Abstract
The ubiquitous Ser/Thr Protein Phosphatase 1 (PP1) regulates diverse, essential cellular processes such as cell cycle progression, protein synthesis, muscle contraction, carbohydrate metabolism, transcription and neuronal signaling. However, the free catalytic subunit of PP1, while an effective enzyme, lacks substrate specificity. Instead, it depends on a diverse set of regulatory proteins (≥200) to confer specificity towards distinct substrates. Here, we discuss recent advances in structural studies of PP1 holoenzyme complexes and summarize the new insights these studies have provided into the molecular basis of PP1 regulation and specificity.
Keywords: protein phosphatase 1, enzyme regulation, enzyme specificity, structural biology
Introduction
Phosphoproteomic studies have suggested that more than ~70% of all eukaryotic cellular proteins are regulated by phosphorylation [1]. The phosphorylation state itself is meticulously controlled by the intricate action of kinases and phosphatases on tyrosine or serine/threonine residues. However, while more than 420 genes in the human genome encode for serine/threonine kinases, which account for 98.2% of all phosphorylation events, less than 40 genes encode for serine/threonine phosphatases (PSPs). PSP themselves have multiple sub-families, with the largest family being the PhosphoProtein Phosphatases (PPP). The PPP family consists of PP1, PP2A, PP3 (PP2B/Calcineurin), PP4, PP5, PP6 and PP7, with PP1, PP2A and PP3 being the best studied enzymes (Figure 1) [2]. In fact, PP1 and PP2A are two of the most abundant enzymes in many cell types. Lastly, also dual-specificity phosphatases (DUSPs) can dephosphorylate p-Ser and p-Thr residues as well as p-Tyr [3].
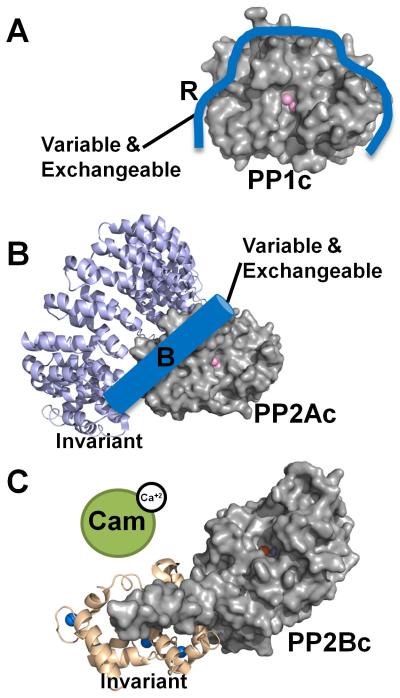
Figure 1. The modular PPP system.
PP1, PP2A and PP2B (Calcineurin, PP3) are metallo-enzymes with a highly similar and conserved catalytic domain structure. However, regulation is achieved via different modes. (A) PP1 is a single catalytic domain (PP1c, gray surface, active site metal ions are depicted as pink spheres; from PDB 3EGG), which can bind to ≥200 biochemically confirmed regulators (blue; indicated by ‘R’). Using one or more of these regulators PP1 is converted into hundreds of highly specific holoenzymes. (B) PP2A (PP2Ac, gray surface, active site metal ions are depicted as pink spheres; from PDB 2NYM) is regulated in a similar manner as PP1, as it binds to a variety of variable and exchangeable B-subunits (blue cylinder). It always binds to an invariant A subunit (magenta ribbon, from PDB 2NYM). The total number of achievable PP2A holoenzymes is ~75. (C) PP2B also consists of a catalytic A and an invariant structural B subunit (PP2Bc, gray surface, active site metal ions are depicted as orange and dark red spheres; B-subunit wheat ribbons, calcium ions are depicted as blue spheres; from PDB 2P6B). However, in order to become activated (release of the auto-inhibitory helix from the active site), calcium activated Calmodulin (green) must bind to PP2B.
Protein Phosphatase 1 (PP1, ~38.5 kDa; Figure 2) is the a highly characterized serine/threonine phosphatase [4]. Mammalian genomes contain three different genes that encode four distinct catalytic subunits of PP1: PP1α, PP1β/δ and the splice variants PP1γ1 and PP1γ2. The sequence identity between corresponding isoforms is between 85% (PP1γ2 and PP1β/δ) and 93% (PP1γ1 and PP1γ2), with the majority of the amino acid differences found in the N- and C-terminal extremities (Figure 2A). PP1α, PP1β/δ and PP1γ1 are ubiquitous, while PP1γ2 is restricted to the testis.
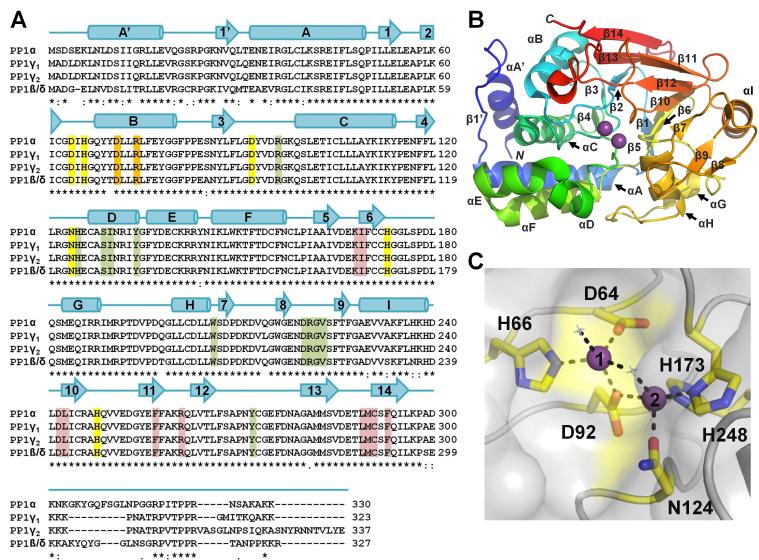
Figure 2. PP1 sequence and structure.
(A) Sequence alignment of PP1 α, β, γ1 and γ2 [all human]. Identical (*), conservative (:) and similar (.) residues are indicated. Sequence identity between corresponding isoforms is 93% (γ1/γ2), 91% (α/γ1), 89% (α/β), 88% (α/γ2), 87% (γ1/β), and 85% (γ2/β). Secondary structure elements of PP1α are indicated above the sequences with α-helices represented as cylinders and β-strands indicated with arrows. Metal coordination residues are highlighted in yellow, residues critical for coordinating molecular toxins are highlight in green (highlighted residues are within 5 Å of the bound toxin in each of the eight PP1:molecular toxin structures determined to date, see Table 1), residues that form the RVxF binding pocket are highlight in red and residues that play a central role in binding targeting proteins/substrates that bind in the PP1 C-terminal groove are highlighted in orange. (B) PP1α (3E7A) illustrated as a cartoon and colored from blue to red from the N- to the C-terminus, respectively. Bound metal ions are shown as spheres (magenta). PP1 secondary structural elements are labeled. (C) PP1α (3E7A) in a surface representation with the residues that coordinate the metals shown as sticks and colored yellow. Metals 1 and 2 are represented as purple spheres and numbered. Metal binding residues are 100% conserved in PP1, PP2A and PP2B.
Compared to protein kinases, the catalytic domains of PSPs appear to have little intrinsic specificity. For example, in contrast to protein kinases, which can select substrates based on local consensus amino acid sequences near a target phosphorylation site, PSPs exhibit only limited preferences. In the case of PP1, the apo-enzyme (apo-PP1 is the catalytic protein including its active site metals) is tightly regulated by its interaction with >200 known targeting proteins [4], proteins that both localize PP1 to distinct regions of the cell and modulate its substrate specificity (Figure 1A). Furthermore, PP1 specific inhibitory proteins, such as inhibitor-2 and DARPP-32, bind and block the PP1 catalytic active site, which contains two metal ions (Mn2+, when heterologously expressed in bacteria [5, 6]) that are required for PP1 activity (Figure 2B,C). The catalytic site of PP1 is at the intersection of three potential substrate binding regions: the hydrophobic, acidic and C-terminal grooves (Figure 3A). Most PP1 regulatory proteins and some PP1 substrates interact with PP1 via a primary PP1-binding motif, the RVxF motif, which generally conforms to the consensus sequence [K/R][K/R][V/I][x][F/W], where x is any residue other than Phe, Ile, Met, Tyr, Asp, or Pro [7-9]. While the RVxF interaction is necessary for regulatory protein binding, it does not influence PP1 enzymatic activity, as it is 20 Å away from the active site (Figure 3B,C). Additional, recently identified docking sites [10-13], such as the SILK and MyPhoNE motifs [9], also play essential roles in the regulation of PP1 activity and substrate specificity. However, a detailed understanding and, more importantly, the ability to predict from sequence alone how the >200 PP1 targeting proteins bind and direct PP1 specificity is still entirely missing. One reason for this lack of understanding of how PP1 achieves substrate specificity is the limited number of PP1 structures and the holoenzyme complexes it forms. In this review, we focus on recent advances in structural studies of PP1 holoenzyme complexes and summarize new insights from these studies.
