Abstract
Thyroid transcription factor 1 (TTF1), a member of the NK family of transcription factors required for basal forebrain morphogenesis, functions in the postnatal hypothalamus as a transcriptional regulator of genes encoding neuromodulators and hypophysiotrophic peptides. One of these peptides is gonadotropin-releasing hormone (GnRH). Here we show that Ttf1 mRNA abundance vary in a diurnal and melatonin-dependent fashion in the preoptic area (POA) of the rat, with maximal Ttf1 expression attained during the dark phase of the light/dark cycle, preceding the nocturnal peak in GnRH mRNA content. GnRH promoter activity oscillates in a circadian manner in GT1-7 cells, and this pattern is enhanced by TTF1 and blunted by siRNA-mediated Ttf1 gene silencing. TTF1 trans-activates GnRH transcription by binding to two sites in the GnRH promoter. Rat GnRH neurons in situ contain key proteins components of the positive (BMAL1, CLOCK) and negative (PER1) limbs of the circadian oscillator, and these proteins repress Ttf1 promoter activity in vitro. In contrast, Ttf1 transcription is activated by CRY1, a clock component required for circadian rhythmicity. In turn, TTF1 represses transcription of Rev-erbα, a heme receptor that controls circadian transcription within the positive limb of the circadian oscillator. These findings suggest that TTF1 is a component of the molecular machinery controlling circadian oscillations in GnRH gene transcription.
Keywords: circadian rhythm, GnRH gene expression, clock genes, TTF1, transcriptional control
Introduction
Thyroid transcription factor 1 (TTF1), also known as Nkx 2.1 and T/ebp (1,2), is a member of the Nkx family of homeobox genes (3,4) that is expressed in the embryonic basal forebrain in a temporally restricted manner. In the absence of TTF1, development of the hypothalamus (as well as other structures of the basal forebrain) is severely impaired (1,5,6).
Notwithstanding the critical importance of TTF1 in basal forebrain embryogenesis, selected postmitotic neurons of this region continue to express the Ttf1 gene indicating that TTF1 not only provides morphogenetic signals to the developing forebrain, but also contributes to regulating mature neuronal functions (2,6,7). After birth, TTF1 can be detected in gonadotropin-releasing hormone (GnRH) neurons of the preoptic area (POA), preproenkephalinergic neurons of the lateral ventromedial nucleus (8,9), kisspeptin neurons of the arcuate nucleus (10), and ependymoglial cells of the third ventricle and median eminence (8,9). In female rats, Ttf1 mRNA abundance increases in the hypothalamus before the natural onset of puberty, and after puberty-inducing hypothalamic lesions, suggesting an involvement of TTF1 in the control of female puberty (8). Using immortalized GnRH producing GT1-7 cells, TTF1 has been show to regulate genes involved in neuronal morphogenesis and differentiation, as well as genes that participate in the neuroendocrine control of reproductive development (11).
An intriguing feature of the GnRH neurosecretory system is the circadian/diurnal periodicity of GnRH release [reviewed in (12,13)]. Such an oscillatory behavior has been long recognized as a critical component of the process by which the GnRH neuronal network is activated at puberty (14–16) and during the estrous cycle (13). It is also clear that the diurnal changes in GnRH neuronal activity are driven by a hypothalamic pacemaker residing in the suprachiasmatic nucleus (SCN) (17,18). At puberty, GnRH secretion begins to increase in the evening (rodents) or at night (primates, including humans), before comparable increases become apparent during the day (14–16). During the rodent estrous cycle, the preovulatory surge of gonadotrophins, driven by changes in GnRH secretion, only occurs in the evening (12,19). Although it is well established that this rhythm is, to a significant extent, determined by a circadian oscillator located in the suprachiasmatic nucleus (13,20), recent studies have unveiled the presence of a clock mechanism intrinsic to GnRH neurons (21–23).
Whether transcription factors regulating GnRH expression, such as TTF1, do so via mechanisms coupled to or interacting with the circadian clock is not known. Suggesting that this may be the case are the observations that Ttf1 expression changes diurnally in the rat medial basal hypothalamus (24) and that TTF1 regulates transcription of another hypothalamic peptide, the pituitary adenylate cyclase-activating polypeptide (PACAP), in a circadian fashion (24). It is thus plausible that the trans-activating effect of TTF1 on GnRH promoter activity has a temporal dimension entrained by the circadian clock. The studies we report here support this concept.
Materials and Methods
Animals and tissue preparation
Two-month-old male Sprague-Dawley rats (Daehan Animal Breeding Company, Chungwon, Chungbuk, South Korea) were housed in a room with a conditioned photoperiod (12 h light/12 h darkness, lights on from 0700 – 1900 h) and temperature (23 −25 °C), and allowed ad libitum access to tap water and pelleted rat chow. To determine daily rhythm of mRNA levels, the animals were acclimated by exposing them for a week to dim red light during the night time. Animals were sacrificed by decapitation at 3 h intervals during the day (0900 h, 1200 h, 1500 h and 1800 h) and night (2100 h, 2400 h, 0300 h and 0600 h). The night experiments were performed under dim red light. Following dissection of the preoptic area (POA) region, the tissues were quickly frozen on dry ice, and stored at −85 °C until RNA isolation. The POA fragments collected consisted of the entire suprachiasmatic region, including the POA proper and the suprachiasmatic nuclei (SCN).
Pinealectomy and administration of melatonin
To remove the pineal gland, the animals were placed in a stereotaxic frame, and the gland was excised with a fine forceps via a 4-mm-wide orifice drilled at the level of the skull’s lambda landmark. After surgery, the orifice was covered with dental cement (Stoelting, Wood Dale, IL). Sham-operated animals underwent an identical procedure, but without removing the gland. Two weeks after the operation, some animals were subcutaneously injected with melatonin (25 mg/kg body weight, Sigma-Aldrich. ST. Louis, MO) at 1800 h and sacrificed 6 h later under dim red light. Following dissection of the POA, the tissues were quickly frozen on dry ice, and stored at −85°C until RNA isolation.
RNA extraction
Total RNA was isolated using Trizol reagent (Sigma-Aldrich, St Louis, MO) as previously reported (24,25).
RNase protection assay (RPA)
RPA Probes: A 331-bp BamHI-ApaI cDNA fragment corresponding to the 5’ end of the Ttf1 coding region was excised from a plasmid (kindly provided by R. DiLauro, Stazione Zoologica A. Dohrn, Naples, Italy) containing the entire Ttf1 coding region (26). The 5’ cDNA fragment was cloned into the BamHI-ApaI sites of the riboprobe vector pGEM-11Z(+) and used for transcription of both sense and antisense RNA strands labelled with 32P-UTP. The GnRH probe used for RPAs was described previously (27). The template cDNA (227 bp size) used to transcribe a MT1 cRNA was cloned by reverse transcription-PCR of total RNA from rat POA using primers designed according to the rat MT1 mRNA sequence (NM_053676). The primers used were: sense, 5’-AAC CTG CTG GTC ATC CTG TCT G-3’; antisense, 5’-CCC GGT GAT GTT GAA TAC CGA G-3’. The PCR product obtained was cloned into pGEM-T vector (Promega, Madison, WI). Antisense and sense RNA probes were generated by T7 and SP6 polymerase-directed transcription, respectively. The cyclophilin probe used was described previously (8,27,28).
The RPA procedure employed has been described in detail elsewhere (8,28). Briefly, sense and antisense 32P-UTP-labeled rat Ttf1, MT1 and GnRH RNA probes were generated and purified using a Fullengther apparatus (Biokey Co., Portland, OR), as reported (8,28). RNA samples (5 µg/tube) or increasing amounts of in vitro synthesized Ttf1, MT1 and GnRH sense mRNA (0.03–2 pg/tube, used to generate standard curves) were hybridized to 500,000 cpm of Ttf1, MT1 and GnRH cRNA probes, respectively, for 18–20 h at 45 °C. The tissue RNA samples were simultaneously hybridized to a cyclophilin cRNA probe (5,000 cpm/tube) to correct for procedural losses (8). At the end of the hybridization, the samples were treated with ribonucleases A and T1 to remove unhybridized RNA species. The protected cRNA fragments were separated on a polyacrylamide-urea gel (5% acrylamide, 7 M urea), and the hybridization signals were detected by exposure of the dried gels to X-Omat X-ray film (Amersham Biosciences, Uppsala, Sweden). The intensity of the signals obtained was quantified as reported (28), using the NIH Image J program.
Electrophoretic mobility shift assays (EMSA)
These assays were carried out using a purified TTF1 homeodomain (TTF1-HD) protein. Procedures for expression, purification, and quantitation of the TTF1-HD were described elsewhere (29). Double-stranded oligodeoxynucleotides, 5’-labeled with λ32P-ATP were the probes used for gel-retardation assays. Their sequences were: GnRH enhancer site 1: 5’-ACA-AAT-TGG-GTT-TAA-GTA-ATT-GGG-CTT-3’, GnRH promoter site 2: 5’- GAT-TTT-AAT-GAC-CAA-GTT-TAA-GAA-AAT-3’. Binding assays (30 min incubation) employed different concentrations (15–150 nM) of TTF1-HD protein and 20,000 cpm of DNA probe in a buffer containing 20 mM, Tris-HCl pH 7.6, 75 mM KCl, 0.25 mg/ml bovine serum albumin (BSA), 5mM dithiothreitol (DTT), 12.5 µg/ml calf thymus DNA, and 10% glycerol. Protein-DNA complexes were separated from unbound DNA on native 7.5% polyacrylamide gels using 0.5× TBE as the running buffer (1.5 h at 4 °C). Gels were fixed and subjected to phosphoimaging analysis using a Bio-Rad GS525 instrument. Protein-DNA complex signals were quantitated using Multi-analyst software (Bio-Rad Laboratories, Hercules, CA). Binding of TTF1-HD to oligodeoxynucleotides derived from the GnRH promoters was expressed as percentage of the TTF1 bound to oligonucleotide C, which contains the core binding motif and flanking sequence of the proximal TTF1 binding site in the thyroglobulin promoter (30).
Cell culture
GT1-7 cells were grown in high glucose DMEM (4.5 mg/ml glucose, Hyclone Laboratories, Inc., Logan UT) supplemented with 10% fetal bovine serum (FBS; Hyclone Laboratories, Inc., Logan UT), and 100 U/ml penicillin/streptomycin (Thermo Fisher Scientific, Inc., Pittsburgh, PA), and maintained at 37°C in 5% CO2. Cells were seeded in 6 well plates (500,000 cells/well) and synchronised by incubation in a serum-free medium for 3 h followed by incubation in DMEM containing 10% FBS (T=00h). Cells were then harvested for RNA extraction or used in promoter assays.
NIH3T3 cells (passages 5 to 10, ATCC, Manassas, VA) were grown in Dulbecco’s modified Eagle medium (DMEM, Sigma) supplemented with 10% fetal bovine serum (FBS, Hyclone), 100 µg/ml penicillin, and 100 µg/ml streptomycin. For promoter assays, the cells were seeded in 12 well plates (200,000 cells/well), and grown in 10% FBS DMEM without antibiotics.
