Abstract
As with many other viruses, the initial cell attachment of rotaviruses, major causative agent of infantile gastroenteritis, is mediated by interactions with specific cellular glycans1–4. The distally located VP8* domain of the rotavirus spike protein VP45 mediates such interactions. The existing paradigm is that ‘sialidase-sensitive’ animal rotavirus strains bind to glycans with terminal sialic acid (Sia), whereas ‘sialidase-insensitive’ human rotavirus (HR) strains bind to glycans with internal Sia such as GM13. Although the involvement of Sia in the animal strains is firmly supported by crystallographic studies1,3,6,7, it is not yet known how VP8* of HRs interacts with Sia and whether their cell attachment necessarily involves sialoglycans. We found that VP8* of a HR strain specifically recognizes A-type histo-blood group antigen (HBGA) using a glycan array screen comprised of 511 glycans, and that virus infectivity in HT-29 cells is abrogated by anti-Atype antibodies as well as significantly enhanced in CHO cells genetically modified to express the A-type HBGA, providing a novel paradigm for initial cell attachment of HR. HBGAs are genetically determined glycoconjugates present in mucosal secretions, epithelial and on red blood cells8, and are recognized as susceptibility and cell attachment factors for gastric pathogens like H. pylori9 and noroviruses10. Our crystallographic studies show that the A-type HBGA binds to the HR VP8* at the same location as the Sia in the VP8* of animal rotavirus, and suggest how subtle changes within the same structural framework allow for such receptor switching. These results raise the possibility that host susceptibility to specific HR strains and pathogenesis are influenced by genetically controlled expression of different HBGAs among the world’s population.
Based on neutralization specificity of the outer capsid proteins VP7 and VP4, rotaviruses are classified into G (VP7) and P (VP4) genotypes following a dual nomenclature system similar to influenza viruses11. The crystallographic structures of VP8* from two sialidase-insensitive human strains, representing P[8] (Wa)1 and P[4] (DS1)12, from two sialidase-sensitive animal strains, representing P[3] (RRV)6,7 and P[7] (CRW-8)1, and the structures of two animal VP8* with bound Sia1,6,12 have been previously reported. NMR, cell binding and neutralization assays, showed that the sialidase-insensitive P[8] Wa strain binds to gangliosides such as GM1 using internal Sia3. These studies suggested that while the sialidase-sensitive strains recognize glycans with terminal Sia such as GD1a, the sialidase-insensitive rotavirus stains bind to gangliosides such as GM1 with an internal Sia moiety, and gave rise to the notion that Sia is the key determinant for host cell recognition in rotaviruses. Our goal was to determine whether all sialidase-insensitive HR genotypes recognize gangliosides with an internal Sia moiety for initial cell attachment or whether they recognize different glycans in a genotype-dependent manner. VP8* (aa 64-224), cloned from a HR strain (HAL1166), first isolated from a child in Finland13, was expressed in E. coli, purified to homogeneity, and crystallized for structural analysis. The sialidase-insensitive HAL1166 strain, phylogenetically and serologically belongs to G8P[14] genotype14. Although not as prevalent as the P[4] and P[8] genotypes, the P[14] genotypes are being increasingly documented by global rotavirus surveillance15–17, and P[14] HRs are thought to be able to jump from animal to human hosts17.
