Abstract
Runx2, best known for its role in regulating osteoblast-specific gene expression, also plays an increasingly recognized role in prostate and breast cancer metastasis. Using the C4-2B/Rx2dox prostate cancer cell line that conditionally expressed Runx2 in response to doxycycline treatment, we identified and characterized G9a, a histone methyltransferase, as a novel regulator for Runx2 activity. G9a function was locus-dependent. Whereas depletion of G9a reduced expression of many Runx2 target genes, including MMP9, CSF2, SDF1, and CST7, expression of others, such as MMP13 and PIP, was enhanced. Physical association between G9a and Runx2 was indicated by co-immunoprecipitation, GST-pulldown, immunofluorescence, and fluorescence recovery after photobleaching (FRAP) assays. Since G9a makes repressive histone methylation marks and is primarily known as a corepressor, we further investigated the mechanism by which G9a functioned as a positive regulator for Runx2 target genes. Transient reporter assays indicated that the histone methyltransferase activity of G9a was not required for transcriptional activation by Runx2. Chromatin immunoprecipitation assays for Runx2 and G9a showed that G9a was recruited to endogenous Runx2 binding sites. We conclude that a subset of cancer-related Runx2 target genes require recruitment of G9a for their expression, but do not depend on its histone methyltransferase activity.
Keywords: coregulator, transcription, chromatin, G9a, Runx2
Introduction
Runx2 together with Runx1 and Runx3 form the mammalian Runt family of transcription factors that share a Runt DNA-binding domain and are responsible for regulating diverse transcriptional programs [Cameron and Neil, 2004; Ducy et al., 1997]. While Runx2 controls osteoblast and chondrocyte development [Ducy et al., 1997; Komori et al., 1997; Otto et al., 1997; Schroeder et al., 2005], Runx1 is important for hematopoiesis [Lo Coco et al., 1997; Woolf et al., 2003], and Runx3 is important for neurogenesis, thymopoiesis and gut epithelium maintenance [Ito, 2004; Levanon et al., 2002; Woolf et al., 2003].
In addition to its important role in skeletal development, Runx2 is expressed in breast and prostate cancer cells where it induces expression of genes involved in epithelial-mesenchymal transition, metastasis, and osteolysis [Akech et al., 2010; Baniwal et al., 2010; Chimge et al., 2011; Little et al., 2011]. Both the androgen receptor (AR) and estrogen receptor α (ERα) bind to and either stimulate or inhibit expression of Runx2 target genes, suggesting a complex relationship [Baniwal et al., 2009; Baniwal et al., 2012; Chimge et al., 2012; Khalid et al., 2008].
The mechanism by which Runx2 activates expression of its target genes is still under intensive study but presumably involves recruitment of a number of coregulator proteins to target genes by Runx2 [Schroeder et al., 2005; Westendorf, 2006]. Coregulator proteins are important effectors of chromatin modification and remodeling, and they also regulate the recruitment and activation of RNA polymerase II and its basal transcription factors. In particular, coregulators and their mechanisms of action have been extensively studied in connection with the action of nuclear receptors [Lonard and O’Malley B, 2007; Lonard and O’Malley, 2005]. Less is known about the influence of these coregulator proteins on Runx2-mediated transcription and their potential to affect the interaction of Runx2 with nuclear receptors. The histone H3 Lys-9 (H3K9) methyltransferase G9a has functional interactions with many DNA-binding transcription factors including Runx3, and we have previously shown that G9a also functions as a coregulator for nuclear receptors, including ERα and AR [Lee et al., 2006; Lee et al., 2009; Purcell et al., 2011]. Surprisingly, G9a expression stimulated transient reporter gene activation by ERα and AR, whereas depletion of endogenous G9a in MCF7 breast cancer cells caused selective positive and negative effects on steroid hormone regulated genes. Since G9a is a major contributor to H3K9 methylation in euchromatin, the role of G9a and its lysine methyltransferase activity in repression of transcription has been studied intensely [Duan et al., 2005; Gyory et al., 2004; Lee, 2011; Nagano et al., 2008; Nishio and Walsh, 2004; Roopra et al., 2004; Tachibana et al., 2002]. G9a also represses transcription through interactions with DNA methyltransferases [Epsztejn-Litman et al., 2008]. In contrast, the extent and mechanism of G9a coactivator function are poorly understood.
Because Runx2 and G9a have common functional interactions with ERα and AR [Baniwal et al., 2009; Baniwal et al., 2012; Chimge et al., 2012; Khalid et al., 2008], G9a influences the intracellular localization of Runx3 [Lee et al., 2009; Lee, 2011], and G9a and Runx2 are co-expressed in prostate cancer cells [Akech et al., 2010; Kelly et al., 2010], we tested the possibility of physical and functional interactions between Runx2 and G9a. We used the cell line C4-2B/Rx2dox, which was engineered to conditionally express Runx2 by treatment with doxycycline (dox). These cells express Runx2 at near physiological levels seen in prostate and breast cancer cells [Baniwal et al., 2010; Chimge et al., 2011]. We found that G9a can dramatically and selectively modulate Runx2-dependent gene expression, both positively and negatively. To further explore the mechanism by which G9a affects Runx2-mediated gene expression, we investigated the physical association between Runx2 and G9a, examined the role of G9a methyltransferase activity in the coactivation of Runx2-induced gene expression, and tested whether Runx2 recruits G9a to Runx2 target genes. Our results suggest that G9a regulates key Runx2 target genes with important roles in prostate cancer progression and metastasis.