Quantitative (q) PCR
GT1-7 cells were cultured as detailed above. After synchronisation, the cells were harvested at 4h intervals for 48 h and total RNA was isolated. Following reverse transcription (50 ng of total RNA in a 15 µl volume) using a Reverse Transcription System (Promega, Madison, WI), segments of Ttf1, MT2 and GnRH mRNA were amplified by real-time PCR using 10 µl of the reverse transcription reaction and the following primer sets: Ttf1 sense, 5’-CGA GCG GCA TGA ATA TGA GT-3’; Antisense, 5’-GCT GTC CTG CTG CAG TTG TT-3’; GnRH sense, 5’-ACT GTG TGT TTG GAA GGC TGC-3’; antisense, 5’-TTC CAG AGC TCC TCG CAG ATC-3’; Mt2 sense, 5’-AGG AAG GCA AAG GCT GAG AG-3’; antisense, 5’-TGG CGA AAA CCA CAA ACA CT-3’; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) sense, 5’-TGT GAA CGG ATT TGG CCG TA-3’; antisense, 5’-ACT TGC CGT GGG TAG AGT CA-3’. Real-time PCR using the QuantiTect SYBR Green PCR Kit (Promega), was carried out in capillaries of a DNA Engine Opticon Continuous Fluorescence Detection System (MJ Research Inc., Waltham, MA) for approximately 40 cycles using the following conditions: at 94°C for 30 sec, 56°C for 30 sec and 72°C for 35 sec.
Secreted alkaline phosphatase (SEAP) assay
To continuously monitor GnRH promoter activity under the influence of TTF1, we used an SEAP assay system. The rat GnRH promoter (−3002 to +88) conjugated with a SEAP reporter sequence (pGnRH-SEAP) was subcloned into a SEAP vector (pSEAP2-Basic, Clontech, Mountain View, CA) after excising it from the construct pGnRH3.0Luc (provided by J.L. Roberts, University of Texas Health Science Center at San Antonio, USA) using XhoI and HindIII restriction enzymes. Twenty-four hours after seeding in 10% FBS DMEM without antibiotics, GT1-7 cells were transiently transfected with pGnRH-SEAP vectors together with TTF1-pcDNA (24) or Tttf1-shRNA vectors (31) using Lipofectamine as the transfection reagent (Invitrogen, Carlsbad, CA). After synchronisation, the medium was collected at 4 h intervals for 48 h. Chemiluminescence detection of SEAP activity was performed as previously reported (24).
Western blots
TTF1 protein was detected in GT1-7 cells by western blot analysis, as previously reported (24). Briefly, GT1-7 cells were transfected with pGnRH-SEAP vectors together with TTF1-pcDNA (32) or Ttf1-shRNA vectors (31) as described above and were harvested for western blot experiment 24 hour after transfection. Thirty ug of protein were loaded on a 10% SDS polyacrylamide gel under denaturing conditions (samples containing a final concentration of 5 % β-mercaptoethanol); after electrophoresis the size-fractionated proteins were transferred to a polyvinylidene difluoride membrane (PVDF, Millipore, Billerica, MA, USA). The membrane was blocked in 5% skim milk and incubated overnight with monoclonal mouse anti-TTF1 antibody (1:1,000, Thermo Fisher Scientific Inc.) or with monoclonal mouse anti-β-actin antibody (1:4,000, Sigma-Aldrich). The membranes were incubated for 2 h with goat anti-mouse IgG-HRP antibodies (1:4,000, Santa Cruz Biotech. Inc., Santa Cruz, CA). The intensity of the signals obtained was quantified using the NIH Image J program.
Site-directed mutagenesis of the GnRH promoter and transcriptional assay
Two putative TTF1-binding sites containing the core recognition site of TTF1 (26) were identified in the promoter region of the mGnRH gene using the Transcription Element Search System (TESS; http://www.cbil.upenn.edu/bin/tess/tess). The first one was identified in the enhancer region (−1622 to −1617, 5’-TTAAGT-3’) and the second one in the proximal promoter region (−101 to −96, 5’-CCAAGT-3’). These putative binding sites were deleted by site-directed mutagenesis using the The QuickChange® XL Site-Directed Mutagenesis Kit (Stratagene, San Diego, CA) and the previously characterized pGL3-LHRHp construct (8), termed here pGL3-GnRHp. As previously described (8,33), GT1-7 cells were plated in 10% FBS DMEM without antibiotics, as reported above, and cells were transiently transfected with the Lipofectamine/PLUS reagent (Invitrogen) following the manufacturer’s instruction. Briefly, 1.5 µl Lipofectamine reagent (Invitrogen, Carlsbad, CA) per µg DNA were premixed for 15 min with the intact or mutated luciferase reporter plasmid pGL3-GnRHp (34), in conjunction with various amounts of pcDNA3.1-Zeo-rTTF1 added to the Plus Reagent (Invitrogen) at a ratio of 1.5 µl/ug DNA. The resulting mixture was incubated for an additional 10 min at RT before it was added to the cells. The transfection medium was replaced after 5 h by fresh 10% FBS DMEM without antibiotics. Transfection efficiency was normalized by cotransfection with the control plasmid pCMV•Sport-βgal (20 ng/well, Invitrogen). The DNA amount per well was kept at 250 ng using an empty pcDNA3.1-Zeo plasmid as the equalizer. Cell lysates were assayed for luciferase and β-gal 48 h after transfection (8,33).
TTF1/Clock gene promoter assays
Twenty-four hours after seeding NIH 3T3 cells, 1.5 µl Lipofectamine reagent (Invitrogen, Carlsbad, CA) per µg DNA were premixed for 15 min with the various reporter gene constructs in Opti-MEM medium (Invitrogen) and added to a mixture of Opti-MEM/Plus Reagent (Invitrogen) at a ratio of 1.5 µl/ug DNA. The resulting mixture was incubated for an additional 10 min at RT before it was added to the cells. Transfection efficiency was normalized by co-transfecting cells with 100 ng/well of a TK-β-galactosidase construct (provided by BS Katzenellebogen, University of Illinois at Urbana-Champaign). The total amount of DNA transfected was kept constant at 1.1 µg/well (Experiment 1) or 1.2 µg/well (Experiment 2) by adding the appropriate amount of empty pcDNA3.1 plasmid to each well. The transfection medium was replaced after 5 h by fresh 10% FBS DMEM without antibiotics. The cells were harvested 48 h after transfection and assayed for luciferase and β-galactosidase, as reported (8,35). Each condition was tested using at least 3 wells/experiment, and each experiment was repeated 2–3 times. Promoter assays using GT1-7 cells were performed as described in the previous section.
To generate a plasmid expressing luciferase under the control of the Bmal1 promoter, we subcloned the mBmal1 promoter from the pd2EGFP-mBmal1prom1 construct (plasmid expressing eGFP under the mBmal1 promoter, provided by N. Cermakian, McGill University, Montreal, Canada) into pGL2 Basic (Promega). Briefly, we excised the promoter from pd2EGFP with BglII/XmaI restriction enzymes (New England Biolabs, Ipswich, MA).The 3’ ends were blunted using the large Klenow fragment of DNA polymerase I (New England Biolabs), and the DNA fragment was subcloned into pGL2 Basic linearized at the SmaI restriction site using the Stratagene DNA ligation kit (Agilent Technologies, Santa Clara, CA). The correct promoter orientation into this vector was verified by restriction enzyme digestion (BamHI, Promega) before amplification of the plasmid for transfection assays. All plasmid DNA preparations were purified using the Qiagen EndoFree Plasmid Maxi kit (Qiagen, Valencia, CA).
In one series of experiments, the long (5kb) form of the Ttf1 rat promoter (500ng/well) inserted in the pSVoa-delta luciferase plasmid (36) (provided by S. Kimura, Laboratory of Metabolism, National Cancer Institute, NIH, Bethesda, MD) was co-transfected with 250ng/well of the following expression vectors: mBMAL1-pcDNA3.1 and mCLOCK-pcDNA3.1 (provided by S. Honma, Hokkaido University Graduate School of Medicine, Sapporo, Japan), mREV-ERBα-pcDNA3 (provided by P. Gos, University of Geneva), mPer1-pcDNAV5 and mPer2-pcDNAV5 (provided by H. Okamura, Kyoto University, Kyoto, Japan) and mCry1V5-pcDNA (provided by S. Reppert, University of Massachusetts Medical School, Boston, MA).
In a second series of experiments, increasing concentrations (100, 250, 500 and 1000ng/well) of TTF1-pcDNA (8) were co-transfected with 100ng of the following promoter constructs: mBmal1-pGL2 basic and hREV-ERBα-pGL2 basic (provided by V. Laudet, University of Lyon, France), mCry1-pGL3 basic and mPer1-pGL3 basic (from S. Reppert, University of Massachusetts Medical School, Boston, MA). Because REV-ERBα has been shown to repress both the Bmal1 promoter (37) and its own promoter (38), we used REV-ERBα (mREV-ERBα-pcDNA3; 250ng/well) as a positive control in experiments involving the effect of TTF1 on the activity of these promoters (mBmal pGL2 basic and hREV-ERBα-pGL2 basic constructs). BMAL1 (mBMAL1-pcDNA3.1) and CLOCK (mCLOCK-pcDNA3.1), previously shown to trans-activate Cry1 and Per1 transcription (39–41) were used (at 250ng each/well) as a positive control when examining the effect of TTF1 on these promoters (mCry1-pGL3 basic and mPer1-pGL3basic constructs).
Immunohistofluorescence detection of clock gene products in GnRH neurons
We stained GnRH neurons with mouse monoclonal antibody 4H3 (42), at a 1:3,000 dilution. CLOCK was detected with rabbit polyclonal antibodies (1:2,000; Millipore, Billerica, MA); PER1 also with rabbit polyclonal antibodies (1:2,000; Millipore), and BMAL1 with guinea pig polyclonal antibodies (1:4,000; Millipore). Prepubertal, 28 day-old female rats were perfused via the left ventricle of the heart between 10:00 and 13:00 h with 4% paraformaldehyde-PBS, pH 7.4. Frozen sections (30 µm) of their brains were prepared using a freezing sliding microtome, mounted on Superfrost glass slides and dried for 2 h under an air stream before staining. The sections were incubated for 48 h at 4 °C with the primary antibodies indicated above. At the end of this period, GnRH immunostaining was developed to a red color using Alexa 568-donkey antimouse IgG (1:500; Invitrogen). CLOCK and PER1 staining was developed to a green color by incubating the sections with biotinylated donkey antirabbit gamma globulin (1:250, Invitrogen), followed by Streptavidin-Alexa 488 (1:500, Invitrogen). BMAL1 staining was also developed to a green color by incubating with biotinylated goat antiguinea pig gamma globulin (1:250, Invitrogen), followed by Streptavidin-Alexa 488. Fluorescent images were acquired as reported (43,44), and images derived from a single 0.5 µm optical section were selected for presentation. Specificity of the primary antibodies used has been reported earlier (42,45,46). In addition, for every reaction we included sections incubated without the first antibodies. All negative controls omitting the BMAL1, CLOCK and PER1 antibodies showed only GnRH neurons. The converse was also true, i. e, omitting the GnRH antibodies resulted in the visualization of only BMAL1, CLOCK and PER1 positive cells. We used 2% normal donkey serum for blocking, which in our experience results in minimum background staining.
Statistical analysis
Data were analyzed using SigmaStat sofware (version 3.11; Systat Software Inc., San Jose, CA). Data passing a normality test were analyzed using the two-tailed Student’s t test to compare two groups or a one-way ANOVA followed by the Student-Newman-Keuls multiple comparison test to compare several groups. When the data failed the normality test, they were analysed using an ANOVA test on ranks followed by the Kruskal-Wallis one-way ANOVA ranks test. A P value of <0.05 was considered statistically significant.