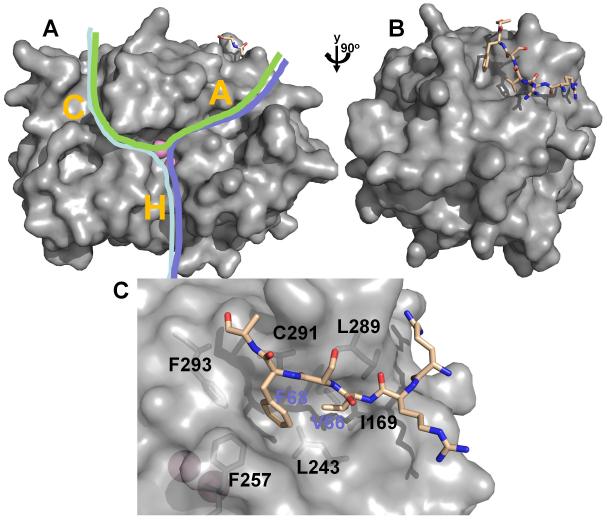
Figure 3. PP1 regulation.
(A) PP1 (from the Gm:PP1 structure [38]) is shown as surface representation. Active site metal ions are depicted as pink spheres and the Gm RVxF peptide is shown in stick representation (beige). The C-terminal (C), hydrophobic (H) and acidic (A) substrate binding grooves are indicated with a letter. Different PP1 substrates (indicated by green, pink and blue lines) can bind via a single groove, or, as shown here, multiple grooves, illustrating that substrates likely bind PP1 via a variety of mechanisms. (B) PP1 is rotated by 90° in order to highlight the RVxF peptide binding site. (C) Detailed view of the RVxF binding site, illustrating the deep hydrophobic pockets which bind the ‘V’ and ‘F’ residues (V66 and F68 for the Gm peptide) of the RVxF motif.
PP1: expression and purification
For many years, the key challenge for PP1 structural biology was access to pure, homogenous, active PP1 in amounts necessary for structural studies. Protocols for the extraction of apo-PP1, apo-PP2A as well as various dimeric and heterotrimeric forms of PP2A, and a dimeric form of PP2B from natural source were established decades ago [14, 15]. These protocols were of tremendous importance in the field as they enabled many key advances in our understanding of these enzymes to be made. In fact, PP1 purified from natural source (typically rabbit muscle) is still the most active enzyme. However, purification from natural source has inherent limitations. For example, these protocols typically result in a mixture of isoforms and studies on PSP mutants are impossible.
To overcome these limitations, protocols for the heterologous expression of PP1 [16-19], PP2A [20, 21] and PP2B [22, 23] in bacteria or insect cells have been established. Bacterial expression of PP2B is readily achieved by using a bicistronic vector for the co-expression of the PP2B A (catalytic) and B (structural) subunits. In contrast, expression of PP1 or PP2A in either bacteria or insect cells is much more difficult. PP1 is readily expressed, but highly insoluble when produced in E. coli. While recent advances in PP2A production have led to a significantly improved protocol and, in turn, yield for the PP2A expressed in insect cells, the yield of PP1 from this same system is, at best, about 0.1 mg/L of Sf9 culture (Nairn laboratory). Moreover, E. coli produced PP1 differs from native PP1 in that it typically, exhibits phosphotyrosine phosphatase activity and is insensitive to regulation by several targeting subunits [17, 19, 24]. Thus, most structural biology efforts has since focused on improving the yield and activity of PP1 produced from E. coli.
One strategy to overcome insolubility in E. coli, is to fuse the insoluble protein (PP1) to a protein solubility tag, e.g. maltose-binding protein (MBP). While this does result in some soluble MBP-PP1 fusion protein, the cleavage of the MBP tag by a protease, e.g. TEV, was very slow and incomplete (Peti laboratory, unpublished). A second strategy is refolding. Multiple refolding protocols were tested, but did not result in significant yields of folded, active PP1 (Peti laboratory, unpublished). Thus, we developed and optimized a novel PP1 bacterial expression protocol [6], which has resulted in: (1) a 10-fold increase in PP1 yield, (2) a more than 100-fold increase in PP1 activity, (3) a possibility to readily form holoenzymes with high affinity and, critically, (4) the highest resolution structures of both toxin:PP1 and regulatory protein:PP1 holoenzyme complexes determined to date.
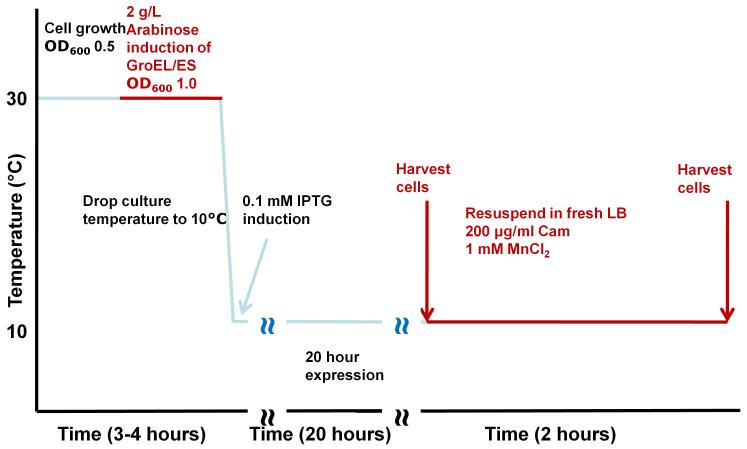
Our protocol, the ‘chaperone-assisted PP1 expression protocol’, relies on the following key features (Figure 4). First, PP1 is subcloned into a pET-28a (Novagen) derived vector RP1B, which encodes a N-terminal Thio6His6 expression/purification tag (MGSDKIHHHHHH) and a TEV (tobacco etch virus) cleavage site (ENLYFQGH) [25]. This allows the purification tag to be cleaved prior to biochemical and structural studies. Second, expression is carried out at a very low temperature (10°C), which increases the yield of soluble protein. Third, PP1 is expressed in the presence of metals (usually MnCl2; however, other metals can be readily substituted as described below), which bind the PP1 active site and stabilize its structure. Fourth, PP1 is co-expressed with chaperones, typically GroEL/ES, as this chaperone system results in the highest yield of soluble, active protein. Fifth, and most importantly, PP1 undergoes ‘in vivo refolding’. Specifically, after expression, the bacterial cells are pelleted, resuspended in fresh medium and the expressed PP1, still in the presence of GroEL/ES, is refolded in vivo during a period of inhibited ribosome activity. It is the co-expression of PP1 with chaperones and the in vivo refolding step that are the most significant advances of this protocol.
Figure 4. Recombinant PP1 expression.
Detailed scheme (timeline) illustrating the steps of our recently developed PP1 expression protocol.
Due to the significant increase in yield, multiple biochemical, biophysical and structural studies on PP1 and its holoenzyme complexes are now possible. For example, the chaperone-assisted PP1 production protocol is highly amenable to modification, e.g. PP1 can be expressed in the presence of a variety of different metals, including Zn2+, Cu2+, Ni2+, Fe2+ and Mn2+. Interestingly, Mn2+-PP1 has always exhibited the highest activity and stability (Peti laboratory, unpublished). In addition, ICP-AES spectroscopy experiments confirmed that two Mn2+ ions bind the PP1 active site in solution [5] (two Mn2+ ions are also present in our toxin:PP1 and regulatory protein:PP1 holoenzyme structures [6, 26]). Interestingly, two Mn2+ ions were also identified in the PP2A active site (as tested by ICP-MS; PP2A expressed in insect cells) [27].
Critically, the PP1 produced using this protocol is much more similar to native PP1. While our recombinant PP1 still possess activity against p-Nitrophenyl Phosphate (p-NPP) [5, 19, 26], the specific activity of chaperone-assisted PP1 is only 8-times lower than native PP1, compared to 1000-fold lower for PP1 produced using other bacterial expression protocols (tested against the most common PP1 substrate, phosphorylase a, a specific substrate of the Gm:PP1 holoenzyme; M. Bollen, KU Leuven, personal communication). In addition, in contrast to PP1 produced using other protocols, the chaperone-assisted PP1 is sensitive to regulation by multiple targeting subunits (i.e., I-2 and spinophilin). Thus, for the first time, a bacterially expressed PP1 closely mimics the behavior and activity of native PP1.
PP1 holoenzyme formation
PP1 is tightly regulated by its interaction with more than 200 targeting and inhibitor proteins, yet very little is understood about how these proteins direct PP1 activity at a molecular level because very few of these holoenzyme structures have been determined to atomic resolution (Figure 5) [26, 28, 29]. One strategy that has been successful is the co-expression of PP1 and a regulatory protein from two separate plasmids in E. coli, which was used for the production and subsequent structure determination of both the MYPT1:PP1 [28] and the Inhibitor-2:PP1 [29] holoenzyme complexes. While successful for these two complexes complex, co-expression has not been optimal for many other complexes tested by both ourselves and others. Thus, we developed a novel route of holoenzyme formation and purification which relies on the individual expression of PP1, via the chaperone-assisted protocol, and its regulatory proteins. This holoenzyme formation protocol is applicable for many PP1 holoenzymes and is also readily amenable to variations in both PP1 (i.e., isoforms; constructs of different lengths, a critical factor for crystal formation) and its regulatory proteins.
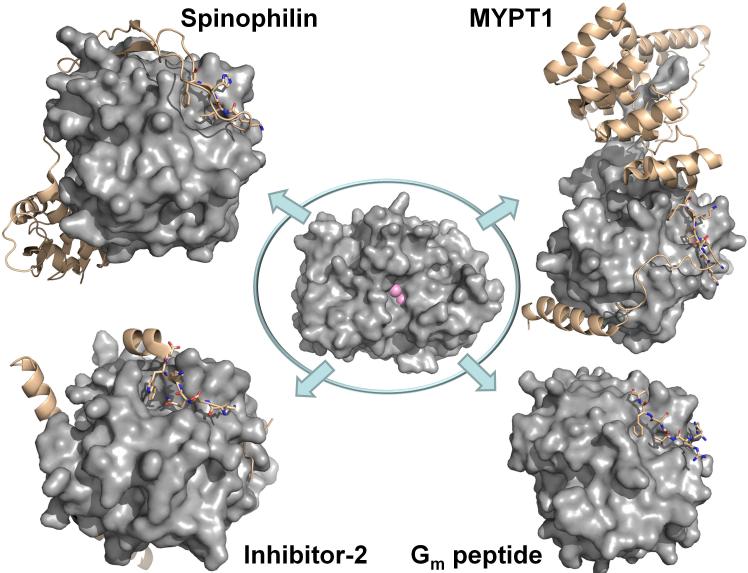
Figure 5. PP1 holoenzyme structures.