Materials and Methods
Cell culture and lentiviral vectors
Cos7 and CV1 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% complete fetal bovine serum (FBS) at 37 °C and 5 % CO2. C4-2B cells were obtained from ViroMed Laboratories (Minneapolis, MN), and maintained in RPMI 1640 medium supplemented with 10% FBS. C4-2B cells are derived from LNCaP cells that were grown in castrated nude mice and metastasized to bone. C4-2B/Rx2dox cells containing a stably integrated dox-inducible FLAG-Runx2 transgene were previously described [Baniwal et al., 2010]. Construction of lentiviral vectors encoding shRNAs that target two distinct regions within G9a open reading frame, and preparation of lentivirus particles for infection of C4-2B/Rx2dox cells were performed as described previously [Ou et al., 2011]. The oligonucleotide sequences for constructing plasmids encoding shRNA specific for G9a (shG9a) and for a random sequence not present in human genome (nonspecific shRNA, shNS) are listed in Table 1. Dox (Calibiochem, La Jolla, CA, USA) was used at 0.25 μg/ml unless otherwise stated, and an equal volume of distilled water was used as vehicle control. Puromycin at the final concentration of 5 ng/ml in cell culture medium was used to select cells expressing G9a or NS shRNA. Immunoblotting was conducted with primary antibodies against hemagglutinin (HA) epitope (3F10 Roche Indianapolis, IN), G9a (Sigma-Aldrich Corp. St. Louis, MO), actin (Santa Cruz Biotechnology, Santa Cruz, CA) and FLAG epitope (M2, Sigma-Aldrich Corp.St. Louis, MO). Secondary antibodies against rat IgG (Santa Cruz Biotechnology, Santa Cruz, CA) and rabbit or mouse IgG (Promega, Fitchburg, WI or LI-COR Lincoln, NE) were used for chemiluminescence detection by film or for quantitative infrared imaging.
Table 1.
Sequences for the primers and oligonucleotides
| Gene | Forward (5′ to 3′) | Reverse (5′ to 3′) |
|---|---|---|
| CSF-2 | ATGTGAATGCCATCCAGGAG | AGGGCAGTGCTGCTTGTAGT |
| CST7 | TCCCAGGACCTTAACTCACG | GCTTCAAGGTGTGGTTGGTT |
| CXCL12/SDF-1 | ATGAACGCCAAGGTCGTG | CTTTAGCTTCGGGTCAATGC |
| GAPDH | GTCATGGGTGTGAACCATGAGA | GGTCATGAGTCCTTCCACGATAC |
| MMP9 | TTGACAGCGACAAGAAGTGG | GCCATTCACGTCGTCCTTAT |
| OC | GGCAGCGAGGTAGTGAAGAG | CTGGAGAGGAGCAGAACTGG |
| PGC | ACAGGCACCTCTCTGCTAACT | AGTAGCCGTTGTTACTGAGGAT |
| PIP | GTACGTCCAAATGACGAAGTCAC | CAGCAGCATCATCAGGGCAGATG |
| RASD1 | GTGTTCAGTCTGGACAACCGC | CTGCTCGATCTCGCGCTGGTC |
| RunX2 | CACGAATGCACTATCCAGCCAC | CGCCAAACAGATTCATCCATTC |
| Primers for qPCR in ChIP | ||
| CSF-2 | GAAGCTTGGCTGAATAGATGC | ACACCAGACATATGAAGCAACATC |
| PGC | TCTCTCTTATCGCTTGCACCTCCT | TAGTCTAATCGCTGCCTCCCTGC |
| Primers for shRNA expression | ||
| shG9a_5 Sequence targeting nucl. 866–888 (286–294 amino acid residues) | ||
|
Sense oligo: 5′-CTTGTGGAAAGGACGAAACACCGACAGCAAGTATGAAGTTAAA GCtcTTCAAGAGAGAGCTTCAACTTCAGACTTGCTGTCTTTTTCTGCAGTTTT | ||
|
Reverse oligo: 5′-3′ AAAACTGCAGAAAAAGACAGCAAGTCTGAAGTTGA AGCTCTCTCTTGAAGAGCTTTAACTTCATACTTGCTGTCGGTGTTTCGTCCTTTCCACAAG | ||
| shG9a_7 Sequence targeting nucl. 280–302 (91–99 amino acid residues) | ||
|
Sense oligo: 5′-CTTGTGGAAAGGACGAAACACCGGATGAATCTAAGAATCTGGA GGgaTTCAAGAGATCCCTCAAGATTCTCAGATTCATCCTTTTTCTGCAGTTTT | ||
|
Reverse oligo: 5′-AAAACTGCAGAAAAAGGATGAATCTGAGAATCTTGAGGGATCTCTTGAAT CCCTCCAGATTCTTAGATTCATCCGGTGTTTCGTCCTTTCCACAAG | ||
| shNS Nonspecific sequence | ||
|
Sense oligo: 5′-CTTGTGGAAAGGACGAAACACCGGGTAGGTTCAACTAGCAAGAC TCTTTCAAGAGAAGAGTCCTGCTAGTCGAACCTACCCTTTTTCTGCAGTTTT | ||
|
Reverse oligo: 5′-AAAACTGCAGAAAAAGGGTAGGTTCGACTAGCAGGACTCTTCTC TTGAAAGAGTCTTGCTAGTTGAACCTACCCGGTGTTTCGTCCTTTCCACAAG | ||
| siG9a sequences from Dharmacon SMART Pool | ||
| siRNA 5 | GGACCUUCAUCUGCGAGUA | |
| siRNA 6 | GAACAUCGAUCGCAACAUC | |
| siRNA 7 | GGAGGUAGCCCGUUACAUG | |
| siRNA 8 | GGAGAGGUGUACUGCAUAG | |
G9a depletion and quantitative reverse-transcriptase PCR (qRT-PCR)
C4-2B/Rx2dox cells infected with lentiviruses encoding shNS or shG9a were grown in phenol red-free DMEM with 5% charcoal-stripped fetal bovine serum (CSS) for 48 h and then grown for an additional 24 h in the presence or absence of 250 ng/ml dox. The CSS was used to ensure the absence of androgenic hormones, which activate AR and cause it to interact with Runx2 and inhibit Runx2-mediated transcription [Baniwal et al., 2009]. Cells were harvested either in TRIZOL (Invitrogen, Carlsbad, CA) for qRT-PCR or RIPA buffer for immunoblot analysis; qRT-PCR was performed as described [Purcell et al., 2011] using primers specified in Table 1. Results shown are mean and range of variation for duplicate PCR reactions performed on cDNA samples from a single experiment. Results are expressed relative to GAPDH mRNA levels and are representative of at least two independent experiments.