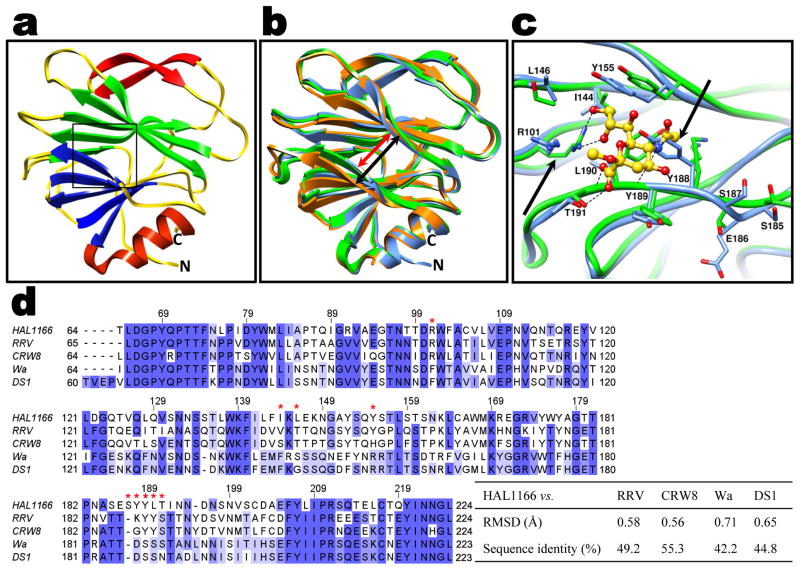
The structure of the HAL1166 VP8* determined to 1.5 Å resolution shows the characteristic galectin-like fold with two twisted β-sheets separated by a shallow cleft as observed in the VP8* structures from other rotavirus strains (Fig. 1a). The structure of P[14] VP8* superimposes well with all of the VP8* structures previously determined. One significant difference between these structures is in the width of the cleft separating the two twisted β-barrel sheets (Fig. 1b). In P[14] VP8*, it is narrower, similar to that in the VP8* of the animal strains, than the cleft in the VP8* of the other two human strains. In the animal VP8* structures, Sia binds near the cleft (Fig. 1c). Although the cleft in the P[14] VP8* structure is of similar dimensions as in the animal VP8* structures, the structural features in this region of P[14] VP8* is not compatible with Sia binding. In addition to changes in the amino acid residues (Fig. 1d) and side-chain orientations, the positioning of the amino acid residues is slightly shifted in this region because of an insertion (aa 187). The side-chain of Y188 is oriented in such a way that it would cause steric hindrance to Sia binding (Fig. 1c). Furthermore, the P[14] VP8* structure with a narrower cleft and several amino acid changes (Fig. 1d) is not compatible with binding of GM1 as suggested, based on computer modeling, for VP8* of the other human strains with a wider cleft3.
Fig. 1. VP8* structure of HAL1166 P[14] HR strain and structural comparison with other VP8* structures.
a. Cartoon representation of the P[14] VP8* structure showing a galectin-like fold with the characteristic two twisted β-sheets (in blue and green) separated by a cleft. The Sia binding site as observed in the animal VP8* is shown by a box. The β-ribbon and the C-terminal α-helix of the structure are shown in red. The N- and C-termini are denoted. b. Structural alignment of P[14] VP8* structure (blue) with VP8* structure of sialidase-sensitive animal rotavirus RRV strain (green, PDB id: 1KQR) and VP8* of sialidase-insensitive HR strain Wa (orange, PDB id: 2DWR) shown in the same orientation as in a. The width of the cleft in the P[14] VP8* is narrower (red arrow) than in the Wa VP8* structure (black arrow). c. P[14] VP8* exhibits changes in the amino acid composition and side chain orientations in the region corresponding to Sia binding site of RRV VP8*. The amino acid residues interacting with Sia in the RRV VP8* structure are shown as green sticks, and bound Sia is shown as yellow sticks with oxygen atoms in red. The residues in this region of the P[14] VP8* structure are shown as blue sticks. The residue numbering corresponds to HAL1166 VP8*. The key amino acid changes in this region between P[14] and RRV VP8* are indicated by black arrows. Noticeable is how Y188 in the P[14] VP8* structure causes steric hindrance if Sia were to bind in this region. Also noticeable is the change in side chain orientation of the conserved R101. d. Alignment of HAL1166 VP8* with other VP8* sequences from sialidase-sensitive animal (RRV, CRW-8) strains, and sialidase-insensitive HR (Wa and DS1) strains. The residues that interact with Sia in the animal VP8* as shown in c are indicated by red stars on the top of the sequence. Highly conserved (>80%), and moderately conserved (>60%) regions are colored in dark and lighter blue respectively. The root mean square deviation (rmsd) of the matching Cα atoms between the P[14] VP8* and other VP8* structures along with percentage of sequence identity are shown in the table on the right.
These observations prompted us to undertake a high throughput screening of a glycan array comprised of 511 different glycans including several glycans with terminal or internal Sia. Such a screening, which has been used to identify cellular glycans for a variety of pathogens including bacterial toxins18, influenza viruses19 and papovaviruses20, unambiguously showed specific binding to glycans with a terminal GalNAcα1-3(Fucα1-2)Galβ1-4GlcNAc sequence, which is a characteristic of A-type HBGA (Table S1, Figs. S1 and S2). None of the sialylated glycans, with either internal or terminal Sia, showed significant binding (Table S1, blue).