Center: PP1 is a single catalytic domain (PP1c, gray surface, active site metal ions are depicted as pink spheres; from PDB 3EGG). Upper left. The spinophilin:PP1 holoenzyme (spinophilin is shown as beige ribbons; RVxF residues are highlighted as sticks. Lower left. The I-2:PP1 holoenzyme (I-2 is shown as beige ribbons; RVxF residues are highlighted as sticks). Upper right. The MYPT1:PP1 holoenzyme (MYPT1 is shown as beige ribbons; RVxF residues are highlighted as sticks). Lower right. The Gm:PP1 RVxF peptide structure (the RVxF peptide is shown as beige sticks).
Apo-PP1, even produced using the chaperone-assisted protocol, is marginally stable. It must be maintained at concentrations ≤5 μM, and, even at these concentrations, is only stable for short periods of time before it begins to form soluble aggregates. These aggregates, which are not detected in activity assays, can be readily identified using size exclusion chromatography (SEC, Superdex 200 26/60). Thus, one of the central steps in our protocol is an on-column formation of the toxin:PP1 or regulatory protein:PP1 complex, followed by up to 6 additional purification steps [26]. Extremely homogeneous, monodisperse PP1 holoenzyme samples are produced using this protocol, which has already allowed for the determination of the highest resolution structures of PP1 complexes to date, including PP1 bound to a small molecular toxin (nodularin-R:PP1, PDB 3E7A, 1.63 Å) [6] and the PP1 bound to a neuronal targeting protein (spinophilin:PP1 holoenzyme, PDB 3EGG, 1.85 Å) [26] (Figure 5). Most importantly, this protocol has also been used to produce multiple PP1 holoenzymes for biochemical and structural studies, including I-2:PP1 [11], spinophilin:I-2:PP1 (a triple complex) [5], spinophilin:nodularin-R:PP1 (a triple complex) [26], neurabin:PP1 [26], tautomycin:PP1 [6], as well as many other currently unpublished PP1 holoenzymes.
The early days of PP1 structural biology
No structure of native PP1 (i.e., purified from natural source) has been determined. Rather, the 15 PP1 structures that have been determined to date (Table 1) have been expressed in E. coli. For these structures, the expression media was typically supplemented with MnCl2 and, in many but not all cases, MnCl2 was also added to all purification buffers. In 1995, the Kuriyan laboratory determined the 3-dimensional structure of PP1α in complex with the natural product toxin microcystin (although PP1α 1-330 was crystallized, only residues 7-300 were observed in the electron density maps, demonstrating that the N- and C-termini of PP1 are disordered) [30]. This structure provided the first detailed insights into the PP1 fold, the local conformation of the PP1α active site and, in turn, its catalytic mechanism (the PP1 fold is shown in Figure 2B). In addition, the electron density maps demonstrated that PP1 contains two metals, positioned about 3.3 Å apart in a shallow pocket at the center of the molecule (Figure 2C). The anomalous signal at these sites ruled out Mg, Ni, Cu or Zn, but was unable to distinguish between Mn, Fe or Co. The six residues that coordinate the metals (Asp64, His66, Asp92, Asn124, His173 and His248 in PP1α, Figure 2A) are strictly conserved in the eukaryote PPP family. This structure also revealed that the active site is located at the intersection of three proposed substrate binding grooves (the acidic, the hydrophobic and the C-terminal binding grooves; Figure 3A).
Table 1. Overview of all currently determined PP1 structures.
| PP1 Complex1 | PP1 isoform | Res (Å) | Metals | PDB | Year | Ref |
|---|---|---|---|---|---|---|
| Ligand bound | ||||||
| tungstate:PP12 | gamma | 2.50 | Mn/Fe | n/a | 1995 | [32] |
| Toxin bound | ||||||
| microcystin:PP13 | alpha | 2.10 | Mn/Mn | 1FJM | 1995 | [30] |
| okadaic acid:PP1 | gamma | 1.90 | Mn/Mn | 1JK7 | 2001 | [34] |
| caluculin A:PP1 | gamma | 2.00 | Mn/Mn | 1IT6 | 2002 | [33] |
| motuporin:PP1 | gamma | 2.10 | Mn/Mn | 2BCD | 2006 | [64] |
| dihydromicrocystin-LA:PP1 | gamma | 2.30 | Mn/Mn | 2BDX | 2006 | [64] |
| *nodularin-R:PP1 | alpha | 1.63 | Mn/Mn | 3E7A | 2009 | [6] |
| *tautomycin:PP1 | alpha | 1.70 | Mn/Mn | 3E7B | 2009 | [6] |
| RVxF peptide bound | ||||||
| Gm(63-75):PP1 | gamma | 3.00 | Mn/Mn | n/a* | 1997 | [38] |
| Rb(870-882):PP1 | alpha | 3.20 | Mn/Mn | 3N5U | 2010 | [65] |
| Regulatory protein bound | ||||||
| MYPT1:PP1 | delta | 2.70 | Mn/Mn | 1S70 | 2004 | [28] |
| Inhibitor-2:PP1 | gamma | 2.50 | Mn | 2O8G | 2006 | [29] |
| *Inhibitor-2:PP14 | alpha | NMR/SAXS | Mn/Mn | 2010 | [11] | |
| *Spinophilin:PP14 | alpha | 1.85 | Mn/Mn | 3EGG | 2010 | [26] |
| *Spinophilin:PP14 | alpha | SAXS | Mn/Mn | 2011 | [66] | |
| *Neurabin:PP1 | alpha | 2.20 | Mn/Mn | 3HVQ | 2010 | [26] |
| Triple Complex | ||||||
| *PP1:Spinophilin:Nodularin-R | alpha | 2.00 | Mn/Mn | 3EGH | 2010 | [26] |
| *PP1:Spinophilin:Inhibitor-24 | Alpha | NMR/SAXS | Mn/Mn | 2011 | [5] |
PP1 protein for all PP1 crystal structures determined to date produced in E. coli. In all cases, expression medium was supplemented with 1 mM MnCl2. In most cases, purification and final protein buffers were also supplemented with MnCl2 (0.2 - 1.0 mM).
Proton induced X-ray emission (PIXE) used to identify 1 Mn and 0.5 Fe in the active site.
Metals listed as Mn2+ in 1FJM; reported that the presence of a strong anomalous signal rules out Mg, Ni, Cu or Zn, but could not discriminate between Mn, Fe or Co.
ICP-AES spectroscopy experiments have confirmed that PP1 holoenzymes prepared using the protocol described in this review, such as PP1:Spinophilin, contain 2 Mn ions at the active site (MnCl2 is used only during expression, not during purification).
Structures of PP1 complexes determined by the authors.
The microcystin:PP1 structure also provided the first direct comparison between different PPP catalytic domains, as the 3-dimensional structure of PP2B was determined at the same time [23, 31]. It was discovered that the structures of PPP catalytic domains were nearly invariant (which was subsequently confirmed with the first structures of PP2A [21, 27]). Shortly thereafter, the Barford laboratory reported the structure of PP1γ in complex with tungstate [32]. The structures of PP1α and PP1γ were discovered to be nearly identical, demonstrating that ligand/inhibitor binding (tungstate/microcystin-LR, respectively) did not lead to significant changes in the conformation of PP1. In this latter study, the authors used proton-induced X-ray emission spectroscopy to show that their PP1 had one Mn2+ and 0.5 Fe (either Fe2+ or Fe3+). In this case, PP1 was expressed in E. coli in the presence of MnCl2 and purified using MnCl2 supplemented buffers; no Fe was added to the medium [18].
Inhibition by small molecular toxins
Many PPPs can be readily and potently inhibited by a large number of active site-directed small molecular toxins, with the most prominent inhibitors being microcystin [30], nodularin [6], calyculin A [33], tautomycin [6] and okadaic acid [34]. Indeed, for many years, a microcystin affinity column was an essential tool for PP1 biochemistry and the identification of novel PP1 regulatory proteins [35]. The toxins bind and occlude the PP1 active site and also extend into the PP1 hydrophobic groove. An overlay of the PP1:toxin structures, in addition to the PP1 residues that mediate toxin binding, is illustrated in Figures 6A,6B.
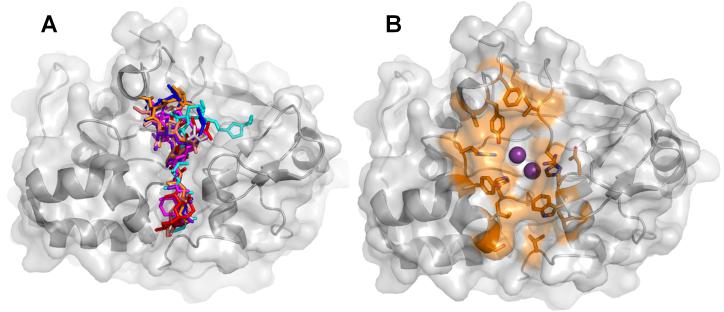
Figure 6. PP1:toxin structures.