Co-immunoprecipitation assay
Cos7 cells were seeded at 300,000 per well in six-well plates and grown in phenol red-free DMEM with 5% CSS for 24 h. Cells were co-transfected with plasmids encoding HA-G9a and Flag-Runx2, and 24 h later they were lysed in a buffer containing 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 1% Protease Inhibitors Cocktail (Sigma-Aldrich Corp. St. Louis, MO). After homogenization by passing ten times through a 1 cc microfine insulin syringe, lysates were cleared of cellular debris by centrifugation at 14,000 rpm for 5 min in a microfuge, and 15% of the lysate solution was set aside to assess input. The remaining lysate was immunoprecipitated with approximately 3 μg of the specified antibody and 30 μl Protein-G beads (Amersham Biosciences, Little Chalfont, Buckinghamshire, England), and then washed three times for 5 min each with the same buffer, followed by centrifugation at 4,000 rpm for 1 min. Bound proteins were analyzed by SDS-PAGE and immunoblotting using the indicated antibodies.
GST pull-down assay
35S-labeled Runx2 was synthesized from a PCR-amplified DNA product by in vitro transcription and translation using TNT 7 Quick for PCR DNA kit (Promega, Fitchburg, WI). As bait, a GST fusion protein with mouse-G9a was immobilized on GSH beads (Amersham Biosciences, Little Chalfont, Buckinghamshire, England) followed by overnight incubation at 4 °C with 35S-labeled Runx2. After six times washing with NETN buffer [Koh et al., 2001] containing 200 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl, pH 7.6, and 0.5 % and 0.01 % Nonidet P40, the beads were boiled in SDS sample buffer for analysis by SDS PAGE and autoradiography.
Immunofluorescence
C4-2B/Rx2dox cells were grown on 18-mm coverslips in six-well plates for 24 h using phenol red-free DMEM containing 5% CSS in the presence or absence of dox. Cells were transfected with 100 ng of pSG5.HA-G9a for 24 h. Cells were then fixed with 95% methanol for 15 min and permeabilized with 1% saponin (Sigma, St. Louis, MO). Proteins were visualized with the respective primary antibodies and secondary antibodies conjugated to either rhodamine or a fluorescein tag. Cells were mounted using Vectashield Hard Set mounting medium (Vector Laboratories, Burlingame, CA) with DAPI which intercalates with the DNA to allow it to be visualized by fluorescence microscopy. Cells were viewed using a LSM 510 Zeiss confocal microscope at 60X magnification.
Fluorescence recovery after photobleaching (FRAP)
To measure FRAP, a nuclear region in cells expressing GFP-G9a protein was photobleached and allowed to recover over time. The intensity of the fluorescence in the bleached area was measured, plotted relative to the pre-bleaching intensity and controlled for background loss in intensity. Cos7 cells were plated at 40,000 cells per well in an 8-well Lab-Tek Chambered coverglass with cover (Thermo Fisher Scientific, Hampton NH). Cells were cultured in phenol red free DMEM containing 5% CSS for 24 h before transfection with plasmid encoding GFP-G9a, with or without plasmid encoding Runx2. After transfection, cells were grown in phenol red free DMEM-5% CSS for 24 h. Cells were viewed using a LSM 510 Zeiss confocal microscope at 60X magnification. The cells were observed at a wavelength of 488 nm at 4% intensity for 15 seconds, prior to bleaching for 5 seconds with 80% intensity. Subsequently images were captured at each second for 200 s at 488 nm (4% intensity) to record the recovery from photobleaching. All data obtained was normalized to the average starting intensity.
Transient reporter gene activity assay
Transient transfections were performed with a luciferase reporter plasmid controlled by six tandem copies of osteoblast specific element 2 (6XOSE2) [Ducy and Karsenty, 1995]. Plasmid expressing mouse Runx2 with a FLAG tag was described previously [Khalid et al., 2008], as were pSG5.HA-G9a expressing full length mouse G9a with an HA tag and pSG5.HA-G9a (H/K) encoding methyltransferase-deficient G9a with the H1166K mutation [Lee et al., 2006]. CV1 cells were seeded 70,000 per well in 12-well plates and grown in phenol red-free DMEM supplemented with 5% CSS for 24 h before transfection with BioT reagent (Bioland Scientific, Paramount, CA) according to the manufacturer’s specification. Molar equivalent amounts of the empty vector pSG5 were transfected to balance promoter number and pCAT-basic promoter-less plasmid was used to balance total mass of DNA transfected for all the wells. At 48 h after transfection, cells were lysed with passive lysis buffer (Promega, Fitchburg, WI) for 15 min, and luciferase assays were performed with a luminometer (BMG Labtech Fluostar Optima, Ortenberg Germany). SDS PAGE and immunoblotting with indicated antibodies were performed on cell lysates.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed as previously described [Purcell et al., 2011]. C4-2B/Rx2dox cells were cultured in phenol red-free DMEM containing 5% CSS for 2 days before addition of 250 ng/ml of dox. ChIP was performed with 10 μl of antibody against G9a (Abcam, Cambridge UK) or 1 μg antibody against FLAG-Runx2 (M2, Sigma-Aldrich Corp., St. Louis, MO), and the immunoprecipitated DNA was analyzed by quantitative PCR (qPCR) using primers specific for the indicated Runx2 binding sites (Table 1). The signal from the immunoprecipitated DNA was normalized to the signal from DNA prepared from the same amount of chromatin before immunoprecipitation (input). Results shown are mean and range of variation for duplicate PCR reactions performed on DNA samples from a single experiment. Results shown are representative of at least two independent experiments.
