Abstract
Decoding post-transcriptional regulatory programs in RNA is a critical step in the larger goal to develop predictive dynamical models of cellular behavior. Despite recent efforts1–3, the vast landscape of RNA regulatory elements remain largely uncharacterized. A longstanding obstacle is the contribution of local RNA secondary structure in defining interaction partners in a variety of regulatory contexts, including but not limited to transcript stability3, alternative splicing4 and localization3. There are many documented instances where the presence of a structural regulatory element dictates alternative splicing patterns (e.g. human cardiac troponin T) or affects other aspects of RNA biology5. Thus, a full characterization of post-transcriptional regulatory programs requires capturing information provided by both local secondary structures and the underlying sequence3,6. We have developed a computational framework based on context-free grammars3,7 and mutual information2 that systematically explores the immense space of small structural elements and reveals motifs that are significantly informative of genome-wide measurements of RNA behavior. The application of this framework to genome-wide mammalian mRNA stability data revealed eight highly significant elements with substantial structural information, for the strongest of which we showed a major role in global mRNA regulation. Through biochemistry, mass-spectrometry, and in vivo binding studies, we identified HNRPA2B1 as the key regulator that binds this element and stabilizes a large number of its target genes. Ultimately, we created a global post-transcriptional regulatory map based on the identity of the discovered linear and structural cis-regulatory elements, their regulatory interactions and their target pathways. This approach can also be employed to reveal the structural elements that modulate other aspects of RNA behavior.
To isolate stability from other aspects of mRNA behavior, we performed whole-genome mRNA stability measurements by incubating MDA-MB-231 cells in the presence of 4-thiouridine (4sU), which is efficiently incorporated into cellular RNA. Subsequently, 4sU-labeled transcripts were captured and quantified at different time-points after the removal of 4sU from the growth medium. We calculated a relative decay rate for each transcript based on the rate at which 4sU-labeled transcripts, in the absence of 4sU in the media, are replaced by newly synthesized unlabeled mRNAs in the population (Supplementary Fig. 1). These measurements were then used to identify the putative cis-regulatory elements (linear and structural) that underlie transcript stability. A number of methods have been previously introduced for discovering structural motifs mainly based on free energy minimization, local sequence alignments or a combination of both alignments and secondary structure predictions 3,6,8. However, the extent to which these in silico predictions reflect stable in vivo molecular conformations has not been fully explored9. In fact, the RNA binding proteins and complexes that interact with their target transcripts may facilitate the formation of secondary structures in vivo. Thus, we sought to bypass the need for predicting thermodynamically stable secondary structures by efficiently enumerating a large space of potential structural motifs. We developed TEISER (Tool for Eliciting Informative Structural Elements in RNA), a framework for identifying the structural motifs that are informative of whole-genome measurements across all the transcripts. In this approach, structural motifs are defined in terms of context-free grammars7 (CFGs) that represent hairpin structures as well as primary sequence information (see Methods and Supplementary Fig. 2). TEISER employs mutual information to measure the regulatory consequences of the presence or absence of each of roughly 100 million different seed CFGs (see Methods). Mutual information is a robust non-parametric measure that reveals general dependencies across discrete or continuous measurements2,10. For example, when applied to the transcript stability data, TEISER captures the dependency between the stability of each mRNA and the presence or absence of a given structural motif in its 5’ and 3’ untranslated regions (UTRs). TEISER, subsequently, uses these measurements to choose and further refine the most informative motifs, and performs a series of statistical tests, e.g. randomization-based statistics and jackknifing tests, to achieve very low (<0.01) false-discovery rates (see Methods and Supplementary Fig. 2).
