SUMMARY
Interactions of the CHMP protein carboxyl terminal tails with effector proteins play important roles in retroviral budding, cytokinesis, and multivesicular body biogenesis. Here we demonstrate that hydrophobic residues at the CHMP4B C-terminal amphipathic α-helix bind a concave surface of Brox, a mammalian paralog of Alix. Unexpectedly, CHMP5 was also found to bind Brox and specifically recruit endogenous Brox to detergent-resistant membrane fractions through its C-terminal 20 residues. Instead of an α-helix, the CHMP5 C-terminal tail adopts a tandem β-hairpin structure that binds Brox at the same site as CHMP4B. Additional Brox:CHMP5 interface is furnished by a unique CHMP5 hydrophobic pocket engaging the Brox residue Y348 that is not conserved among the Bro1 domains. Our studies thus unveil a novel conformation of the CHMP protein C-terminal tails, and provide new insights into the overlapping but distinct binding profiles of ESCRT-III and the Bro1 domain proteins.
INTRODUCTION
The endosomal sorting complex required for transport (ESCRT) machinery plays crucial roles in membrane fission and remodeling events during retroviral budding (Dordor et al., 2011), cytokinesis (Caballe and Martin-Serrano, 2011), multivesicular body biogenesis (Henne et al., 2011; Hurley, 2010), and autophagy (Rusten and Stenmark, 2009). In mammalian cells, it consists of several protein complexes such as ESCRT-0, ESCRT-I, ESCRT-II and ESCRT-III, plus the VPS4-LIP5 complex, and several associated proteins such as the Bro1 domain-containing proteins Alix, HD-PTP and Brox (Peel et al., 2011). Among these, ESCRT-III and VPS4 are the most highly conserved and essential components, as the former assembles into detergent-resistant polymers to induce membrane scission, and the latter recycles ESCRT-III into soluble monomers for the next ESCRT cycle (Hurley and Hanson, 2010; Lata et al., 2008b; Shim et al., 2008).
ESCRT-III family is composed of seven protein families named charged multivesicular body proteins (CHMP) 1–7 and IST-1 (increased sodium tolerance-1), all of which possess characteristic bipartite sequences containing basic N-terminal and acidic C-terminal fragments. Interactions between these two polarized segments maintain CHMP proteins in self-inhibited states as soluble monomers (Bajorek et al., 2009; Lata et al., 2008a; Shim et al., 2007; Zamborlini et al., 2006). Binding of acidic lipids to the CHMP N-terminal region induces conformational changes followed by polymerization and exposure of their C-terminal fragments, which in turn recruit VPS4 and other effector proteins (Bajorek et al., 2009; Lata et al., 2008a; Shim et al., 2007). CHMP proteins are known to bind the microtubule-interacting and transport (MIT) domains of VPS4, Vta1/LIP5 and other effector proteins through their MIT-interaction motifs (MIMs) with different affinities (Azmi et al., 2008; Kieffer et al., 2008; Row et al., 2007; Ward et al., 2005). These interactions generally involve the MIT domain three-helix bundles bound to MIMs in either amphipathic α-helices such as those from CHMP1A (Stuchell-Brereton et al., 2007), CHMP1B (Yang et al., 2008), Did2 (yeast homolog of CHMP1)(Xiao et al., 2009), Vps2 (yeast homolog of human CHMP2)(Obita et al., 2007), and CHMP3 (Solomons et al., 2011), or in an extended conformation such as those from CHMP6 (Kieffer et al., 2008) and its archaea homolog Saci1372 (Samson et al., 2008). Notably, all of the α-helical MIMs consist of the C-terminal tails of the respective CHMP proteins. In addition, the C-terminal tails of CHMP4A, B and C isoforms adopt α-helical conformations and bind a conserved hydrophobic pocket at the Bro1 domain of Alix, which is involved in retroviral budding (Fisher et al., 2007; McCullough et al., 2008; Usami et al., 2007). The C-terminal tail of CHMP5 does not have a distinctive amphipathic feature and has not been reported to be involved in protein complex formation. The α4-α5 helices of CHMP5, predicted based on the core structure of CHMP3 (Muziol et al., 2006), is associated with the Vta1/LIP5 N-terminal MIT domains (Azmi et al., 2008; Bowers et al., 2004; Ward et al., 2005; Xiao et al., 2008). This interaction co-localizes CHMP5 with Vta1 to the endosomes and indirectly potentiates VPS4 activity to disassemble the ESCRT-III complex (Nickerson et al., 2010; Shiflett et al., 2004).
Brox was initially identified as a Bro1 domain-containing protein that binds CHMP4B and contains a C-terminal CAAX farnesylation motif (Ichioka et al., 2008). Recently, the Bro1 domain of Brox was demonstrated to adopt a typical boomerang structure similar to that of Alix, and binds HIV-1 Gag NC domain in a similar manner, but did not appear to promote HIV-1 release (Sette et al., 2011; Zhai et al., 2011). Consistent with previous reports of CHMP4B binding to Bro1 domain-containing proteins (Doyotte et al., 2008; Ichioka et al., 2008; Ichioka et al., 2007; Katoh et al., 2003; Strack et al., 2003), structural comparison and modeling suggested that the Bro1 domains from Brox and HD-PTP could bind CHMP4B at the same conserved hydrophobic pockets as Alix (Sette et al., 2011; Zhai et al., 2011). As Brox is a relatively new member of the Bro1 domain family, little is known about its partner proteins other than CHMP4B.
To investigate the interaction of Brox with other components of the ESCRT-III machinery, we tested binding of Brox to all known CHMP proteins. Surprisingly, we found that both CHMP5 and CHMP4B associated with Brox, and the CHMP5 C-terminal tail was required for its specific recruitment of Brox to membrane fractions. To understand the molecular basis of the Brox:CHMP5 and Brox:CHMP4B interaction, we determined the crystal structures of Brox in complex with the C-terminal tails of CHMP5 and CHMP4B. Structural analysis revealed that the overall character of the Brox:CHMP5 and Brox:CHMP4B interface is similar, with dominant contribution by hydrophobic contacts. Unexpectedly, the CHMP5 C-terminal tail adopts a tandem β-hairpin structure with its second β-hairpin anchored its interface with Brox, whereas the C-terminal tail of CHMP4B adopts a canonical α-helical structure representative of all other CHMP C-terminal tail structures determined to date. The structures demonstrate that the C-terminal tails of the CHMP proteins can adopt distinctive conformations that interact with Bro1 domain proteins in versatile modes.
RESULTS
Brox Interacts with Both CHMP4B and CHMP5
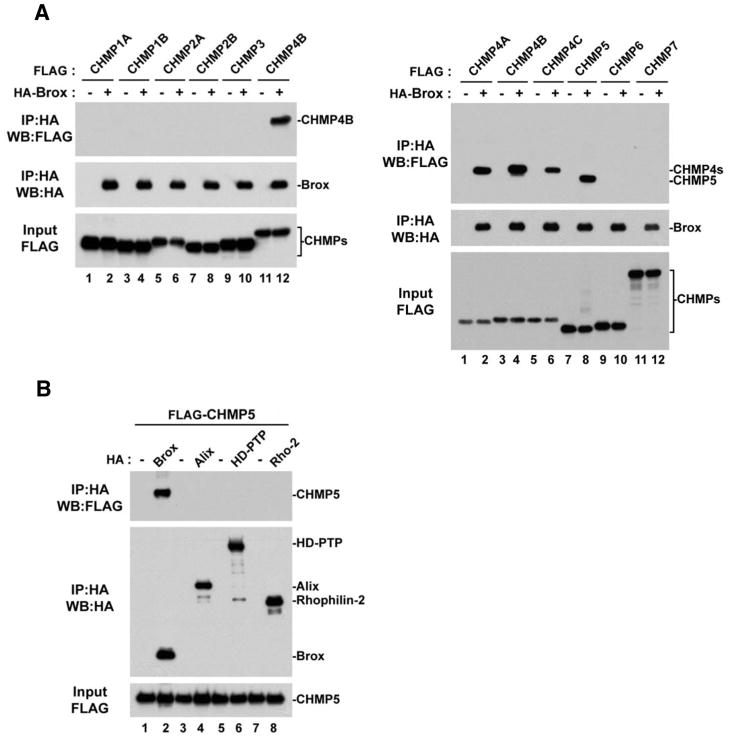
Among the seven CHMP proteins, only the CHMP4 isoforms are known to recruit the Bro1 domain containing proteins to the ESCRT-III machinery (Ichioka et al., 2008; Ichioka et al., 2007; Katoh et al., 2003). It is unclear whether other CHMP proteins also play a role in this process. To investigate this possibility, we used Brox in a binding study with all known CHMP proteins. To our surprise, although no interactions were detected with CHMP1A, 1B, 2A, 2B, 3, 6 and 7 (Figure 1A, left panel), Brox was found to capture CHMP5 as efficiently as the three CHMP4 isoforms (Figure 1A, right panel, compare lanes 2, 4, 6 and 8). Because the ability to bind CHMP4 proteins is one of the main characteristic that defines Bro1 domain-containing proteins such as the yeast Bro1p and human Alix and HD-PTP (Ichioka et al., 2007; Katoh et al., 2003; Kim et al., 2005; Odorizzi et al., 2003), we next asked whether this new Brox:CHMP5 interaction was specific to Brox or a general property of Bro1-domain containing proteins. To answer this, we tested the four known human Bro1-domain containing proteins for their ability to bind CHMP5. In contrast to Alix, HD-PTP and Rhophilin-2, only Brox was found in complex with CHMP5 (Figure 1B, lane 2) demonstrating that the Brox:CHMP5 association is specific.