(A) Overlay of the eight PP1:toxin complexes; toxins are illustrated as sticks (3E7A, grey; 3E7B, red; 1IT6, cyan; 1JK7, magenta; 2BCD, salmon; 3EGH, purple, 1FMJ, blue; 2BDX, orange). (B) The PP1 residues that mediate toxin binding. Highlighted residues (shown as sticks and colored orange) are within 5 Å of the bound toxin in each of the PP1:molecular toxin structures determined to date.
Because the active sites of PPPs, especially PP1 and PP2A, are so similar, there are often only very small differences in the ability of these toxins to inhibit PPP activity. Okadaic acid has the largest difference in IC50 between PP2A and PP1, with the IC50 of PP1>PP2A. Thus okadaic acid is often used as a reagent to selectively inhibit PP2A in cell biological studies. The molecular basis of this difference is subtle. The crystal structures of the PP1 [34] and PP2A [27] bound to okadaic acid (PDBs 1JK7 and 2IE4) showed that the toxin:PP1/PP2A interactions are largely conserved between the two complexes. The most significant difference was the position of α7-α8 loop, which, in PP2A, was closed about okadaic acid while in PP1 it was open, and thus ~4-5 Å further away. Thus, okadaic acid appears to binds slightly more deeply into the hydrophobic groove in PP2A and this likely explains why it inhibits PP2A activity more effectively than that of PP1.
While most toxins inhibit PP2A and PP1 with nearly equivalent IC50 values, all toxins have a significantly decreased (1000-10000 fold) IC50’s for PP2B/Calcineurin, despite all three PPPs having nearly identical residues and structures at their active sites. One potential explanation for this difference in inhibition susceptibility is the nature of the metals bound in the active site (Mn2+ for PP1 expressed in E. coli, Mn2+ for PP2A expressed in insect cells and Fe/Zn for calcineurin expressed in E. coli; the identity of the endogenous metals in PP1 is still unknown). A second explanation is the differences in sequence between the proteins in regions outside the metal binding residues. To this end, kinetic and crystallographic studies revealed a role for the Cys273 in toxin binding by PP1. Specifically, mutation of Cys273 to Ala, Ser or Leu (the corresponding residue in PP2B) results in an 11-20-fold increase in the IC50 values for microcystin-LR toxin inhibition of PP1 [36, 37].
First insights to PP1 targeting
In 1997, the Barford laboratory reported the structure of PP1 in complex with the RVxF peptide from Gm (Gm residues 63-75; only residues 64-69 were observed in the electron density map) [38] (Figures 3B,C and 5). More than 90% of all PP1 regulators contain an identifiable RVxF sequence [4] and thus, this structure provided the first atomic resolution insights into how multiple PP1 regulatory subunits bind and direct PP1 specificity. The Gmpeptide:PP1 peptide structure revealed that the RVxF binding site is ~20 Å away from the PP1 active site. It is characterized by two deep hydrophobic pockets that are comprised of residues which are either invariant or highly conserved among all PP1 isoforms from yeast to humans. Critically, these residues are not conserved in PP2A and PP2B, which explains why PP1 regulatory proteins with RVxF motifs do not bind other PPPs [39]. The residues buried in the PP1 hydrophobic pockets, which results in a loss of nearly 1000 Å2 of solvent accessible surface area upon complex formation, are ‘V’ and ‘F’ of the RVxF motif. The importance of these residues was confirmed by mutagenesis studies which showed that the mutation of the ‘F’ residue abolished the ability of Gm peptides to disrupt the PP1-GL complex (mutation of the corresponding ‘F’ residue in other PP1 regulatory proteins is now regularly used to disrupt regulatory protein binding to PP1) [38, 40]. Unexpectedly, in spite of the extensive interactions between PP1 and the Gm RVxF peptide, the structure of PP1 was largely unchanged. Thus, PP1 conformation appeared to be largely invariant independent of the molecular entity (ligand/toxin/peptide) to which it was bound.
Collectively, these studies showed that the RVxF motif, which is present in >90% (>180) of all PP1 regulators [4], functions primarily as an anchoring motif, as RVxF peptide binding does not influence the overall structure nor alter the substrate specificity of PP1.
PP1 holoenzyme structural biology
Despite the importance of PP1 in multiple biological processes, the first PP1 holoenzyme structure (MYPT1:PP1) was reported only seven years ago, in 2004, from the Dominguez laboratory (PDB 1S70) [28] (Figure 5). This was clearly a milestone for the field. This structure showed that MYPT1 binds to PP1 at multiple anchor points, only one of which is the RVxF motif, and was the first indication that multiple grooves on PP1 function to bind and interact with PP1 regulatory proteins. The PP1 interaction domain of MYPT1, like most PP1 regulatory PP1 binding domains, is largely unstructured in solution [41]. In the PP1 holoenzyme, this domain becomes ordered and wraps around the outer edge of PP1, positioning the C-terminus of the MYPT1 ankyrin repeats to optimally bind the PP1 C-terminus (residues 301-309; this is the only structure in which PP1 residues C-terminal to ~300 have ever been observed) [28, 41]. None of the proposed PP1 substrate binding grooves (Figure 3A) are apparently occupied by MYPT1; i.e., they are still largely accessible for substrate binding. Thus, due to the fact that binding of MYPT1 to PP1 leads to a reshaping of the catalytic cleft in PP1 the authors hypothesized that this structural change, as well as a change in the overall electrostatic potential, leads to a change in substrate specificity. However, an earlier structure-function study indicated that a distinct sub-region of MYPT1 was responsible for the positive effects of MYPT1 on myosin phosphatase activity (residues 1-38), while a region after the RVXF motif and extending into the ankyrin repeats (residues 39-150) was responsible for inhibition of phosphorylase dephosphorylation [42].
More recently, the Inhibitor-2:PP1 (I-2) structure was determined [11, 29] (Figure 5). We have shown that I-2 is largely unstructured in solution, except for a single strongly populated α-helix (residues 127-156), and that very limited regions of I-2 become ordered upon PP1 binding [11, 43] (Figure 7). This was also shown by the I-2:PP1 crystal structure, in which only 59 of the 206 residues of I-2 were sufficiently ordered that they could be modeled into the electron density [29]. I-2 interacts at three sites on PP1. First, I-2 residues 44-56 bind the PP1 RVxF motif interaction site via an interaction nearly identical to that observed in the Gm:PP1 and MYPT1:PP1 structures. Second, I-2 residues 130-169 forms a long ordered α-helix, as predicted by NMR studies, which lies directly over the PP1 active site and blocks its access to other substrates. This helix is anchored in the acidic and hydrophobic grooves of PP1. Third, I-2 residues 12-17 interact at a third site, now known as the SILK interaction site [44]. Although the RVxF motif is the most commonly identified PP1 binding motif, recent studies have revealed that many PP1 regulatory proteins contain additional motifs and that each contributes to PP1 binding [4]. The SILK interaction site is located on the opposite side of the PP1 active site, below the RVxF interaction site (Figure 5). The importance of this site in the formation of PP1 triple complexes [5] will be described in more detail below.
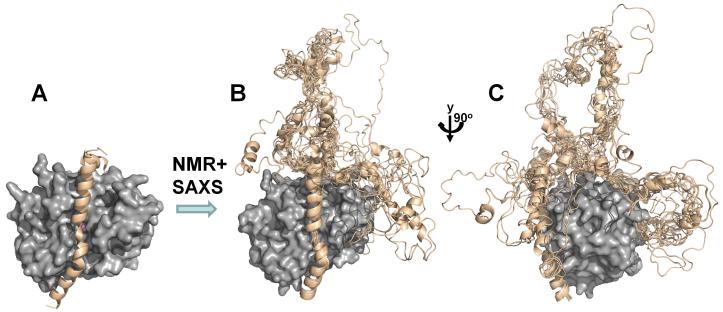
Figure 7. PP1:Inhibitor-2 structure.
(A) I-2:PP1 holoenzyme X-ray structure (PP1c, gray surface, I-2 beige ribbons, PDB 2O8G); (B) the use of NMR and SAXS allowed for the determination of the I-2:PP1 ensemble structure; (C) PP1 is rotated by 90° in order to highlight the I-2 RVxF and SILK peptide binding sites.
PP1 conformation is invariant
To date, 15 PP1 structures have been reported, including tungstate:PP1 (1), toxin:PP1 (7), peptide:PP1 (2), regulatory protein:PP1 holoenzyme (4) and toxin:regulatory protein:PP1 holoenzyme (1) complexes. Remarkably, and as predicted by the Gmpeptide:PP1 structure, the conformation of PP1 is essentially unchanged between these complexes, i.e., its conformation is independent of the interaction partner. This is shown in Figure 8, where the backbones of the PP1 catalytic domains from 15 complexes are superimposed. The average root mean square deviation (RMSD) is ~0.5 Å, with a maximum RSMD of 0.9 Å (between the microcystin:PP1 and okadaic acid:PP1 complexes; microcystin is covalently attached to PP1 at Cys273, leading to a small conformational change in the β12-β13 loop). This shows that the multitude of binding pockets for various toxins and targeting proteins are largely preformed in PP1. Moreover, this also suggests that any changes associated with ligand/toxin/regulatory protein binding are subtle, often involving small changes in loops and even changes in rotomer conformations of the sidechains. The most significant conformational changes between the PP1 structures are found in loops or single secondary structural elements, including the β12-β13 loop, the αA’-β2 loop, the β11-β12 loop, helix αI and β-strand β14 (the C-terminal strand of the structured catalytic domain).