Results
Ttf1 and GnRH mRNA levels vary in a diurnal fashion in the rat POA during the normal day-night cycle
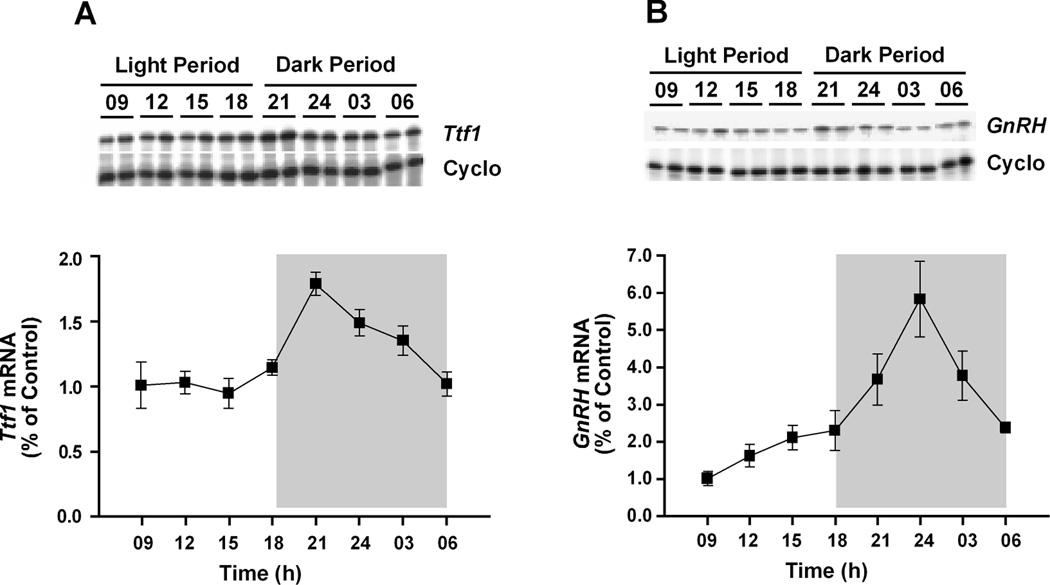
Diurnal changes in Ttf1 and GnRH mRNA levels were determined by RPA in the POA of 2-month-old male rats. Both Ttf1 and GnRH mRNA abundance varied in a diurnal manner (Fig. 1). Ttf1 mRNA levels were relatively constant during day time, but increased sharply after the lights were turned off at 1900 h, peaking at 2100 h. Levels decreased steadily thereafter, reaching day time values by 0600 h (Fig. 1A). GnRH mRNA levels showed a similar pattern of expression, with peak levels occurring at midnight, i.e., 3 h after the peak in Ttf1 mRNA levels (Fig. 1B). Levels decreased between midnight and 0600 h, but without reaching day time values until later in the day. By 0600 h (1 h before the lights are turned on), GnRH mRNA levels were about 2.5-fold higher than at 0900 h. These results indicate that expression of the Ttf1 and GnRH genes in the POA oscillates in a diurnal fashion, and that the changes in Ttf1 mRNA expression precede those in GnRH mRNA levels.
Figure 1. Daily rhythms in Ttf1 and GnRH mRNA levels in the rat POA as assessed by RPA.
A, Diurnal changes in Ttf1 mRNA abundance; A representative autoradiogram illustrating these changes is shown on top of a graph depicting changes in mean Ttf1 mRNA levels (n = 8/time point). B, Diurnal changes in GnRH mRNA abundance (n = 6/time point). The shaded areas denote the dark period of the day. Each value represents the mean ± SEM for each time period and is calculated using individual values normalized according to the cyclophillin (Cyclo) mRNA content detected in each sample. Values at 0900 h are used as the reference point to express subsequent fluctuations in mRNA levels.
Pinealectomy prevents, and melatonin restores, the diurnal changes of Ttf1 and GnRH mRNA abundance in the POA
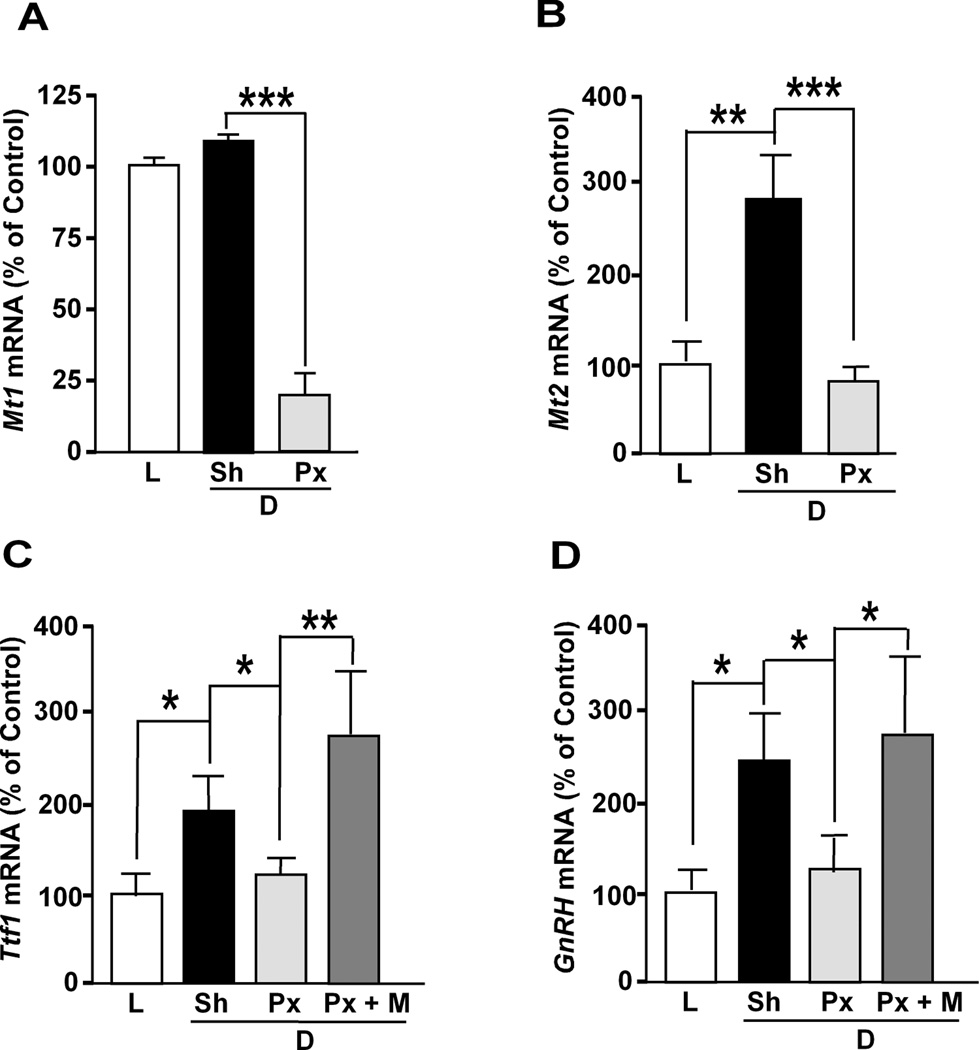
One major extracellular input to the neuroendocrine hypothalamus directed by the day-night cycle is melatonin released from pineal gland (47). Melatonin release increases during the night and induces a nocturnal increase of hypothalamic GnRH expression in rats (48,49). This effect is in all likelihood mediated by the high-affinity melatonin receptors MT1 and MT2, as these receptors are not only expressed in the rat hypothalamus as a whole (50), but also in GnRH neurons in situ (MT1) (50) and GT1-7 cells in vitro (MT1 and MT2) (51). Based on these observations, we reasoned that the nocturnal increase of Ttf1 expression in the POA may also be a melatonin-dependent phenomenon. Mt1 mRNA abundance in the POA was not significantly different between the day (1200 h, L) and night (2400 h, D), but 2 weeks after Px, the mRNA levels (measured at midnight) were strikingly decreased (Fig. 2A). This result is consistent with previous findings showing that Mt1 mRNA content decreases in both the SCN and pars tuberalis of PX rats (52). In contrast to Mt1 mRNA, Mt2 mRNA levels were significantly increased during the night (Fig. 2B), and this increase was obliterated by Px (Fig. 2B). Though expression of Mt2 in rat GnRH neurons in vivo is uncertain (50), the nocturnal increase in Mt2 mRNA abundance we observed is strikingly similar to that of Ttf1 and GnRH mRNA levels, suggesting that the MT2 receptor may also be involved in the diurnal regulation of Ttf1 and GnRH expression, perhaps indirectly through associated neuronal systems.
Figure 2. Effect of pinealectomy and melatonin on the diurnal change of Ttf1 and GnRH mRNA levels in the rat POA.
Two-month old male rats were pinealectomized (Px) or sham-operated (Sh) and kept under a normal light and dark cycle (12 h light/12 h dark, lights on 0700 – 1900 h) for 2 weeks. Then, groups of animals were euthanized at either 1200 h or 2400 h to determine effect of light (L) and dark (D) on Mt1 mRNA with RNase protection assays and Mt2, Ttf1 and GnRH mRNA levels by using qPCR. Some animals were subcutaneoulsy injected with melatonin (M) 6 h before sacrifice. A, Mt1 mRNA abundance is slightly greater at midnight (Sh/D) than at noon (L), and decreased markedly after Px, in animals sacrificed at 2400 h (Px/D). B, Mt2 mRNA content significantly increases at midnight (Sh/D) as compared to noon (L), and decreases after Px (Px/D). C, Ttf1 mRNA abundance increases significantly at midnight (Sh/D) as compared to noon (L), and Px obliterates this increase (Px/D); in contrast, melatonin increases Ttf1 mRNA abundance (Px+M/D). D, GnRH mRNA content changes in a similar fashion. Bars are mean ± SEM (n=4–6/group). One-way ANOVA followed by Student-Newman-Keuls multiple comparison test * = P<0.05, ** = P<0.01, *** = P<0.001.
As before (Fig. 1A), Ttf1 mRNA levels in the POA showed a diurnal variation (Fig. 2C), with low values at 1200 h (L) and high values at 2400 h (D); this diurnal fluctuation was significantly attenuated in PX rats (D/PX, Fig. 2C), and enhanced by a single dose of melatonin (25 mg/Kg BW) given 6 h earlier (at 18:00 h). In agreement with earlier observations (49), the diurnal fluctuation in GnRH mRNA levels was also prevented by PX, and restored by melatonin (Fig. 2D), as previously shown by others (48,49). Our results indicate that, as is the case of GnRH mRNA, the nocturnal increase of Ttf1 mRNA abundance in the POA appears to be due, to a significant extent, to melatonin released from the pineal gland.