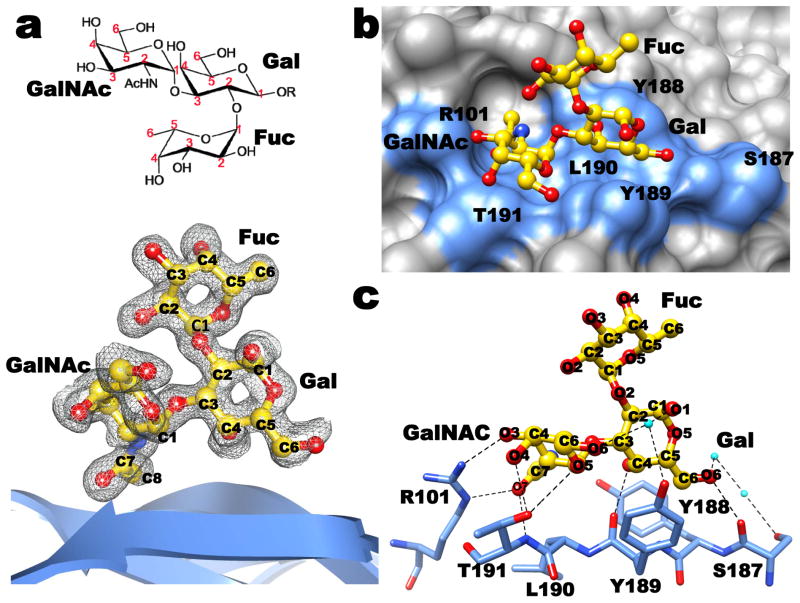
To understand the structural interactions between P[14] VP8* and A-type HBGA, we co-crystallized VP8* with tri- and tetrasaccharides that correspond to the terminal structure in the Atype HBGA. The structure of the complex, determined to a similar resolution of 1.5 Å as the unliganded structure, clearly showed density for the bound ligand (Fig. 2a). The HBGA binds near the cleft region at the same location as the Sia in the VP8* structures of the animal rotavirus strains (Fig. 2b and Fig. S3). The binding of HBGA, which does not cause any conformational changes in the VP8*, involves a network of both direct and solvent-mediated hydrogen bond interactions, in addition to several stabilizing hydrophobic contacts (Fig. S4). The terminal GalNAc and Gal of the HBGA participate in all of the interactions (Fig. 2c). The GalNAc participates in direct hydrogen bonding interactions involving the side chains of R101 and T191, and hydrophobic interactions involving L190 and T191, whereas Gal participates in hydrogen bonding interactions with the main chain carbonyl groups of Y189 and S187, and hydrophobic interactions with Y189 and Y188. The proximal sugar moieties project out from the surface of VP8* without making any direct contacts with VP8* (Figs. 2 and S3).
Fig. 2. Structural analysis of P[14] VP8*-A-type HBGA interactions.
a. The chemical structure of the A-type trisaccharide (above) and simulated annealing omit difference map (below), contoured at 3σ level, showing the binding of A-type trisaccharide to P[14] VP8*. Bound A-type trisaccharide (GalNAc: N-acetylgalactosamine; Gal: galactose; Fuc: fucose) is shown in a ball-and-stick representation (yellow) inside the map with its carbon atoms numbered following the standard convention. The nitrogen and the oxygen atoms in the trisaccharide are colored in blue and red, respectively. b. Surface representation of the P[14] VP8* structure (grey) with the bound A-trisaccharide shown in stick representation (with same color scheme as in a). The acetamido group of GalNAc inserts into a well-defined pocket in the VP8* structure. The amino acid residues in the P[14] VP8* which participate in hydrogen bond and hydrophobic interactions with the trisaccharide are indicated in blue. c. Network of hydrogen bond interactions (dashed lines) between the VP8* residues (light blue) and A-type trisaccharide (colored as in a). Participating water molecules are shown as small spheres (cyan). More detailed interactions between VP8* and the ligand are given in Fig. S4. The terminal two saccharide moieties of the A-type tetrasaccharide (GalNAcα1-3(Fucα1-2)Galβ1-4GlcNAc) also show similar interactions with the VP8* (Fig. S3).
The structure of P[14] VP8* with HBGA shows how subtle amino acid changes result in altered ligand specificity. The only positionally conserved common residue between P[14] and animal VP8* that participates in the ligand interactions is R101. Although the position matches well, its side-chain orientation differs significantly between the two structures (Fig. 1c). Without this change, the side-chain of R101 in the P[14] VP8* structure would cause steric hindrance to the GalNAc moiety of the HBGA. Most other residues that participate in the ligand interactions are upstream of position 187 in the VP8* sequence, where an insertion occurs in the P[14] VP8*. This insertion along with sequence variation alters the configuration of the binding site in the P[14] VP8* to allow specific interaction with HBGA. Insertion at position 187 causes a localized change which makes the side-chain of Y188 clash with Sia when placed in the structure of P[14] VP8* (Fig. 1c). Following this residue, the polypeptide chain reverts back to the same course as in the animal VP8* structures such that Y189 and T191, which interact with HBGA, are now positionally equivalent to Y188 and S190 which interact with Sia in the animal VP8* (Figs. 1c and 2c).