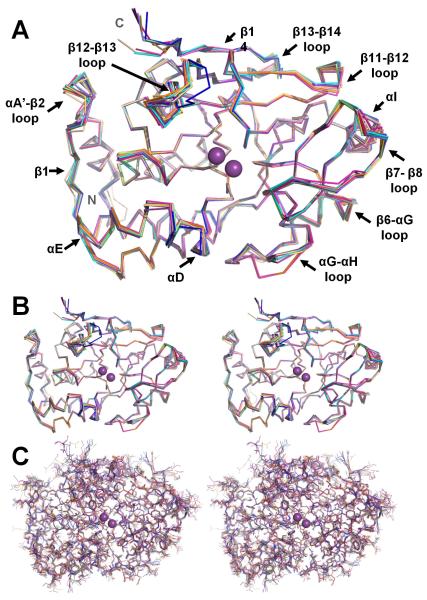
Figure 8. PP1 conformation is invariant.
(A) Overlay of the PP1 structures, illustrated as backbone ribbons (1FMJ, dark blue; 1IT6, cyan; 1JK7, light magenta; 1S70, beige; 2BCD, salmon; 2BDX, orange; 2O8G, slate; 3E7A, grey; 3E7B, lime green; 2EGG teal; 3HVQ, pink; 3N5U, yellow-orange; 3EGH, purple-blue; Gm, violet-purple). The bound metals are represented as spheres and colored magenta. The regions of PP1 that undergo the changes in conformation between the structures are labeled. (B) Stereo view of the overlay in A. (C) Same stereo view as in B except all atoms are shown.
The spinophilin:PP1 holoenzyme; structure defines function
Spinophilin [45] is a 817-residue neuronal regulatory protein that targets PP1 to neuronal synapses, where it controls essential neuronal processes such as AMPA receptor activity and cytoskeletal reorganization and thus plays a central role in learning and memory [40, 45, 46]. Furthermore, spinophilin has been shown to have an important role in the actions of drugs of abuse and changes in PP1 function have been associated with Parkinson’s disease [47].
As observed for most PP1 regulators, the spinophilin PP1-binding domain (residues 424-494) is an intrinsically disordered polypeptide/protein (IDP) [26]. IDPs lack bulky hydrophobic residues (Val, Leu, Ile, Met, Phe, Trp and Tyr) that typically form the hydrophobic core of folded proteins [48, 49] and, in turn, lack a single, stable fold. Instead, they are enriched in charged and often short side chain amino acids (Gln, Ser, Pro, Glu, Lys and Gly). The significantly increased flexibility of IDPs allows them to form novel unexpected complexes that are essential for a multitude of vital biological processes [50, 51].
Although the spinophilin PP1-binding domain is an IDP, it is still able to interact tightly and specifically with PP1 (Kd ~8 nM) [26]. In 2010, we determined the 1.85 Å crystal structure of the spinophilin:PP1α holoenzyme (Figure 5). As discussed above, the overall structure of PP1 is largely identical to that of all previously reported PP1 structures [30, 52]. In contrast, the intrinsically unstructured PP1-binding domain of spinophilin becomes restricted to a single conformation in the holoenzyme complex. We also determined the 3-dimensional structure of the neurabin:PP1 holoenzyme, with neurabin being the neuron specific isoform of spinophilin; they have ~80% sequence identity and no major differences were observed between the two structures.
Although cell biological and biochemical studies had shown that residues 417-494, which includes the RVxF motif (spinophilin residues RKIHF447-451), were critical for PP1 binding, it was not known how these additional residues interacted with PP1 outside of the RVxF interaction site. Our structure showed that spinophilin residues 476-492 are alpha helical, and that residues 430-434 and 456-460 form two β-strands (spinophilin β-strands 1/2), which extend the PP1 β-sheet 1 (PP1 β-strands 14/13/2/3/4) into an 7-strand β-sheet. In addition, spinophilin residues 462-469 bind directly into the PP1 C-terminal substrate binding groove. The conformational changes in PP1 that accompany occupation of this groove are subtle. First, there is a small shift of the end of PP1 β-strand β14, which slightly narrows the top of the C-terminal groove so the β14 backbone atoms can hydrogen bond with spinophilin. Second, the side chain of PP1 residue Arg74, which in every other PP1 structure lies across the C-terminal groove and forms a salt bridge with PP1 residue Asp71, rotates up out of the groove, resulting in a ~5 Å displacement of the guanidinium group. This movement opens up access to the C-terminal groove, enabling spinophilin to bind (Figure 9). Here, spinophilin residue Arg469 is the driving force for this interaction, as it forms backbone and sidechain hydrogen-bond interactions (intra-molecularly with Tyr462 and Asn464 and inter-molecularly with Asp71) with PP1 and, in turn, primes spinophilin residues Tyr462 and Tyr467 to form extensive hydrophobic interactions with PP1. A detailed mutagenesis study of the spinophilin:PP1 interaction showed that residues in not only the spinophilin RVxF motif, but also in the C-terminal groove binding are both critical for PP1 binding [26].
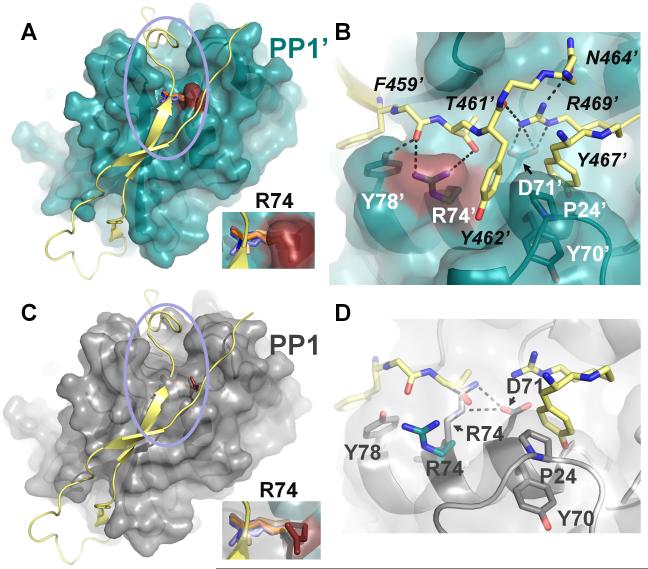
Figure 9. The PP1 C-terminal groove accessibility and binding is regulated by R74 (PP1).
(A) PP1:spinophilin complex (3EGG). Spinophilin (yellow) is shown in cartoon representation. PP1 (teal) is illustrated as a transparent surface, with R74 colored in red. Residue R74 from all PP1 structures determined to date are shown in stick representation (with the exception of the PP1:spinophilin:nodularin and PP1:neurabin complexes, which are identical to the PP1:spinophilin complex and not included for clarity). PP1 structures were superimposed as in Figure 1. In all cases, with the exception of the PP1:spinophilin complex, R74 lies across the C-terminal groove. Inset, close-up of R74. (B) Close-up of the area enclosed in the pink circle in (A). Colors identical to those in (A) with residues numbers for PP1 (white) and spinophilin (black) indicated (spinophilin residues 465 and 466 were removed for clarity, as were some carbonyls). Polar interactions between PP1 side chains and spinophilin are indicated by black dashed lines. Spinophilin residue R469 makes multiple electrostatic contacts with PP1 residue Asp71 and other spinophilin residues. (C) Same as (A) except PP1 (grey) is from the PP1:nodularin complex (3E7A). (D) Same as (B) except PP1 residues (grey) are now from PP1:nodularin complex. The polar interactions between PP1 residues R74 and D71 are illustrated by dashed lines (black). R74 (teal) from the PP1:spinophilin complex is also shown to illustrate the large conformational change of this residue between the two complexes. Finally, a subset of the spinophilin residues in (B) show that the side chain of spinophilin T461 sterically clashes that of PP1:nodularin complex PP1 residue R74, explaining why R74 must change conformation in order for spinophilin to bind PP1.
How does spinophilin direct PP1 specificity?
Spinophilin directs PP1 substrate specificity in a manner independent of its ability to target PP1 to distinct subcellular locations. In order to understand how this is achieved at a molecular level, the ability of the spinophilin:PP1 holoenzyme to dephosphorylate both spinophilin:PP1 holoenzyme specific and non-specific substrates was assayed [26]. Unexpectedly, spinophilin does not bind near nor alter the structure of the PP1 active site (the latter being confirmed by determining the structure of the spinophilin:nodularin-R:PP1 triple complex). This suggested that any change in substrate specificity of the spinophilin:PP1 holoenzyme complex was likely due to direct modification of a PP1 substrate binding surface. A detailed mutagenesis and activity study revealed that spinophilin directs PP1 specificity by a novel mechanism, steric inhibition of substrate binding sites. Because spinophilin occupies the PP1 C-terminal substrate binding groove, substrates, e.g. phosphorylase a, that require this groove for binding are excluded and thus unable to be dephosphorylated.
The importance of this mechanism is illustrated by the following study. Mutation of Asp71, a PP1 residue located in the center of the C-terminal substrate binding pocket which forms a hydrogen bond with spinophilin Arg469, severely impairs this steric inhibition. First, mutation of Asp71 reduced spinophilin binding efficiency by ~60%. Second, it resulted in no change in the de-phosphorylation efficiency of PP1 for a specific spinophilin:PP1 substrate (GluR1). This demonstrates that the catalytic activity of the PP1 mutant is not impaired and that GluR1 does require either spinophilin nor binding to the PP1 C-terminal substrate binding groove for dephosphorylation. Finally, this mutation resulted in a substantial increase in the KM for PP1 dephosphorylation of phosphorylase a [53], demonstrating that, in contrast to GluR1, phosphorylase a requires the C-terminal substrate binding pocket for dephosphorylation. Finally, this is the only PP1 interaction region in which the bound sequences between spinophilin and neurabin have 100% primary sequence identity, further highlighting the importance of this interaction.