Figure 1. CHMP5 is a novel binding partner of Brox.
(A) Interactions between Brox and ESCRT-III. FLAG-tagged ESCRT-III proteins were expressed in HEK293T cells in the presence or absence of HA-tagged Brox, and immunoprecipitation was performed using anti-HA antibody conjugated beads. Both input and immunoprecipitated complexes were visualized by western blot using the indicated antibodies.
(B) FLAG-tagged CHMP5 was expressed in HEK293T cells in the presence or absence of the indicated HA-tagged Bro1-containing proteins.
Immunoprecipitation was performed using anti-HA antibody conjugated beads.. Both input and immunoprecipitated complexes were analyzed by western blot using the indicated antibodies.
CHMP5 Recruits Endogenous Brox to Cellular Membranes
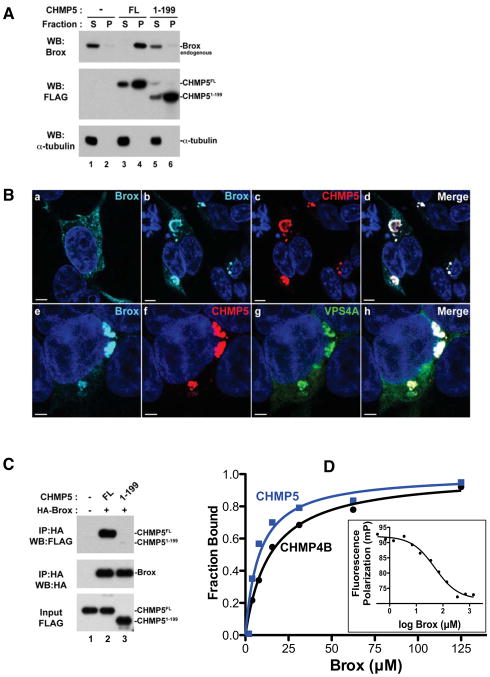
ESCRT-III proteins are known to cycle between cytoplasmic inactive forms (closed conformation) and membrane-bound active forms (open conformation) where they assemble into polymers that facilitate membrane fission (Bajorek et al., 2009; Bodon et al., 2011; Fabrikant et al., 2009; Hanson et al., 2008; Lata et al., 2008b; Pincetic et al., 2010; Shim et al., 2008). Overexpression of CHMP proteins triggers the formation of detergent-resistant complexes that can capture binding partners on cellular membranes (Pincetic et al., 2010; Shim et al., 2007). To test whether cellular Brox is recruited to membranes by CHMP5, we performed sedimentation assays, in which cellular extracts are separated into Triton-soluble (S) and Triton-insoluble (P) fractions (membrane-enriched) as previously described (Pincetic et al., 2010; Shim et al., 2007). As expected, endogenous Brox distributed mainly to the cytoplasmic soluble fraction in the absence of exogenously expressed ESCRT-III factors (Figure 2A, lanes 1–2). However, expression of CHMP5 dramatically redistributed Brox to the insoluble membrane-enriched fraction (Figure 2A, lanes 3–4). This suggests that specific interaction between CHMP5 and Brox recruited the latter to detergent-resistant membrane fractions. In contrast, CHMP4B was able to recruit both Brox and Alix to the membrane fractions (Figure S1A), consistent with its ability to interact with both proteins (Ichioka et al., 2008). To further characterize the interaction of CHMP5 and Brox in the cellular context, we performed immunofluorescence studies of CHMP5 and Brox. In contrast to Brox alone that exhibited a diffuse cytoplasmic staining, co-expression with CHMP5 redistributed Brox to endosomal-like structures where both proteins colocalized (Figure 2B, panels ad). The accumulation of GFP-VPS4A together with Brox and CHMP5 identified these structures as class E compartments (Figure 2B, panels e–h). These results are consistent with our sedimentation assays, and suggest that the Brox:CHMP5 complex is an integral component of the ESCRT machinery.
Figure 2. CHMP5 recruits Brox to VPS4A-positive endosomes and the CHMP5 C-tail is essential for Brox:CHMP5 interaction.
(A) CHMP5 recruits Brox to cellular membranes. HEK293T cells were transfected with empty vector (lanes 1–2), FLAG-tagged CHMP5 FL (lanes 3–4) or 1–199 (lanes 5–6). Forty-eight hours post transfection, cells were lysed and fractionated into soluble (S) and pellet (P) fractions as described in Experimental Procedures. Distribution of FLAG-tagged ESCRT-III and endogenous Brox was analyzed by western blot using the indicated antibodies. α-tubulin was used as a control protein for the soluble fraction.
(B) Brox and CHMP5 colocalize to VPS4A-positive endosomes. HA-tagged Brox was expressed in HEK293T cells alone (a), with FLAG-tagged CHMP5 (b–d) or in combination with both FLAG-tagged CHMP5 and GFP-tagged VPS4A (e–h). Cells were immunostained as described in Experimental Procedures. Brox, CHMP5, GFP-VPS4A are stained turquoise, red and green, respectively. Merge panels show colocalization of Brox and CHMP5 (d) or Brox, CHMP5 and VPS4A (h) in white. Scale bar = 5μm.
(C) The Brox binding site maps to the last 20 residues of CHMP5. FLAG-tagged CHMP5 full-length (FL, 1–219) or the C-terminal truncated fragment (1–199) was expressed in HEK293T cells in the presence or absence of the HA-tagged Brox. Examination of the protein interactions was performed similar to (A).
(D) Both CHMP4B and CHMP5 C-tails bind Brox. The C-tails of CHMP4B and CHMP5 bind Brox with dissociation constants of 14.3±1.4 μM (black) and 8.1±2.5 μM (blue), respectively, as measured by the FP assay. CHMP5 competes with CHMP4B for binding to Brox (insert) with an IC50 of 49.9 μM.
See also Figure S1.
The Brox:CHMP5 Association Requires the C-terminal Tail of CHMP5
To map the Brox binding region in CHMP5, we tested their interaction using immunoprecipitation assays. The C-terminal tails of the CHMP4 isoforms were previously shown to recapitulate the binding of the full-length CHMP4 proteins to the Alix Bro1 domain through hydrophobic residues at successive turns of their α-helices (McCullough et al., 2008), but it was unclear whether the CHMP5 C-terminal tail functions similarly to those from CHMP4s in Brox binding. We therefore generated truncation mutants from the C-terminus of CHMP5 and tested their ability to interact with Brox. Interestingly, removing the last 20 residues of CHMP5 (CHMP5 1-199) was sufficient to abrogate its interaction with Brox (Figure 2C, lane 3). This suggests that CHMP5 contains a new Brox-binding motif within its last 20 amino acid residues.
Since the full-length CHMP5 recruited Brox to detergent-insoluble membrane fractions, we next tested whether ectopic expression of the CHMP5 1-199 mutant affected the cellular distribution of Brox. In contrast to the full-length CHMP5, co-expression of the CHMP5 1-199 had no effect on Brox distribution in the Triton-soluble fraction (Figure 2A, compare lanes 3–4 and 5–6), even though both the CHMP5 full-length and the truncation mutant retained their propensity to sediment in the membrane-enriched pelletable fraction. Therefore, recruitment of Brox to the membrane-enriched fraction requires specific interaction with the CHMP5 C-terminal tail. In agreement, the CHMP5 C-tail binds Brox with an affinity comparable to that for the Brox:CHMP4B interaction (Figure 2D) as measured by a fluorescence polarization (FP) assay, and CHMP5 competed with CHMP4B for association with Brox. This is in contrast to the Alix Bro1 domain, which binds CHMP4B but has no detectable affinity for CHMP5 (Figure S1B).