Results
G9a activates or represses Runx2-mediated transcription in a gene-specific manner in prostate cancer cells
The C4-2B prostate cancer cell line is an important model for studying prostate cancer that metastasizes to bone [Wu et al., 1994]. Under normal growth conditions C4-2B cells hardly express Runx2. In order to characterize regulatory effects of Runx2 in prostate cancer, we previously generated C4-2B/Rx2dox cells by transducing C4-2B cells with lentiviruses encoding a dox-inducible Runx2 expression system [Baniwal et al., 2009]. As observed by the immunoblot analysis, dox treatment induced Runx2 expression without affecting G9a protein or mRNA levels (Fig. 1A). As expected, qRT-PCR analysis demonstrated that Runx2 induction by dox increased expression of its target genes (Fig. 1B, C). To examine the influence of G9a on Runx2-induced transcription, we transduced C4-2B/Rx2dox cells with lentiviral particles encoding shRNA against G9a (shG9a_5) or against a non-specific sequence (shNS). shG9a but not shNS efficiently reduced G9a mRNA and protein levels, while Runx2 expression was essentially equal in the two cell populations (Fig. 1A). G9a depletion dramatically reduced the Runx2-induced mRNA levels for most of the tested target genes, including MMP9, PGC, CSF2, SDF-1, and CST7 (Fig. 1B), suggesting a requirement of G9a for full response to Runx2 in these cases. In contrast, the Runx2-induced expression of two other target genes, MMP13 and PIP, was higher in the G9a-depleted cells compared with the control cells expressing shNS (Fig. 1C), suggesting antagonism of Runx2 in these cases. Similar results were obtained when Runx2-induced expression of selected target genes was examined after depletion of G9a with shRNA targeting a different region of the G9a mRNA (shG9a_7, data not shown). The contrasting positive and negative effects of G9a depletion on different Runx2 target genes serve as mutual controls against possible differences in the two cell populations expressing shNS versus shG9a. Thus, G9a may function as a coactivator or a corepressor for Runx2 in a gene-specific manner.
Fig. 1. Selective positive and negative coregulator effects by G9a on Runx2 target genes.
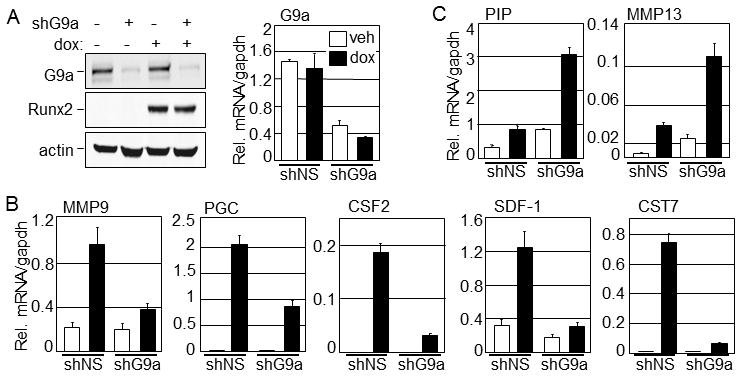
C4-2B/Rx2dox cells were infected with lentiviral vectors encoding shRNA targeting G9a mRNA (shG9a) or a non-specific sequence (shNS), and the infected populations were selected with puromycin. The two infected cell populations were cultured in medium supplemented with charcoal-stripped serum (CSS) containing dox (250 ng/ml) to induce Runx2 expression or equal volume of vehicle (distilled water) for 24 h before harvest. A: Immunoblots (left) were performed using antibodies against G9a, FLAG epitope (to detect Runx2), and actin as a control. mRNA levels were assessed by qRT-PCR (right). B & C: The mRNA levels for the indicated target genes of Runx2 were analyzed by qRT-PCR as described in Materials and Methods. All experiments were repeated at least three times and the representative results are shown. Abbreviations used: PIP, Prolactin-induced protein; MMP13 and MMP9, Matrix metalloproteinsase-13 and -9, respectively; PGC, Progastricsin-C; CSF-2, Colony-stimulating factor-2; SDF-1, Stromal differentiating factor-1; CST7, Cystatin-7.
G9a interacts and colocalizes with Runx2
To begin exploring how G9a may exert its effect on Runx2-mediated transcription, we tested whether G9a associates with Runx2 in cultured cells. Cos-7 cells were co-transfected with expression plasmids encoding FLAG-tagged Runx2 and HA-tagged G9a, and the whole cell lysates were subjected to immunoprecipitation using Flag-specific antibodies or non-specific IgG. Immunoblot analysis of the immunoprecipitates indicated that G9a co-precipitated specifically with Runx2 in amounts similar to the 15% input, while no detectable G9a co-precipitated with non-specific IgG (Fig. 2A). This result suggests a robust association between G9a and Runx2. Next, we assessed whether Runx2 bound G9a in a cell free system using GST pull-down assays. Runx2 was transcribed and translated in a cell free system in the presence of [35S]methionine, and was incubated with bacterially expressed GST-G9a or GST alone as the control. Autoradiography of the proteins bound to GST-G9a and GST indicated that Runx2 associated specifically with G9a (Fig. 2B), suggesting that G9a directly interacts with Runx2.
Fig. 2. G9a associates with Runx2.
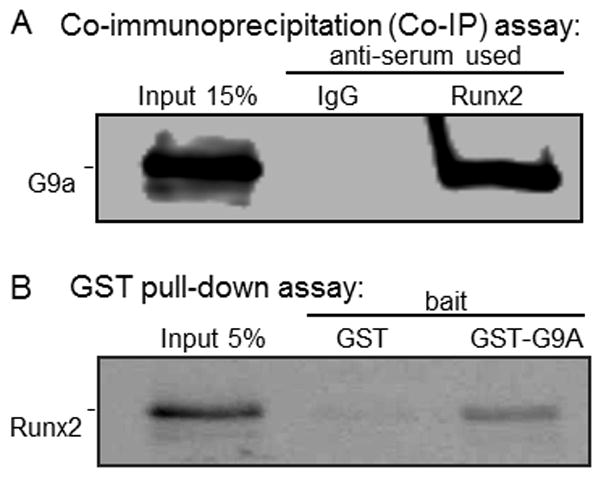
A: Cos-7 cells were transfected with expression vectors for FLAG-Runx2 and HA-G9a (500 ng each in 6-well dishes). Cell extracts were immunoprecipitated with FLAG antibodies, and the immunoprecipitates were analyzed by immunoblotting using antibodies against HA epitope. B: GST or GST-G9a on glutathione agarose beads was incubated with in vitro transcribed and translated 35S-labeled Runx2. The bound protein fraction was analyzed by SDS-PAGE and autoradiography.
To further explore the association of Runx2 with G9a in cultured cells, and to study changes in their intracellular localization patterns when co-expressed, we performed immunofluorescence studies in the C4-2B/Rx2dox cells. In the absence of dox-induced Runx2, endogenous G9a was broadly localized in the nucleus and formed discrete small, punctate foci (Fig. 3A, bottom row). After 24 h of dox treatment, Runx2 was primarily detectable in discrete nuclear foci (Fig. 3A, top row). More importantly, overlay of the G9a (green) and Runx2 (red) patterns in the same cells indicated their co-localization in many of the nuclear foci (Fig. 3A, top row, yellow color). However, there also remained distinct foci of Runx2 or G9a that did not overlap. These foci with overlapping and non-overlapping patterns of G9a and Runx2 occupancy may be indicative of differential local chromatin landscapes. Thus, co-localization of G9a and Runx2 in specific nuclear foci suggests that Runx2 and G9a interact physically in discrete sub-nuclear compartments.
