Abstract
Like eukaryotes, bacteria must coordinate division with growth to ensure cells are the appropriate size for a given environmental condition or developmental fate. As single-celled organisms, nutrient availability is one of the strongest influences on bacterial cell size. Classic physiological experiments conducted over four decades ago first demonstrated that cell size is directly correlated with nutrient source and growth rate in the Gram-negative bacterium Salmonella typhimurium. This observation subsequently served as the basis for studies revealing a role for cell size in cell cycle progression in a closely related organism, Escherichia coli. More recently, the development of powerful genetic, molecular, and imaging tools has allowed us to identify and characterize the nutrient-dependent pathway responsible for coordinating cell division and cell size with growth rate in the Gram-positive model organism B. subtilis. Here, we discuss the role of cell size in bacterial growth and development and propose a broadly applicable model for cell size control in this important and highly divergent domain of life.
INTRODUCTION
Coordinating growth with division is essential to ensure that cells are the appropriate size for a given environmental condition or developmental fate. This is true not only for multicellular plants and animals, but also for single-celled organisms that need to adapt quickly to rapid changes in environmental conditions. Like their eukaryotic counterparts, in the absence of environmental or internal pressure to increase size, exponentially growing bacteria cultured under a constant set of parameters exhibit little size variation between cells. Maintenance of cell size within a narrow band indicates that cells have mechanisms to transiently adjust the timing of division to correct aberrations in cell size generated through stochastic events. Similarly, although bacterial cell size is essentially constant under steady state conditions, environmental challenges and developmental programs frequently require changes in cell size just as they do for other single and multicellular organisms.
Here we review what is known about bacterial cell size control during steady state growth, in response to changes in nutrient availability, and during development. For reasons of brevity we will focus on two well-studied model systems, Escherichia coli and Bacillus subtilis. Where appropriate we will also include information gleaned from work in other organisms.
Before we delve into the discussion of bacterial cell size control in mesophilic model systems, it is important to note that bacteria occupy habitats that include thermal vents where temperatures are well over 100°C, 5M saline salt pools, and environments where ionizing radiation levels are 1000X times the lethal dose for humans. Bacteria also exhibit a vast array of morphologies ranging from rods and filaments to cocci, spirals and amoeboid-like forms. The diversity of bacteria is mirrored in the size of individual species which range from ~0.3μm for obligate intracellular pathogenic members of the genus Mycoplasma, to ~600μm for Epulopiscium fishelsoni, a Gram-positive commensal inhabitant of Surgeonfish guts, and 750μm for Thiomargarita namibiensis, a chemilithotrophic Gram-negative bacterium native to coastal Namibia [3–5]. While T. namibiensis is essentially a large gas vesicle surrounded by a thin layer of cytoplasm, Epulopiscium has managed to overcome diffusion-dependent limitations on cell size in part by increasing genome number along with cell size. These tens-of-thousands of genomes are arranged around the periphery of the Epulopiscium cell, where they are thought to facilitate responses to local stimuli and thereby contribute to maintenance of the extremely large cell size [6].
Similarly, although it is not the focus of this review, it is important to note that cell size and shape are, not surprisingly, sensitive to changes in the morphogenesis of the bacterial cell wall. In particular, enzymes involved in synthesizing the peptidoglycan material that constitutes the bacterial cell wall, as well as the Mre proteins which recent data suggest help coordinate peptidoglycan synthesis, all play an important role in cell size control by maintaining cell shape and width within normal parameters. For excellent reviews on this topic see [7–9].
BINARY FISSION: A DECEPTIVE SIMPLE MODE OF REPRODUCTION
Bacteria exhibit many forms of reproduction including binary fission, budding (Planctomycetes), hyphal growth (Actinomycetes), daughter cell formation (Epulopiscium), and the formation of multicellular baeocytes (the cyanobacterium Stanieria). Of these, binary fission is one of the most common, and is by far the best understood.
Binary fission in B. subtilis and E. coli is deceptively simple. During exponential growth, cells double in mass and then divide in the middle to produce equivalently sized daughter cells. Despite its apparent simplicity, binary fission is in fact the culmination of a complex, elaborately orchestrated series of events. Binary fission requires cells to double in mass, initiate and terminate at least one round of chromosome replication, decatenate and segregate sister chromosomes (also referred to as nucleoids), assemble the division machinery precisely at midcell, and coordinate membrane invagination with cell wall synthesis to form a complete septum (Figure 1).
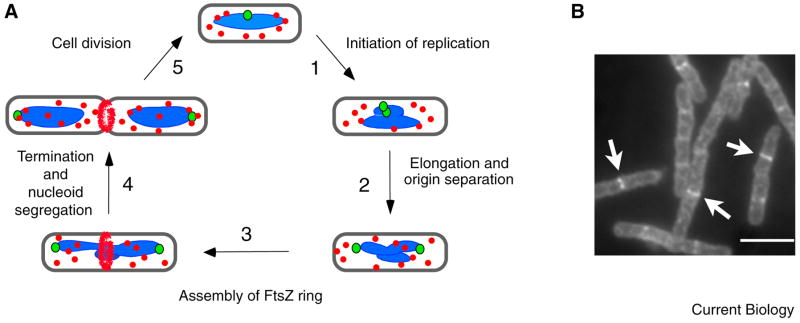
Figure 1. The bacterial division cycle.