The C-terminal Tails of the CHMP4B and CHMP5 Adopt Entirely Different Structures
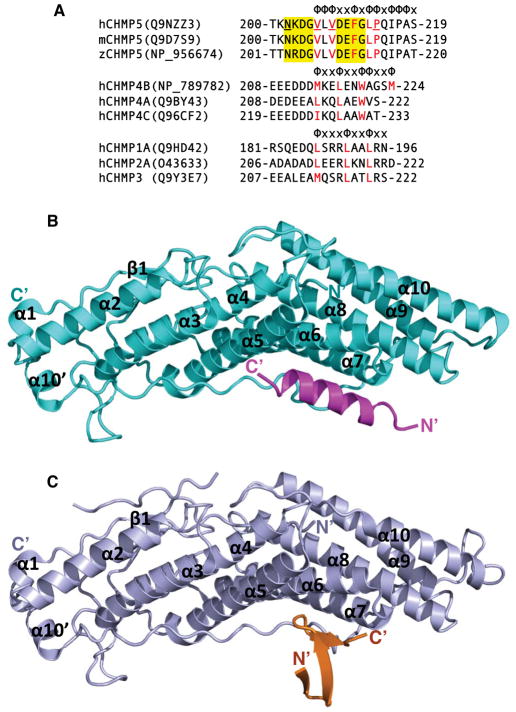
The CHMP4 C-tails were shown previously to engage the Alix Bro1 domain through hydrophobic residues (McCullough et al., 2008), but the spacing of the hydrophobic residues is highly divergent among the CHMP C-terminal tails (Figure 3A), To illustrate the molecular basis of the Brox interaction with CHMP5 and CHMP4B, we co-crystallized the Bro1 domain of Brox in complex with the C-terminal fragments of CHMP5 or CHMP4B and determined their structures (Table 1). As expected, the Brox:CHMP4B crystal structure shows that the C-terminal tail of CHMP4B adopts an amphipathic α-helical conformation bound to a pocket at the concave side of the Brox structure (Figure 3B and S2A). Similar to the structure of Alix:CHMP4B complex (McCullough et al., 2008), the CHMP4B hydrophobic residues M214, L217, W221, and M224 at successive turns of the α-helix contact Brox residues from the α5, α6, and α7 helices, and the long C-terminal loop (Figure 4A and S3). Comparison of the Brox structures in the presence and absence of CHMP4B reveals a large conformational change near residue Y348, with its Cα atom shifted ~6 Å (Figure S3B). This is largely due to the presence of the CHMP4B residue M214 that protrudes from the α helix and interacts with hydrophobic residues L208, L212 and Y348 in Brox. This in effect pushes the Y348 loop away from the body of the boomerang structure. Both residues L208 and L212 of Brox were previously reported to be important for CHMP4B binding by Popov and colleagues (Popov et al., 2009), and similar mode of interaction was observed between the same CHMP4B residue M214 and the Alix Bro1 domain residues I212 and L216 (McCullough et al., 2008) (Figure 4B).
Figure 3. Brox binds the C-terminal tails of CHMP4B and CHMP5.
(A) The C-terminal tail sequences for selected CHMP proteins. The sequences for human, mouse and zebrafish CHMP5, human CHMP4B, 4A, and 4C, and human CHMP1A, 2A, and 3 C-terminal tails are shown in three separate groups. Patterns of hydrophobicity are indicated above each group with “Φ” denotes hydrophobic residues and “x” denotes other residues. The hydrophobic residues involved in interaction with Brox, Alix, and VPS4 are marked in red for each group. Residues at the two β-turns of CHMP5 are shown in yellow shade, and those involved in interaction with Brox residue Y348 are underlined.
(B) Brox and CHMP4B are shown as cyan and magenta ribbons, respectively. The secondary structures of the Bro1 domain and the N- and C-termini of the CHMP4B are labeled.
(C) Brox and CHMP5 are shown as lightblue and orange ribbons, respectively. The secondary structures of the Bro1 domain and the N- and C-termini of the CHMP5 are labeled.
See also Figure S2.
Table 1.
X-ray Diffraction Data Collection and Structural Refinement
| Broxl-CHMP5 a | Broxl-5P b | Broxs-CHMP5 c | Broxs-5P | Broxl-CHMP4B | |
|---|---|---|---|---|---|
| Data Collection | |||||
| Spacegroup | C2 | C2 | P212121 | P212121 | P6122 |
| Unit cell (a, b, c) (Å) (α, β, γ) (°) |
172.4,46.0,68.6 90,104.5,90 |
173.3,46.2,68.8 90,105.0,90 |
68.9,111.4,116.3 90,90,90 |
69.2,111.4,116.0 90,90,90 |
138.2,138.2,373.8 90,90,120 |
| Wavelength (Å) | 1.075 | 1.075 | 1.075 | 1.075 | 1.075 |
| Resolution (Å) (Last shell) | 50.0–2.6 (2.7–2.6) | 50.0–3.1 (3.2–3.1) | 50.0–2.7 (2.8–2.7) | 50.0–2.6 (2.7–2.6) | 50.0–3.8 (3.9–3.8) |
| No of reflections (total/unique) | 54848/15918 | 34035/9731 | 97052/23032 | 116969/27011 | 95231/21693 |
| Completeness (%) | 97.4 (83.9) d | 99.2 (94.8) d | 91.6 (93.6) d | 94.0 (89.2) d | 99.3 (98.4) d |
| I/σ(I) | 8.7 (1.8) d | 6.1 (1.9) d | 7.1 (2.4) d | 9.0 (2.8) d | 18.3 (2.2) d |
| Rmerge (%) e | 8.45 (57.0) d | 12.5 (63.0) d | 13.3 (65.9) d | 9.8 (39.2) d | 3.61 (55.0) d |
| Refinement | |||||
| No. of protein atoms | 3237 | 3244 | 6238 | 6238 | 3155 |
| No. of solvent/hetero-atoms | 84/6 | 22/6 | 157/6 | 183/18 | 0 |
| Rmsd bond lengths (Å) | 0.005 | 0.007 | 0.005 | 0.004 | 0.006 |
| Rmsd bond angles (°) | 0.969 | 1.142 | 0.895 | 0.863 | 1.294 |
| Rwork (%) f | 16.6 | 19.6 | 18.6 | 17.7 | 23.3 |
| Rfree (%) g | 21.1 | 25.1 | 25.1 | 24.1 | 24.4 |
| Ramachandranplot favored/disallowed h | 97.0%/0.0 | 95.3%/0.0 | 96.3%/0.0 | 97.3%/0.0 | 96.7%/0.0 |
| PDB code | 3ULY | 3UM0 | 3UM1 | 3UM2 | 3UM3 |
Broxl is a long form of the human Brox protein (residues 2–411), and CHMP5 represents the C-terminal tail of CHMP5 (residues 151–219).
5P represents a peptide of the CHMP5 C-terminal 20 residues.
Broxs is a short form of the human Brox protein (residues 2–377).
Asterisked numbers correspond to the last resolution shell.
Rmerge = Σh Σi |Ii(h) -<I(h)> |/ΣhΣi Ii(h), where Ii(h) and <I(h)> are the ith and mean measurement of the intensity of reflection h.
Rwork = Σh||Fobs (h)|-|Fcalc (h)||/Σh|Fobs (h)|, where Fobs (h) and F calc (h) are the observed and calculated structure factors, respectively. No I/σ cutoff was applied.
Rfree is the R value obtained for a test set of reflections consisting of a randomly
selected 5% subset of the data set excluded from refinement.
Values from Molprobity server (http://molprobity.biochem.duke.edu/).