Fig. 3. Runx2 colocalizes with G9a and enhances its intranuclear mobility.
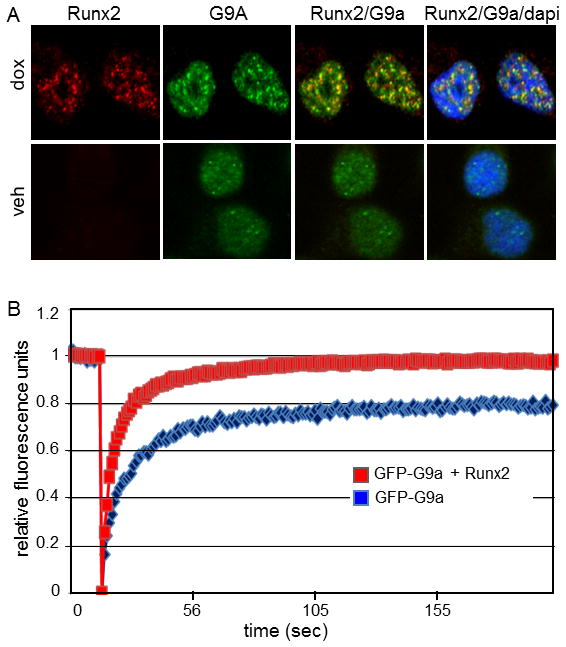
A: C4-2B/Rx2dox cells were treated for 24 h with dox to induce Runx2 expression or vehicle (veh) as control, and immunofluorescence analysis was performed to detect G9a (green) or Runx2 (red) proteins. DAPI staining (blue) indicates dense chromatin organization in the cell nuclei. Runx2/G9a, overlay of red and green images (yellow) indicates overlap; Runx2/G9a/dapi, overlay of red, green, and blue images. B: Fluorescence recovery after photobleaching of G9a-GFP was assessed as described in Materials and Methods in Cos7 cells transfected with plasmid encoding GFP-G9a alone (blue curve) or together with plasmid encoding Runx2 (red curve).
Runx2 affects intranuclear mobility of G9a
Intranuclear mobility of G9a was assessed by fluorescence recovery after photobleaching (FRAP) of dox- or vehicle-treated C4-2B/Rx2dox cells transiently expressing GFP-G9a (Fig. 3B). The initial photobleaching is indicated by the precipitous drop in green fluorescence, whereas the kinetics and extent of fluorescence recovery in the photobleached area shows the rate and extent of GFP-G9a mobility. The half-time (i.e. the rate) of fluorescence recovery was similar in the presence and absence of Runx2, indicating that there was no global change in the mobility of GFP-G9a. However, the extent of fluorescence recovery was only 80% in the absence of Runx2 but approached 100% when it was present. The incomplete recovery of fluorescence indicates that a portion of GFP-G9a is relatively immobile, such that the photobleached GFP-G9a is not replaced by unbleached GFP-G9a during the recovery period. After Runx2 induction, this immobile fraction of GFP-G9a gains mobility, resulting in near complete fluorescence recovery. Thus, Runx2 alters the mobility of a discrete fraction of GFP-G9a moving it from an environment where it is immobile to a new environment where it regains mobility. Together with our studies that showed strong interaction between G9a and Runx2 (Fig. 2), we conclude that their hetero-oligomerization results in enhanced intranuclear mobility of GFP-G9a.
Methyltransferase activity of G9a is not required for its coactivator function
G9a has been extensively characterized as a corepressor that is recruited by repressive transcription factors in order to achieve transcriptionally inert chromatin states. In this setting the corepressor function of G9a often involves its C-terminal SET domain, which methylates the lysine-9 residue of histone H3 (H3K9) resulting in a repressive chromatin structure. However, we observed that G9a supports or opposes transcriptional activation by Runx2 in a gene-specific manner (Fig. 1). Similar behavior of G9a was found for its role as a coregulator for ERα [Purcell et al., 2011]. Because the coactivator function of G9a (i.e. its positive effect on gene expression) is less well characterized than its corepressor function, we established a transient reporter gene assay to test whether the methyltransferase activity of G9a is involved in the coactivator function of G9a. CV-1 cells were transiently transfected with the 6XOSE2-luciferase reporter plasmid, which contains six tandem Runx2-binding sites (Fig. 4A), along with plasmids encoding Runx2 and G9a. Runx2 levels were titrated to attain minimal reporter gene activity in the absence of the co-transfected G9a expression plasmid (data not shown). In this system, increasing levels of G9a co-expression caused a dramatic enhancement of reporter gene activation by Runx2 (Fig. 4B). To examine the influence of the histone methyltransferase activity of G9a on Runx2 transactivation potential, we used the G9a(H/K) mutant, where the H1166K point mutation inactivates the histone methyltransferase activity [Lee et al., 2006]. Expression of the G9a H/K mutant at levels similar to wild type G9a (Fig. 4C) produced a similar stimulatory effect on Runx2-mediated reporter activity (Fig. 4B). In the absence of Runx2, G9a over-expression had no effect on reporter gene activity (data not shown). Therefore, the methyltransferase activity of G9a is not obligatory for its ability to function as a Runx2 coactivator.
Fig. 4. Enhancement of Runx2-mediated transcription by G9a in a transient reporter assay.