FtsZ assembly is coordinated with DNA replication and segregation. A. (1) In newborn cells, FtsZ (red) is unassembled. A circular chromosome (blue) with a single origin of replication (green) is located at mid-cell. (2–4) After chromosome replication initiates, the origins of replication separate and move to opposite poles of the cell. Once replication is complete, the condensed chromosomes separate, leaving a nucleoid free space. (3) FtsZ ring formation coincides with chromosome segregation. Assembly starts from a single point at mid-cell and extends bidirectionally. (5) During cytokinesis, the FtsZ ring constricts at the leading edge of the invaginating membrane. B. Immunofluorescence micrograph of exponentially growing B. subtilis cells with FtsZ rings. Arrows indicate examples of cells with rings. Bar = 5μm.
In contrast to eukaryotes, the bacterial cell cycle is not divided into discrete stages. Instead cell growth, DNA replication, chromosome segregation, and even the initial assembly of the division machinery can overlap with one another, a physically challenging proposition at faster growth rates. Due to its overlapping nature, the nomenclature used for describing stages of the eukaryotic cell cycle (G1, S, G2, and M) is not useful when describing the bacterial cell cycle. The alternative nomenclature includes three discrete periods: B, the time between cell birth and the initiation of DNA replication, C, the period required for chromosome replication and D period, the time between the termination of replication and division.
Under steady state conditions, B. subtilis and E. coli cells exhibit little variation in cell size beyond the requirements of binary fission [1, 2]. Maintaining cell size within these parameters suggests cells precisely control both the timing and position of cell division and can compensate for stochastic events that lead to a reduction in cell size or an increase in cell size, by transiently altering the length of their cell cycle (Figure 2). Although changes in the duration of any cell cycle event can theoretically impact cell size, in E. coli and B. subtilis, only two, the initiation of DNA replication and cell division, have been implicated as important control points in the homeostatic regulation of cell size. Below we discuss the role of initiation and division in the spatial and temporal control of cell size under steady state conditions.
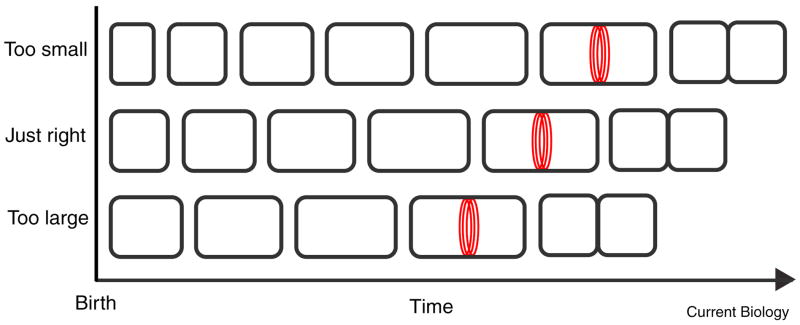
Figure 2. Transient changes in the length of the cell cycle are required for cell size homeostasis under steady state conditions.
B. subtilis and E. coli cells exhibit little variation in cell size beyond the requirements of binary fission during steady state growth. Individual cells that are born too short transiently increase the length of their cell cycle to increase size while cells that are too long experience a transient reduction in the length of their cell cycle to reduce the daughter cell size. At the time of division (red rings), the size of all three cells is the same resulting in the production of appropriately sized daughter cells.
Cell size and the initiation of DNA replication
The initiation of DNA replication is tightly correlated with achievement of a particular cell size in both E. coli and B. subtilis [10, 11]. Merging data from the seminal physiological studies of Moselio Schaechter, Ole Maaløe, and Neils Kjeldgaard working in Salmonella [12] and Helmstetter and Cooper, working in E. coli [13], William Donachie deduced that the mass of bacterial cells at the time of replication initiation is constant, regardless of growth rate [10]. Donachie interpreted this data to mean that attainment of a specific cell size is required to trigger DNA replication. On the basis of this interpretation he proposed existence of a positive regulator that accumulates in a growth-dependent manner, reaching critical levels only when cells attain a specific size. As a model for cell size control, Donachie’s proposal was intuitively appealing; as the first step in the cell cycle, changes in the timing of replication initiation should theoretically impact the entire cell cycle, and with it, cell size.