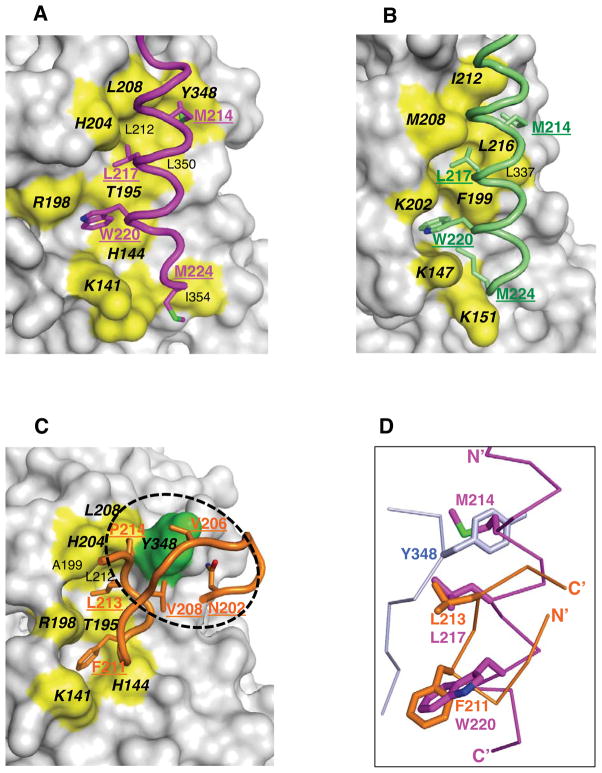
Figure 4. Brox employs similar surface for CHMP4B and CHMP5 binding.
(A) The CHMP4B C-terminal tail is shown in magenta with the four Brox-contacting hydrophobic residues in stick models and labeled. The Brox residues in contact with CHMP4B are shown as yellow colored surface and labeled, of which identical interface residues in both Brox:CHMP4B and Brox:CHMP5 (C) structures are in bold and italic.
(B) The Alix Bro1 domain in complex with CHMP4B (3C3Q) is shown as a reference for (A) and (C), with the CHMP4B colored lime. The conserved CHMP4B-binding residues between Alix and Brox (A) are labeled in bold and italic.
(C) The CHMP5 C-terminal tail is shown in orange with Brox-contacting residues in stick models and labeled. The Brox residues in contact with CHMP5 are shown similar to those in (A), except for residue Y348 shown in green. The second Brox:CHMP5 interface centered at Y348 is indicated with a dotted circle. Identical interface residues in both Brox:CHMP4B (A) and Brox:CHMP5 structures are in bold and italic. For clarity, the CHMP5 residues T200, K201, and I216 are omitted.
(D) The Brox:CHMP4B and Brox:CHMP5 structures are superimposed, and the CHMP4B and CHMP5 are represented as magenta and orange Cα traces. The three hydrophobic residues M214, L217 and W220 from CHMP4B, and the two hydrophobic residues F211 and L213 from CHMP5, as well as residue Y348 from Brox in the Brox:CHMP5 structure, are shown in magenta, orange, and lightblue sticks, respectively.
See also Figure S3.
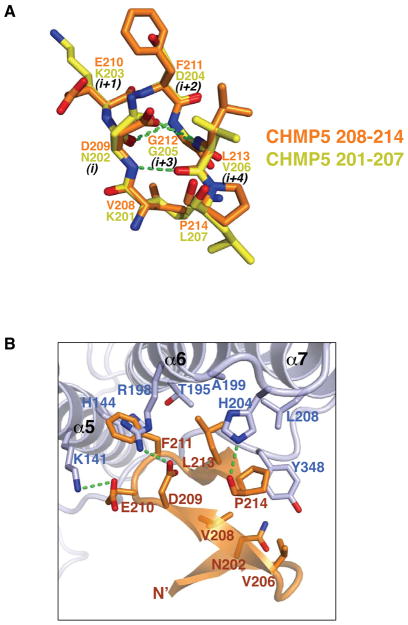
As the structures for all CHMP C-terminal tails reported to date adopt α-helical conformation, we were surprised to find that the C-terminal tail of CHMP5 adopts a tandem β-hairpin structure with three short β strands (Figure 3C and S2C). Both of the β-hairpins belong to a common 3:5 type composed of a classic type I β-turn and a G1 β-bulge at residues G205 and G212, respectively (Pantoja-Uceda et al., 2006; Richardson, 1981; Sibanda et al., 1989) (Figure 5A and S4). The patterns of the main-chain hydrogen bonds are identical for the two β-hairpins, with one hydrogen bond between the carbonyl oxygen of residue i and the amide nitrogen of residue i+3, and the other hydrogen bond between the amide nitrogen of residue i and the carbonyl oxygen of residue i+4. The overall structure of the tandem β-hairpins are stabilized by four additional main-chain:main-chain and main-chain:side-chain hydrogen bonds. These include hydrogen bonds between the carbonyl oxygen of L207 and amide nitrogen of Q215, and the amide nitrogen of V208 and carbonyl oxygen of T200 (Figure S4A), as well as the N202 (residue i) side-chain to G205 (residue i+3) main-chain amide hydrogen bond and D209 (residue i) side-chain to G212 (residue i+3) main-chain amide hydrogen bond (Figure 5A). The seven Cα atoms of the two β-hairpins can be superimposed with an rmsd of 0.15 Å, indicating essentially identical main-chain conformation.
Figure 5. The Brox:CHMP5 interface.
(A) The two β-hairpins from CHMP5 are superimposed and represented as yellow (residues 201–207) and orange (residues 208–214) stick models. Hydrogen bonds are shown with green dotted lines. Residues in the β-turns are denoted with i to i+4, according to conventions by Sibanda and colleagues (Sibanda et al., 1989).
(B) Brox and CHMP5 are shown as lightblue and orange ribbons with the interface residues represented in sticks. Hydrogen bonds are shown with green dotted lines. For clarity, the CHMP5 residues Q215 and I216 are omitted. See also Figure S4.
During structural determination of the Brox-CHMP5 complex, a second region of positive electron density was observed for crystals containing the longer “Broxl” form at the convex surface of the Brox structure (Figure S2B). Residues T384 to P394 were then built in the current model for the Broxl-CHMP5 and Broxl-5P structures (Table 1) based on the close proximity of the density to the last residue P379 of the Bro1 domain, and the characteristic bulged side chains of P386 and P388 in an extended Brox C-terminal region. Nonetheless, the current resolution (2.6 Å) does not allow for unambiguous determination of the amino acid side chains, and thus we can not formally exclude the possibility of alternative models.
The Brox:CHMP5 Complex Employs Two Adjacent Interfaces
Despite the drastic structural differences, CHMP5 binds Brox at a similar surface as CHMP4B, but engages two adjacent hydrophobic surfaces with its second β-hairpin. At the first interface, the CHMP5 residues F211 and L213 dock onto a pocket formed by Brox α5 helix (residues K141 and H144), α6 helix and the following linker (residues T195, R198, A199 and H204), and α7 helix (residue L208). Most of these same Brox residues are also involved in binding of CHMP4B residues W220 and L217 (Figure 4A, 4C and 5B). The third CHMP4B hydrophobic residue M214 has no equivalence in CHMP5, instead, the Brox residue Y348 occupies the same equivalent position (Figure 4D). It binds a unique pocket in CHMP5 formed by residues N202, V206, V208 and P214, constituting an adjacent second Brox:CHMP5 interface (Figure 4C and 5B). Remarkably, these Brox-binding residues are identical among the CHMP5 proteins from zebrafish to human, but are not conserved in other CHMPs (Figure 3A). In addition, the Brox residue Y348 is not conserved among other Bro1 domain proteins (Sette et al., 2011; Zhai et al., 2011; Ichioka et al., 2008), which is consistent with the specific interaction between Brox and CHMP5 (Figure 1A and S1A). The Brox loop containing Y348 has the most divergent sequences among the Bro1 domain-containing proteins, transverses the entire length of the Bro1 domain structures, and is longer in Brox than in Alix or HD-PTP (Sette et al., 2011; Zhai et al., 2011). These observations suggest that this long C-terminal loop of the Bro1 domains may be adaptable to different partner proteins and confer unique functions to each protein.
The peptide directions are reversed between CHMP4B and CHMP5 in reference to Brox, such that the major hydrophobic residues of comparable sizes (F211 from CHMP5 and W220 from CHMP4B, and L213 from CHMP5 and L217 from CHMP4B) occupy the same pockets in Brox (Figure 4D). In contrast to the Brox:CHMP4B structure, CHMP5 binding does not induce major structural changes in the Brox C-terminal loop with less than 2 Å shifts of the Cα atoms, suggesting a rigid-body fit between Brox and CHMP5 (Figure S3D). In total, there are 1100 Å2 of solvent accessible surface are buried at the Brox:CHMP5 C-terminal tail interface, compared with 1300 Å2 buried at the Brox:CHMP4B C-terminal tail interface.