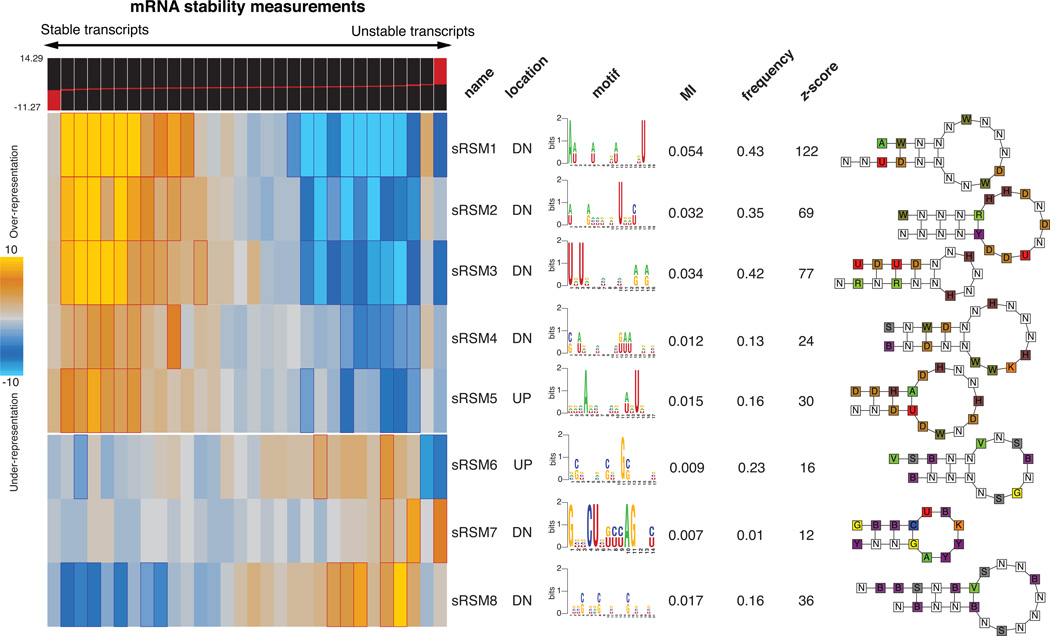
Application of TEISER to the mRNA stability measurements in MDA-MB-231 cells revealed eight strong structural motif predictions that passed our statistical tests aimed at finding the most likely elements causally involved in mRNA stability (Fig. 1 and Supplementary Fig. 3). Apart from being highly informative of mRNA stability measurements, these putative regulatory elements show a variety of other characteristics that support their functionality. For example, four of the discovered motifs are also informative of transcript stability measurements in mouse11 (Supplementary Fig. 4a). Furthermore, these motifs are highly conserved between human and mouse genomes (see Methods and Supplementary Fig. 3) and are also informative of co-expression clusters discovered across independent whole-genome datasets (Supplementary Fig. 4b).
Figure 1. Discovery of RNA structural motifs informative of genome wide transcript stability.
Each RNA structural motif is shown along with its pattern of enrichment/depletion across the range of mRNA stability measurements throughout the genome. The transcripts are partitioned into equally populated bins based on their stability measures, going from left (highly stable) to right (unstable). In the heatmap representation, a gold entry marks the enrichment of the given motif in its corresponding stability bin (measured by log-transformed hypergeometric p-values), while a light-blue entry indicates motif depletion in the bin. Red and blue borders mark highly significant motif enrichments and depletions, respectively. Included are the motif names, their location (UP for 5’UTR and DN for 3’UTR), their sequence information (in the form of a logo) and their frequency (the fraction of transcripts that carry at least one instance of the motif). Also shown are the associated mutual information values. Each mutual information (MI) value is used to calculate a z-score, which is the number of standard-deviations of the actual MI relative to MI's calculated for 1.5 million randomly shuffled stability profiles. A structural illustration of each motif is also presented using the following single letter nucleotide code: Y=[UC], R=[AG], K=[UG], M=[AC], S=[GC], W=[AU], B=[GUC], D=[GAU], H=[ACU], V=[GCA] and N=any nucleotide.