Later work subsequently identified DnaA, a highly conserved AAA+ ATPase, as a good candidate for Donachie’s positive, growth-dependent regulation of replication initiation [14–16]. In its active, ATP bound form DnaA binds cooperatively to sequence specific DnaA boxes within the chromosomal origin of replication (oriC) and drives open complex formation, facilitating loading of the replication machinery [17–20]. DnaA binds to its own promoter and autoregulates its production [21–23]. Following initiation in E. coli, DnaA is inactivated through a variety of mechanisms, including sequestration of its promoter, titration by chromosomal binding sites, and conversion to the inactive ADP-bound form, to ensure that only one round of replication is initiated per division cycle [16, 24]. The ratio of free DnaAATP to oriC then increases in a growth rate dependent manner until it is high enough to support initiation, a point that is coincident with achievement of a specific mass in wild type E. coli [16, 24].
Consistent with a role for DnaA in the size-dependent regulation of replication initiation, significantly reducing DnaA expression delays initiation and increases cell size, while overexpressing DnaA leads to premature initiation and a reduction in cell size in both E. coli and B. subtilis [14, 23]. Similarly, short E. coli mutants delay initiation until they reach a size that is approximately equivalent to wild type cells [25]. This delay is alleviated following a modest increase in DnaA expression, supporting the idea that growth-dependent accumulation of active DnaA to critical levels is the primary trigger for initiation in E. coli.
Although it is easy to imagine how normalizing size at initiation might be a conserved strategy to ensure that cell size is maintained under steady state conditions, several lines of evidence argue against this possibility. First and foremost, altering initiation timing in E. coli leads to compensatory changes in the timing and duration of downstream cell cycle events, particularly the length of time available for chromosome replication. Cells that initiate replication early exhibit an extended C period, while those that delay initiation, increase the rate of DNA replication, reducing C period by as much as 30% [25–30]. Thus, cell size control is unlikely to be solely a product of initiation mass. Moreover, although B. subtilis superficially appears to maintain a constant initiation mass [31], data from short mutants suggests the initiation of replication in B. subtilis is independent of cell size [2, 25], a finding consistent with recent work on the regulation of DnaA activity in this organism [32–40]. Together, these data argue for the presence of homeostatic mechanisms responsible for maintaining cell size that can, depending on the circumstance, override the effect of changes in initiation mass.
Cell size and assembly of the division machinery
As the last step in the cell cycle, the bacterial equivalent of M phase, the precise spatial and temporal regulation of cell division is a fundamental part of cell size control. Dividing before doubling in mass or mislocalizing the division machinery both lead to aberrations in daughter cell size and potentially fatal defects in chromosome segregation. For most bacteria, cell division is initiated by assembly of the tubulin homolog FtsZ into a ring-like structure at mid-cell, the future site of cell division [41, 42] (Figure 1). The FtsZ ring serves as a scaffold for assembly of the cell division machinery and constricts at the leading edge of the invaginating membrane during binary fission. The nature of the signals initiating FtsZ assembly at the beginning of the division process and stimulating constriction of the ring at the end of the process are not known. In E. coli and B. subtilis, FtsZ levels are constant across a wide range of growth rates, a finding that suggests FtsZ ring formation is controlled at the level of FtsZ assembly, rather than by altering FtsZ levels over the course of the cell cycle [43]. A multitude of factors function collectively to ensure that FtsZ ring formation and constriction are coordinated, both temporally and spatially, with DNA replication and chromosome segregation [41]. (In contrast to B. subtilis and E. coli, FtsZ levels, like those of many other proteins, are regulated in a cell cycle dependent manner in Caulobacter [44].)
The positional regulation of FtsZ assembly
Binary fission by definition requires the division machinery to be precisely positioned at mid-cell. While factors that promote FtsZ assembly at mid-cell have yet to be indentified, inhibitors that prevent FtsZ assembly at aberrant positions play an important role in the positional regulation of cell division in E. coli and B. subtilis. Two sets of inhibitors in particular, function in concert to help restrict assembly of FtsZ and the division machinery to the DNA-free space at mid-cell. Components of the Min system, the first regulators of cell division to be characterized at the molecular level [45], inhibit FtsZ assembly at the cell poles while the DNA-associated proteins, SlmA in E. coli and Noc in B. subtilis, help prevent assembly of the division machinery over unsegregated chromosomal DNA (for reviews on both sets of inhibitors see: [46, 47]). Defects in components of the Min system result in a high frequency of aberrant FtsZ assembly at sites immediately adjacent to cell poles and the formation of tiny anucleate minicells—the product of polar division events. In contrast, although single mutations in noc or slmA have little impact on either the temporal or spatial control of cell division, they are synthetically lethal when combined with mutations in min due to the formation of FtsZ rings at anomalous positions [48, 49].
The study of the double mutants supports a model in which Min and Noc in B. subtilis and SlmA in E. coli function together to help prevent FtsZ assembly at cell poles and over unsegregated DNA. The completion of chromosome segregation reveals a division inhibition free zone at mid-cell that is then utilized by FtsZ. Both the Noc and SlmA chromosomal binding sites are concentrated toward the origin of replication and absent in the terminus. Based on their chromosomal location, regions enriched with SlmA or Noc binding sites should move from the future division site prior to termination of replication [50, 52–54]. While SlmA interacts directly with FtsZ to inhibit assembly, the primary target of Noc-mediated division inhibition is not known [50, 51].