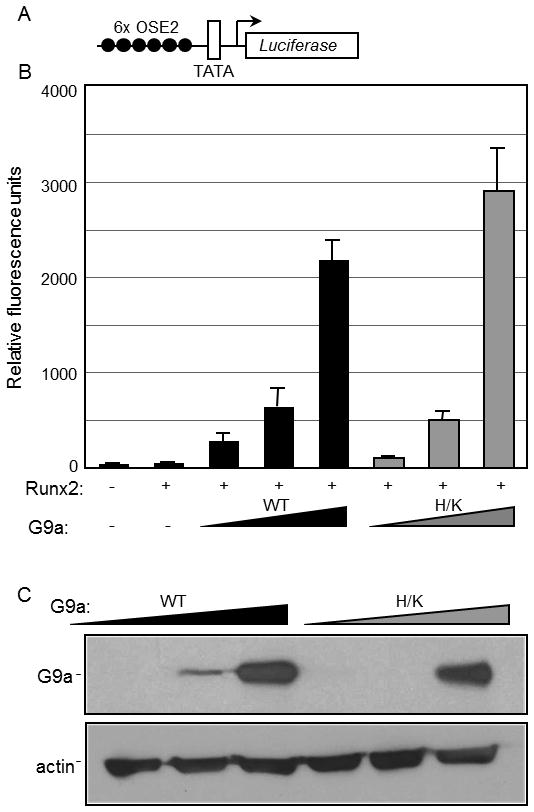
CV1 cells in 12-well plates were transfected with the 6XOSE2-Luciferase reporter plasmid illustrated in A (200 ng/well) alone or together with expression vectors for Runx2 (1 ng) and either HA-tagged G9a full length (G9a) or HA-G9a(H/K) methyltransferase-deficient mutant (H/K) (50, 100 and 200 ng). After transfection the cells were grown for 48 h before they were subjected to luciferase assays (B) and immunoblot analysis using antibodies against HA and actin (C).
Runx2-induced G9a occupancy at the regulatory sites of target genes
In order to enhance the transcription of endogenous Runx2-target genes, G9a could either act directly on the target gene due to its interaction with Runx2 or indirectly by inducing or repressing the expression of an independent gene/s which in turn influences expression of Runx2 target genes. Our initial observations indicated a strong physical and functional interaction between G9a and Runx2 (Figs. 2–4) suggesting that G9a may be a novel coregulator for Runx2 that remodels chromatin and/or directs the assembly of an active transcription complex. We therefore performed ChIP assays in the C4-2B/Rx2dox cells to test whether Runx2 facilitated G9a recruitment to the chromatin at Runx2-binding sites in the vicinity of their target genes. C4-2B/Rx2dox cells treated with dox or vehicle were subjected to ChIP analysis using antibodies against Runx2 or G9a, and the immunoprecipitated DNA was analyzed by qPCR using primers specific for Runx2-binding sites located 4 and 0.1 kb upstream of the CSF2 and PGC gene transcription start sites, respectively. These genes were chosen because of their dependence on G9a for stimulation by Runx2 (Fig. 1), and because they display well-defined Runx2-occupied regions [Little et al., 2011]. ChIP assays showed that dox-induced Runx2 occupied the expected sites (Fig. 5A), and more importantly, enhanced the co-recruitment of endogenous G9a (Fig. 5B). Thus, during transcriptional activation, both G9a and Runx2 are present at the regulatory elements of their target genes. Taken together, these findings indicate that G9a strongly associates with Runx2 in vitro and in vivo, and that Runx2 recruits G9a to function as a gene-specific coactivator.
Fig. 5. Runx2 recruits G9a to the regulatory elements of its target genes.
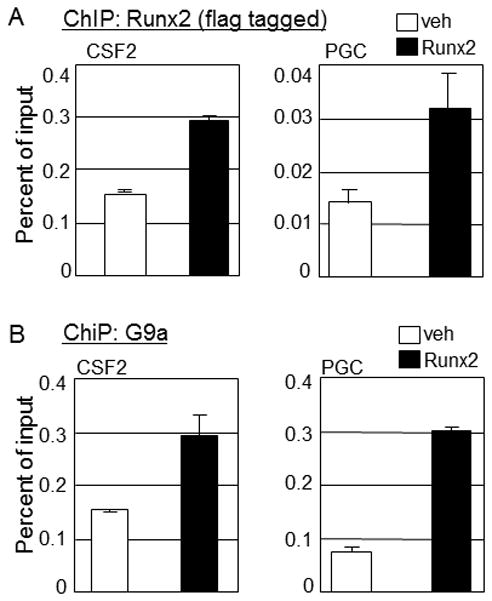
C4-2B/Rx2dox cells were plated in 15-cm dishes and cultured in media supplemented with CSS for two days and then treated with dox or vehicle for an additional 16 h before ChIP analysis using antibodies against FLAG to detect Runx2 (A) or against G9a (B). Immunoprecipitated DNA was analyzed with primers (Table 1) designed to amplify the Runx2-occupied regions of the indicated target genes.
Discussion
G9a, a well known corepressor, employs its methyltransferase activity to deposit repressive histone marks in chromatin regions following its recruitment by repressive transcription factors [Duan et al., 2005; Gyory et al., 2004; Nagano et al., 2008; Nishio and Walsh, 2004; Roopra et al., 2004; Tachibana et al., 2002]. However, we have previously shown that G9a also can act as a powerful coactivator for steroid hormone receptors in transient reporter gene assays, and as a selective activator and repressor for a subset of endogenous target genes of steroid hormone receptors [Lee et al., 2006; Purcell et al., 2011]. Here we show that G9a regulates Runx2-mediated transcription in a similar fashion, that is, as a coactivator in transient reporter assays and a locus specific activator or repressor of endogenous Runx2 target genes. While G9a coactivator function for steroid receptors required the coexpression of additional coactivators such as GRIP1 [Lee et al., 2006], G9a alone enhanced Runx2 regulated expression in CV1 cells. shRNA-mediated depletion of G9a in C4-2B/Rx2dox prostate cancer cells affected the expression of several well characterized endogenous Runx2 target genes. As was the case for steroid hormone receptors [Purcell et al., 2011], the requirement for G9a varied with different Runx2-target genes, such that G9a depletion inhibited expression of some but enhanced the expression of others. In contrast to modest effects of G9a on the expression of endogenous target genes of steroid hormone receptors [Purcell et al., 2011], G9a had remarkable effects on the expression of several Runx2 target genes. This suggests that G9a functions as a critical coregulator for Runx2-regulated gene expression in cancer cells. Alterations of the endogenous expression levels, availability, or activity of G9a protein, e.g. by post-translational modifications or interaction with other proteins, could modulate the pattern of Runx2-regulated gene expression in a developmental and cell type specific manner. For example, G9a dependent Runx2-target genes including MMP9, MMP13, PGC, CSF2, SDF-1, CST7, and PIP are important regulators of tumor growth, invasion, and/or metastasis [Baniwal et al., 2010; Baniwal et al., 2012; Barnes et al., 2004; Blyth et al., 2005; Dai et al., 2010; Kos and Lah, 1998; Little et al., 2011; Taichman et al., 2002]. Thus, modulation of G9a coregulator activity could alter the regulation of these genes by Runx2 and modify prostate cancer disease progression.