Formation of the spinophilin:PP1 holoenzyme
NMR spectroscopy has proven to be an excellent tool for deriving atomic resolution parameters in order to understand the structural preferences of IDP ensemble structures. Thus, we used NMR-derived local and long range probes for secondary and tertiary structure [43] to investigate the unbound ensemble structure(s) of spinophilin. Furthermore, in collaboration with Dr. Julie Forman-Kay (University of Toronto, Ca), we used the ENSEMBLE calculation program [54, 55] to gain further insight into the structure of unbound spinophilin. ENSEMBLE detected two dominant populated clusters for free spinophilin. In both clusters, a ~30% populated helix was detected at the C-terminus, which is the conformation observed in the spinophilin:PP1 holoenzyme structure. In addition, in the lower, 18% populated spinophilin cluster, tertiary interactions were detected that clearly resemble β-strand contacts also observed in the spinophilin:PP1 holoenzyme structure. Taken together, the structures of the unbound spinophilin ensemble reveal that many of the intramolecular structural features observed in PP1-bound spinophilin are pre-formed in unbound spinophilin.
Comparison of PP1 and PP2B interactions
The observation the spinophilin extends the PP1 central β-sheet into a 7-strand β-sheet was unexpected, yet extremely exciting, as spinophilin binds PP1 near the structurally homologous substrate binding surface of PP2B: the PxIxIT:PP2B interaction site (the PxIxIT motif is present in many PP2B substrates and is critical for PP2B binding) [56]. This led us to investigative the structural basis of the differential interaction(s) between PP1/PP2B interacting proteins and their respective phosphatases. Specifically, small structural changes in PP1/PP2B have a significant effect on the surface complimentary structure of the enzymes (Figure 10). First, there is a ‘collapse’ of the RVxF binding site in PP2B and second, the creation of the PxIxIT binding site for PP2B substrates. This provides specific binding sites for specific substrates and regulators of PP1 and PP2B, respectively. The change of these regulatory and substrate binding sites is due to an increase in length of the β11-β12 loop, which is 6 amino acids (residues 258-263) long in PP1, but 12 amino acids (residues 291-302) in PP2B. This increased loop structure enables Arg260 in PP1 to adopt an entirely different sidechain conformation than the PP2B counterpart, Arg292. Furthermore, Asp253 and Thr255 (PP1, β10-β11 loop and β11 sheet) adopt a different conformation than residues Thr288 and Met290, which become part of the PxIxIT binding pocket in PP2B. This is the first molecular insight into the differential mode of specificity regulation between the highly similar (by primary sequence) enzymes PP1 and PP2B.
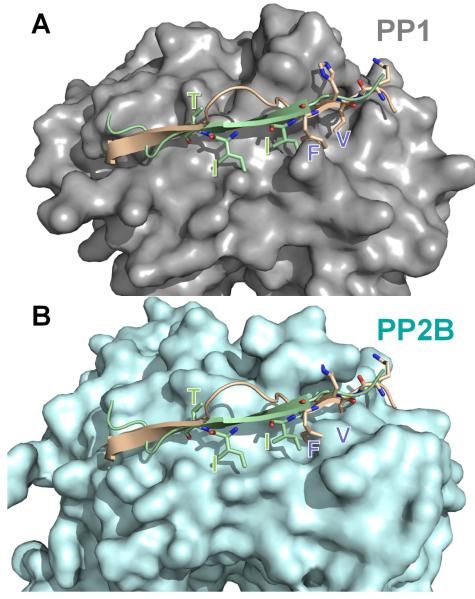
Figure 10. Overlapping, but distinct regulatory and substrate binding sites in PP1/PP2B.
The spinophilin:PP1 (PDB 3EGG) holoenzyme was superimposed onto PP2B:PIVIIT (PDB 2P6B) using PP1 and PP2B. In (A), PP1 (grey) is shown in surface representation with spinophilin (residues 446-461 of spinophilin, which includes the RVxF motif and the β14 binding residues) in beige and the PIVIIT peptide in green. As can be seen, both the PP1 regulator spinophilin, as well as the PP2B substrate mimicking PIVIIT peptide bind to the β14 sheet of PP1/PP2B. (B) Same as in ‘A’ except PP2B (light blue) is now shown in surface representation. The residues that are essential for mediating phosphatase specific interactions are shown in stick representation and labeled in both panels. Comparison of panels ‘A’ and ‘B’ illustrates that the deep hydrophobic pockets that are present in PP1 (RVxF binding pockets) or PP2B (PIVIIT binding pockets) are not present in the other phosphatase. This structural data provides the first molecular insight into the mechanism of specificity regulation between the highly homologous enzymes PP1 and PP2B.
PP1 triple complexes: dynamic regulation of the spinophilin:PP1 holoenzyme by inhibitor-2
Given that many PP1 regulatory proteins are IDPs, it is not surprising that many of them stay, at least to some extent, dynamic even when in a complex with PP1 (i.e., I-2 in the I-2:PP1 complex [11, 29]). Recently, it was shown that pairs of targeting and inhibitor proteins can bind to PP1 simultaneously, forming PP1 triple complexes [57-59]. This leads to an additional layer of complexity in PP1 regulation. Since regulatory protein binding at the RVxF site is mutually exclusive (PP1 has only one RVxF interaction site), which of the two regulatory proteins will be bound at this site (i.e., is the ‘stronger’ binder)? A second related question is which additional PP1 interaction sites, e.g. the SILK site of I-2, are occupied and what is their relative importance for triple complex formation. Understanding the relative importance of all PP1 regulatory protein interaction sites are critical for understanding how PP1 is regulated in the cell.
The only PP1 heterotrimeric PP1 complex that has been biochemically and structurally investigated is the PP1:spinophilin:I-2 (PSI) holoenzyme [5] (Figure 11). The PSI complex is extremely stable and can be purified to homogeneity using size exclusion chromatography. However, structural results have shown that both regulatory proteins interact with PP1 via the RVxF motif. Thus, one or both regulatory proteins must change conformation in order for the triple complex to form. Because this complex is highly dynamic in solution, NMR spectroscopy and small angle X-ray scattering (SAXS) were the only experimental tools that could be used to provide atomic level information on this complex. These studies showed that in the PSI complex (~60 kDa) I-2 is bound to PP1 via the SILK motif and the long I-2 inhibitory helix, which binds and blocks the PP1 active site [5], while spinophilin binds PP1 via its RVxF interaction site and C-terminal substrate binding groove. Thus, for I-2, the inhibitory α-helix and especially the SILK motif are sequences that mediate PP1 binding in PSI, a critical result that illustrates that binding motifs outside the well characterized RVxF site can be the key interaction site for PP1 regulatory proteins.
Figure 11. PP1:Spinophilin:Inhibitor-2 structure.
(A) SAXS-based ensemble structure for the PP1:spinophilin:I-2 (PSI) complex, which highlights the increased flexibility of I-2 in the PSI complex versus the I-2:PP1 complex (compare with Fig. 5). (PP1c, gray surface; I-2, wheat ribbon, spinophilin, slate ribbons); (B) PP1 is rotated by 90° in order to highlight the spinophilin RVxF peptide binding site (spinophilin residues I449 and F451, as part of the RVxF motif, are highlighted as spheres); (C) PP1 is rotated by additional 90° in order to highlight the I-2 SILK peptide binding site (I-2 residues 13-16, the SILK motif, are highlighted as spheres).
Combined biochemical, NMR spectroscopy and SAXS analysis allowed for the structural model of PSI complex. Release of the I-2 RVxF motif leads to an increase in the size of the flexible I-2 loop in the PSI complex as when compared to the loop in the I-2:PP1 complex (Figure 7). Conversely, while the RVxF motif of spinophilin remains bound to PP1 in the PSI complex, the shorter α-helix that connects the spinophilin PP1 binding and PDZ domains dissociates from PP1 in order to accommodate bound I-2. As these are the first structural, molecular insights into PP1 triple complexes, it is not possible to draw detailed conclusions as to whether these observations will hold for all other potential PP1 triple complexes. However, these studies have certainly highlighted the importance of PP1 binding motifs outside the RVxF site for PP1 regulation and activity.
Future outlook for PP1: mechanisms, motifs, metals and drugs
Structural biology research on all PPPs [2, 60], but especially PP1 and PP2A, has significantly enhanced our understanding of the regulation of PPP activity. An initial specific mode of PP1 regulation by the PP1 targeting protein spinophilin, which directs PP1 specificity via steric inhibition of alternative substrate binding sites [4, 6, 11, 12, 43] has been discovered. This is a novel method for creating enzyme specificity. Nevertheless, as PP1 is regulated by a huge number of protein (~200 confirmed; ~650 predicted) [4], it is likely, and has already been experimentally observed in some cases (unpublished results), that PP1 employs a multitude of mechanisms to create specific holoenzymes. Detailed structural analysis of these holoenzymes will lead to a more comprehensive understanding of PP1 interaction motifs outside of the canonical RVxF motif or the active site [61]. Currently, there is only extremely limited overlap of PP1 binding sites between the structurally analyzed PP1 holoenzymes (Figure 5), suggesting multiple surfaces of PP1 are potential protein:protein interaction sites. Therefore, it is only through the structural analysis of additional PP1 holoenzymes that common interaction sites will be identified. This, in turn, will allow new common interaction motifs to be defined and thus, ultimately, enable the mode of PP1 interaction and regulation of each confirmed and new PP1 regulator to be identified via its primary sequence alone. It will be interesting to determine if additional common interaction sites between the families of PPP’s, are detected. Finally, an unresolved question is the nature of the metals bound at the active site of PP1 in the native enzyme. Here future biochemical, structural and computational analysis will ultimately allow this point to be clarified.