Although Noc/SlmA and Min help prevent aberrant FtsZ ring formation and division at cell poles and across unsegregated nucleoids, they do not appear to impact the timing of divison under normal conditions nor do they function as the bacterial equivalent of a G2-M phase checkpoint. First and foremost, as mentioned above, single mutations in noc or slmA have no significant impact on the timing or position of cell division [48, 49]. Moreover, the severe cell division defect associated with the loss of both noc/slmA and min appears to be due to titration of FtsZ away from the medial division site, rather than a consequence of aberrent division events across unsegregated DNA [48, 49]. Finally, FtsZ ring formation and division can take place over unsegregated chromosomes in sporulating B. subtilis cells, short B. subtilis mutants, and in B. subtilis cells in which replication is artificially blocked prior to termination [2, 55–57]. In the two former cases, disaster is averted through the actions of the SpoIIIE DNA translocase which pumps chromosomal DNA out of the way of the invaginating septum [2, 55].
Noc is also found in S. aureus, which does not have a Min system. In contrast to E. coli and B. subtilis, single mutations in S. aureus noc lead to mislocalization of FtsZ, division over unsegregated DNA and DNA breaks, suggesting it plays a much more central role in coordinating division with chromosome segregation during normal growth in this organism [58]. In C. crescentus, an unrelated chromosome-associated protein MipZ contributes to the spatial regulation of division by helping coordinate chromosome segregation with FtsZ assembly [59].
While a positive acting factor directing FtsZ to mid-cell has yet to be identified in B. subtilis or E. coli, there are hints that the initiation of DNA replication may play a role in establishing the FtsZ assembly site at mid-cell [60, 61]. Blocking replication initiation through the use of conditional mutants, leads to the formation of an asymmetrically positioned FtsZ ring adjacent to a medially positioned bacterial chromosome in both E. coli and B. subtilis [61, 62]. However, permitting initiation, but blocking the first steps in elongation in B. subtilis, using outgrowing spores to synchronize cells, leads to the formation of medial FtsZ rings [61]. Similar findings have also been reported in C. crescentus, suggesting the role of replication initiation in division site selection may be conserved [63].
Coordinating FtsZ assembly with cell size under steady state conditions
At its most fundamental level, maintaining cell size under steady state conditions requires cell division to be precisely coordinated with growth rate and mass doubling time. Dividing before doubling in mass reduces cell size while dividing too late increases cell size. In bacteria, cell division is dependent on assembly, maturation and constriction of the cytokinetic ring. FtsZ and other components of the division machinery assemble at the nascent division site early in the cell cycle, at a time that coincides with, but is not dependent on chromosome segregation [2, 64].
Once formed, the cytokinetic ring is present for a significant period of time influenced by growth rate and mass doubling time. In fast growing B. subtilis cells (mass doubling time ~ 26 minutes) the Z period (the time the FtsZ ring is present) is ~22 minutes. In slow growing cells from the same strain (mass doubling time ~80 minutes), the Z period is ~40 minutes. Notably, the Z period is significantly longer than the time required for the cell to physically divide. In E. coli, the Z period is ~50 minutes for K-12 cells cultured under conditions supporting 85 minutes of mass doubling time, while the time between the first evidence of constriction and physical separation of daughter cells is ~ 22 minutes [65]. Why the Z period is so long is something of a conundrum. Although FtsZ contributes to cell elongation in some species, including E. coli and C. crescentus, in others with similarly long Z periods, it is required solely for cross wall synthesis [66, 67]. One possibility is that an extended Z period provides the cell with ample opportunity to modify the timing of division in response to changes in nutrient availability, DNA damage, and aberrations in prior events in the cell cycle.
FtsZ levels are constant over a wide range of mass doubling times in both E. coli and B. subtilis, a finding that strongly argues against a model in which cell division is controlled by oscillations in FtsZ concentration [43]. Instead, it is likely that the timing of FtsZ ring formation, and with it maturation and constriction of the cytokinetic ring, are governed through finely graded changes FtsZ assembly dynamics over the course of the cell cycle. This model is consistent with the observation that mutations that alter the efficiency of FtsZ ring formation and division significantly increase average cell size [46, 68–71]. Importantly, under steady state growth conditions, a small reduction in FtsZ expression leads to a transient delay in division. After increasing size to accommodate the reduction in FtsZ levels, cells resume growth with mass doubling times indistinguishable from wild type [72]. In other words, growth rate, not cell size, appears to be the overriding mechanism governing the timing of bacterial cell division.