Circadian oscillations of GnRH promoter activity are facilitated by TTF1
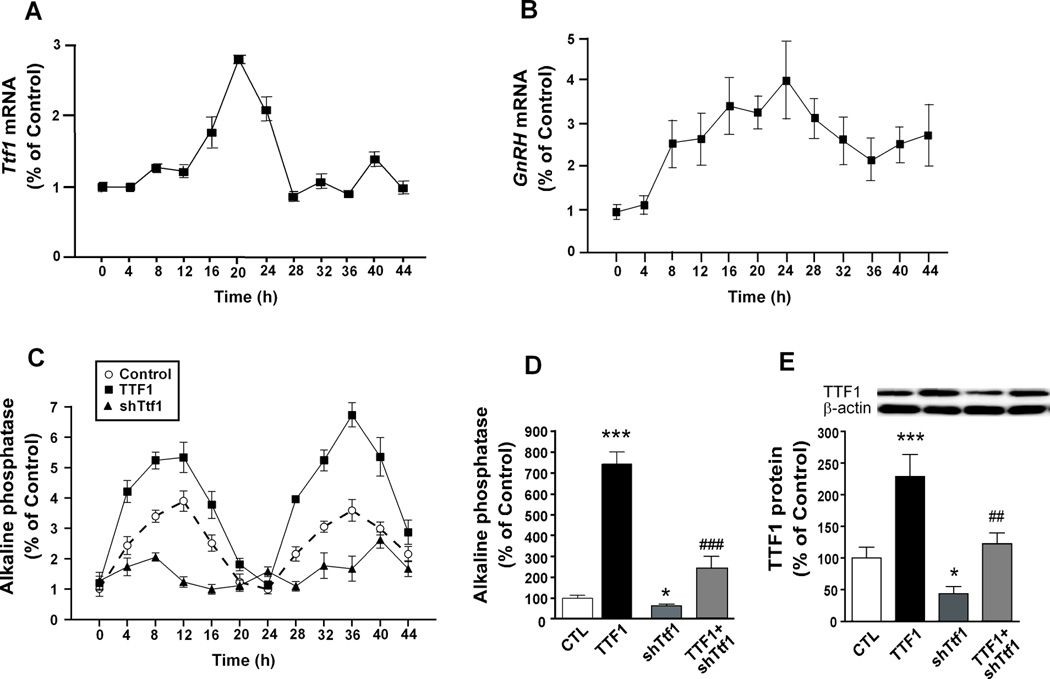
To study the effect of TTF1 on the circadian activity of the GnRH promoter in vitro, we used GT1-7 cells, as they were previously found to express GnRH mRNA in a circadian fashion (23). We first determined if GT1-7 cells also display an endogenously circadian rhythm of Ttf1 mRNA expression. We collected the cells at 4 h intervals for 48 h after synchronising them by serum deprivation, and measured their Ttf1 and GnRH mRNA content. The results showed that, consistent with the in vivo oscillation in expression (Fig. 1A),Ttf1 mRNA levels vary in a circadian fashion in GT1-7 cells during the first 24 h after synchronization (Fig. 3A), with peak values being observed about 4 h before the circadian peak in GnRH mRNA values (Fig. 3B). Neither circadian oscillation was sustained during the second 24 h period examined after synchronisation.
Figure 3. Ttf1 mRNA abundance varies in a circadian fashion in GT1-7 cells and TTF1 enhances the circadian increase in GnRH promoter activity as assessed by a SEAP reporter assay.
A, Ttf1 mRNA levels measured by real-time PCR fluctuate in a circadian manner in GT1-7 cells after synchronisation of the cells with serum-free medium. B, GnRH mRNA content also varies in a circadian manner in GT1-7 cells. C, Amplification of the diurnal changes in GnRH promoter activity by TTF1 and blunting of these changes by shRNA-mediated Ttf1 knock-down. An SEAP reporter vector containing a 3 kb-long GnRH promoter was cotransfected (500 ng/well) into GT1-7 cells along with a TTF1 expression vector (TTF1, 500 ng) or a Ttf1 shRNA vector (shTtf1, 500 ng). Starting 24 h after transfection, the medium was collected for SEAP assay at 4 h intervals for 48 h. Each point represents the mean ± SEM (n=6 for control and TTF1; n=4 for shTtf1). D, Quantification and statistical analysis of the peak values achieved by each group shown in C; bars are mean ± SEM. E, Changes in the content of TTF1 protein after transfecting GT1-7 cells with either the TTF1 expression vector or shTtf1, or both, as determined by Western blotting. β-actin was used as a normalizing control. One-way ANOVA followed by Student-Newman-Keuls multiple comparison test * = P<0.05, *** = P<0.001 vs. control; ## = P<0.01, ### = P<0.001 vs. TTF1.
We subsequently set out to determine if the diurnal changes of GnRH expression observed in vivo in the rat POA depend on circadian variations in TTF1 levels. To answer this question, we monitored changes of GnRH promoter activity for 2 days in cultured GT1-7 cells that were transfected with a construct in which the coding region of a secreted alkaline phosphatase gene is controlled by the GnRH promoter (pGnRH-SEAP). TTF1 was overexpressed by co-transfecting the cells with a TTF1 expression vector; endogenous TTF1 production was reduced by RNA interference using a Ttf1 siRNA previously described (31). Under the culture conditions employed, GnRH promoter activity exhibited an autonomous 24 h rhythm (Fig. 3C). Increasing the cellular content of TTF1 enhanced the peak amplitude of GnRH promoter rhythmic activity, without affecting nadir values of activity. In contrast, shRNA-mediated Ttf1 knock-down blunted these fluctuations (Fig. 3C). These data suggest that TTF1 facilitates the circadian rhythm of GnRH transcription, by increasing the amplitude but not the frequency of the rhythm.
In addition to the increased peak amplitude, and as previously observed with the PACAP promoter (24), total GnRH promoter activity was also enhanced by TTF1 (Fig. 3D). Conversely, knocking-down TTF1 expression blunted both basal GnRH promoter activity and the TTF1-induced peak amplitude (Fig. 3D). Measurement of TTF1 protein revealed that, as expected, cells transfected with the TTF1 expression vector had increased TTF1 protein levels; Ttf1 shRNA reduced the increase in TTF1 protein content seen in cells transfected with the TTF1 expression vector (Fig. 3E). It was also evident that Ttf1 shRNA decreased basal TTF1 levels in cells not transfected with the TTF1 expression vector (Fig. 3E). This finding indicates that, as seen before in rat GnRH neurons in situ (8), GT1-7 cells also express the Ttf1 gene.
TTF1 transactivates the GnRH promoter via specific binding sites
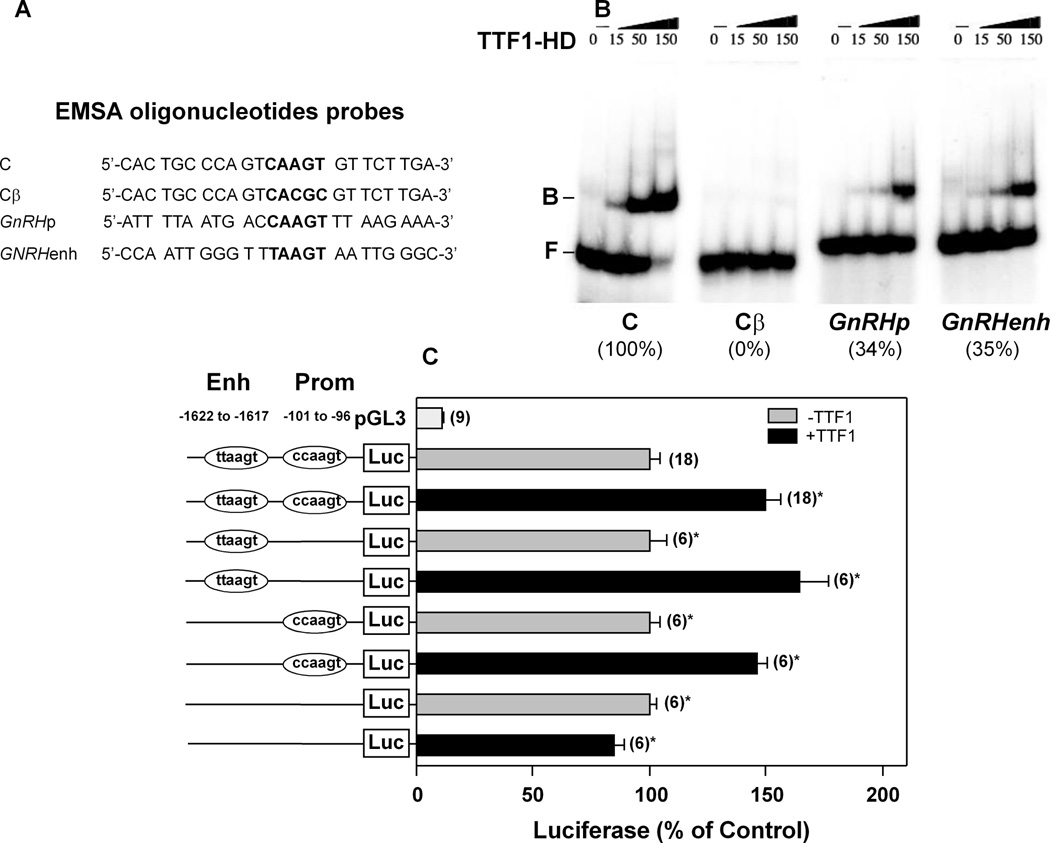
We previously showed that TTF1 increases GnRH promoter activity (8). Because the 5’ flanking region of the mGnRH gene contains sequences that conform the consensus TTF1 binding motif (26), we sought to determine if the purified TTF1 homeodomain (TTF1-HD) could bind to these regions. Computer analysis of the 5’ flanking region of the mGnRH gene using a search program (www.cbil.upenn.edu.tess/) verified the existence of two putative core TTF1 motifs. One of these sites is located within the proximal GnRH promoter (−101 to −96) and the other in the GnRH enhancer region (−1622 to −1617). This latter region targets expression of the GnRH gene to GnRH neurons in the basal forebrain and enhances GnRH promoter activity (53,54).
To determine if TTF1 binds to these sites, we used increasing amounts of the purified TTF1 homedomain (TTF1-HD) protein in EMSAs and 24-mer oligodeoxynucleotide probes containing the putative TTF1-HD binding sites and flanking sequences present in either the enhancer (GnRHenh) or proximal promoter (GnRHp) region of the mGnRH gene (Fig 4A). The oligodeoxynucleotide “C” containing the sequence of a high-affinity TTF1-HD binding site found in the thryroglobulin promoter (30) was used both as a positive control and as a reference to estimate the affinity of TTF1-HD to the TTF1 sites in the GnRH promoter. Cβ is a mutant of the C oligodeoxynucleotide in which the core binding site for TTF1-HD was deleted (30). When comparing the binding affinity of both GnRH probes to that of the C probe (considered as 100%), we found that the GnRHp and GnRHenh probes also bound TTF1-HD, although with less (~35%) affinity (Fig. 4B). As expected, a mutated C probe (Cβ) exhibited no binding (Fig. 4B). Promoter assays using a GnRH promoter construct in which the enhancer region is placed directly upstream from the proximal GnRH promoter (55) demonstrated that TTF1 trans-activates this construct (Fig. 4C). Neither deletion of the distal TTF1 binding site (TTAAGT) nor the proximal site (CCAAGT) affected the ability of TTF1 to transactivate the promoter. However, deletion of both sites obliterated the TTF1 effect (Fig. 4C). These results indicate that TTF1 binds to specific binding sites in both the GnRH enhancer and proximal promoter to stimulate GnRH transcription. Furthermore the ability of TTF1 to enhance circadian GnRH promoter activity, and the circadian pattern of Ttf1 mRNA expression in both the POA and GT1-7 cells, suggests the involvement of clock genes in this relationship.
Figure 4. TTF1-HD binds to putative TTF1 binding sites in the GnRH promoter and trans-activates GnRH transcription.