Mutation of the Brox:CHMP5 and Brox:CHMP4B Interface Residues Compromises their Interactions
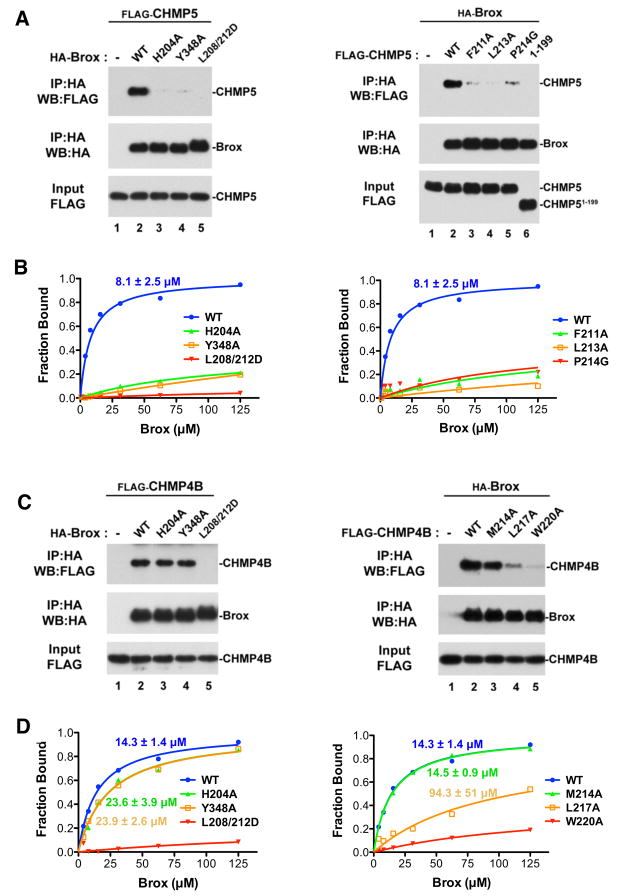
To examine the significance of the Brox:CHMP4B and Brox:CHMP5 interfaces observed in the crystal structures, we mutated the interface residues and performed immunoprecipitation and in vitro binding assays to evaluate the interactions (Figure 6). The Brox residues L208 and L212 are located at its interface with both CHMP4B and CHMP5, and were previously reported to be important for Brox:CHMP4B binding (Popov et al., 2009). In agreement, mutation of these residues resulted in undetectable binding to both CHMP5 (Figure 6A left panel, lane 5 and Figure 6B left panel) and CHMP4B (Figure 6C left panel, lane 5, Figure 6D left panel). Similarly, the Brox residues H204 and Y204 are essential for CHMP5 binding (Figure 6A left panel, lanes 3 and 4 and Figure 6B left panel), as H204 form a crucial hydrogen bond with the main-chain carbonyl oxygen of CHMP5 P214 (Figure 5B), and Y348 engages a largely hydrophobic pocket formed by four CHMP5 residues (Figure 4C). Neither H204A nor Y348A appears to be necessary for CHMP4B binding (Figure 6C left panel, lanes 3 and 4 and Figure 6D left panel), because both are located at the periphery of the Brox:CHMP4B interface devoid of main-chain contacts or significant hydrophobic interactions (Figure 4A). Importantly, mutation of the CHMP5/CHMP4B-binding surface residues in Brox did not compromise the proper folding of the mutants, as shown by their similar CD spectra to the wild type Brox protein (Figure S5).
Figure 6. Characterization of the Brox:CHMP5 and Brox:CHMP4B interfaces in the cell.
(A) Identification of Brox (left panel) and CHMP5 (right panel) residues important for the Brox:CHMP5 interaction in the cell. FLAG-tagged CHMP5 proteins (WT or the indicated mutant) were expressed in HEK293T cells in the presence or absence of HA-tagged Brox (WT or the indicated mutant). Forty-eight hours post transfection, cells were lysed in RIPA buffer and cleared lysates were incubated with anti-HA antibody conjugated beads. Both input and immunoprecipitated complexes were analyzed by western blot using the indicated antibodies.
(B) Fluorescence polarization assay for the Brox (left panel) and CHMP5 (right panel) mutants. The binding isotherms and the dissociation constants are marked and color-coded.
(C) Identification of Brox (left panel) and CHMP4B (right panel) residues important for the Brox:CHMP4B interaction in the cell. FLAG-tagged CHMP4B proteins (WT or the indicated mutant) were expressed in HEK293T cells in the presence or absence of HA-tagged Brox (WT or the indicated mutant). Examination of the protein interactions was performed similar to (A).
(D) Fluorescence polarization assay for the Brox (left panel) and CHMP4B (right panel) mutants. The binding isotherms and the dissociation constants are marked and color-coded.
See also Figure S5.
On the CHMP5 side of the interface, mutation of residues F211 and L213 at its first interface with Brox is deleterious to binding, as well as mutation of P214 at the second interface (Figure 6A right panel, lanes 3–5 and Figure 6B right panel). In comparison, the CHMP4B residues W220 and L217, binding at the equivalent Brox surface as residues F211 and L213 from CHMP5, play essential roles in Brox:CHMP4B association (Figure 6C right panel, lanes 4–5 and Figure 6D right panel). By contrast, M214 at the peripheral of the interface appears not essential for this interaction (Figure 6C right panel, lane 3 and Figure 6D right panel). In summary, our observations of the Brox:CHMP5/CHMP4B interactions are consistent with our analysis of their crystal structures.
DISCUSSION
Our structural and biochemical studies show that the C-terminal tails of both CHMP4B and CHMP5 bind Brox at the same site, but employ entirely different conformations to posit two equivalent hydrophobic residues at the interface. Re-organization of the Brox loop near residue Y348 accommodates the third hydrophobic residue M214 from CHMP4B, whereas a unique CHMP5 hydrophobic pocket binds the Brox residue Y348 at an equivalent location as the CHMP4B residue M214. Importantly, interaction of Brox with the CHMP5 C-terminal tail is essential for recruitment of Brox to detergent-resistant membrane fractions, which may be relevant to its function during membrane remodeling events. This work not only provides novel insights into the ESCRT-III and Bro1 domain interactions, but also lays the groundwork for future investigation of the unique functions of the CHMP4 and CHMP5 proteins through their overlapping but distinct binding profiles to the Bro1 domain proteins.
The most novel finding of this work is that CHMP5 is a new partner for Brox, adopts a tandem β-hairpin structure at its C-terminal tail, and binds Brox at a similar site as CHMP4B but with reversed peptide directions. β-hairpins are the simplest super-secondary structures that are abundant in globular protein structures (Sibanda and Thornton, 1985). Because of their frequent occurrence at the hotspots of protein:protein interface, there is intense interest in the design of β-hairpins or their mimetics as potential therapeutic agents (Pantoja-Uceda et al., 2006; Robinson, 2008). In fact, the first example of a designed β-hairpin has a sequence of NPDG very similar to the first β-turn of the CHMP5 C-terminal tail (Blanco et al., 1993). Analysis of the CHMP5 C-terminal tail sequence suggests that it is in keeping with previous statistical analysis of β-turn preferences (Hutchinson and Thornton, 1994). In addition, the CHMP5 C-terminal fragment is dominated by branched/hydrophobic residues that favor β-instead of α-secondary structures (Minor and Kim, 1994). Therefore, the tandem β-hairpin structure of the CHMP5 C-terminal tail is dictated by its amino acid sequence. Single and tandem β-hairpin structures have been identified previously in the nuclear receptor PPARγ, methyltransferase, γ-chymotrypsin, the sindbisvirus capsid protein, and the WW domains (Figure S4B-G) (Efimov, 1992; Ilsley et al., 2002). Notably, the WW domains are known to be one of the smallest and most adaptable mediators of protein-protein interactions (Ilsley et al., 2002), which parallels the critical involvement of the β-hairpins at the Brox:CHMP5 interface.