THE NUTRIENT-DEPENDENT CONTROL OF BACTERIAL CELL SIZE
In landmark work Schaechter, Maaløe, and Kjeldgaard first noted that the size of bacterial cells corresponds with growth rate, which itself is dependent on the nutrient condition in which they are cultured [12, 73]. They observed that Salmonella cells were approximately twice as large when cultured at fast growth rates in a rich nutrient source than the same cells cultured at slower growth rates in a poor nutrient source. The nutrient-dependent control of cell size was subsequently shown to apply to other evolutionarily similar and distant bacteria including B. subtilis [74, 75] and S. aureus, where long-term glucose limitation leads to a heterogeneously sized population in which the average diameter is reduced by ~40% [76]. Single-celled eukaryotes, including the classic cell cycle model system, Schizosaccharomyces pombe, also modulate size in response to changes in nutrient availability [77]. While B. subtilis cells increase exclusively in length, E. coli cells cultured in nutrient-rich medium are both longer and wider than those cultured in nutrient-poor medium [1, 78, 79].
The ability to coordinate growth rate with size require cells to 1) sense nutrient availability and 2) transmit this information to the division apparatus to alter size accordingly (Figure 3A). In B. subtilis the nucleotide sugar UDP-glucose (UDP-glc) appears to function as an intracellular proxy for nutrient availability in the signal transduction pathway governing cell size. Mutations in pgcA or gtaB, genes required for the synthesis of UDP-glc, reduce the length of B. subtilis cells by 35% and 25% respectively during growth in nutrient-rich medium without a significant impact on growth or DNA replication [2, 80]. E. coli cells defective in pgm (the pgcA homolog) are also ~30% shorter than wild type cells during growth in LB, suggesting UDP-glc may also serve as a proxy for nutrient availability in this evolutionarily divergent bacterium [25, 81].
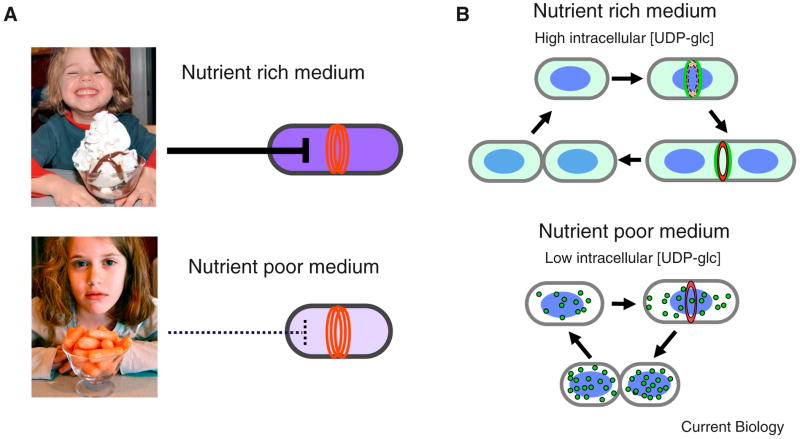
Figure 3. A carbon-dependent inhibitor of cell division.
A. Carbon source has the largest impact on the size of E. coli and B. subtilis cells at division. A rich carbon source, depicted here as an ice cream sundae, ensures cells are large enough to accommodate extra DNA generated by multifork replication via carbon-dependent inhibition of cell division (red rings). Conversely, a poor carbon source, depicted here as carrots, has little effect on cell division, resulting in a reduction in average cell size. UDP-glc, purple, serves as the intracellular proxy for carbon and is thus at a higher concentration in cells cultured in carbon-rich conditions than in carbon-poor conditions. B. The glucosyltransferase UgtP inhibits division in a carbon-dependent manner. (Top) In a rich carbon source, high intracellular concentrations of UDP-glc stimulate UgtP (green) localization to mid-cell where it interacts directly with FtsZ to inhibit assembly and/or maturation of the FtsZ ring and increase cell size. (Bottom) During growth in a poor carbon source, UgtP expression levels are reduced and the remaining protein is sequestered in randomly positioned foci, permitting division to proceed unimpeded and reducing average cell size.
Specifically why UDP-glc would be conserved as an intracellular proxy for nutrient availability in the growth-dependent regulation of cell size is not entirely clear. In contrast to its precursor glucose-6-phosphate, UDP-glc appears to be required exclusively for non-essential processes in B. subtilis and E. coli, including generation of glucosylated lipids, modification of cell wall polymers, and synthesis of periplasmic carbohydrates (e.g. [82–84]). Like glycogen synthesis, cells may shunt glucose through the UDP-glc biosynthesis pathway only when carbon and other nutrients required for biosynthesis are in excess, the same conditions that support rapid growth and multifork replication. (ADP-glc rather than UDP-glc is used as the precursor for glycogen synthesis in bacteria [85]). Intriguingly, in E. coli UDP-glc also appears to be part of the signal transduction cascade controlling activation of the stationary phase transcription factor σS, a phenomenon that is also associated with carbon limitation [86]. Given the significant evolutionary distance between E. coli and B. subtilis (they are evolutionarily more divergent than humans and bakers yeast), it will be interesting to determine if UDP-glc functions as a proxy for nutrient availability in other bacterial, and even eukaryotic, systems.