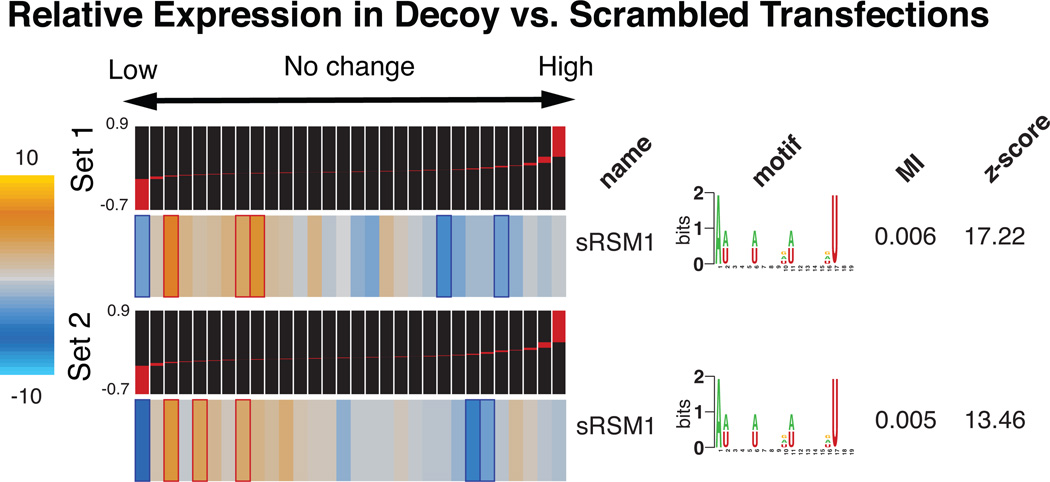
Among the putative structural motifs discovered by TEISER, we chose sRSM1 (structural RNA Stability Motif-1)—the most statistically significant 3’ UTR element (z-score=122)—for further analysis. In order to probe the functionality of sRSM1 instances across the genome, we performed in vivo titration experiments using synthetic oligonucleotides10,12. Upon transfecting MDA-MB-231 cells with decoy RNA molecules harboring sRSM1 instances (Supplementary Fig. 5), we observed a notable reduction in the level of endogenous transcripts that carried this motif, in comparison to their level in the control cells transfected with scrambled RNA molecules (Fig. 2). This global down-regulation points to the presence of a trans-acting factor that, upon interaction with sRSM1, stabilizes its target transcripts. The decoy (synthetic) sRSM1 elements compete with endogenous mRNAs for the putative trans-acting factor, which results in the observed reduction in the level of its target mRNAs. Furthermore, reporter constructs carrying instances of sRSM1 showed a marked decrease in transcript decay rate in comparison to scrambled controls, further suggesting a direct role for this structural element in transcript stability (Supplementary Fig. 6).
Figure 2. The regulatory role of sRSM1.
Whole-genome expression levels were measured in decoy-transfected samples relative to the controls transfected with scrambled RNA molecules (see Methods). The measurements were performed in duplicate, for two independent decoy/scrambled sets (the relative transcript levels were subsequently averaged across the two replicates in each set). Genes were sorted and quantized into equally populated bins based on the average log-ratio of their expression levels in the decoy samples relative to the scrambled controls. TEISER was used to show the enrichment/depletion patterns of transcripts harboring sRSM1 in their 3’ UTRs. Mutual information values and the associated z-scores are also presented.