G9a could modulate the expression of Runx2-target genes through its recruitment to the regulatory elements in the promoter or enhancer regions or by an indirect mechanism whereby G9a regulates other yet unknown genes, which in turn regulate Runx2 target genes. While we cannot completely rule out indirect mechanisms of G9a action, our data strongly indicate that G9a directly associates with Runx2 and is co-recruited to the regulatory elements of at least some Runx2 target genes. In addition to the association indicated by co-immunoprecipitation and GST pull-down assays, immunofluorescence microscopy of C4-2B/Rx2dox cells revealed that dox-induced Runx2 co-localized with G9a. Additionally, FRAP analysis showed that about 20% of G9a is immobile in the absence of Runx2, and Runx2 expression mobilizes this G9a fraction. One attractive interpretation of the FRAP data is that in the absence of Runx2, some G9a is tightly associated with proteins located in an immobile fraction within the nucleus (e.g., the nuclear matrix); and Runx2 binding to G9a diminishes its interaction with components in the immobile fraction. Based on these results we conclude that G9a through its physical interaction with Runx2 is recruited to the regulatory elements of Runx2 target genes.
Runx2 stimulation of transient reporter gene expression responded in a directly proportional manner to G9a expression levels, suggesting a direct regulation of Runx2-mediated transcription by G9a. Additionally, a G9a mutant that lacked methyltransferase activity retained the ability to enhance Runx2-mediated reporter activity, supporting the hypothesis that positive effects of G9a on Runx2-mediated transcription do not involve an indirect mechanism that requires the corepressor function of G9a. This is consistent with our previous findings that the methyltransferase activity of G9a was not required for the ability of G9a to enhance transient reporter gene activation by steroid hormone receptors [Lee et al., 2006; Purcell et al., 2011]. Therefore, the positive action of G9a presumably involves interaction of G9a with other proteins, e.g. components of the chromatin remodeling machinery or with other coregulators that facilitate the assembly of an active transcription complex on Runx2 target genes. More elaborate studies must be performed to confirm the requirement and identity of the specific proteins that are recruited by G9a to activate transcription.
In conclusion, although G9a has primarily been characterized for its function as a corepressor, we have shown here that G9a is recruited by Runx2 to a subset of its target genes to activate transcription. Since G9a functions as a coactivator for both Runx2 and steroid hormone receptors, two very different classes of transcription factors, it is likely that G9a has the dual ability to function as a coactivator or a corepressor for a diverse array of transcription factors.
Acknowledgments
Grant sponsor: NIH-NIDDK, DK055274 (to MRS) and DK071122 (to BF)
Immunofluorescence and FRAP were performed at the microscopy core facility of the USC Research Center for Liver Diseases. We thank Dr. G. Karsenty (Columbia University) for providing the 6XOSE2 plasmid. BF holds the J. Harold and Edna L. LaBriola Chair in Genetic Orthopedic Research at USC.
Footnotes
Work performed at the University of Southern California, Los Angeles, California
All authors declare no conflict of interest.
References
- Akech J, Wixted JJ, Bedard K, van der Deen M, Hussain S, Guise TA, van Wijnen AJ, Stein JL, Languino LR, Altieri DC, Pratap J, Keller E, Stein GS, Lian JB. Runx2 association with progression of prostate cancer in patients: mechanisms mediating bone osteolysis and osteoblastic metastatic lesions. Oncogene. 2010;29:811–21. doi: 10.1038/onc.2009.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniwal SK, Khalid O, Gabet Y, Shah RR, Purcell DJ, Mav D, Kohn-Gabet AE, Shi Y, Coetzee GA, Frenkel B. Runx2 transcriptome of prostate cancer cells: insights into invasiveness and bone metastasis. Mol Cancer. 2010;9:258. doi: 10.1186/1476-4598-9-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniwal SK, Khalid O, Sir D, Buchanan G, Coetzee GA, Frenkel B. Repression of Runx2 by androgen receptor (AR) in osteoblasts and prostate cancer cells: AR binds Runx2 and abrogates its recruitment to DNA. Mol Endocrinol. 2009;23:1203–14. doi: 10.1210/me.2008-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniwal SK, Little GH, Chimge NO, Frenkel B. Runx2 controls a feed-forward loop between androgen and prolactin-induced protein (PIP) in stimulating T47D cell proliferation. J Cell Physiol. 2012;227:2276–82. doi: 10.1002/jcp.22966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes GL, Hebert KE, Kamal M, Javed A, Einhorn TA, Lian JB, Stein GS, Gerstenfeld LC. Fidelity of Runx2 activity in breast cancer cells is required for the generation of metastases-associated osteolytic disease. Cancer Res. 2004;64:4506–13. doi: 10.1158/0008-5472.CAN-03-3851. [DOI] [PubMed] [Google Scholar]
- Blyth K, Cameron ER, Neil JC. The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer. 2005;5:376–87. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- Cameron ER, Neil JC. The Runx genes: lineage-specific oncogenes and tumor suppressors. Oncogene. 2004;23:4308–14. doi: 10.1038/sj.onc.1207130. [DOI] [PubMed] [Google Scholar]