In B. subtilis, the glucosyltransferase UgtP serves as the UDP-glc dependent effector responsible for coordinating cell size with nutrient availability [2]. As expected for a nutrient-dependent regulator of division, defects in UgtP reduce cell size by approximately 20% during growth in nutrient-rich medium but do not significantly impact cell size under nutrient limiting conditions. ugtP mutants do not exhibit any apparent defects in cell growth or viability, suggesting their primary defect is in cell size homeostasis. In vitro UgtP interacts directly with FtsZ to inhibit FtsZ assembly.
In the cell, UgtP-mediated division inhibition is coordinated with nutrient availability through UDP-glc dependent changes in UgtP localization (Figure 3B). UgtP is distributed throughout the cytoplasm of rapidly growing B. subtilis cells cultured in a rich nutrient source and concentrated at the cytokinetic ring where it interacts directly with FtsZ to inhibit division, thereby increasing cell size. Conversely, during growth in nutrient-poor medium or in cells defective for UDP-glc biosynthesis, UgtP is sequestered away from the cytokinetic ring in small punctate foci that are randomly distributed within the cell. The precise nature of the UgtP foci (i.e. storage units for inactive protein, UgtP oligomers, etc.) has yet to be determined. UgtP-mediated division inhibition is further repressed during growth in nutrient-poor medium via nutrient-dependent reductions in either its expression or stability [2]. Under these conditions, FtsZ ring formation and division proceed unimpeded, reducing average cell size.
ugtP null mutants are wild type for cell growth, mass doubling time, and Z period [2]. Thus, UgtP-mediated division inhibition is likely to be transient, taking place only when cells must increase size following an increase in nutrient availability, or in the event that a cell is too small for a particular growth condition. Because division inhibition is transient, data from steady state cultures does not clarify whether UgtP delays division by inhibiting FtsZ ring formation or by preventing maturation of an already extant FtsZ ring.
UgtP is a bifunctional protein whose glucosyltransferase activity is required for generating di-glucosyl-diacylglycerol, the membrane anchor for lipoteichoic acid, a major component of the Gram-positive bacterial cell wall. Although the di-glucosyl modification is generally dispensable for growth and viability, ugtP null mutants can exhibit condition-dependent defects in cell wall morphology [87, 88]. UgtP may thus serve as a link between cell wall synthesis and division under conditions supporting rapid growth. A UDP-glc-dependent effector responsible for coordinating nutrient availability with cell size has yet to be identified in E. coli, which does not have a UgtP homolog.
Cell size and chromosome segregation
While it is clear that B. subtilis and E. coli coordinate size with nutrient availability, precisely why they do so is less clear. The most likely possibility is that the increase in size permits cells to accommodate extra DNA generated by multifork replication. Multifork replication makes it possible for certain bacteria to sustain mass doubling times shorter than the period required to initiate and complete chromosome replication and division. Although initiation is still limited to once per division cycle, multifork replication permits the initiation of a new round of DNA replication prior to completion of the previous round and rapidly growing cells are thus born with multiple active replication forks [13]. B. subtilis and E. coli cells grown in a complex, nutrient-rich medium supplemented with glucose, can have twelve or more replication forks proceeding simultaneously.
Consistent with growth rate dependent increases in size being a mechanism for dealing with the excess DNA generated by multifork replication, both E. coli and B. subtilis increase size at faster growth rates such that the cell mass to DNA content is maintained under conditions supporting multifork replication [1, 31, 75, 89, 90]. While it is formally possible that the increase in size is a result of the increased biosynthesis due to the additional DNA, data from short B. subtilis and E. coli mutants argue against this idea. In the mutant cells, the DNA content and origin to cell mass ratio is elevated under conditions supporting multifork replication, despite a ~35% reduction in size [25, 43]. Instead we favor the idea that growth rate dependent increases in size are the result of nutrient-dependent changes in FtsZ assembly, as discussed below.
The pressure to maintain a constant DNA to cell mass ratio over a range of growth rates may reflect a physical constraint of chromosome segregation. Theoretical work suggests that reductions in the concentration of DNA are more amenable to chromosome segregation [91]. Hence, increasing cell size during multifork replication may be required to ensure that DNA concentration remains within ideal parameters. Consistent with this idea, E. coli and B. subtilis both appear to require achievement of a critical length prior to the initiation of chromosome segregation [11, 92]. Moreover, mutations that reduce B. subtilis cell size lead to an increased frequency of FtsZ assembly and cell division across unsegregated nucleoids, regardless of the presence of Noc [2].
BACTERIAL CELL SIZE AND DEVELOPMENT
As it is in many eukaryotes, bacterial cell size is frequently tied to developmental fate. For example, members of the photosynthetic genus Anabaena typically grow as long chains of vegetative cells. Under nitrogen limiting conditions, however, approximately one-in-ten cells differentiates into a much larger, nitrogen fixing heterocyst [93]. Similarly, upon entering the root hairs of leguminous plants, Rhizobia increase in size as they differentiate into nitrogen fixing bacteroids [94]. Sporulation in the filamentous soil bacterium Streptomyces coelicolor is preceded by the formation of evenly spaced septa that divide syncytial aerial filaments into coccoid spores, each containing a single copy of the genome. Although it is not known how spore size is determined, recent studies in S. coelicolor have identified the factors required for stimulating FtsZ assembly and division at precisely spaced positions thereby ensuring the production of uniformly sized spores [95]. Two of the best studied examples of developmentally regulated cell size control are asymmetric division during the C. crescentus cell cycle, and the polar cell division that is the first morphologically distinct step in B. subtilis spore formation.