We used streptomycin-binding RNA aptamer immobilization coupled with mass spectrometry13 to discover candidates that bind, in vitro, to the decoy instances of sRSM1, but not to the scrambled versions (Supplementary Fig. 7). After isolation under stringent conditions and in-solution digestion of RNA-bound proteins followed by nanoliquid chromatography-tandem mass spectrometry, we identified HNRPA2B1 as a promising candidate (Supplementary Table 1). This RNA-binding protein is a member of the A/B subfamily of heterogeneous nuclear ribonucleoproteins (hnRNPs)14 and carries two repeats of quasi-RRM RNA binding domains (Supplementary Fig. 8). Moreover, the established roles of other members of this family, namely HNRNPD and HNRNA1, in regulating RNA stability15 and binding terminal stem-loops16 further suggest HNRPA2B1 as a functional regulator. Also, more than 4,000 transcripts carry potentially functional instances of sRSM1 (see Methods), implicating this motif as a major global regulator of mRNA stability. The HNRPA2B1 transcript, at the same time, is highly abundant in the cell (one standard deviations higher than average17), thus making it a promising candidate for global modulation of mRNA stability through sRSM1.
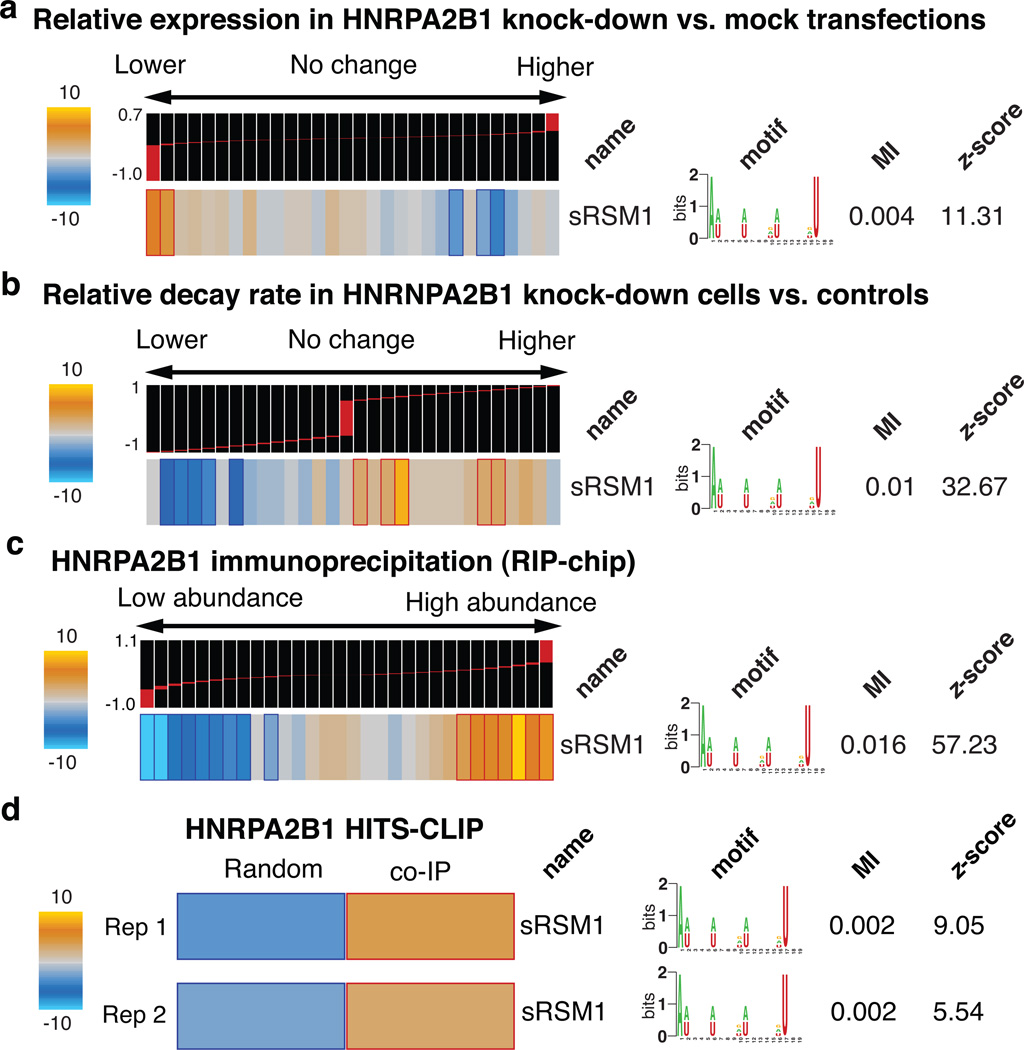
In order to directly assess the regulatory consequences of modulating HNRPA2B1, we performed knock-down experiments followed by gene expression profiling. Consistent with our prior observations, HNRPA2B1 knock-down caused a significant decrease in the expression level of transcripts carrying sRSM1 (Fig. 3a). Stability measurements in the knock-down cells confirmed that the observed down-regulation of these transcripts was in fact due to changes in stability (see Methods), with the transcripts carrying sRSM1 elements showing a marked increase in their corresponding relative decay rates (Fig. 3b).