A, Sequences of the oligodeoxynucleotides used for EMSA. B, Autoradiograms of electrophoretic mobility shift assays (EMSA) demonstrating that increasing concentrations of TTF1-HD (15, 50 and 150 nM) results in increasing TTF1-HD binding to the indicated oligodeoxynucleotides described in A. The relative affinity of each binding site for TTF1-HD is expressed as percentage of the binding intensity of TTF1-HD to the canonical TTF1 recognition site in the positive control thyroglobulin promoter (C, considered as 100%) (30). Cβ, a mutant form in which the core binding site for TTF1-HD was deleted (26), was used as a negative control. F = free DNA; B= protein-bound DNA. C, A pGL-3 plasmid carrying the intact enhancer/promoter regions of the GnRH gene (Enh/Prom) or DNA fragments bearing either 6 bp deletions of TTF1 core binding sites present in each of these regions (Del-GnRH enh, Del GnRH prom) or a double deletion (Del-GnHRH enh/prom) were cotransfected (250 ng/well) into GT1-7 cells with 100 ng/well pcDNA-Zeo-rTTF-1 and the resulting changes in luciferase activity were measured 48 h later. The positions of the deleted TTF-1 binding sites (enh −1622 to −1617; prom −101 to −96) are indicated. Bars represent mean ± SEM and figures above bars are number of wells per group. Student t test, * = P<0.05 vs. groups not treated with TTF1.
GnRH neurons in situ express core components of the circadian clock
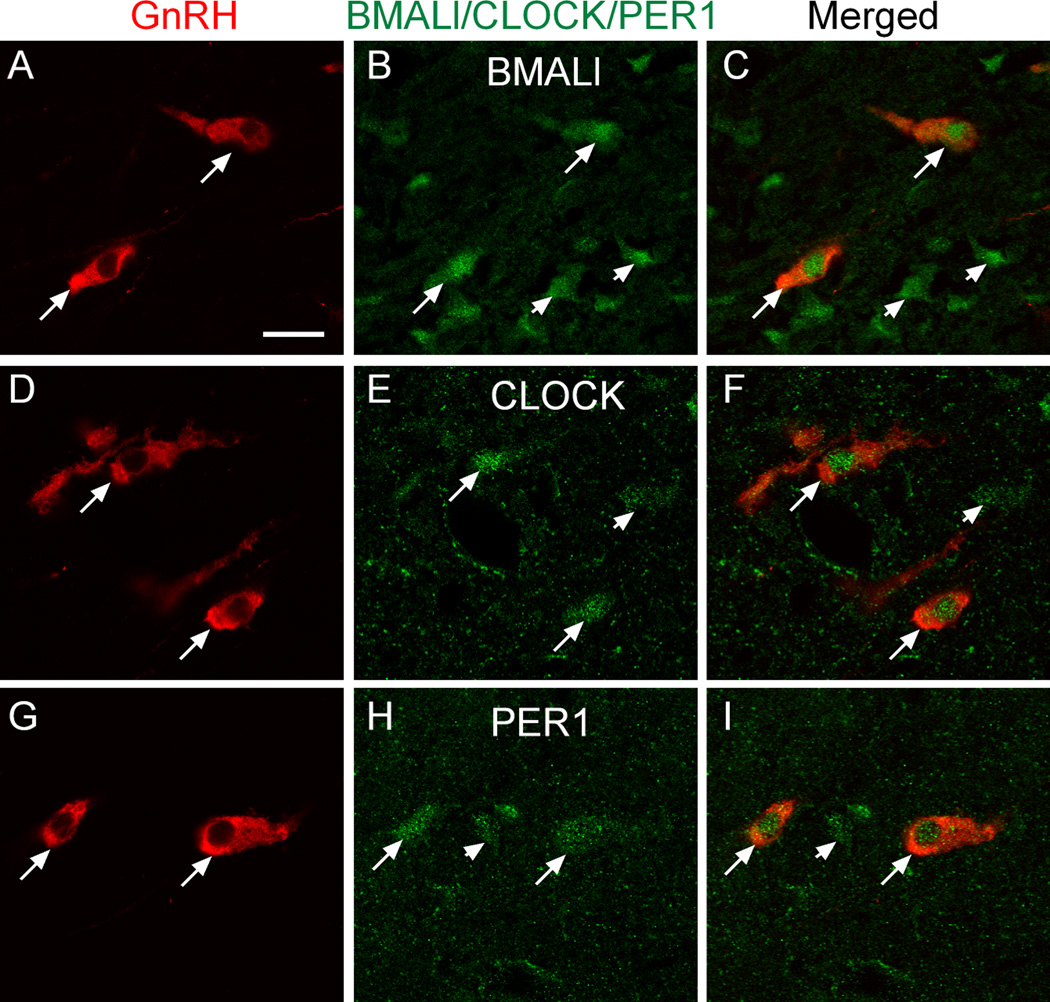
To determine if GnRH neurons in vivo contain core clock proteins, we detected these proteins by double immunofluorescent staining. It is known that GT1-7 cells express core clock proteins (22,23), and that at least three of these proteins (BMAL1, PER1, PER2) can also be detected in mouse GnRH neurons in vivo (21,56). By confocal imaging, we found that GnRH neurons of the rat POA express CLOCK, in addition to BMAL1 and PER1, and that all three of them have a nuclear localization (Fig. 5). Although the content of these clock proteins would be expected to display a diurnal variation, no attempts were made to verify this assumption.
Figure 5. GnRH neurons of the rat POA contain the core clock proteins BMAL1, CLOCK and PER1 as detected by double immunohistofluorescence.
A–C, BMAL1; D–F, CLOCK; G–I, PER1. Arrows point to examples of colocalization. Arrowhead in H points to a PER1 positive cell lacking GnRH immunoreactivity. Notice the nuclear localization of BMAL1, CLOCK and PER1. Bar = 10 µm.
Clock proteins and TTF1 trans-activate the GnRH promoter in a non-synergistic, non-additive manner
The GnRH 5’-flanking region contains several E-boxes (Supplementary Fig.1A), which are recognition sequences typically mediating the trans-activational effects of CLOCK/BMAL1 heterodimers on clock-controlled genes, like Per1, Per2, Cry1 and Cry2 (17,57,58). Promoter assays carried out in GT1-7 cells showed that CLOCK/BMAL1 increase GnRH transcription and that this effect is neither additive nor synergistic to that of TTF1 (Supplementary Fig. 1B). These results suggest that core clock genes and TTF1 trans-activate the enhancer/proximal region of the GnRH promoter along a common pathway.
The Ttf1 promoter is regulated by core clock proteins and TTF1 protein modulates the promoter activity of hREV-ERBα and mPer1 in vitro
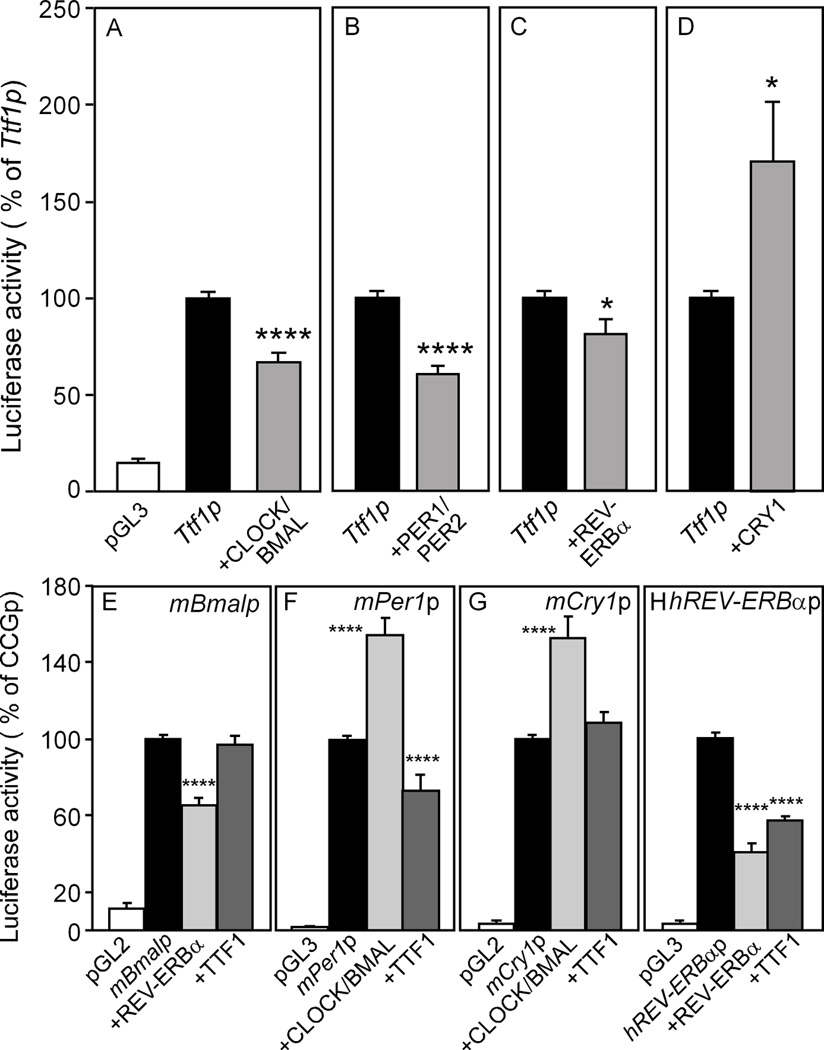
The aforementioned findings and the presence of several E-boxes in the 5’-flanking region of the rat Ttf1 gene (Supplementary Fig. 2) prompted us to investigate the effect of core clock proteins on Ttf1 promoter activity using gene reporter assays in GT1-7 (Supplementary Fig 3A and B) and NIH-3T3 cells (Fig 6 and Supplementary Fig. 3 C–F). Ttf1 promoter activity was slightly reduced by the transient expression of both mBMAL/mCLOCK proteins (Fig 6A, Supplementary Fig 3A) and also by mPER1/mPER2 (Fig 6B). A modest decrease was observed in cells over-expressing the transcriptional repressor REV-ERBα (Fig 6C). In contrast, mCRY1 unambiguously increased Ttf1 promoter activity (Fig 6D). These results suggest that clock genes mainly affect Ttf1 transcription via a CRY1-mediated trans-activational effect.
Figure 6. Effect of clock proteins on Ttf1 promoter activity and effect of TTF1 on the transcription of core clock genes (CCGs) in NHI 3T3 cells.
A–D, mBMAL/mCLOCK, mPER1/mPER2 and mREV-ERBα repress the Ttf1 promoter, but CRY1 increases Ttf1 transcription. The rat Ttf1 promoter was used at 500ng/well. The expression vectors (mBMAL1-pcDNA3.1, mCLOCK-pcDNA3, mREV-ERBα-pcDNA3, mPer1-pcDNAV5, mPer2-pcDNAV5 and mCry1V5-pcDNA) were used at 250ng/well. The total DNA amount/well was kept constant at 1.1 µg/well by adding the appropriate amount of pcDNA3.1 to each well. Results (mean±SEM) are expressed as percentage of Ttf1 promoter-driven luciferase activity alone. T test, Ttf1p vs CCG, *, p<0.05; ****, p<0.0001. n=16/condition (3 different experiments). E–H, TTF1 represses the Per1 and Rev-erbα promoters (F,H), without affecting the transcriptional activity of Bmal1 and Cry1 (E,G). The responsiveness of each CCG gene promoter (CCGp) was verified by co-transfecting plasmids encoding clock proteins known to modify the transcriptional activity of each promoter being tested, i.e. mREV-ERBα for the mBmal and hRev-erbα promoters (E, H) and mBMAL/mCLOCK for the mCry1 and mPer1 promoters (F, G). Results (mean±SEM) are expressed as percentage of each promoter-driven luciferase activity alone. One-way ANOVA, experimental condition vs control (CCGp) **, p<0.01; ***, p<0.001;. n=18–24/condition (4–7 different experiments) except for (F), n=6–12, (2 different experiments). TTF1-pcDNA was used at 500 ng/well; mBmal1-pGL2 basic, hREV-ERBα-pGL2, mCry1-pGL3 and mPer1-pGL3 were used at 100ng/well; mREV-ERBα-pcDNA3, mBMAL1-pcDNA3.1 and mCLOCK-pcDNA3.1 at 250ng/well. Total DNA amount/well was kept constant at 1.2 µg/well by adding the appropriate amount of pcDNA3.1 to each well.