In C. crescentus each round of the cell cycle produces two cell types, the larger stalked cell and the smaller, flagellated swarmer cell [44]. In addition to their distinct morphologies, these cells have very different developmental fates: stalked cells are capable of initiating new rounds of DNA replication and division while the swarmer cells remain in G1 phase until they differentiate into stalked cells. Although it was initially thought that the size difference was established by preferential growth on the stalk side of a medially positioned FtsZ ring, recent data suggest that the cell size asymmetry is established by positioning the FtsZ slightly closer to the swarmer cell pole [96]. This occurs through interactions between DNA proximal to the chromosomal origin of replication, the polarity determinant TipN, and the division inhibitor MipZ [59, 96–99]. Another intriguing, size-related aspect of Caulobacter development, is the ability of the stalk to increase in size in response to changes in extracellular phosphate. The stalk, which is an extension of the cell body rather than an extra appendage, exhibits an up to a 30-fold increase in length upon phosphate starvation [100]. The increase in stalk length increases the surface area to volume ratio of the cell and is thought to increase the ability of Caulobacter cells to take up phosphate.
Endospore formation in B. subtilis requires an even more dramatic asymmetric division event that divides the cell into two compartments, the large mother cell and the tiny forespore [101] (Figure 4). The switch from medial to asymmetric division is mediated by the transcription factor Spo0A. As a response regulator protein, Spo0A is activated by phosphorylation as part of a signal transduction cascade initiated in response to starvation and crowding [102]. Once activated, Spo0A induces the expression of genes that suppress FtsZ assembly at mid-cell and activate FtsZ assembly at both cell poles [103, 104] (Figure 4). In particular, Spo0A-dependent induction of the SpoIIE phosphatase stimulates re-localization of FtsZ from a spiral intermediate that extends the length of the cell to ring structures at both poles [103]. Only one of the polar rings matures into a septum, through an apparently stochastic process.
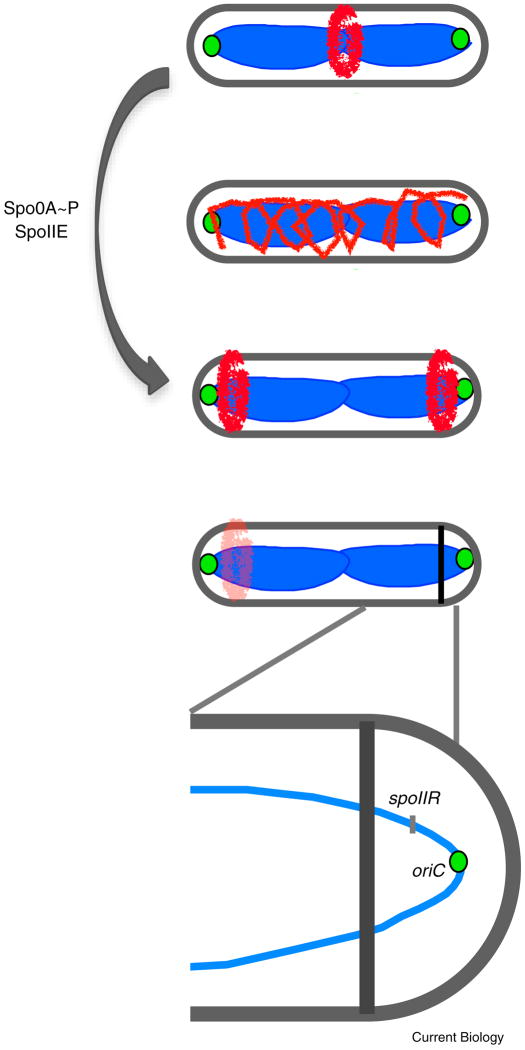
Figure 4. Developmentally regulated changes in division site selection establish cell type specific gene expression during sporulation in B. subtilis.
FtsZ (red), DNA (blue), origin of replication (green). Activation of the transcription factor Spo0A in response to nutrient limitation and cell crowding induces expression of genes, including spoIIE, required for relocalization of FtsZ from mid-cell to both poles via a spiral intermediate. Through a stochastic process, one FtsZ ring is used for polar septation while the other one is disassembled in response to the onset of mother cell-specific gene expression. (Bottom inset) Activation of forespore-specific gene expression is controlled in part via transient genetic asymmetry. The asymmetrically positioned septum bisects the chromosome, such that only the origin proximal ~30% is in the forespore immediately following septation. Forespore-specific gene expression immediately following septation is thus limited to origin proximal loci until the remainder of the chromosome is pumped into the forespore through the actions of the DNA translocase, SpoIIIE.