Figure 3. HNRPA2B1 stabilizes transcripts through direct in vivo binding to sRSM1 structural motifs.
a, Genome-wide expression levels were measured in HNRPA2B1 siRNA-transfected samples relative to mock-transfected controls. TEISER was used to capture the enrichment/depletion pattern of transcripts carrying sRSM1 across the relative expression values. Experiments were performed in triplicate, each with an independent siRNA targeting HNRPA2B1 and the resulting log ratios were averaged for each transcript. b, Transcript decay rates were compared in HNRPA2B1 knock-downs versus mock-transfected controls. These measurements were then analyzed by TEISER to visualize the extent to which the decay rates of transcripts carrying sRSM1 elements were increased following HNRPA2B1 knock-down. c, Using UV-crosslinking followed by immunoprecipitation, mRNAs that bind HNRPA2B1 were extracted and compared against the input mRNA population (RIP-chip). The log ratio calculated for each mRNA denotes its abundance in the immunoprecipitated sample relative to the input control. Bins to the right contain the mRNAs that were captured as interacting partners with HNRPA2B1. Similar to the prior examples, TEISER was used to show the enrichment/depletion pattern of transcripts carrying sRSM1 in their 3’ UTRs. The values associated with each transcript were calculated as the average of log ratios from biological replicates. d, HNRPA2B1 binding sites were identified using immunoprecipitation followed by high-throughput sequencing (HITS-CLIP). Instances of the sRSM1 element are significantly enriched in these sites relative to a population of random sequences from 3’ UTRs that are not represented in the sequenced population.
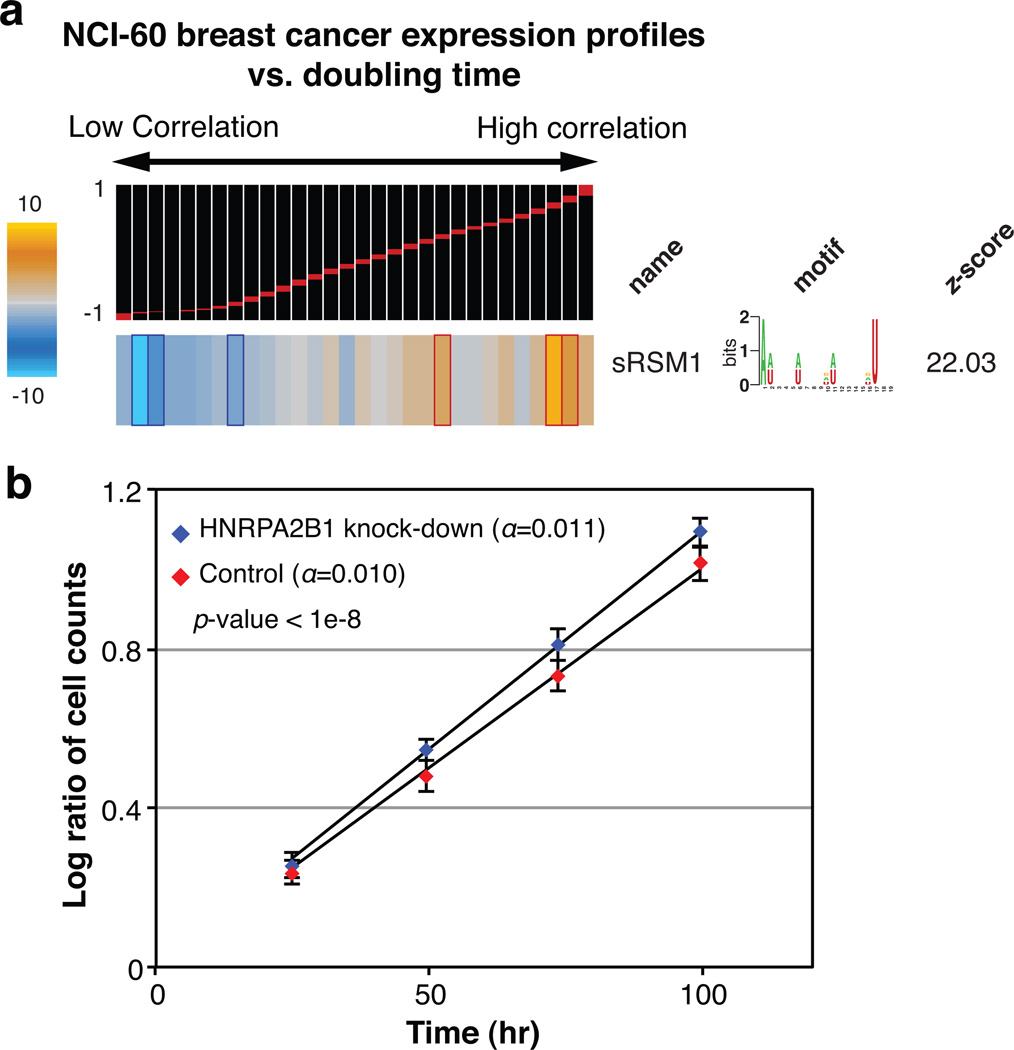