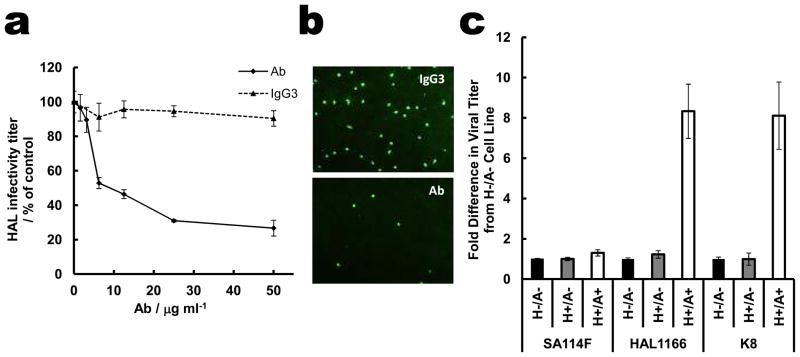
The remarkable overlap of the HBGA binding site in the P[14] VP8* with that of the Sia in the animal VP8* structure strongly suggests that A-type HBGA is a cell attachment factor for P[14] rotavirus strains. To examine the biologic relevance of HBGA binding to P[14] VP8*, virus infectivity assays were performed. Using intestinal HT29 cells isolated from a type A individual, dose-dependent abrogation of HAL1166 infectivity was observed, with a greater than 75% reduction at the highest concentration of anti-A-type HBGA antibody as compared to an isotype control antibody (Figs. 3a and 3b). In contrast, this antibody did not inhibit the sialidase-sensitive SA11 rotavirus strain (Fig. 3a). To further ascertain the specificity of the HAL1166 to A-type HBGA, infectivity assays were performed using parental CHO cells, which do not express any HBGA, and genetically-engineered CHO cells expressing either A- or H-type HBGA. CHO cells expressing type A HBGA showed a large increase in infectivity with HAL1166, but not SA11, compared to parental CHO cells or those expressing type H alone (Fig. 3c). Similarly, low infectivity was observed in Caco-2 cells, isolated from a blood type O individual, compared to HT29 cells that express type A HBGA (data not shown). Specificity to A-type HBGA was further confirmed by performing hemagglutination assays using P[14] GST-VP8*. Type A blood cells, but not type O or B, were hemagglutinated by soluble VP8* (Fig. S5).
Fig. 3. HAL1166 rotavirus specifically recognizes A-type HBGA.
a. Dose-dependent inhibition of HAL1166 infection in HT29 cells by anti-A-type antibody (Ab, white bars for HAL1166, and black bars for SA11). Isotype control IgG3 did not inhibit HAL1166 infectivity (grey bars). Error bars (also in Fig. 3c) represent standard deviation and the p-values were determined by student t-test, n=3. All concentrations of anti-A-type antibody reduced infectivity compared to control with p < 0.05. b. Representative immunofluorescence microscopy images of HT29 cells infected with HAL1166 rotavirus in the presence of 50 μg/ml of IgG3 (top) and anti-A-type antibody (bottom). c. Infectivity of SA11 P[1], HAL1166 P[14], and K8 P[9] rotavirus strains in the parental CHO cells (H-/A-), the single transfectant with the Fut2 enzyme (H+/A-), and the double transfectant with both Fut2 and A type histo-blood group glycosyltranferase (H+/A+). The fold difference in infectivity was determined compared to parental cells. For HAL1166 and K8 HR increase in the infectivity in CHO (A+/H+) cells compared to parental CHO, and CHO (H+/A-), the p-values were < 0.01.
This is the first study describing the structural interactions between a human VP8* and a cellular glycan. Importantly, it shows that binding of sialylated glycans is not obligatory among sialidase-insensitive HR strains. Our finding that P[14] VP8* specifically recognizes HBGA raises important questions such as whether other HR strains interact with similar or other HBGAs in a serotype-dependent manner like in human noroviruses10, and whether genetically controlled differential expression of HBGAs among world’s population plays a role in susceptibility to HRs. In a recently published paper, G8P[14] rotavirus was identified in the stool samples from two adults with diarrhea, who lived in the same geographical area in Denmark21. The blood type of one of these patients and of another patient infected with a G6P[14] virus was type A (B. Böttiger, personal communication). Although this is a small sample size, these findings warrant further epidemiological studies to determine whether HBGA is a susceptibility factor for rotaviruses. Based on sequence comparisons, our prediction that VP8* of the K8 HR strain (P[9] genotype) would also recognize A-type HBGA (Fig. S6) is firmly supported by infectivity assays with parental and derivative CHO cells (Fig. 3c).