This study identified Brox as the second interacting partner for CHMP5 described to date, and provides a starting point to address the functional significance of the Brox:CHMP5 interaction. Our understanding of CHMP5/Vps60 function in eukaryotic cells has been limited to its role in regulating VPS4 function in a number of cellular processes via binding its other cellular partner, LIP5/Vta1 (Azmi et al., 2008; Shiflett et al., 2004; Ward et al., 2005). Even though CHMP5 is not strictly essential for the function of ESCRT-III in membrane scission and MVB biogenesis, it plays an essential role in regulating the function of late endosomes and lysosomes: CHMP5 deficiency caused accumulation of late endosome compartments, and enhanced cell surface receptor signaling due to diminished receptor turnover, and was embryonically lethal (Shim et al., 2006). In addition, CHMP5 was shown to be essential for spindle formation during mitosis and was found to localize at the midbody during cytokinesis, suggesting its broad participation in cell division (Morita et al., 2010; Morita et al., 2007). Furthermore, CHMP5 was reported to play a key role in the initiation of innate immune response against retroviral infection by mediating post-translational modification of ESCRT-III proteins and blockade of retroviral release (Kuang et al., 2011). It remains to be determined whether some of the defects observed in CHMP5-depleted mammalian cells were a consequence of interference with Brox function. Since CHMP5 and CHMP4B compete for the same binding site in Brox, an attractive model would be that CHMP5 participates in dissociating Brox from CHMP4B once the membrane scission is complete. By using a novel and distinct mode of binding, Brox appears to have acquired a unique way to regulate and/or disassemble the CHMP4 proteins by recruiting CHMP5 and masking a functional interface. This is in contrast to the Alix Bro1 domain, which binds CHMP4B but has no detectable affinity for CHMP5, and might be utilizing other modes of regulation for CHMP4 binding and function. For example, the V or PRD domain of Alix could mask the CHMP4 binding site at the Alix Bro1 domain in an autoinhibited state (Zhou et al., 2009). Studies are underway to further understand these processes.
EXPERIMENTAL PROCEDURES
A full description of the methods is in the Supplemental Information.
Immunoprecipitation Assay
The mammalian expression vectors for N-terminal HA-tagged human Alix, Brox, HD-PTP and Rhophilin-2 were reported previously (Dussupt et al., 2009). Expression vectors for ESCRT-III proteins were generated by PCR amplification from cDNA previously described (Dussupt et al., 2009) or purchased from Open Biosystems (Huntsville, AL) and subcloned into p3XFLAG-myc-CMV-26 (Sigma, St. Louis, MO). Point mutations were generated using the Quik-change mutagenesis kit (Stratagene, Santa Clara, CA). HEK293T cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) with the indicated expression vectors encoding HA- and FLAG-tagged proteins. Forty-eight hours post-transfection, cell lysates were precipitated with agarose beads covalently linked to anti-HA mouse monoclonal antibody (Sigma, St. Louis, MO). The beads were then extensively washed and eluted with HA peptide (100 mg/ml, Sigma, St. Louis, MO). Immunoprecipitates and cell lysates (input fractions) were visualized by SDS-PAGE and western blot with anti-FLAG or anti-HA antibody (Sigma, St. Louis, MO).
Sedimentation Assay
The sedimentation assay was adapted from previously described protocols (Pincetic et al., 2010; Shim et al., 2007). HEK293T cells were transfected with the indicated plasmids and 48 hours post-transfection, cells were harvested and resuspended in a lysis buffer containing 10 mM Tris-HCl pH 7.5, 10 % sucrose, 1 mM EDTA, 0.1 % Triton X-100 and protease inhibitor cocktail. The soluble (S) and pellet (P) fractions were separated by centrifugation at 10,000 × g for 15 min at 4°C. Pellets were resuspended into the same volume of lysis buffer as the supernatant and sonicated to shear DNA. Equal volumes of fraction S and P were analyzed by SDS-PAGE and western blot using anti-FLAG antibody (M2, Sigma, St. Louis, MO), anti-α-tubulin antibody (DM1A, Sigma, St. Louis, MO) and rabbit antisera to Alix and Brox produced in the Bouamr lab.
Immunofluorescence microscopy
HEK293T cells were seeded on glass coverslips and transfected with the indicated plasmids using Lipofectamine 2000. Twenty-four hours post transfection, the cells were fixed in 3.7 % paraformaldehyde in PBS, quenched with 100 mM Glycine in PBS, permeabilized with 0.5% Triton X-100 (Sigma, St. Louis, MO) and blocked in 1% BSA in PBS. Cells were stained with mouse anti-HA and rabbit anti-FLAG primary antibodies (Sigma, St. Louis, MO) followed by Alexa 633-conjugated anti-mouse and Alexa 594-conjugated anti-rabbit secondary antibodies (Invitrogen, Carlsbad, CA). Nuclei were counterstained with DAPI. Coverslips were mounted with ProLong Gold (Invitrogen, Carlsbad, CA) and sequential Z-sections (about 0.25 μm each) were obtained by confocal microscopy using a Leica SP5 inverted confocal microscope (Leica Microsystems, Exton, PA). Micrographs were generated using Imaris software (version 7.3, Bitplane AG, Zurich, Switzerland).
Protein Expression and Purification
The C-terminal fragments of human CHMP5 (residues 151–219) and human CHMP4B (residues 121–224) were cloned in a modified pET30a(+) vector (EMD Biosciences, San Diego, CA). The expression vector were transformed into E. coli BL21 (DE3) cells and protein expression was induced with 0.5 mM isopropyl-β-d-thiogalactoside (IPTG) at OD600 = 0.6 and grew overnight at 15°C. Cells were harvested and lysed in a buffer containing 25 mM Tris-HCl, pH 8.0, 125 mM NaCl and 10 mM imidazole. Cleared cell lysates were purified by nickel affinity chromatography with a HisPrep FF 16/10 affinity column (GE Healthcare Biosciences, Piscataway, NJ). This was followed by TEV digestion and size-exclusion chromatography, and a second nickel affinity chromatography in a buffer containing 25 mM Tris-HCl pH 8.0 and 150 mM NaCl. Two forms of human Brox proteins were used in this study: residues 2–411 and denoted “Broxl” in Table 1, and residues 2–377 and denoted “Broxs” in Table 1. The Brox and Alix Bro1 domains were expressed and purified as reported previously (Sette et al., 2011) and similar to the above for the CHMP proteins. Both the “Broxl” and “Broxs” forms were used in co-crystallization with CHMP5, and only the “Broxl” form was used in co-crystallization with CHMP4B.
Crystallization and X-ray Diffraction Data Collection
Crystals for the Brox:CHMP4B complex was obtained by mixing purified “Broxl” form and the CHMP4B C-terminal fragment at a 1:1.2 molar ratio and crystallizing with a well solution containing 0.1 M imidazole buffer at pH 7.2, 15% glycerol and 1 M sodium citrate at 4°C. The Brox:CH MP5 crystals were obtained by mixing the Brox proteins and the CHMP5 C-terminal fragment at a 1:1.5 molar ratio and crystallizing with a well solution containing 0.2 M magnesium acetate, 0.1M sodium cacodylate, pH 6.5, and 20% polyethylene glycol 8000 at 18°C. A synthesized peptide (TKNKDGVLVDEFGLPQIPAS, United peptides, Rockville, MD) corresponding to the CHMP5 C-terminal 20 residues (denoted “-5P” in Table 1) was used to obtain co-crystals with Brox that are essentially identical to those using the 69 residue CHMP5 C-terminal fragment (denoted “-CHMP5” in Table 1). The crystals were flash frozen in liquid nitrogen with 20% glycerol as cryoprotectant. X-ray diffraction data were collected at beamline 23-ID at the Argonne National Laboratory (Argonne, IL) or beamline X29 at the Brookhaven National Laboratory (Upton, NY). The diffraction data were processed with the HKL2000 package (Zbyszek Otwinowski, 1997).
Structure Determination and Refinement
Structures for the Brox:CHMP4B and Brox:CHMP5 complexes were determined by molecular replacement with program Phaser (McCoy, 2007) using the crystal structure of Brox (3R9M) (Sette et al., 2011) as a search model. Electron density maps calculated with the molecular replacement solutions showed clear positive densities at the concave surface of the Brox boomerang structure (Figure S2). The identity of the CHMP4B C-terminal tail was determined based on the prominent density for the CHMP4B W220 side-chain, the strong cylinder-shaped density for a helical structure, and the overall similarity between the Alix:CHMP4B (3C3Q) and Brox:CHMP4B structures (Figure S2A). The registry of the CHMP5 C-terminal tail was unambiguously determined based on the identical positive densities calculated with the CHMP5 C-terminal fragment (“-CHMP5”) and the 20 residue peptide (“-5C”), the prominent density for the F211 side-chain, no side-chain density for the adjacent G212, and the following LP sequence has a characteristic ring structure at the Proline main-chain (Figure S2C).