In principle, our observations are consistent with a possible indirect role for HNRPA2B1—brought about, for instance, by a common partner that binds both HNRPA2B1 and sRSM1 sites. The direct interaction between HNRPA2B1 and its potential target genes can be tested through cross-linking and immunoprecipitation of HNRPA2B1, which, through local UV photoreactivity of bases and amino-acids, can detect direct physical interactions18. We expressed a tagged clone of HNRPA2B1 in MDA-MB-231 cells, and after UV-crosslinking, immunoprecipitated this protein and the target mRNA molecules that were bound to it. We then labeled the isolated RNA population and hybridized it to microarrays with the input total RNA as control (a method called RIP-chip19). We observed a highly significant enrichment of sRSM1 in the immunoprecipitated population (Fig. 3c). In order to reduce the background and better pinpoint the HNRPA2B1 binding sites, we treated the samples with nuclease prior to immunoprecipitation under denaturing conditions and sequenced the HNRPA2B1-bound RNA population (HITS-CLIP20). We observed that sRSM1 elements were significantly enriched in the identified putative binding sites, in comparison with randomly selected sequences21 (Fig. 3d). These observations demonstrate that HNRPA2B1 directly interacts with sRSM1 in vivo and functions to stabilize its target transcripts through this regulatory element. These transcripts, in turn, modulate a variety of cellular processes and pathways. For example, we observed a significant positive correlation between sRSM1 target transcripts and doubling-time in NCI-60 breast cancer cell-lines (Fig. 4a). Indeed, knocking-down HNRPA2B1 resulted in a slight but significant increase in growth rate (by 10%, p-value<10−8) further highlighting the regulatory role of this global modulator in a key cellular process (Fig. 4b).