To determine if TTF1 is involved in the transcriptional control of core clock genes, we tested the effect of TTF1 on Bmal1, Per1, Cry1 and REV-ERBα promoter activities in NIH 3T3 cells. TTF1 had no effect on mBmal1 promoter activity (Fig 6E), a minimal repressive effect on mPer1 transcriptional activity (Fig 6F), and no effect on mCry1 promoter activity (Fig. 6G). In contrast, TTF1 was very effective in repressing the REV-ERBα promoter (Fig 6H). These assays were conducted using conditions that allowed us to reproduce known effects of clock proteins on clock gene expression. For instance, REV-ERBα has been reported to repress the Bmal1 promoter (37), and inhibited mBmal1 transcription in our study (Fig.6E). CLOCK/BMAL1 transactivate Per1 (40) and Cry1 transcription (41) and this is the effect we observed (Fig. 6F, G). Lastly, REV-ERBα repressed transcription of its own gene (Fig. 6H), in agreement with results reported by others (38). The repressive effects of TTF1 on Per1 and REV-ERBα were maximal at the concentration of 100 ng, remaining at this level at concentrations as high as 500–1,000 ng (Supplementary Fig. 3D and F).
Discussion
The present study provides evidence for the concept that TTF1, a transcriptional activator of GnRH transcription (8,10), plays a role in facilitating circadian changes in GnRH expression. Ttf1 mRNA abundance in the rat POA area oscillates in a diurnal fashion; peak levels of Ttf1 mRNA, attained during the early night hours, precede the nocturnal peak in GnRH mRNA. The latter rhythm, previously shown by others (21), is confirmed by our observations. Our results show that TTF1 enhances GnRH transcription by binding to at least two recognition sites, one located within the GnRH enhancer region and the other in the proximal GnRH promoter. The fact that both of these regions are required for cell-type specific expression of the GnRH gene (53,55), suggests that TTF1 contributes to controlling circadian GnRH expression in the POA by targeting core regulatory components of the GnRH transcriptional machinery for trans-activation.
The nocturnal changes in both Ttf1 and GnRH expression were obliterated by PX, which also reduced drastically Mt1 and Mt2 receptor mRNA levels in the POA. Conversely, melatonin administration restored the nocturnal increase in both GnRH and Ttf1 mRNA levels. It would appear that this effect is exerted via MT1 receptors, because most actions of melatonin in the CNS are mediated by these receptors (59). Their loss compromises the ability of melatonin to both inhibit SCN neuronal firing (59) and to act as a zeitgeber throughout the brain (47). Previous reports have shown that PX results in decreased MT1 receptor density in the SCN (52) and lowers GnRH mRNA levels in GnRH neurons of the rat POA (49). Other authors have shown that melatonin administration increases GnRH mRNA abundance in the aging rat brain (48). Our findings indicate that melatonin is not only required for the diurnal oscillation in GnRH mRNA levels, but is also essential for the nocturnal increase in Ttf1 expression in the rat POA. These in vivo findings are at odds with in vitro results showing that melatonin decreases GnRH mRNA content in GT1-7 cells (51). The discrepancy may be due to heterogeneity among GT1-7 subcultures as Provenzano et al did not find detectable levels of TTF1 in these cells (11) whereas we did (Fig. 3E). It is also possible that GT1-7 cells behave differently from GnRH neurons in situ or that melatonin requires cellular networks other than GnRH neurons to facilitate in vivo diurnal changes in GnRH expression.
The mechanism(s) underlying this supportive effect of melatonin is unknown. Because MT1 receptors are widely expressed throughout the brain (47), melatonin may support circadian oscillations in TTF1 and GnRH mRNA abundance by not only acting on the SCN pacemaker (59), but also on cellular subsets functionally and anatomically connected to GnRH neurons. Melatonin may also activate MT1 receptors located on GnRH neurons, because MT1 mRNA (but not MT2 mRNA) is expressed in some GnRH neurons in situ (50).
GnRH promoter activity varies in a circadian manner in GT1-7 cells with peak levels attained in mid-subjective night. This pattern of activity closely resembles the in vivo changes in GnRH mRNA abundance, and its amplitude is enhanced by TTF1. It was, however, surprising that neither the Ttf1 nor the GnRH mRNA rhythm persisted beyond the first 24 h after in vitro synchronization. This could be due to the synchronization protocol we used, to the absence of an initial TTF1 peak of sufficient amplitude to sustain the circadian activity of the GnRH promoter, and to the absence of melatonin supplementation to entrain these rhythms. We believe that the most parsimonious explanation resides with the latter two possibilities, because a) Increasing TTF1 levels in GT1-7 cells does result in robust and sustained circadian changes in exogenous GnRH promoter activity (Fig. 3c), and b) In vivo melatonin administration restores the diurnal rhythm in hypothalamic Ttf1 and GnRH expression lost after PX (Fig. 2). A requirement for melatonin to sustain the in vitro circadian rhythms of both Ttf1 and GnRH expression is also suggested by the results of Roy et al (51) who demonstrated that melatonin does entrain a circadian rhythm in GnRH mRNA levels in GT1-7 cells. Because RNAi-mediated suppression of endogenous Ttf1 mRNA levels blunts the circadian oscillations in GnRH promoter activity, it is likely that the ability of TTF1 to trans-activate the GnRH promoter is tied to a clock mechanism that at least in part operates within the GnRH neuronal network. The presence of CLOCK, BMAL1 and PER1 in the nucleus of GnRH neurons in situ indicates that these cells have some of the basic molecular components required for this mechanism to operate in vivo.
TTF1 may amplify the circadian peaks of GnRH promoter activity by modifying the expression and/or activity of clock genes known to regulate GnRH circadian output (22,23). Alternatively, Ttf1 expression itself may be under the control of clock genes. The diurnal pattern of Ttf1 mRNA abundance observed in vivo suggests that Ttf1 expression is regulated by clock proteins. At least part of this regulation is exerted directly at the transcriptional level because: a) The Ttf1 5’flanking region contains eight recognizable E-box sites (Supplementary Fig. 1), and b) in vitro Ttf1 promoter activity is regulated by clock genes.
Ttf1 transcription was modestly inhibited by CLOCK/BMAL1, two protein components of the circadian oscillator’s positive limb (60–62), and also by PER1/PER2, which are components of the negative limb (60–62). It is likely that the CLOCK/BMAL1-mediated Ttf1 transcriptional inhibition is not a direct effect, but is mediated by an increase in PER1/PER2 transcription as both conditions result in similar inhibitory effects. REV-ERBα, a negative regulator of Bmal1 expression that controls the period length of rhythms in gene expression within the positive limb of the circadian oscillator (37) had a negligible inhibitory effect. In contrast, Ttf1 promoter activity was increased by CRY1, a component of the negative feedback loop that inhibits CLOCK/BMAL1-induced transcription (41), but that also enhances Bmal1 expression (62) by preventing ubiquitination of PER2 (63), a positive regulator of Bmal1 expression. According to the basic transcriptional mechanisms regulating core clock networks {7283,7250}, it is expected that overexpressing CLOCK/BMAL1 would produce a synchronous increase in PER1/2 and CRY1 which would subsequently affect Ttf1 transcription in a similar fashion. Although the results we observed are contradictory to these expectations, there are reasonable interpretations that can be entertained. A potential explanation is that overexpression of proteins able to regulate clock controlled genes disrupts the circadian expression of other elements of the clock network, as was reported in the case of CRY1 and REV-ERBα {5331}. It is also possible that the activating effect of CRY1 may be due to direct trans-activation of the Ttf1 promoter. This latter effect is consistent with the reported stimulatory effect of CRY1 on GnRH release (22), the evolutionary conserved direct transactivation of downstream promoters by Cry1 (64,65), and the emerging concept that different components of the mammalian circadian clock can act directly on clock associated genes to directly activate or repress transcription (66).
TTF1 trans-activates GnRH transcription by directly binding to specific recognition sites in the GnRH promoter. In a cellular context this effect is manifested as an increase in the amplitude – but not the period – of the diurnal changes in GnRH promoter activity. This feature suggests that TTF1 increases GnRH transcription by interacting with clock proteins as they bind to E-boxes in the GnRH 5’flanking region (Supplementary Fig. 2), and/or by directly modulating clock gene transcription. In the present study we only addressed the later possibility by assessing the ability of TTF1 to affect clock gene transcription. Our results showed that TTF1 markedly represses the REV-ERBα promoter without affecting Bmal1 or Cry1 promoter activity, and modestly repressing Per1 transcription. The lack of effect of TTF1 on the Bmal1 promoter is surprising, because it would be expected that following repression of REV-ERBα, Bmal1 expression would rise. Such an effect may no longer detected 48 h after TTF1 transfection, or alternatively, it may be discerned only in a circadian context. It is also plausible that it may have been obliterated due to a disruption of circadian gene networks caused by TTF1 overexpression [as previously observed following Cry1 overexpression (58)]. Further studies are required to resolve this issue.
Within a broader context, our results suggest that, instead of acting directly on core components of the primary feedback loop controlling the circadian oscillator (CLOCK/BMAL1), TTF1 modulates clock gene circuit activity by repressing one of the interlocking associated loops, driven by the retinoic acid-related orphan receptor Rev-erbα (37). By doing so, TTF1 would act in a circadian context to reduce Rev-erbα accumulation, resulting in activation of Bmal1 expression (37) and/or Cry de-repression (67).