A second region of positive electron density was observed at the convex surface of the Brox structure for crystals containing the “Broxl” form only, and Brox residues T384 to P394 were built in the current model for the Broxl-CHMP5 and Broxl-5P structures based on the close proximity of the density to the last residue P379 of the Bro1 domain, and the characteristic bulged side chains of P386 and P388 in an extended Brox C-terminal region. Nonetheless, we can not formally exclude the possibility of alternative models because the current resolution does not allow for unambiguous determination of the amino acid side chains. The Brox:CHMP5 structural models were built and refined using programs COOT (Emsley and Cowtan, 2004) and PHENIX (Adams et al., 2010), respectively. The Brox:CHMP4 structure was refined with the deformable elastic network (DEN) approach incorporated in program CNS (Brünger et al., 1998; Schroder et al., 2010). Calculation of the solvent accessible surface area was carried out using program areaimol from the CCP4 program suite (Lee and Richards, 1971; Potterton et al., 2003). All structure figures were prepared with program Pymol (Schrödinger, LLC.).
Fluorescence Polarization Assay
5-Fluorescein (FAM) labeled peptides (Table S1) (United Biosystems, Rockville, MD) were dissolved in 25 mM Tris-Cl, pH 8.0 and 150 mM NaCl. Purified wild type or mutant “Broxl” protein samples were diluted with the above buffer and mixed with the 2 nM FAM labeled peptides. The mixtures were assayed in black 96 well plates with a Molecular Devices Paradigm spectrometer (Molecular Devices, Sunnyvale, CA). For the CHMP4B and CHMP5 competition assay, 2 nM FAM-labeled CHMP4B peptide and increasing concentrations of unlabeled CHMP5 peptide were mixed with 50 μM Brox protein and the fluorescence polarization was measured. Data were analyzed and plotted using program GraphPad Prism version 5.0 (GraphPad Software, San Diego, CA).
Circular Dichroism (CD) Spectroscopy
A J-715 spectrometer (Jasco, Easton, MD) was used to measure the CD spectra of the wild-type and mutant Brox protein samples in PBS buffer (pH 7.4) at 0.5 mg/ml. The spectra were acquired from 198 to 260 nm with a 1 nm stepsize at 20 °C and the buffer only spectrum was subtracted from the Brox sample spectra.
Supplementary Material
Highlights.
CHMP5 binds and recruits Brox to membrane fractions through its C-terminal tail
The CHMP5 C-terminal tail adopts a tandem β-hairpin structure
The CHMP4B C-terminal tail α-helix binds at a concave surface of Brox
The CHMP5 β-hairpins and CHMP4B α-helix bind the same surface at Brox
Acknowledgments
The authors would like to thank the X29A beam line scientists at the Brookhaven National Laboratory for their assistance with X-ray diffraction data collection. We thank Masatoshi Maki for reagents and Alicia Buckler-White and her team at LMM for DNA sequencing and the Biological Imaging team at the Research Technologies Branch, NIAID for assistance with confocal microscopy. We are grateful to Tengchuan Jin for help with the FP assay, Grzegorz Piszczek at the Biochemistry and Biophysics Center of NHLBI for assistance with CD spectroscopy, and D. Eric Anderson at the Mass Spectrometry facility of NIDDK for technical support. F. B. and T.S.X. are supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH. This project was also supported by funds from the Office of AIDS Research (OAR, NIH) to FB and TSX.
Footnotes
ACCESSION NUMBERS
Coordinates and structure factor files have been deposited in the RCSB Protein Data Bank with the accession codes 3ULY, 3UM0, 3UM1, 3UM2, and 3UM3.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi IF, Davies BA, Xiao J, Babst M, Xu Z, Katzmann DJ. ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Developmental cell. 2008;14:50–61. doi: 10.1016/j.devcel.2007.10.021. [DOI] [PubMed] [Google Scholar]
- Bajorek M, Schubert HL, McCullough J, Langelier C, Eckert DM, Stubblefield WM, Uter NT, Myszka DG, Hill CP, Sundquist WI. Structural basis for ESCRT-III protein autoinhibition. Nat Struct Mol Biol. 2009;16:754–762. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco FJ, Jimenez MA, Herranz J, Rico M, Santoro J, Nieto JL. Nmr Evidence of a Short Linear Peptide That Folds into a Beta-Hairpin in Aqueous-Solution. J Am Chem Soc. 1993;115:5887–5888. [Google Scholar]
- Bodon G, Chassefeyre R, Pernet-Gallay K, Martinelli N, Effantin G, Lutje Hulsik D, Belly A, Goldberg Y, Chatellard-Causse C, Blot B, et al. Charged multivesicular body protein-2B (CHMP2B) of the endosomal sorting complex required for transport-III (ESCRT-III) polymerizes into helical structures deforming the plasma membrane. The Journal of biological chemistry. 2011;286:40276–40286. doi: 10.1074/jbc.M111.283671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers K, Lottridge J, Helliwell SB, Goldthwaite LM, Luzio JP, Stevens TH. Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic. 2004;5:194–210. doi: 10.1111/j.1600-0854.2004.00169.x. [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, et al. Crystallography and NMR system (CNS): A new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Caballe A, Martin-Serrano J. ESCRT machinery and cytokinesis: the road to daughter cell separation. Traffic. 2011;12:1318–1326. doi: 10.1111/j.1600-0854.2011.01244.x. [DOI] [PubMed] [Google Scholar]
- Dordor A, Poudevigne E, Gottlinger H, Weissenhorn W. Essential and supporting host cell factors for HIV-1 budding. Future Microbiol. 2011;6:1159–1170. doi: 10.2217/fmb.11.100. [DOI] [PubMed] [Google Scholar]
- Doyotte A, Mironov A, McKenzie E, Woodman P. The Bro1-related protein HD-PTP/PTPN23 is required for endosomal cargo sorting and multivesicular body morphogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6308–6313. doi: 10.1073/pnas.0707601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussupt V, Javid MP, Abou-Jaoude G, Jadwin JA, de La Cruz J, Nagashima K, Bouamr F. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog. 2009;5:e1000339. doi: 10.1371/journal.ppat.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov AV. A novel super-secondary structure of beta-proteins. A triple-strand corner. FEBS letters. 1992;298:261–265. doi: 10.1016/0014-5793(92)80072-o. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fabrikant G, Lata S, Riches JD, Briggs JA, Weissenhorn W, Kozlov MM. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput Biol. 2009;5:e1000575. doi: 10.1371/journal.pcbi.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128:841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. The Journal of cell biology. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Developmental cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Hurley JH. The ESCRT complexes. Crit Rev Biochem Mol Biol. 2010;45:463–487. doi: 10.3109/10409238.2010.502516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nat Rev Mol Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson EG, Thornton JM. A revised set of potentials for beta-turn formation in proteins. Protein science : a publication of the Protein Society. 1994;3:2207–2216. doi: 10.1002/pro.5560031206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichioka F, Kobayashi R, Katoh K, Shibata H, Maki M. Brox, a novel farnesylated Bro1 domain-containing protein that associates with charged multivesicular body protein 4 (CHMP4) Febs J. 2008;275:682–692. doi: 10.1111/j.1742-4658.2007.06230.x. [DOI] [PubMed] [Google Scholar]
- Ichioka F, Takaya E, Suzuki H, Kajigaya S, Buchman VL, Shibata H, Maki M. HD-PTP and Alix share some membrane-traffic related proteins that interact with their Bro1 domains or proline-rich regions. Arch Biochem Biophys. 2007;457:142–149. doi: 10.1016/j.abb.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Ilsley JL, Sudol M, Winder SJ. The WW domain: linking cell signalling to the membrane cytoskeleton. Cell Signal. 2002;14:183–189. doi: 10.1016/s0898-6568(01)00236-4. [DOI] [PubMed] [Google Scholar]
- Katoh K, Shibata H, Suzuki H, Nara A, Ishidoh K, Kominami E, Yoshimori T, Maki M. The ALG-2-interacting protein Alix associates with CHMP4b, a human homologue of yeast Snf7 that is involved in multivesicular body sorting. The Journal of biological chemistry. 2003;278:39104–39113. doi: 10.1074/jbc.M301604200. [DOI] [PubMed] [Google Scholar]
