that were most informative based on visual inspection of a scree plot. To perform an analysis adjusted for population stratification, we included the top principal components as covariates in the original analysis.
Supplementary Material
Acknowledgments
This work has been supported by the intramural program of the National Human Genome Research Institute and grants from: Glenn Award for Research in Biological Mechanisms of Aging (N.B.), the NIH PO1 AG027734 (N.B.), the NIH/NIA Longevity Consortium U19 AG023122 (G.A., T.P), the NIH/NIA R56 AG027216 (C.B.), and NIH/NHLBI R01 HL87681-01 (N.S.). We thank Drs. Gilles Thomas, Helene Blanche, and Valerie Morel for genotyping the French samples analyzed in this study, and Narisu Narisu for providing technical assistance with the genotyping of the Stage 1 samples.
References
- Anselmi CV, Malovini A, Roncarati R, Novelli V, Villa F, Condorelli G, Bellazzi R, Puca AA. Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 2009;12:95–104. doi: 10.1089/rej.2008.0827. [DOI] [PubMed] [Google Scholar]
- Armitage P. Tests for linear trends in proportions and frequencies. Biometrics. 1955;11:375–386. [Google Scholar]
- Cao K, Blair CD, Faddah DA, Kieckhaefer JE, Olive M, Erdos MR, Nabel EG, Collins FS. Progerin and telomere dysfunction collaborate to trigger cellular senescence in normal human fibroblasts. J Clin Invest. 2011;121:2833–2844. doi: 10.1172/JCI43578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K, Capell BC, Erdos MR, Djabali K, Collins FS. A lamin A protein isoform overexpressed in Hutchinson-Gilford progeria syndrome interferes with mitosis in progeria and normal cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4949–4954. doi: 10.1073/pnas.0611640104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capell BC, Collins FS. Human laminopathies: nuclei gone genetically awry. Nat Rev Genet. 2006;7:940–952. doi: 10.1038/nrg1906. [DOI] [PubMed] [Google Scholar]
- Capell BC, Collins FS, Nabel EG. Mechanisms of cardiovascular disease in accelerated aging syndromes. Circ Res. 2007;101:13–26. doi: 10.1161/CIRCRESAHA.107.153692. [DOI] [PubMed] [Google Scholar]
- Castro E, Edland SD, Lee L, Ogburn CE, Deeb SS, Brown G, Panduro A, Riestra R, Tilvis R, Louhija J, Penttinen R, Erkkola R, Wang L, Martin GM, Oshima J. Polymorphisms at the Werner locus: II. 1074Leu/Phe, 1367Cys/Arg, longevity, and atherosclerosis. Am J Med Genet. 2000;95:374–380. [PubMed] [Google Scholar]
- Christensen K, Johnson TE, Vaupel JW. The quest for genetic determinants of human longevity: challenges and insights. Nat Rev Genet. 2006;7:436–448. doi: 10.1038/nrg1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran WG. Some methods for strengthening the common χ2 tests. Biometrics. 1954;10:417–451. [Google Scholar]
- de Lorgeril M, Renaud S, Mamelle N, Salen P, Martin JL, Monjaud I, Guidollet J, Touboul P, Delaye J. Mediterranean alpha-linolenic acid-rich diet in secondary prevention of coronary heart disease. Lancet. 1994;343:1454–1459. doi: 10.1016/s0140-6736(94)92580-1. [DOI] [PubMed] [Google Scholar]
- Deelen J, Beekman M, Uh HW, Helmer Q, Kuningas M, Christiansen L, Kremer D, van der Breggen R, Suchiman HE, Lakenberg N, van den Akker EB, Passtoors WM, Tiemeier H, van Heemst D, de Craen AJ, Rivadeneira F, de Geus EJ, Perola M, van der Ouderaa FJ, Gunn DA, Boomsma DI, Uitterlinden AG, Christensen K, van Duijn CM, Heijmans BT, Houwing-Duistermaat JJ, Westendorp RG, Slagboom PE. Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging Cell. 2011;10:686–698. doi: 10.1111/j.1474-9726.2011.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flachsbart F, Caliebe A, Kleindorp R, Blanche H, von Eller-Eberstein H, Nikolaus S, Schreiber S, Nebel A. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2700–2705. doi: 10.1073/pnas.0809594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geesaman BJ, Benson E, Brewster SJ, Kunkel LM, Blanche H, Thomas G, Perls TT, Daly MJ, Puca AA. Haplotype-based identification of a microsomal transfer protein marker associated with the human lifespan. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:14115–14120. doi: 10.1073/pnas.1936249100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- Herskind AM, McGue M, Holm NV, Sorensen TI, Harvald B, Vaupel JW. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900. Hum Genet. 1996;97:319–323. doi: 10.1007/BF02185763. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium'. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Kuningas M, Magi R, Westendorp RG, Slagboom PE, Remm M, van Heemst D. Haplotypes in the human Foxo1a and Foxo3a genes; impact on disease and mortality at old age. Eur J Hum Genet. 2007;15:294–301. doi: 10.1038/sj.ejhg.5201766. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang WJ, Cao H, Lu J, Wu C, Hu FY, Guo J, Zhao L, Yang F, Zhang YX, Li W, Zheng GY, Cui H, Chen X, Zhu Z, He H, Dong B, Mo X, Zeng Y, Tian XL. Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum Mol Genet. 2009;18:4897–4904. doi: 10.1093/hmg/ddp459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungquist B, Berg S, Lanke J, McClearn GE, Pedersen NL. The Effect of Genetic Factors for Longevity: A Comparison of Identical and Fraternal Twins in the Swedish Twin Registry. The Journals of Gerontology Series A: Biological Sciences and Medical Sciences. 1998;53A:M441–M446. doi: 10.1093/gerona/53a.6.m441. [DOI] [PubMed] [Google Scholar]