- Chimge NO, Baniwal SK, Little GH, Chen YB, Kahn M, Tripathy D, Borok Z, Frenkel B. Regulation of Breast Cancer Metastasis by Runx2 and Estrogen Signaling: Role of SNAI2. Breast Cancer Res. 2011;13:R127. doi: 10.1186/bcr3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimge NO, Baniwal SK, Luo J, Coetzee S, Khalid O, Berman BP, Tripathy D, Ellis MJ, Frenkel B. Opposing Effects of Runx2 and Estradiol on Breast Cancer Cell Proliferation: In Vitro Identification of Reciprocally Regulated Gene Signature Related to Clinical Letrozole Responsiveness. Clin Cancer Res. 2012;18:901–11. doi: 10.1158/1078-0432.CCR-11-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Lu Y, Yu C, Keller JM, Mizokami A, Zhang J, Keller ET. Reversal of chemotherapy-induced leukopenia using granulocyte macrophage colony-stimulating factor promotes bone metastasis that can be blocked with osteoclast inhibitors. Cancer Res. 2010;70:5014–23. doi: 10.1158/0008-5472.CAN-10-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Zarebski A, Montoya-Durango D, Grimes HL, Horwitz M. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol Cell Biol. 2005;25:10338–51. doi: 10.1128/MCB.25.23.10338-10351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy P, Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol Cell Biol. 1995;15:1858–69. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–54. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, Deplus R, Fuks F, Shinkai Y, Cedar H, Bergman Y. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008;15:1176–83. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyory I, Wu J, Fejer G, Seto E, Wright KL. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat Immunol. 2004;5:299–308. doi: 10.1038/ni1046. [DOI] [PubMed] [Google Scholar]
- Ito Y. Oncogenic potential of the RUNX gene family: ‘overview’. Oncogene. 2004;23:4198–208. doi: 10.1038/sj.onc.1207755. [DOI] [PubMed] [Google Scholar]
- Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2010;28:1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalid O, Baniwal SK, Purcell DJ, Leclerc N, Gabet Y, Stallcup MR, Coetzee GA, Frenkel B. Modulation of Runx2 activity by estrogen receptor-alpha: implications for osteoporosis and breast cancer. Endocrinology. 2008;149:5984–95. doi: 10.1210/en.2008-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh SS, Chen D, Lee YH, Stallcup MR. Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J Biol Chem. 2001;276:1089–98. doi: 10.1074/jbc.M004228200. [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–64. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Kos J, Lah TT. Cysteine proteinases and their endogenous inhibitors: target proteins for prognosis, diagnosis and therapy in cancer (review) Oncol Rep. 1998;5:1349–61. doi: 10.3892/or.5.6.1349. [DOI] [PubMed] [Google Scholar]
- Lee DY, Northrop JP, Kuo MH, Stallcup MR. Histone H3 lysine 9 methyltransferase G9a is a transcriptional coactivator for nuclear receptors. J Biol Chem. 2006;281:8476–85. doi: 10.1074/jbc.M511093200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Kim J, Kim WH, Lee YM. Hypoxic silencing of tumor suppressor RUNX3 by histone modification in gastric cancer cells. Oncogene. 2009;28:184–94. doi: 10.1038/onc.2008.377. [DOI] [PubMed] [Google Scholar]
- Lee YM. Control of RUNX3 by histone methyltransferases. J Cell Biochem. 2011;112:394–400. doi: 10.1002/jcb.22969. [DOI] [PubMed] [Google Scholar]
- Levanon D, Bettoun D, Harris-Cerruti C, Woolf E, Negreanu V, Eilam R, Bernstein Y, Goldenberg D, Xiao C, Fliegauf M, Kremer E, Otto F, Brenner O, Lev-Tov A, Groner Y. The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J. 2002;21:3454–63. doi: 10.1093/emboj/cdf370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little GH, Noushmehr H, Baniwal SK, Berman BP, Coetzee GA, Frenkel B. Genome-wide Runx2 occupancy in prostate cancer cells suggests a role in regulating secretion. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Coco F, Pisegna S, Diverio D. The AML1 gene: a transcription factor involved in the pathogenesis of myeloid and lymphoid leukemias. Haematologica. 1997;82:364–70. [PubMed] [Google Scholar]
- Lonard DM, O’Malley BW. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol Cell. 2007;27:691–700. doi: 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Lonard DM, O’Malley BW. Expanding functional diversity of the coactivators. Trends Biochem Sci. 2005;30:126–32. doi: 10.1016/j.tibs.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–20. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- Nishio H, Walsh MJ. CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proc Natl Acad Sci U S A. 2004;101:11257–62. doi: 10.1073/pnas.0401343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–71. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Ou CY, LaBonte MJ, Manegold PC, So AY, Ianculescu I, Gerke DS, Yamamoto KR, Ladner RD, Kahn M, Kim JH, Stallcup MR. A coactivator role of CARM1 in the dysregulation of beta-catenin activity in colorectal cancer cell growth and gene expression. Mol Cancer Res. 2011;9:660–70. doi: 10.1158/1541-7786.MCR-10-0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell DJ, Jeong KW, Bittencourt D, Gerke DS, Stallcup MR. A distinct mechanism for coactivator versus corepressor function by histone methyltransferase G9a in transcriptional regulation. J Biol Chem. 2011;286:41963–71. doi: 10.1074/jbc.M111.298463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roopra A, Qazi R, Schoenike B, Daley TJ, Morrison JF. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol Cell. 2004;14:727–38. doi: 10.1016/j.molcel.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Schroeder TM, Jensen ED, Westendorf JJ. Runx2: a master organizer of gene transcription in developing and maturing osteoblasts. Birth Defects Res C Embryo Today. 2005;75:213–25. doi: 10.1002/bdrc.20043. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, Shinkai Y. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–91. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taichman RS, Cooper C, Keller ET, Pienta KJ, Taichman NS, McCauley LK. Use of the stromal cell-derived factor-1/CXCR4 pathway in prostate cancer metastasis to bone. Cancer Res. 2002;62:1832–7. [PubMed] [Google Scholar]
- Westendorf JJ. Transcriptional co-repressors of Runx2. J Cell Biochem. 2006;98:54–64. doi: 10.1002/jcb.20805. [DOI] [PubMed] [Google Scholar]
- Woolf E, Xiao C, Fainaru O, Lotem J, Rosen D, Negreanu V, Bernstein Y, Goldenberg D, Brenner O, Berke G, Levanon D, Groner Y. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci U S A. 2003;100:7731–6. doi: 10.1073/pnas.1232420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HC, Hsieh JT, Gleave ME, Brown NM, Pathak S, Chung LW. Derivation of androgen-independent human LNCaP prostatic cancer cell sublines: role of bone stromal cells. Int J Cancer. 1994;57:406–12. doi: 10.1002/ijc.2910570319. [DOI] [PubMed] [Google Scholar]