Figure 4. HNRPA2B1 regulates growth rate.
a, Whole genome expression levels across five breast cancer cell lines (MCF7, MDA-MB-231, HS578T, BT-549 and T47D) were correlated against their doubling times17. The resulting values, ranging from −1 to 1, were analyzed by TEISER to probe the enrichment/depletion pattern of transcripts carrying sRSM1. b, The growth of HNRPA2B1 siRNA-transfected samples was compared to those of mock-transfected controls. For each time-point, the number of cells in four independent samples was counted in duplicates (n=8), yielding an estimated growth-rate (α). Shown are the average log-ratios, their standard deviation at each time-point, and the statistical significance of the observed difference in growth-rate.
Revealing the detailed post-transcriptional regulatory code relies on the discovery of all the cis-regulatory elements that contribute to changes in transcript abundance. In addition to the sRSMs identified through TEISER, we also discovered a large diverse set of lRSMs (linear RNA Stability Motifs), including six known miRNA recognition sites, that are informative of transcript stability measurements (Supplementary Fig. 9). These motifs were identified by FIRE2, a framework for discovering informative linear motifs. Combining these two approaches provided us with an extensive set of putative regulatory elements that cover both structural and primary sequence components. The next step in deciphering the post-transcriptional regulatory program involves the identification of target pathways that are potentially modulated by each element. Using iPAGE10, for pathway analysis of gene expression, we showed that our discovered elements likely target a diverse array of cellular processes and pathways (Supplementary Fig. 10). For example, the sRSM1 structural element is significantly enriched in the 3’ UTRs of the genes involved in “Notch signaling”, while avoiding the UTRs of other pathways such as “nucleosome assembly” (Supplementary Fig. 11). These results demonstrate that while post-transcriptional regulatory mechanisms are poorly characterized, they have potentially far-reaching impact on specific cellular processes.
Regulatory programs often employ combinatorial interactions between various cis-regulatory elements to modulate gene expression2,22. We utilized mutual information to reveal such potential interactions in the post-transcriptional regulatory programs governing mRNA stability (Supplementary Fig. 12 and 13). For example, sRSM1 showed significant interactions with a number of structural and linear motifs, including sRSM8 and sRSM3 (Supplementary Fig. 11). These observed interactions might reflect cross talk, or insulation, between the underlying regulatory processes that act upstream of these elements. The full map of such interactions (Supplementary Fig. 14 and 15) reveals a complex network of motif-pathway relationships that set the stage for molecular dissection and predictive modeling of post-transcriptional regulation from sequence.
While we have studied mRNA stability under normal and static conditions in a single cell line, the full regulatory program that governs mRNA stability likely involves a much richer repertoire of cis-regulatory elements operating within a more complex regulatory network. Also, while we have focused on transcript stability, our framework is general in concept and can be employed to study complex regulatory programs governing other aspects of RNA biology. For example, the established role of local secondary structures in shaping the splicing code4,23 suggests alternative splicing as a prominent area for analysis using this framework. The large repertoire of publicly available whole-genome expression datasets similarly offers a rich resource for identifying the post-transcriptional regulatory modules that underlie steady-state measurements.
Methods Summary
TEISER relies on calculating mutual information values between whole-genome measurements and millions of predefined structural motifs. The statistically significant motifs are then optimized and elongated through a greedy algorithm. The mRNA stability measurements were performed using a previously published method1. The decoy/scrambled experiments and siRNA knock-downs were performed using lipofectamin 2000 reagent (Invitrogen). For hybridizations, we used human 4×44k whole-genome human arrays (Agilent). Isolation and identification of RNA-binding proteins were based on previously published protocols13,24. HNRPA2B1 target transcripts were isolated based on the CLIP protocol18.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Acknowledgements
We thank the members of the Tavazoie laboratory for helpful comments on the project and manuscript. We are also grateful to Nora Pencheva, Bambi Tsui, Sohail Tavazoie and Lars Dölken for their intellectual and technical contributions. S.T. was supported by grants from NHGRI (2R01HG003219) and the NIH Director's Pioneer Award.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions HG, HSN and ST conceived and designed the study. HG and HSN developed TEISER. RS contributed to the execution of the study. HG, HSN, TMG, PO, IMC and ST designed the experiments. HG, PO, LF and TMG performed the experiments. HG, HSN and TMG analyzed the results. HG, HSN and ST wrote the paper.