- Malovini A, Illario M, Iaccarino G, Villa F, Ferrario A, Roncarati R, Anselmi CV, Novelli V, Cipolletta E, Leggiero E, Orro A, Rusciano MR, Milanesi L, Maione AS, Condorelli G, Bellazzi R, Puca AA. Association study on long-living individuals from Southern Italy identifies rs10491334 in the CAMKIV gene that regulates survival proteins. Rejuvenation Res. 2011;14:283–291. doi: 10.1089/rej.2010.1114. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- McClintock D, Ratner D, Lokuge M, Owens DM, Gordon LB, Collins FS, Djabali K. The mutant form of lamin A that causes Hutchinson-Gilford progeria is a biomarker of cellular aging in human skin. PLoS One. 2007;2:e1269. doi: 10.1371/journal.pone.0001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merideth MA, Gordon LB, Clauss S, Sachdev V, Smith AC, Perry MB, Brewer CC, Zalewski C, Kim HJ, Solomon B, Brooks BP, Gerber LH, Turner ML, Domingo DL, Hart TC, Graf J, Reynolds JC, Gropman A, Yanovski JA, Gerhard-Herman M, Collins FS, Nabel EG, Cannon RO, 3rd, Gahl WA, Introne WJ. Phenotype and course of Hutchinson-Gilford progeria syndrome. N Engl J Med. 2008;358:592–604. doi: 10.1056/NEJMoa0706898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebel A, Kleindorp R, Caliebe A, Nothnagel M, Blanche H, Junge O, Wittig M, Ellinghaus D, Flachsbart F, Wichmann HE, Meitinger T, Nikolaus S, Franke A, Krawczak M, Lathrop M, Schreiber S. A genome-wide association study confirms APOE as the major gene influencing survival in long-lived individuals. Mechanisms of Ageing and Development. 2011;132:324–330. doi: 10.1016/j.mad.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Novelli V, Viviani Anselmi C, Roncarati R, Guffanti G, Malovini A, Piluso G, Puca AA. Lack of replication of genetic associations with human longevity. Biogerontology. 2008;9:85–92. doi: 10.1007/s10522-007-9116-4. [DOI] [PubMed] [Google Scholar]
- Park SK, Kim K, Page GP, Allison DB, Weindruch R, Prolla TA. Gene expression profiling of aging in multiple mouse strains: identification of aging biomarkers and impact of dietary antioxidants. Aging Cell. 2009;8:484–495. doi: 10.1111/j.1474-9726.2009.00496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genetics. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlikowska L, Hu D, Huntsman S, Sung A, Chu C, Chen J, Joyner AH, Schork NJ, Hsueh WC, Reiner AP, Psaty BM, Atzmon G, Barzilai N, Cummings SR, Browner WS, Kwok PY, Ziv E. Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell. 2009;8:460–472. doi: 10.1111/j.1474-9726.2009.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- Renaud S, de Lorgeril M. Wine, alcohol, platelets, and the French paradox for coronary heart disease. Lancet. 1992;339:1523–1526. doi: 10.1016/0140-6736(92)91277-f. [DOI] [PubMed] [Google Scholar]
- Rodriguez S, Coppede F, Sagelius H, Eriksson M. Increased expression of the Hutchinson-Gilford progeria syndrome truncated lamin A transcript during cell aging. Eur J Hum Genet. 2009;17:928–937. doi: 10.1038/ejhg.2008.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez S, Eriksson M. Low and high expressing alleles of the LMNA gene: implications for laminopathy disease development. PLoS ONE. 2011;6:e25472. doi: 10.1371/journal.pone.0025472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science (New York, N.Y. 2006;312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani P, Solovieff N, DeWan A, Walsh K, Puca A, Hartley SW, Melista E, Andersen S, Dworkis DA, Wilk JB, Myers RH, Steinberg MH, Montano M, Baldwin CT, Hoh J, Perls TT. Genetic signatures of exceptional longevity in humans. PLoS ONE. 2012 doi: 10.1371/journal.pone.0029848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skytthe A, Pedersen NL, Kaprio J, Stazi MA, Hjelmborg JV, Iachine I, Vaupel JW, Christensen K. Longevity studies in GenomEUtwin. Twin Res. 2003;6:448–454. doi: 10.1375/136905203770326457. [DOI] [PubMed] [Google Scholar]
- Soerensen M, Dato S, Christensen K, McGue M, Stevnsner T, Bohr VA, Christiansen L. Replication of an association of variation in the FOXO3A gene with human longevity using both case-control and longitudinal data. Aging Cell. 2010;9:1010–1017. doi: 10.1111/j.1474-9726.2010.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Zhao JH, Zhang D, Kruse TA, Christensen K. Power for genetic association study of human longevity using the case-control design. Am J Epidemiol. 2008;168:890–896. doi: 10.1093/aje/kwn205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Plenge RM, Ransom M, Lee A, Villoslada P, Selmi C, Klareskog L, Pulver AE, Qi L, Gregersen PK, Seldin MF. Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genetics. 2008;4:e4. doi: 10.1371/journal.pgen.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.