Lastly, because of the essential role of PP1 in a vast array of fundamental biological regulatory signaling cascades, specific PP1 inhibitors or activators hold tremendous clinical potential for the treatment of a wide variety of diseases. Critical milestone publications by the Yuan [62] and more recently by the Bertolotti laboratory [63] showed that one can develop ‘regulatory protein’-specific inhibitors for PP1. Clearly, structural biology will also play a key role for providing a detailed understanding of how these inhibitors interact with PP1 holoenzymes and the molecular insights that will enable them to be improved. Collectively, these studies will provide the necessary molecular details required to establish PP1 as a highly specific drug target against numerous diseases such as Parkinson’s disease, drug addiction and cancer.
Acknowledgement
The authors thank all current and previous members of the Page and Peti laboratory for their contribution to this ongoing research during the last 7 years. The authors thank Dr. Mathieu Bollen for stimulating input into the extensive biology of PP1 and Dr. Julie Forman-Kay for support with the ENSEMBLE method. The work was supported by grant R01NS056128 from the National Institute of Neurological Disorders and Stroke to W.P., grant R01GM098482 from the National Insitute of General Medicine and an American Cancer Society research scholar grant (RSG-08-067-01-LIB) to R.P.
Abbreviations
- IDP
Intrinsically disordered protein
- NMR
nuclear magnetic resonance
- SAXS
small angle X-Ray scattering
- PP1
protein phosphatase 1
- I-2
Inhibitor-2
- PSI
PP1:spinophilin:I-2 complex
- MYPT1
myosin phosphatase targeting subunit 1
- Gm
skeletal muscle glycogen targeting subunit
References
- 1.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 2.Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–84. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Patterson KI, Brummer T, O’Brien PM, Daly RJ. Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem J. 2009;418:475–89. doi: 10.1042/bj20082234. [DOI] [PubMed] [Google Scholar]
- 4.Bollen M, Peti W, Ragusa MJ, Beullens M. The extended PP1 toolkit: designed to create specificity. Trends Biochem Sci. 2010;35:450–8. doi: 10.1016/j.tibs.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dancheck B, Ragusa MJ, Allaire M, Nairn AC, Page R, Peti W. Molecular investigations of the structure and function of the protein phosphatase 1- spinophilin-inhibitor 2 heterotrimeric complex. Biochemistry. 2011;50:1238–46. doi: 10.1021/bi101774g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelker MS, Page R, Peti W. Crystal structures of protein phosphatase-1 bound to nodularin-R and tautomycin: a novel scaffold for structure-based drug design of serine/threonine phosphatase inhibitors. J Mol Biol. 2009;385:11–21. doi: 10.1016/j.jmb.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wakula P, Beullens M, Ceulemans H, Stalmans W, Bollen M. Degeneracy and Function of the Ubiquitous RVXF Motif That Mediates Binding to Protein Phosphatase-1. Journal of Biological Chemistry. 2003;278:18817–18823. doi: 10.1074/jbc.M300175200. [DOI] [PubMed] [Google Scholar]
- 8.Meiselbach H, Sticht H, Enz R. Structural analysis of the protein phosphatase 1 docking motif: molecular description of binding specificities identifies interacting proteins. Chem Biol. 2006;13:49–59. doi: 10.1016/j.chembiol.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 9.Hendrickx A, Beullens M, Ceulemans H, Den Abt T, Van Eynde A, Nicolaescu E, Lesage B, Bollen M. Docking Motif-Guided Mapping of the Interactome of Protein Phosphatase-1. Chemistry & Biology. 2009;16:365–371. doi: 10.1016/j.chembiol.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Hurley T, Yang J, Zhang L, Goodwin K, Zou Q, Cortese M, Dunker A, DePaoli-Roach A. Structural basis for regulation of protein phosphatase 1 by inhibitor-2. J Biol Chem. 2007;282:28874–28883. doi: 10.1074/jbc.M703472200. [DOI] [PubMed] [Google Scholar]
- 11.Marsh JA, Dancheck B, Ragusa MJ, Allaire M, Forman-Kay JD, Peti W. Structural diversity in free and bound states of intrinsically disordered protein phosphatase 1 regulators. Structure. 2010;18:1094–103. doi: 10.1016/j.str.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ragusa MJ, Dancheck B, Critton DA, Nairn AC, Page R, Peti W. Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites. Nat Struct Mol Biol. 2010;17:459–464. doi: 10.1038/nsmb.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terrak M, Kerff F, Langsetmo K, Tao T, Dominguez R. Structural basis of protein phosphatase 1 regulation. Nature. 2004;429:780–784. doi: 10.1038/nature02582. [DOI] [PubMed] [Google Scholar]
- 14.Cohen P, Alemany S, Hemmings BA, Resink TJ, Stralfors P, Tung HY. Protein phosphatase-1 and protein phosphatase-2A from rabbit skeletal muscle. Methods Enzymol. 1988;159:390–408. doi: 10.1016/0076-6879(88)59039-0. [DOI] [PubMed] [Google Scholar]
- 15.Stewart AA, Cohen P. Protein phosphatase-2B from rabbit skeletal muscle: a Ca2+-dependent, calmodulin-stimulated enzyme. Methods Enzymol. 1988;159:409–16. doi: 10.1016/0076-6879(88)59040-7. [DOI] [PubMed] [Google Scholar]
- 16.Zhang AJ, Bai G, Deans-Zirattu S, Browner MF, Lee EY. Expression of the catalytic subunit of phosphorylase phosphatase (protein phosphatase-1) in Escherichia coli. J Biol Chem. 1992;267:1484–90. [PubMed] [Google Scholar]
- 17.Watanabe T, da Cruz e Silva EF, Huang HB, Starkova N, Kwon YG, Horiuchi A, Greengard P, Nairn AC. Preparation and characterization of recombinant protein phosphatase 1. Methods Enzymol. 2003;366:321–38. doi: 10.1016/s0076-6879(03)66024-6. [DOI] [PubMed] [Google Scholar]
- 18.Alessi DR, Street AJ, Cohen P, Cohen PT. Inhibitor-2 functions like a chaperone to fold three expressed isoforms of mammalian protein phosphatase-1 into a conformation with the specificity and regulatory properties of the native enzyme. Eur J Biochem. 1993;213:1055–66. doi: 10.1111/j.1432-1033.1993.tb17853.x. [DOI] [PubMed] [Google Scholar]
- 19.MacKintosh C, Garton AJ, McDonnell A, Barford D, Cohen PT, Tonks NK, Cohen P. Further evidence that inhibitor-2 acts like a chaperone to fold PP1 into its native conformation. FEBS Lett. 1996;397:235–8. doi: 10.1016/s0014-5793(96)01175-1. [DOI] [PubMed] [Google Scholar]
- 20.Myles T, Schmidt K, Evans DR, Cron P, Hemmings BA. Active-site mutations impairing the catalytic function of the catalytic subunit of human protein phosphatase 2A permit baculovirus-mediated overexpression in insect cells. Biochem J. 2001;357:225–32. doi: 10.1042/0264-6021:3570225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho US, Xu W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature. 2007;445:53–7. doi: 10.1038/nature05351. [DOI] [PubMed] [Google Scholar]
- 22.Jin L, Harrison SC. Crystal structure of human calcineurin complexed with cyclosporin A and human cyclophilin. Proc Natl Acad Sci U S A. 2002;99:13522–6. doi: 10.1073/pnas.212504399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kissinger CR, Parge HE, Knighton DR, Lewis CT, Pelletier LA, Tempczyk A, Kalish VJ, Tucker KD, Showalter RE, Moomaw EW, et al. Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex. Nature. 1995;378:641–4. doi: 10.1038/378641a0. [DOI] [PubMed] [Google Scholar]
- 24.Endo S, Connor JH, Forney B, Zhang L, Ingebritsen TS, Lee EY, Shenolikar S. Conversion of protein phosphatase 1 catalytic subunit to a Mn(2+)-dependent enzyme impairs its regulation by inhibitor 1. Biochemistry. 1997;36:6986–92. doi: 10.1021/bi970418i. [DOI] [PubMed] [Google Scholar]
- 25.Peti W, Page R. Strategies to maximize heterologous protein expression in Escherichia coli with minimal cost. Protein Expr Purif. 2007;51:1–10. doi: 10.1016/j.pep.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 26.Ragusa MJ, Dancheck B, Critton DA, Nairn AC, Page R, Peti W. Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites. Nat Struct Mol Biol. 2010;17:459–64. doi: 10.1038/nsmb.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xing Y, Xu Y, Chen Y, Jeffrey PD, Chao Y, Lin Z, Li Z, Strack S, Stock JB, Shi Y. Structure of protein phosphatase 2A core enzyme bound to tumor-inducing toxins. Cell. 2006;127:341–53. doi: 10.1016/j.cell.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 28.Terrak M, Kerff F, Langsetmo K, Tao T, Dominguez R. Structural basis of protein phosphatase 1 regulation. Nature. 2004;429:780–4. doi: 10.1038/nature02582. [DOI] [PubMed] [Google Scholar]
- 29.Hurley TD, Yang J, Zhang L, Goodwin KD, Zou Q, Cortese M, Dunker AK, DePaoli-Roach AA. Structural basis for regulation of protein phosphatase 1 by inhibitor-2. J Biol Chem. 2007;282:28874–83. doi: 10.1074/jbc.M703472200. [DOI] [PubMed] [Google Scholar]