The dsRNA rotaviruses, accounting for ~500,000 deaths annually in children worldwide22, exhibit enormous genetic and strain diversity. In addition to point mutations and gene rearrangements, genetic reassortment between co-circulating strains, similar to influenza viruses, contribute to the expanding diversity of rotaviruses23,24. Current evidence indicates that many of the HR strains including the P[14] HAL1166 strain17, originated from animal reservoirs through reassortment and inter-species transmission23,24. Although effective vaccines are currently available, whether they will remain effective with such expanding virus diversity is an open question25,26. Discovery that a HR strain with host-switching capabilities binds to a nonsialylated but novel glycan receptor opens new approaches to better understand the molecular basis of critical HR-host interactions, which likely influences host-specificity, cell specificity, pathogenesis, and virus evolution.
Methods Summary
Expression, purification, and crystallization of P[14] VP8* and its complex with A-type oligosaccharides, and structure determination, using RRV VP8* structure (PDB id: 1KQR) as a molecular replacement model, and refinement were carried out as described in the Supplementary Methods section. Diffraction data were collected at Baylor College of Medicine using Rigaku FR-E+ SuperBright rotating anode. See Table S2 for data collection and refinement statistics. The carbohydrate-binding specificity of P[14] VP8* was investigated using glycan array v4.2 with 511 glycans in replicates of 6 (Consortium for Functional Glycomics Protein-Glycan Interaction Core (H) (http://www.functionalglycomics). GST-tagged VP8* bound on the glycan array was detected using a fluorescent-labeled anti-GST monoclonal antibody. Infectivity assays were performed as previously described3,27.
Methods
Protein Expression and Purification
VP8* (aa 64-224) of HAL1166 rotavirus strain (P[14] genotype) was cloned into an expression vector pGEX-2T (GE healthcare) with an N-terminal GST tag and a thrombin cleavage site. The recombinant GST-VP8* was expressed in E. coli BL21 (DE3) (Novagen) and purified by Glutathione Sepharose 4 Fast Flow (GE healthcare). The GST tag was cleaved by using thrombin before rebinding the protein mixtures onto a Glutathione Sepharose column to remove the GST, leaving Gly-Ser at the N terminus. The VP8* was then filtered and further purified by size exclusion chromatography on a Superdex-75 (GE healthcare) column with 10 mM Tris, pH7.4, 100 mM NaCl, 1 mM DTT. The concentration of the purified VP8* was determined by measuring absorbance at 280 nm and using an absorption coefficient of 43,010 M−1cm−1 calculated using Vector NTI 11 software (Invitrogen).
Crystallization
Crystallization conditions for P[14] VP8* (13.5 mg/ml) were screened by hanging-drop vapor diffusion using the Mosquito crystallization robot (TTP LabTech) and visualized using Rock Imager (Formulatrix) at 20°C. The crystals from one of the conditions (30% PEG 1500, sodium acetate trihydrate, pH 4.5) were harvested with the screen condition containing 18% glycerol. To obtain crystals of VP8*-HBGA complex, VP8* was co-crystallized with A-type trisaccharide or tetrasaccharide (purchased from Dextra labs), with a 1:52 or 1:46 excess molar ratio of ligand under similar condition as the unliganded P[14] VP8*.
Data Collection and Processing
Diffraction data for both unliganded and liganded VP8* crystals were collected at Baylor College of Medicine using Rigaku FR-E+ SuperBright rotating anode. These data were processed with DTREK28 or IMOSFLM as implemented in the CCP4 suite29. Space group was confirmed using POINTLESS30. The unliganded and liganded VP8* structures in the P21 space group, with one molecule in the asymmetric unit, at ~1.5Å resolution were determined. For initial phasing, the RRV VP8* structure (PDB id:1KQR ) was used as a search model for molecular replacement using Phaser31. Following automated model building and solvent addition using ARP/wARP32, the structure was refined using PHENIX33. The oligosaccharide moieties of the HBGAs were generated using the SWEET 2 package34 of the Glycosciences.de server (http://www.glycosciences.de) and modeled into the electron density using COOT35 and validated by computing simulated annealing omit maps using PHENIX33. The stereochemistry of the oligosaccharides including the allowed conformational angles was checked using the CARP36 package in the Glycosciences.de server (http://www.glycosciences.de). Data collection and refinement statistics are given in Table S2. Ligand interactions were analyzed using COOT and LIGPLOT37 with donor to acceptor distances between 2.6 Å and 3.2 Å for hydrogen bonding interactions, and C-C distances between 3.4 Å and 4.5 Å for hydrophobic interactions. The structural alignments and calculations of rmsd were carried out using the PyMOL (http://pymol.sourceforge.net). Figures were prepared by using Chimera38.