Reprints and permissions information is available at www.nature.com/reprints The microarray and high-throuput sequencing data are deposited at GEO under the umbrella accession number GSE35800.
References
- 1.Dolken L, et al. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. Rna. 2008;14:1959–1972. doi: 10.1261/rna.1136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elemento O, Slonim N, Tavazoie S. A universal framework for regulatory element discovery across all Genomes and data types. Mol Cell. 2007;28:337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rabani M, Kertesz M, Segal E. Computational prediction of RNA structural motifs involved in posttranscriptional regulatory processes. P Natl Acad Sci USA. 2008;105:14885–14890. doi: 10.1073/pnas.0803169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barash Y, et al. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 5.Wan Y, Kertesz M, Spitale RC, Segal E, Chang HY. Understanding the transcriptome through RNA structure. Nat Rev Genet. 2011;12:641–655. doi: 10.1038/nrg3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pavesi G, Mauri G, Stefani M, Pesole G. RNAProfile: an algorithm for finding conserved secondary structure motifs in unaligned RNA sequences. Nucleic Acids Res. 2004;32:3258–3269. doi: 10.1093/nar/gkh650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Searls DB. The language of genes. Nature. 2002;420:211–217. doi: 10.1038/nature01255. [DOI] [PubMed] [Google Scholar]
- 8.Hofacker IL, Fekete M, Stadler PF. Secondary structure prediction for aligned RNA sequences. J Mol Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 9.Kertesz M, et al. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodarzi H, Elemento O, Tavazoie S. Revealing global regulatory perturbations across human cancers. Mol Cell. 2009;36:900–911. doi: 10.1016/j.molcel.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 12.Cutroneo KR, Ehrlich H. Silencing or knocking out eukaryotic gene expression by oligodeoxynucleotide decoys. Crit Rev Eukaryot Gene Expr. 2006;16:23–30. doi: 10.1615/critreveukargeneexpr.v16.i1.20. [DOI] [PubMed] [Google Scholar]
- 13.Windbichler N, Schroeder R. Isolation of specific RNA-binding proteins using the streptomycin-binding RNA aptamer. Nat Protoc. 2006;1 doi: 10.1038/nprot.2006.95. 638-U634. [DOI] [PubMed] [Google Scholar]
- 14.Biamonti G, Ruggiu M, Saccone S, Della Valle G, Riva S. Two homologous genes, originated by duplication, encode the human hnRNP proteins A2 and A1. Nucleic Acids Res. 1994;22:1996–2002. doi: 10.1093/nar/22.11.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 16.Michlewski G, Caceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat Struct Mol Biol. 2010;17:1011–1018. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross DT, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 18.Jensen KB, Darnell RB. CLIP: crosslinking and immunoprecipitation of in vivo RNA targets of RNA-binding proteins. Methods Mol Biol. 2008;488:85–98. doi: 10.1007/978-1-60327-475-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- 20.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456 doi: 10.1038/nature07488. 464-U422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giannopoulou EG, Elemento O. An integrated ChIP-seq analysis platform with customizable workflows. BMC Bioinformatics. 2011;12:277. doi: 10.1186/1471-2105-12-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beer MA, Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, et al. RNA secondary structure in mutually exclusive splicing. Nat Struct Mol Biol. 2011;18:159–168. doi: 10.1038/nsmb.1959. [DOI] [PubMed] [Google Scholar]
- 24.Greco TM, Yu F, Guise AJ, Cristea IM. Nuclear import of histone deacetylase 5 by requisite nuclear localization signal phosphorylation. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.004317. M110 004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 26.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.