- 30.Goldberg J, Huang HB, Kwon YG, Greengard P, Nairn AC, Kuriyan J. Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1. Nature. 1995;376:745–53. doi: 10.1038/376745a0. [DOI] [PubMed] [Google Scholar]
- 31.Griffith JP, Kim JL, Kim EE, Sintchak MD, Thomson JA, Fitzgibbon MJ, Fleming MA, Caron PR, Hsiao K, Navia MA. X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex. Cell. 1995;82:507–22. doi: 10.1016/0092-8674(95)90439-5. [DOI] [PubMed] [Google Scholar]
- 32.Egloff MP, Cohen PT, Reinemer P, Barford D. Crystal structure of the catalytic subunit of human protein phosphatase 1 and its complex with tungstate. J Mol Biol. 1995;254:942–59. doi: 10.1006/jmbi.1995.0667. [DOI] [PubMed] [Google Scholar]
- 33.Kita A, Matsunaga S, Takai A, Kataiwa H, Wakimoto T, Fusetani N, Isobe M, Miki K. Crystal structure of the complex between calyculin A and the catalytic subunit of protein phosphatase 1. Structure (Camb) 2002;10:715–24. doi: 10.1016/s0969-2126(02)00764-5. [DOI] [PubMed] [Google Scholar]
- 34.Maynes JT, Bateman KS, Cherney MM, Das AK, Luu HA, Holmes CF, James MN. Crystal structure of the tumor-promoter okadaic acid bound to protein phosphatase-1. J Biol Chem. 2001;276:44078–82. doi: 10.1074/jbc.M107656200. [DOI] [PubMed] [Google Scholar]
- 35.Damer CK, Partridge J, Pearson WR, Haystead TA. Rapid identification of protein phosphatase 1-binding proteins by mixed peptide sequencing and data base searching. Characterization of a novel holoenzymic form of protein phosphatase 1. J Biol Chem. 1998;273:24396–405. doi: 10.1074/jbc.273.38.24396. [DOI] [PubMed] [Google Scholar]
- 36.Maynes JT, Perreault KR, Cherney MM, Luu HA, James MN, Holmes CF. Crystal structure and mutagenesis of a protein phosphatase-1:calcineurin hybrid elucidate the role of the beta12-beta13 loop in inhibitor binding. J Biol Chem. 2004;279:43198–206. doi: 10.1074/jbc.M407184200. [DOI] [PubMed] [Google Scholar]
- 37.MacKintosh RW, Dalby KN, Campbell DG, Cohen PT, Cohen P, MacKintosh C. The cyanobacterial toxin microcystin binds covalently to cysteine-273 on protein phosphatase 1. FEBS Lett. 1995;371:236–40. doi: 10.1016/0014-5793(95)00888-g. [DOI] [PubMed] [Google Scholar]
- 38.Egloff MP, Johnson DF, Moorhead G, Cohen PT, Cohen P, Barford D. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. The EMBO journal. 1997;16:1876–87. doi: 10.1093/emboj/16.8.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe T, Huang HB, Horiuchi A, da Cruze Silva EF, Hsieh-Wilson L, Allen PB, Shenolikar S, Greengard P, Nairn AC. Protein phosphatase 1 regulation by inhibitors and targeting subunits. Proc Natl Acad Sci U S A. 2001;98:3080–5. doi: 10.1073/pnas.051003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsieh-Wilson LC, Allen PB, Watanabe T, Nairn AC, Greengard P. Characterization of the neuronal targeting protein spinophilin and its interactions with protein phosphatase-1. Biochemistry. 1999;38:4365–73. doi: 10.1021/bi982900m. [DOI] [PubMed] [Google Scholar]
- 41.Pinheiro AS, Marsh JA, Forman-Kay JD, Peti W. Structural signature of the MYPT1-PP1 interaction. J Am Chem Soc. 2011;133:73–80. doi: 10.1021/ja107810r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson DF, Moorhead G, Caudwell FB, Cohen P, Chen YH, Chen MX, Cohen PT. Identification of protein-phosphatase-1-binding domains on the glycogen and myofibrillar targetting subunits. Eur J Biochem. 1996;239:317–25. doi: 10.1111/j.1432-1033.1996.0317u.x. [DOI] [PubMed] [Google Scholar]
- 43.Dancheck B, Nairn AC, Peti W. Detailed structural characterization of unbound protein phosphatase 1 inhibitors. Biochemistry. 2008;47:12346–56. doi: 10.1021/bi801308y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hendrickx A, Beullens M, Ceulemans H, Den Abt T, Van Eynde A, Nicolaescu E, Lesage B, Bollen M. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem Biol. 2009;16:365–71. doi: 10.1016/j.chembiol.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 45.Sarrouilhe D, di Tommaso A, Metaye T, Ladeveze V. Spinophilin: from partners to functions. Biochimie. 2006;88:1099–113. doi: 10.1016/j.biochi.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 46.Allen PB, Ouimet CC, Greengard P. Spinophilin, a novel protein phosphatase 1 binding protein localized to dendritic spines. Proc Natl Acad Sci U S A. 1997;94:9956–61. doi: 10.1073/pnas.94.18.9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown AM, Baucum AJ, Bass MA, Colbran RJ. Association of protein phosphatase 1 gamma 1 with spinophilin suppresses phosphatase activity in a Parkinson disease model. J Biol Chem. 2008;283:14286–94. doi: 10.1074/jbc.M801377200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 49.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–31. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 50.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–82. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 51.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 52.Kita A, Matsunaga S, Takai A, Kataiwa H, Wakimoto T, Fusetani N, Isobe M, Miki K. Crystal structure of the complex between calyculin A and the catalytic subunit of protein phosphatase 1. Structure. 2002;10:715–24. doi: 10.1016/s0969-2126(02)00764-5. [DOI] [PubMed] [Google Scholar]
- 53.Zhang J, Zhang Z, Brew K, Lee EY. Mutational analysis of the catalytic subunit of muscle protein phosphatase-1. Biochemistry. 1996;35:6276–82. doi: 10.1021/bi952954l. [DOI] [PubMed] [Google Scholar]
- 54.Marsh JA, Forman-Kay JD. Structure and disorder in an unfolded state under nondenaturing conditions from ensemble models consistent with a large number of experimental restraints. J Mol Biol. 2009;391:359–74. doi: 10.1016/j.jmb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 55.Choy WY, Forman-Kay JD. Calculation of ensembles of structures representing the unfolded state of an SH3 domain. J Mol Biol. 2001;308:1011–32. doi: 10.1006/jmbi.2001.4750. [DOI] [PubMed] [Google Scholar]
- 56.Li H, Zhang L, Rao A, Harrison SC, Hogan PG. Structure of calcineurin in complex with PVIVIT peptide: portrait of a low-affinity signalling interaction. J Mol Biol. 2007;369:1296–306. doi: 10.1016/j.jmb.2007.04.032. [DOI] [PubMed] [Google Scholar]
- 57.Connor JH, Weiser DC, Li S, Hallenbeck JM, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 assembles a novel signaling complex containing protein phosphatase 1 and inhibitor 1. Mol Cell Biol. 2001;21:6841–50. doi: 10.1128/MCB.21.20.6841-6850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lesage B, Beullens M, Pedelini L, Garcia-Gimeno MA, Waelkens E, Sanz P, Bollen M. A complex of catalytically inactive protein phosphatase-1 sandwiched between Sds22 and inhibitor-3. Biochemistry. 2007;46:8909–19. doi: 10.1021/bi7003119. [DOI] [PubMed] [Google Scholar]
- 59.Terry-Lorenzo RT, Elliot E, Weiser DC, Prickett TD, Brautigan DL, Shenolikar S. Neurabins recruit protein phosphatase-1 and inhibitor-2 to the actin cytoskeleton. J Biol Chem. 2002;277:46535–43. doi: 10.1074/jbc.M206960200. [DOI] [PubMed] [Google Scholar]
- 60.Roy J, Cyert MS. Cracking the phosphatase code: docking interactions determine substrate specificity. Sci Signal. 2009;2:re9. doi: 10.1126/scisignal.2100re9. [DOI] [PubMed] [Google Scholar]
- 61.Honkanen RE, Golden T. Regulators of serine/threonine protein phosphatases at the dawn of a clinical era? Curr Med Chem. 2002;9:2055–75. doi: 10.2174/0929867023368836. [DOI] [PubMed] [Google Scholar]
- 62.Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D, Yuan J. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–9. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- 63.Tsaytler P, Harding HP, Ron D, Bertolotti A. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science. 2011;332:91–4. doi: 10.1126/science.1201396. [DOI] [PubMed] [Google Scholar]
- 64.Maynes JT, Luu HA, Cherney MM, Andersen RJ, Williams D, Holmes CF, James MN. Crystal structures of protein phosphatase-1 bound to motuporin and dihydromicrocystin-LA: elucidation of the mechanism of enzyme inhibition by cyanobacterial toxins. J Mol Biol. 2006;356:111–20. doi: 10.1016/j.jmb.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 65.Hirschi A, Cecchini M, Steinhardt RC, Schamber MR, Dick FA, Rubin SM. An overlapping kinase and phosphatase docking site regulates activity of the retinoblastoma protein. Nat Struct Mol Biol. 2010;17:1051–7. doi: 10.1038/nsmb.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ragusa MJ, Allaire M, Nairn AC, Page R, Peti W. Flexibility in the PP1:spinophilin holoenzyme. FEBS Lett. 2011;585:36–40. doi: 10.1016/j.febslet.2010.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]