Glycan Array Screening
The carbohydrate-binding specificity of HAL1166 VP8* was investigated on glycan array v4.2 comprised of 511 glycans (Consortium for Functional Glycomics, Protein-Glycan Interaction Core-H) (http://www.functionalglycomics). Recombinant GST-tagged VP8* at decreasing concentrations in binding buffer (20 mM Tris-HCl pH 7.4, 150 mM sodium chloride, 2 mM calcium chloride, 2 mM magnesium chloride, 0.05% Tween 20, 1% BSA) was applied to separate glycan arrays, and bound protein was detected using a fluorescent-labeled anti-GST monoclonal antibody. Summary of the glycan array results is given in Table S1. Concentration dependent binding at 20 μg/ml and 2 μg/ml is shown in Fig. S1a and S1b, where the glycans are ranked according to their relative binding strengths (Fig. S1c) as described previously39.
Inhibition and Infectivity Assays
Inhibition assays were performed on HT29 (human intestinal epithelial) cells following previously described protocols40. The monoclonal antibody (MAb) against blood group A antigen (BG-2) was purchased from Covance. The isotype control antibody (MG3-35) was purchased from Abcam. HAL1166 virus was grown as previously described41. Confluent HT29 cell monolayers were grown on 96-well plates. Increasing concentrations of antibodies were allowed to bind to the cells at 4°C for 1 hour before 400 fluorescent focus units (FFU) of virus were added per well. Recombinant VP8* protein competition was determined by treating the cells with 62.5 or 31 μg/ml VP8* at 4 °C for 30 min before virus inoculation as above. After allowing virus attachment to HT29 cells for 1 hour on ice, the inoculum was removed, and the cells were washed with cold DMEM and incubated for 16 hours at 37°C in 95% (vol/vol) air with 5% (vol/vol) CO2. Virus titers in methanol-fixed cell monolayers were determined by staining cells with rabbit anti-RV antibody and Alexa 488-labeled donkey anti-rabbit secondary antibody (Invitrogen). Virus infectivity was expressed as the percentage of focus-forming units in control wells incubated with the same concentrations of bovine serum albumin. Data are given as the mean of three replicates, and the bar indicates the standard deviation. Data were analyzed by Student's t test (two-tailed test).
For infectivity assays, SA11, HAL1166 or K8 viruses were serially diluted and incubated on confluent monolayers in 96 wells plates of HT29, Caco-2, and CHO cells (parental cells or cells expressing type H HBGA, or type H and type A HBGA prepared as previously described42). Following attachment for 1 hour at 37°C, the cells were washed, incubated for 16 hours and processed as described above for immunofluorescence.
Hemagglutination Assay
Pooled human red blood cells (RBCs) were purchased from Immucor (Atlanta, GA). The RBCs were packed via centrifugation at 500 × g for ten minutes and 0.5% suspensions of each RBC type were prepared in 0.85% saline (pH 6.2). GST (negative control) or GST-tagged VP8* were serially diluted via doubling dilution in PBS (0.01 M sodium phosphate, 0.15 M NaCl, pH 5.5; passed through a 0.2-μm-pore-size filter) on 96-well V-bottom plates (Nunc, Naperville, Ill.) in triplicate. The HA activity of the VP8 dimers was tested by mixing 50 uL of the prep (starting dilution of 10 nM) with an equal volume of the RBC suspensions. Recombinant Norovirus virus-like particles (Norwalk virus, genogroup GI.1 and Houston virus, genogroup GII.4) were included as positive controls (5 ug/ml and 10 ug/ml starting dilutions, respectively) because they have well-characterized HA activity that is known to be mediated by interaction with histo-blood group antigens on the surface of the RBCs43. The reaction was allowed to proceed for one hour at 4°C before results were recorded. The titer recorded was the highest dilution of sample that prevented the complete sedimentation of RBCs to the bottom of the well as compared to the negative controls. A commercial blood typing antibody specific for the B antigen was purchased from Immucor and tested at a starting dilution of 1:100 as another positive control.