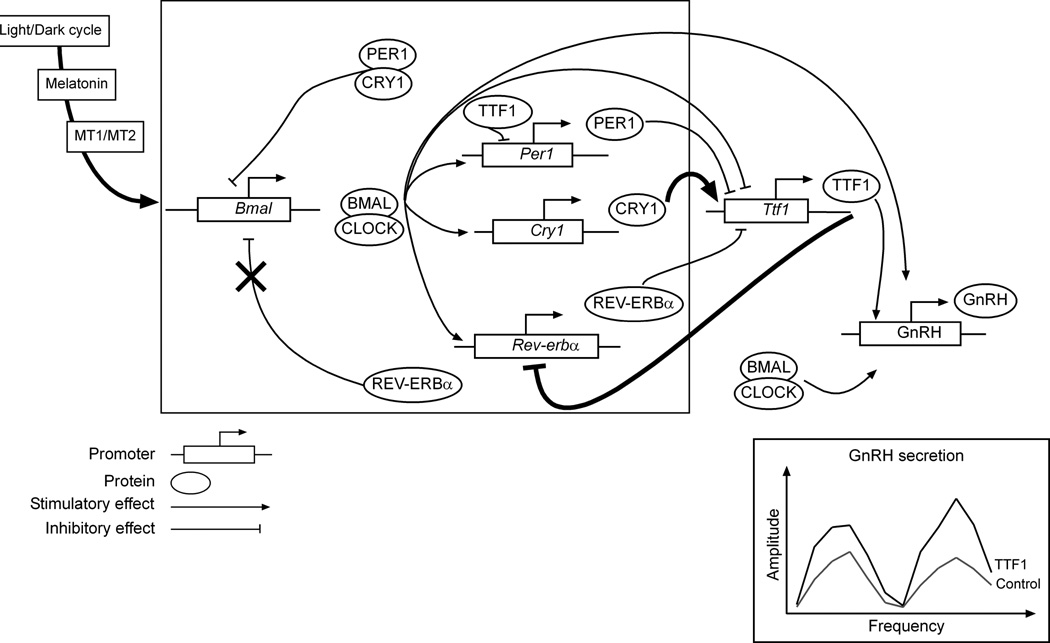
Though complex, these interactions are amenable to interpretations that can be integrated into a transcriptional working model (Fig. 7). According to this model, daily cues such as the light/dark cycle and diurnal fluctuations in melatonin secretion would entrain periodical oscillation in Ttf1 and GnRH expression. Ttf1 can be considered as a clock-controlled gene that regulates GnRH transcription both directly by binding to its promoter and indirectly through interaction with core clock components (Rev-erbα, Cry1, thick lines). Although high-throughput approaches coupled with systems biology strategies have revealed an astounding complexity of circadian oscillators (66), the basic interlocking transcriptional regulatory loops underlying the circadian clock remain firmly established (framed box, Fig. 7). BMAL1 dimerizes with CLOCK to stimulate the transcription of PER1/2, CRY1/2 and REV-ERBα (17,66). While PER1/2 and CRY1/2 negatively regulate CLOCK/BMAL-induced transcription (62), REV-ERBα inhibits the transcription of BMAL1 (37), which subsequently leads to their own transcriptional downregulation. Our data indicate that TTF1 may be part of a secondary regulatory loop involving an increase in Ttf1 transcription driven by CRY1 and a decrease in Rev-erbα transcription (Fig 7, thick lines). This is of particular interest as CRY1 expression, similarly to that of TTF1, peaks at evening phases in the SCN (58,67). In addition, Cry1 is under positive and negative transcriptional regulation by CLOCK/BMAL (41) and REV-ERBα (58,67), respectively. We therefore envision a mechanism by which the nocturnal increase in CRY1 stimulates Ttf1 transcription that will, in turn, decrease REV-ERBα transcription. The latter would delay the transcriptional inhibition of Bmal1, lengthening the time during which CRY1 is sufficiently elevated (41) to increase Ttf1 transcription. This TTF1-mediated regulatory loop would eventually result in increased amplitude of the circadian oscillations in GnRH promoter activity, as revealed by secreted alkaline phosphatase assays measuring GnRH promoter-driven transcription. The idea that CRY1 is positively coupled to the circadian control of GnRH secretion is supported by previous studies showing that CRY1 overexpression in GT1-7 cells leads to an increase in peak amplitude of GnRH secretion (22). Our results also show that PER1 has an inhibitory effect on Ttf1 transcription, which could be part of an inhibitory sub-loop counterbalancing the stimulatory effect of CRY1 on Ttf1 transcription. It is important to recognize that clock proteins may also act directly on the GnRH promoter to modify its activity and elicit GnRH secretion (22,23). The lack of synergism or additivity between TTF1 and CLOCK/BMAL1 effects on GnRH promoter activity we observed in our study may not hold true for TTF1 binding sites and E-box recognition sequences present in regions of the GnRH 5’-flanking region other than the enhancer and proximal promoter we examined here.
Figure 7. A draft model of TTF1 interactions with core clock genes to regulate circadian changes in GnRH promoter activity within GnRH neurons.
The (simplified) transcriptional model includes the well established rhythmic secretion of BMAL1 that, upon dimerization with CLOCK, stimulates the transcription of REV-ERBα PER1/2 and CRY1/2. For the sake of clarity, and according to our own experiments, only Per1 and Cry1 promoters are represented). In turn, these factors inhibit the transcription of BMAL1, which then leads to their own transcriptional downregulation in what is known as the inhibitory limb of the circadian clock (17,66). In GnRH neurons, this rhythmic expression of clock gene is thought to be involved in the manifestation of circadian GnRH secretion (21–23). Our data indicate that daily changes in melatonin secretion entrain the diurnal expression of Ttf1 and GnRH mRNA, acting via MT1 and perhaps also MT2 receptors. In addition of a direct effect of TTF1 and CLOCK/BMAL on GnRH transcription, TTF1 may be part of a secondary regulatory interloop (thick lines) in which, on the one hand, CRY1 stimulates the transcription of Ttf1 while, on the other hand, TTF1 inhibits REV-ERBα transcription. It is speculated that this regulatory loop underlies the ability of TTF1 to increase GnRH transcription in a circadian fashion by increasing the amplitude, and not the period, of clock-driven circadian GnRH promoter activity.
Altogether our results are compatible with the interpretation that TTF1 is part of a regulatory transcriptional sub-network that, acting within the POA, contributes to the circadian oscillator controlling GnRH secretion. By showing entrainment to the light-dark cycle, dependence on melatonin, periodicity and robustness of its daily fluctuations in expression, trans-regulatory strength, and functional connectivity to the molecular machinery underlying the circadian oscillator controlling GnRH secretion, Ttf1 emerges as a tissue-specific clock-associated gene. Furthermore, our results indicate that TTF1 is a modifier of circadian amplitude, and not of clock period. As such, it appears to belong to a handful of clock-related genes regulating the amplitude of circadian oscillations (66). Derangements of these circadian interactions may play a role in the genesis of disorders affecting GnRH secretion both at puberty and during adult reproductive life.
Supplementary Material
Acknowledgements
We thank Ms. Maria Costa for performing the immunohistofluorescence studies and Dr. Anda Cornea (Director of the Imaging Core at the ONPRC) for her assistance with confocal microscopy imaging. This work was supported by research grant (2011K000275) from the Brain Research Center of the 21st Century Frontier Research Program (BJL) and NIH grants HD25123, the Eunice Kennedy Shriver NICHD/NIH through cooperative agreement [U54 HD18185] as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research, and 8P51OD011092-53 for the operation of the Oregon National Primate Research Center (SRO).
References
- 1.Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox CH, Ward JM, Gonzalez FJ. The T/ebp null mouse: Thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- 2.Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126:3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- 3.Price M, Lazzaro D, Pohl T, Mattei M-G, Rüther U, Olivo J-C, Duboule D, Di Lauro R. Regional expression of the homeobox gene Nkx-2.2 in the developing mammalian forebrain. Neuron. 1992;8:241–255. doi: 10.1016/0896-6273(92)90291-k. [DOI] [PubMed] [Google Scholar]
- 4.Pera EM, Kessel M. Demarcation of ventral territories by the homeobox gene NKX2.1 during early chick development. Dev Genes Evol. 1998;208:168–171. doi: 10.1007/s004270050170. [DOI] [PubMed] [Google Scholar]
- 5.Takuma N, Sheng HZ, Furuta Y, Ward JM, Sharma K, Hogan BL, Pfaff SL, Westphal H, Kimura S, Mahon KA. Formation of Rathke's pouch requires dual induction from the diencephalon. Development. 1998;125:4835–4840. doi: 10.1242/dev.125.23.4835. [DOI] [PubMed] [Google Scholar]
- 6.Marin O, Baker J, Puelles L, Rubenstein JL. Patterning of the basal telencephalon and hypothalamus is essential for guidance of cortical projections. Development. 2002;129:761–773. doi: 10.1242/dev.129.3.761. [DOI] [PubMed] [Google Scholar]
- 7.Marin O, Anderson SA, Rubenstein JL. Origin and molecular specification of striatal interneurons. J Neurosci. 2000;20:6063–6076. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee BJ, Cho GJ, Norgren R, Junier M-P, Hill DF, Tapia V, Costa ME, Ojeda SR. TTF-1, a homeodomain gene required for diencephalic morphogenesis, is postnatally expressed in the neuroendocrine brain in a developmentally regulated and cell-specific fashion. Mol Cell Neurosci. 2001;17:107–126. doi: 10.1006/mcne.2000.0933. [DOI] [PubMed] [Google Scholar]
- 9.Davis AM, Seney ML, Stallings NR, Zhao L, Parker KL, Tobet SA. Loss of steroidogenic factor 1 alters cellular topography in the mouse ventromedial nucleus of the hypothalamus. J Neurobiol. 2004;60:424–436. doi: 10.1002/neu.20030. [DOI] [PubMed] [Google Scholar]
- 10.Mastronardi C, Smiley GG, Raber J, Kusakabe T, Kawaguchi A, Matagne V, Dietzel A, Heger S, Mungenast AE, Cabrera R, Kimura S, Ojeda SR. Deletion of the Ttf1 gene in differentiated neurons disrupts female reproduction without impairing basal ganglia function. J Neurosci. 2006;26:13167–13179. doi: 10.1523/JNEUROSCI.4238-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Provenzano C, Pascucci B, Lupari E, Civitareale D. Large scale analysis of transcription factor TTF-1/NKX2.1 target genes in GnRH secreting cell line GT1-7. Mol Cell Endocrinol. 2010;323:215–223. doi: 10.1016/j.mce.2010.02.038. [DOI] [PubMed] [Google Scholar]
- 12.Khan AR, Kauffman AS. The Role of Kisspeptin and RFRP-3 Neurons in the Circadian-Timed Preovulatory Luteinizing Hormone Surge. J Neuroendocrinol. 2011 doi: 10.1111/j.1365-2826.2011.02162.x. Accepted Article- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turek FW, Van Cauter E. Rhythms in reproduction. In: Knobil E, Neill JD, editors. The Physiology of Reproduction. 2nd Edition. New York: Raven Press; 1994. pp. 487–540. [Google Scholar]
- 14.Boyar R, Finkelstein J, Roffwarg H, Kapen S, Weitzman E, Hellman L. Synchronization of augmented luteinizing hormone secretion with sleep during puberty. N Engl J Med. 1972;287:582–586. doi: 10.1056/NEJM197209212871203. [DOI] [PubMed] [Google Scholar]
- 15.Terasawa E, Bridson WE, Nass TE, Noonan JJ, Dierschke DJ. Developmental changes in the luteinizing hormone secretory pattern in peripubertal female rhesus monkeys: Comparisons between gonadally intact and ovariectomized animals. Endocrinology. 1984;115:2233–2240. doi: 10.1210/endo-115-6-2233. [DOI] [PubMed] [Google Scholar]
- 16.Urbanski HF, Ojeda SR. Gonadal-independent activation of enhanced afternoon luteinizing hormone release during pubertal development in the female rat. Endocrinology. 1987;121:907–913. doi: 10.1210/endo-121-3-907. [DOI] [PubMed] [Google Scholar]
- 17.Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 18.Saper CB, Lu J, Chou TC, Gooley J. The hypothalamic integrator for circadian rhythms. Trends Neurosci. 2005;28:152–157. doi: 10.1016/j.tins.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 19.Freeman ME. The neuroendocrine control of the ovarian cycle of the rat. In: Knobil E, Neill JD, editors. The Physiology of Reproduction. 2nd Edition. New York: Raven Press, Ltd.; 1994. pp. 613–658. [Google Scholar]