- Kieffer C, Skalicky JJ, Morita E, De Domenico I, Ward DM, Kaplan J, Sundquist WI. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Developmental cell. 2008;15:62–73. doi: 10.1016/j.devcel.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Kim J, Sitaraman S, Hierro A, Beach BM, Odorizzi G, Hurley JH. Structural basis for endosomal targeting by the Bro1 domain. Developmental cell. 2005;8:937–947. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang Z, Seo EJ, Leis J. Mechanism of inhibition of retrovirus release from cells by interferon-induced gene ISG15. Journal of virology. 2011;85:7153–7161. doi: 10.1128/JVI.02610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Roessle M, Solomons J, Jamin M, Gottlinger HG, Svergun DI, Weissenhorn W. Structural basis for autoinhibition of ESCRT-III CHMP3. Journal of molecular biology. 2008a;378:818–827. doi: 10.1016/j.jmb.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008b;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Fisher RD, Whitby FG, Sundquist WI, Hill CP. ALIX-CHMP4 interactions in the human ESCRT pathway. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7687–7691. doi: 10.1073/pnas.0801567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor DL, Jr, Kim PS. Measurement of the beta-sheet-forming propensities of amino acids. Nature. 1994;367:660–663. doi: 10.1038/367660a0. [DOI] [PubMed] [Google Scholar]
- Morita E, Colf LA, Karren MA, Sandrin V, Rodesch CK, Sundquist WI. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12889–12894. doi: 10.1073/pnas.1005938107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. The EMBO journal. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muziol T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, Gottlinger H, Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Developmental cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Nickerson DP, West M, Henry R, Odorizzi G. Regulators of Vps4 ATPase activity at endosomes differentially influence the size and rate of formation of intralumenal vesicles. Mol Biol Cell. 2010;21:1023–1032. doi: 10.1091/mbc.E09-09-0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, Emr SD, Williams RL. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Katzmann DJ, Babst M, Audhya A, Emr SD. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J Cell Sci. 2003;116:1893–1903. doi: 10.1242/jcs.00395. [DOI] [PubMed] [Google Scholar]
- Pantoja-Uceda D, Santiveri CM, Jimenez MA. De novo design of monomeric beta-hairpin and beta-sheet peptides. Methods Mol Biol. 2006;340:27–51. doi: 10.1385/1-59745-116-9:27. [DOI] [PubMed] [Google Scholar]
- Peel S, Macheboeuf P, Martinelli N, Weissenhorn W. Divergent pathways lead to ESCRT-III-catalyzed membrane fission. Trends in biochemical sciences. 2011;36:199–210. doi: 10.1016/j.tibs.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Pincetic A, Kuang Z, Seo EJ, Leis J. The interferon-induced gene ISG15 blocks retrovirus release from cells late in the budding process. Journal of virology. 2010;84:4725–4736. doi: 10.1128/JVI.02478-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov S, Popova E, Inoue M, Gottlinger HG. Divergent Bro1 domains share the capacity to bind human immunodeficiency virus type 1 nucleocapsid and to enhance virus-like particle production. Journal of virology. 2009;83:7185–7193. doi: 10.1128/JVI.00198-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nature chemical biology. 2006;2:584–590. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- Richardson J. The anatomy and taxonomy of protein structure. Advances in Protein Chemistry. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Robinson JA. Beta-hairpin peptidomimetics: design, structures and biological activities. Acc Chem Res. 2008;41:1278–1288. doi: 10.1021/ar700259k. [DOI] [PubMed] [Google Scholar]
- Row PE, Liu H, Hayes S, Welchman R, Charalabous P, Hofmann K, Clague MJ, Sanderson CM, Urbe S. The MIT domain of UBPY constitutes a CHMP binding and endosomal localization signal required for efficient epidermal growth factor receptor degradation. The Journal of biological chemistry. 2007;282:30929–30937. doi: 10.1074/jbc.M704009200. [DOI] [PubMed] [Google Scholar]
- Rusten TE, Stenmark H. How do ESCRT proteins control autophagy? J Cell Sci. 2009;122:2179–2183. doi: 10.1242/jcs.050021. [DOI] [PubMed] [Google Scholar]
- Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder GF, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464:1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette P, Mu R, Dussupt V, Jiang J, Snyder G, Smith P, Xiao TS, Bouamr F. The Phe105 Loop of Alix Bro1 Domain Plays a Key Role in HIV-1 Release. Structure. 2011;19:1485–1495. doi: 10.1016/j.str.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiflett SL, Ward DM, Huynh D, Vaughn MB, Simmons JC, Kaplan J. Characterization of Vta1p, a class E Vps protein in Saccharomyces cerevisiae. The Journal of biological chemistry. 2004;279:10982–10990. doi: 10.1074/jbc.M312669200. [DOI] [PubMed] [Google Scholar]
- Shim JH, Xiao C, Hayden MS, Lee KY, Trombetta ES, Pypaert M, Nara A, Yoshimori T, Wilm B, Erdjument-Bromage H, et al. CHMP5 is essential for late endosome function and down-regulation of receptor signaling during mouse embryogenesis. The Journal of cell biology. 2006;172:1045–1056. doi: 10.1083/jcb.200509041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S, Kimpler LA, Hanson PI. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- Shim S, Merrill SA, Hanson PI. Novel interactions of ESCRT-III with LIP5 and VPS4 and their implications for ESCRT-III disassembly. Mol Biol Cell. 2008;19:2661–2672. doi: 10.1091/mbc.E07-12-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibanda BL, Blundell TL, Thornton JM. Conformation of β-hairpins in protein structures. A systematic classification with applications to modelling by homology, electron density fitting and protein engineering. J Mol Biol. 1989;206:759–777. doi: 10.1016/0022-2836(89)90583-4. [DOI] [PubMed] [Google Scholar]
- Sibanda BL, Thornton JM. β-hairpin families in globular proteins. Nature. 1985;316:170–174. doi: 10.1038/316170a0. [DOI] [PubMed] [Google Scholar]
- Solomons J, Sabin C, Poudevigne E, Usami Y, Hulsik DL, Macheboeuf P, Hartlieb B, Gottlinger H, Weissenhorn W. Structural basis for ESCRT-III CHMP3 recruitment of AMSH. Structure. 2011;19:1149–1159. doi: 10.1016/j.str.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- Usami Y, Popov S, Gottlinger HG. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. Journal of virology. 2007;81:6614–6622. doi: 10.1128/JVI.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward DM, Vaughn MB, Shiflett SL, White PL, Pollock AL, Hill J, Schnegelberger R, Sundquist WI, Kaplan J. The role of LIP5 and CHMP5 in multivesicular body formation and HIV-1 budding in mammalian cells. The Journal of biological chemistry. 2005;280:10548–10555. doi: 10.1074/jbc.M413734200. [DOI] [PubMed] [Google Scholar]
- Xiao J, Chen XW, Davies BA, Saltiel AR, Katzmann DJ, Xu Z. Structural basis of Ist1 function and Ist1-Did2 interaction in the multivesicular body pathway and cytokinesis. Mol Biol Cell. 2009;20:3514–3524. doi: 10.1091/mbc.E09-05-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Xia H, Zhou J, Azmi IF, Davies BA, Katzmann DJ, Xu Z. Structural basis of Vta1 function in the multivesicular body sorting pathway. Developmental cell. 2008;14:37–49. doi: 10.1016/j.devcel.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Rismanchi N, Renvoise B, Lippincott-Schwartz J, Blackstone C, Hurley JH. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat Struct Mol Biol. 2008;15:1278–1286. doi: 10.1038/nsmb.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamborlini A, Usami Y, Radoshitzky SR, Popova E, Palu G, Gottlinger H. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19140–19145. doi: 10.1073/pnas.0603788103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbyszek Otwinowski WM. Processing of X-ray Diffraction Data Collected in Oscillation Mode In Methods Enzymol. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Zhai Q, Landesman MB, Robinson H, Sundquist WI, Hill CP. Structure of the Bro1 Domain Protein BROX and Functional Analyses of the ALIX Bro1 Domain in HIV-1 Budding. PLoS One. 2011;6:e27466. doi: 10.1371/journal.pone.0027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Pan S, Sun L, Corvera J, Lee YC, Lin SH, Kuang J. The CHMP4b- and Src-docking sites in the Bro1 domain are autoinhibited in the native state of Alix. Biochem J. 2009;418:277–284. doi: 10.1042/BJ20081388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.