Supplementary Material
Acknowledgments
We acknowledge the support from NIH grants AI36040 (to BVV), AI 080656 and P30 DK56338 (to MKE), GM62116 (to the Consortium for Functional Glycomics), and the Robert Welch foundation (Q1279) to BVV. We thank Robert Atmar and Sreejesh Shanker for helpful discussions and BCM X-ray core facility for data collection.
Footnotes
Author Contribution: LH carried out expression, purification, crystallization, diffraction data collection, structure determination. LH, SC, RC, and NC contributed to virus infectivity assays in HT29, CHO cells and hemagglutination assays and data analyses. DS contributed to glycan array experiments and analysis. JL provided parental and genetically modified CHO cells and advice. MKE provided supervision and advice on cell infectivity assays and analysis. LH and BVV analyzed and interpreted the structural data. BVV contributed to the overall direction of the project and wrote the manuscript with input from other authors.
Author Information: The coordinates and structure factors for the P[14] VP8* structures are deposited in Protein Data Bank under accession numbers 4DRR (apo), 4DRV(with A-type trisaccharide) and 4DSO (with A-type tetrasaccharide). Raw glycan array data are available at www.functionalglycomics.org/glycomics/publicdata/selectedScreens.jsp. Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Blanchard H, Yu X, Coulson BS, von Itzstein M. Insight into host cell carbohydrate-recognition by human and porcine rotavirus from crystal structures of the virion spike associated carbohydrate-binding domain (VP8*) J Mol Biol. 2007;367:1215–1226. doi: 10.1016/j.jmb.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 2.Dormitzer PR, et al. Specificity and affinity of sialic acid binding by the rhesus rotavirus VP8* core. J Virol. 2002;76:10512–10517. doi: 10.1128/JVI.76.20.10512-10517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haselhorst T, et al. Sialic acid dependence in rotavirus host cell invasion. Nat Chem Biol. 2009;5 :91–93. doi: 10.1038/nchembio.134. [DOI] [PubMed] [Google Scholar]
- 4.Lopez S, Arias CF. Early steps in rotavirus cell entry. Curr Top Microbiol Immunol. 2006;309:39–66. doi: 10.1007/3-540-30773-7_2. [DOI] [PubMed] [Google Scholar]
- 5.Settembre EC, Chen JZ, Dormitzer PR, Grigorieff N, Harrison SC. Atomic model of an infectious rotavirus particle. EMBO J. 2011;30:408–416. doi: 10.1038/emboj.2010.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dormitzer PR, Sun ZY, Wagner G, Harrison SC. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 2002;21:885–897. doi: 10.1093/emboj/21.5.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kraschnefski MJ, et al. Effects on sialic acid recognition of amino acid mutations in the carbohydrate-binding cleft of the rotavirus spike protein. Glycobiology. 2009;19:194–200. doi: 10.1093/glycob/cwn119. [DOI] [PubMed] [Google Scholar]
- 8.Marionneau S, et al. ABH and Lewis histo-blood group antigens, a model for the meaning of oligosaccharide diversity in the face of a changing world. Biochimie. 2001;83:565–573. doi: 10.1016/s0300-9084(01)01321-9. [DOI] [PubMed] [Google Scholar]
- 9.Ilver D, et al. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- 10.Glass RI, Parashar UD, Estes MK. Norovirus gastroenteritis. N Engl J Med. 2009;361:1776–1785. doi: 10.1056/NEJMra0804575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matthijnssens J, et al. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG) Arch Virol. 2011 doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monnier N, et al. High-resolution molecular and antigen structure of the VP8* core of a sialic acid-independent human rotavirus strain. J Virol. 2006;80:1513–1523. doi: 10.1128/JVI.80.3.1513-1523.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerna G, et al. Identification of a new VP4 serotype of human rotaviruses. Virology. 1994;200:66–71. doi: 10.1006/viro.1994.1163. [DOI] [PubMed] [Google Scholar]
- 14.Ciarlet M, Estes MK. Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J Gen Virol. 1999;80 ( Pt 4):943–948. doi: 10.1099/0022-1317-80-4-943. [DOI] [PubMed] [Google Scholar]