- 20.Maywood ES, O'Neill JS, Chesham JE, Hastings MH. Minireview: The circadian clockwork of the suprachiasmatic nuclei--analysis of a cellular oscillator that drives endocrine rhythms. Endocrinology. 2007;148:5624–5634. doi: 10.1210/en.2007-0660. [DOI] [PubMed] [Google Scholar]
- 21.Hickok JR, Tischkau SA. In vivo circadian rhythms in gonadotropin-releasing hormone neurons. Neuroendocrinology. 2010;91:110–120. doi: 10.1159/000243163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chappell PE, White RS, Mellon PL. Circadian gene expression regulates pulsatile gonadotropin-releasing hormone (GnRH) secretory patterns in the hypothalamic GnRH-secreting GT1-7 cell line. J Neurosci. 2003;23:11202–11213. doi: 10.1523/JNEUROSCI.23-35-11202.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillespie JMA, Chan BPK, Roy D, Cai F, Belsham DD. Expression of circadian rhythm genes in gonadotropin-releasing hormone-secreting GT1-7 neurons. Endocrinology. 2003;144:5285–5292. doi: 10.1210/en.2003-0802. [DOI] [PubMed] [Google Scholar]
- 24.Kim MS, Hur MK, Son YJ, Park J-I, Chun SY, D'Elia AV, Damante G, Cho S, Kim K, Lee BJ. Regulation of pituitary adenylate cyclase-activating polypeptide gene transcription by TTF-1, a homeodomain-containing transcription factor. J Biol Chem. 2002;277:36863–36871. doi: 10.1074/jbc.M206443200. [DOI] [PubMed] [Google Scholar]
- 25.Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. BioTechniques. 1993;15:532–534. [PubMed] [Google Scholar]
- 26.Damante G, Fabbro D, Pellizzari L, Civitareale D, Guazzi S, Polycarpou-Schwartz M, Cauci S, Quadrifoglio F, Formisano S, Di Lauro R. Sequence-specific DNA recognition by the thyroid transcription factor-1 homeodomain. Nucleic Acids Res. 1994;22:3075–3083. doi: 10.1093/nar/22.15.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Junier M-P, Wolff A, Hoffman GE, Ma YJ, Ojeda SR. Effect of hypothalamic lesions that induce precocious puberty on the morphological and functional maturation of the luteinizing hormone-releasing hormone neuronal system. Endocrinology. 1992;131:787–798. doi: 10.1210/endo.131.2.1639024. [DOI] [PubMed] [Google Scholar]
- 28.Ma YJ, Dissen GA, Rage F, Ojeda SR. RNase protection assay Methods: A Companion to Methods in Enzymology. 1996;10:273–278. doi: 10.1006/meth.1996.0102. [DOI] [PubMed] [Google Scholar]
- 29.Damante G, Pellizzari L, Esposito G, Fogolari F, Viglino P, Fabbro D, Tell G, Formisano S, Di Lauro R. A molecular code dictates sequence-specific DNA recognition by homeodomains. EMBO J. 1996;15:4992–5000. [PMC free article] [PubMed] [Google Scholar]
- 30.Civitareale D, Lonigro R, Sinclair AJ, Di Lauro R. A thyroid-specific nuclear protein essential for tissue-specific expression of the thyroglobulin promoter. EMBO J. 1989;8:2537–2542. doi: 10.1002/j.1460-2075.1989.tb08391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JG, Park BS, Yun CH, Kim HJ, Kang SS, D'Elia AV, Damante G, Lee KU, Park JW, Kim ES, Namgoong IS, Kim YI, Lee BJ. Thyroid transcription factor-1 regulates feeding behavior via melanocortin pathway in the hypothalamus. Diabetes. 2011;60:710–719. doi: 10.2337/db10-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim JG, Son YJ, Yun CH, Kim YI, Nam-Goong IS, Park JH, Park SK, Ojeda SR, D'Elia AV, Damante G, Lee BJ. Thyroid transcription factor-1 facilitates cerebrospinal fluid formation by regulating aquaporin-1 synthesis in the brain. J Biol Chem. 2007;282:14923–14931. doi: 10.1074/jbc.M701411200. [DOI] [PubMed] [Google Scholar]
- 33.Ojeda SR, Hill J, Hill DF, Costa ME, Tapia V, Cornea A, Ma YJ. The Oct-2 POU-domain gene in the neuroendocrine brain: A transcriptional regulator of mammalian puberty. Endocrinology. 1999;140:3774–3789. doi: 10.1210/endo.140.8.6941. [DOI] [PubMed] [Google Scholar]
- 34.Guo ZS, Wang L-H, Eisensmith RC, Woo SLC. Evaluation of promoter strength for hepatic ce of autoregulation Endocrinology 1998; 139: 1999–2006. gene expression in vivo following adenovirus-mediated gene transfer. Gene Ther. 1996;3:802–810. [PubMed] [Google Scholar]
- 35.Matagne V, Mastronardi C, Shapiro RA, Dorsa DM, Ojeda SR. Hypothalamic expression of Eap1 is not directly controlled by ovarian steroids. Endocrinology. 2008;150:1870–1878. doi: 10.1210/en.2008-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oguchi H, Kimura S. Multiple transcripts encoded by the thyroid-specific enhancer-binding protein (T/EBP)/thyroid-specific transcription factor-1 (TTF-1) gene: eviden. doi: 10.1210/endo.139.4.5933. [DOI] [PubMed] [Google Scholar]
- 37.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 38.Adelmant G, Begue A, Stehelin D, Laudet V. A functional Rev-erb alpha responsive element located in the human Rev-erb alpha promoter mediates a repressing activity. Proc Natl Acad Sci U S A. 1996;93:3553–3558. doi: 10.1073/pnas.93.8.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang DC, Reppert SM. The circadian clocks of mice and men. Neuron. 2001;29:555–558. doi: 10.1016/s0896-6273(01)00230-6. [DOI] [PubMed] [Google Scholar]
- 40.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 41.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 42.Urbanski HF. Monoclonal antibodies to luteinizing hormone-releasing hormone: Production, characterization, and immunocytochemical application. Biol Reprod. 1991;44:681–686. doi: 10.1095/biolreprod44.4.681. [DOI] [PubMed] [Google Scholar]
- 43.DeFazio RA, Heger S, Ojeda SR, Moenter SM. Activation of A-type g-aminobutyric acid receptors excites gonadotropin-releasing hormone neurons. Mol Endocrinol. 2002;16:2872–2891. doi: 10.1210/me.2002-0163. [DOI] [PubMed] [Google Scholar]
- 44.Heger S, Mastronardi C, Dissen GA, Lomniczi A, Cabrera R, Roth CL, Jung H, Galimi F, Sippell W, Ojeda SR. Enhanced at puberty 1 (EAP1) is a new transcriptional regulator of the female neuroendocrine reproductive axis. J Clin Invest. 2007;117:2145–2154. doi: 10.1172/JCI31752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 46.Hastings MH, Field MD, Maywood ES, Weaver DR, Reppert SM. Differential regulation of mPER1 and mTIM proteins in the mouse suprachiasmatic nuclei: new insights into a core clock mechanism. J Neurosci. 1999;19:RC11. doi: 10.1523/JNEUROSCI.19-12-j0001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simonneaux V, Ribelayga C. Generation of the melatonin endocrine message in mammals: a review of the complex regulation of melatonin synthesis by norepinephrine, peptides, and other pineal transmitters. Pharmacol Rev. 2003;55:325–395. doi: 10.1124/pr.55.2.2. [DOI] [PubMed] [Google Scholar]
- 48.Li S, Givalois L, Pelletier G. Effects of aging and melatonin administration on gonadotropin-releasing hormones (GnRH) gene expression in the male and female rat. Peptides. 1997;18:1023–1028. doi: 10.1016/s0196-9781(97)00076-4. [DOI] [PubMed] [Google Scholar]
- 49.Li S, Pelletier G. Effects of pinealectomy and melatonin on gonadotropin-releasing hormone (GnRH) gene expression in the male rat brain. Endocrine. 1995;3:533–536. doi: 10.1007/BF02738829. [DOI] [PubMed] [Google Scholar]
- 50.Ishii H, Tanaka N, Kobayashi M, Kato M, Sakuma Y. Gene structures, biochemical characterization and distribution of rat melatonin receptors. J Physiol Sci. 2009;59:37–47. doi: 10.1007/s12576-008-0003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roy D, Angelini NL, Fujieda H, Brown GM, Belsham DD. Cyclical regulation of GnRH gene expression in GT1-7 GnRH-secreting neurons by melatonin. Endocrinology. 2001;142:4711–4720. doi: 10.1210/endo.142.11.8464. [DOI] [PubMed] [Google Scholar]
- 52.Gauer F, Masson-Pevet M, Pevet P. Differential regulation of melatonin receptors by short-versus long-term pinealectomy in the rat suprachiasmatic nuclei and pars tuberalis. J Pineal Res. 1994;16:73–76. doi: 10.1111/j.1600-079x.1994.tb00085.x. [DOI] [PubMed] [Google Scholar]
- 53.Whyte DB, Lawson MA, Belsham DD, Eraly SA, Bond CT, Adelman JP, Mellon PL. A neuron-specific enhancer targets expression of the gonadotropin-releasing hormone gene to hypothalamic neurosecretory neurons. Mol Endocrinol. 1995;9:467–477. doi: 10.1210/mend.9.4.7659090. [DOI] [PubMed] [Google Scholar]
- 54.Lawson MA, MacConell LA, Kim J, Powl BT, Nelson SB, Mellon PL. Neuron-specific expression in vivo by defined transcription regulatory elements of the GnRH gene. Endocrinology. 2002;143:1404–1412. doi: 10.1210/endo.143.4.8751. [DOI] [PubMed] [Google Scholar]
- 55.Nelson SB, Lawson MA, Kelley CG, Mellon PL. Neuron-specific expression of the rat gonadotropin-releasing hormone gene is conferred by interactions of a defined promoter element with the enhancer in GT1-7 cells. Mol Endocrinol. 2000;14:1509–1522. doi: 10.1210/mend.14.9.0521. [DOI] [PubMed] [Google Scholar]
- 56.Olcese J, Domagalski R, Bednorz A, Weaver DR, Urbanski HF, Reuss S, Middendorff R. Expression and regulation of mPer1 in immortalized GnRH neurons. NeuroReport. 2003;14:613–618. doi: 10.1097/00001756-200303240-00018. [DOI] [PubMed] [Google Scholar]
- 57.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15(Spec No 2):R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 58.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 59.Liu C, Weaver DR, Jin X, Shearman LP, Pieschi RL, Gribkoff VK, Reppert SM. Molecular dissection of two distinct actions of melatonin on the suprachiasmatic circadian clock. Neuron. 1997;19:91–102. doi: 10.1016/s0896-6273(00)80350-5. [DOI] [PubMed] [Google Scholar]
- 60.Jin X, Shearman LP, Weaver DR, Zylka MJ, De Vries GJ, Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- 61.Bunger Mk, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. MOP3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, Reppert SM. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 63.Yagita K, Tamanini F, Yasuda M, Hoeijmakers JH, van der Horst GT, Okamura H. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 2002;21:1301–1314. doi: 10.1093/emboj/21.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thum KE, Kim M, Christopher DA, Mullet JE. Cryptochrome 1, cryptochrome 2, and phytochrome a co-activate the chloroplast psbD blue light-responsive promoter. Plant Cell. 2001;13:2747–2760. doi: 10.1105/tpc.010345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bose S, Boockfor FR. Episodes of prolactin gene expression in GH3 cells are dependent on selective promoter binding of multiple circadian elements. Endocrinology. 2010;151:2287–2296. doi: 10.1210/en.2009-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang EE, Kay SA. Clocks not winding down: unravelling circadian networks. Nat Rev Mol Cell Biol. 2010;11:764–776. doi: 10.1038/nrm2995. [DOI] [PubMed] [Google Scholar]
- 67.Ukai-Tadenuma M, Yamada RG, Xu H, Ripperger JA, Liu AC, Ueda HR. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell. 2011;144:268–281. doi: 10.1016/j.cell.2010.12.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.