- 15.Chitambar SD, Arora R, Kolpe AB, Yadav MM, Raut CG. Molecular characterization of unusual bovine group A rotavirus G8P[14] strains identified in western India: emergence of P[14] genotype. Vet Microbiol. 2011;148:384–388. doi: 10.1016/j.vetmic.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 16.Fukai K, Saito T, Inoue K, Sato M. Molecular characterization of novel P[14],G8 bovine group A rotavirus, Sun9, isolated in Japan. Virus Res. 2004;105:101–106. doi: 10.1016/j.virusres.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 17.Matthijnssens J, et al. Are human P[14] rotavirus strains the result of interspecies transmissions from sheep or other ungulates that belong to the mammalian order Artiodactyla? J Virol. 2009;83:2917–2929. doi: 10.1128/JVI.02246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Byres E, et al. Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin. Nature. 2008;456:648–652. doi: 10.1038/nature07428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stevens J, et al. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J Mol Biol. 2006;355:1143–1155. doi: 10.1016/j.jmb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Neu U, et al. Structure-function analysis of the human JC polyomavirus establishes the LSTc pentasaccharide as a functional receptor motif. Cell Host Microbe. 2010;8:309–319. doi: 10.1016/j.chom.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Midgley SE, Hjulsager CK, Larsen LE, Falkenhorst G, Bottiger B. Suspected zoonotic transmission of rotavirus group A in Danish adults. Epidemiol Infect. 2011:1–5. doi: 10.1017/S0950268811001981. [DOI] [PubMed] [Google Scholar]
- 22.Parashar UD, Gibson CJ, Bresse JS, Glass RI. Rotavirus and severe childhood diarrhea. Emerg Infect Dis. 2006;12:304–306. doi: 10.3201/eid1202.050006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Estes MK, Kapikian AZ. In: Fields Virology. Knipe DM, Howley PM, editors. Vol. 2. Lippincott Williams & Wilkins; 2007. pp. 1917–1974. [Google Scholar]
- 24.Gray J, Iturriza-Gomara M. Rotaviruses. Methods Mol Biol. 2011;665:325–355. doi: 10.1007/978-1-60761-817-1_18. [DOI] [PubMed] [Google Scholar]
- 25.Angel J, Franco MA, Greenberg HB. Rotavirus vaccines: recent developments and future considerations. Nat Rev Microbiol. 2007;5:529–539. doi: 10.1038/nrmicro1692. [DOI] [PubMed] [Google Scholar]
- 26.Gentsch JR, et al. Serotype diversity and reassortment between human and animal rotavirus strains: implications for rotavirus vaccine programs. J Infect Dis. 2005;192 (Suppl 1):S146–159. doi: 10.1086/431499. [DOI] [PubMed] [Google Scholar]
- 27.Guillon P, et al. Inhibition of the interaction between the SARS-CoV spike protein and its cellular receptor by anti-histo-blood group antibodies. Glycobiology. 2008;18:1085–1093. doi: 10.1093/glycob/cwn093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr Sect D. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 29.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 31.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris RJ, Perrakis A, Lamzin VS. ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 2003;374:229–244. doi: 10.1016/S0076-6879(03)74011-7. [DOI] [PubMed] [Google Scholar]
- 33.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bohne A, Lang E, von der Lieth CW. SWEET - WWW-based rapid 3D construction of oligo- and polysaccharides. Bioinformatics. 1999;15:767–768. doi: 10.1093/bioinformatics/15.9.767. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lutteke T, Frank M, von der Lieth CW. Carbohydrate Structure Suite (CSS): analysis of carbohydrate 3D structures derived from the PDB. Nucleic Acids Res. 2005;33:D242–246. doi: 10.1093/nar/gki013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 38.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 39.Smith DF, Song X, Cummings RD. Use of glycan microarrays to explore specificity of glycan-binding proteins. Methods Enzymol. 2010;480:417–444. doi: 10.1016/S0076-6879(10)80033-3. [DOI] [PubMed] [Google Scholar]
- 40.Haselhorst T, et al. Sialic acid dependence in rotavirus host cell invasion. Nat Chem Biol. 2009;5 :91–93. doi: 10.1038/nchembio.134. [DOI] [PubMed] [Google Scholar]
- 41.Crawford SE, et al. Rotavirus viremia and extraintestinal viral infection in the neonatal rat model. J Virol. 2006;80:4820–4832. doi: 10.1128/JVI.80.10.4820-4832.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guillon P, et al. Inhibition of the interaction between the SARS-CoV spike protein and its cellular receptor by anti-histo-blood group antibodies. Glycobiology. 2008;18:1085–1093. doi: 10.1093/glycob/cwn093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hutson AM, Atmar RL, Marcus DM, Estes MK. Norwalk virus-like particle hemagglutination by binding to h histo-blood group antigens. J Virol. 2003;77:405–415. doi: 10.1128/JVI.77.1.405-415.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.