Activation of forespore specific gene expression appears to be dependent in part on the transient genetic asymmetry generated by formation of the polar septum which bisects one end of the chromosome such that only the origin-proximal ~30% region is in the forespore immediately following septation (for review see [105]). The remainder of the chromosome is later pumped into the forespore via the SpoIIIE DNA translocase [57, 106–108]. The subsequent activation of mother cell specific gene expression “deactivates” the remaining FtsZ ring [109]. As sporulation progresses, the mother cell engulfs the forespore through an endocytosis-like mechanism, synthesizes a protective coat for it, and finally releases the mature spore through the ultimate act of bacterial altruism, cell lysis [102].
It is still an open question if transient genetic asymmetry is sufficient to explain cell-type-specific activation of gene expression, or if the diminutive size of the forespore also plays a role. Intriguingly, in the endospore forming coccus Sporosarcina ureae, which like B. subtilis is a member of the order Bacillales, differential gene expression does not involve formation of a polar septum. Instead, S. ureae is able to achieve differential gene expression following what appears to be binary fission [110]. Why asymmetric division is not required for compartment specific gene expression in S. ureae is unclear, although this observation suggests there may be more than one way to generate differential gene expression in this group of organisms.
A MODEL FOR THE CONTROL OF BACTERIAL CELL SIZE
In an elegant paper published in 1974, Teather et al. suggested the existence of a diffusible factor required for the initiation of cell division in E. coli [68]. Accumulation of this factor to critical levels was proposed to trigger the initiation of division at mid-cell, in much the same way that accumulation of active DnaA to critical levels was proposed to trigger the initiation of chromosome replication. A key aspect of this model was that only enough of this diffusible factor accumulated per mass doubling to initiate one round of division. (Not coincidentally the senior author on this paper was William Donachie, the same person who proposed the existence of DnaA long well before it was identified molecularly [10].) Subsequent work suggests the diffusible factor proposed by Teather et al. is in fact FtsZ [46, 111].
In an extension of Teather’s model, we propose that cell size under steady state conditions is dictated in large part by the amount of FtsZ available for assembly into the cytokinetic ring (Figure 5). In this model, cell division is dependent on the accumulation of sufficient FtsZ to support assembly, maturation and/or constriction of the cytokinetic ring. For organisms like E. coli and B. subtilis where FtsZ concentration remains constant regardless of growth rate, this means that cells need to reach a minimal size to ensure there is sufficient FtsZ to support division (Figure 5A). For organisms that vary FtsZ concentration over the course of the cell cycle, such as Caulobacter, this means that FtsZ levels would need to increase until there was sufficient FtsZ to support division (not shown). This model is supported by work on the dose-dependent effect of partial FtsZ depletion on cell size [70], FtsZ’s extraordinary conservation [64, 112, 113], as well as the identification of an FtsZ inhibitor responsible for the nutrient-dependent control of cell size in B. subtilis [2].
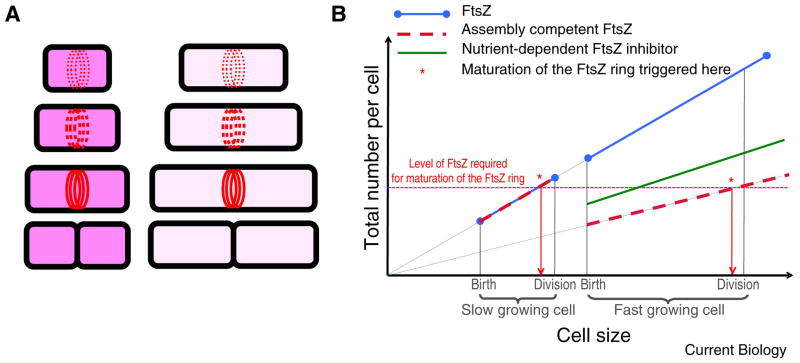
Figure 5. The concentration of assembly-competent FtsZ dictates cell size at division.
A. (Left) Cytoplasmic FtsZ concentration (dark pink background) is constant throughout the cell cycle, however the total amount of FtsZ increases with cell size. Growth-dependent accumulation of FtsZ to critical levels dictates cell size at division. (Right) Reducing the intracellular concentration of assembly-competent FtsZ (light pink background) increases the size at which cells accumulate sufficient FtsZ to support division. B. Graphic model for the growth rate-dependent control of cell size. Asterisks mark the initiation of constriction in response to accumulation of FtsZ to critical levels. In the slower growing cell, on the left, assembly-competent FtsZ accumulates in direct proportion to cell size, reaching critical levels near the end of the cell cycle and stimulating maturation of the FtsZ ring and division. The faster growing cell, on the right, is born larger than its slow growing counterpart due to the presence of a growth-dependent inhibitor of FtsZ assembly (green). Due to the presence of the inhibitor, the faster growing cell is significantly larger when sufficient assembly-competent FtsZ has accumulated to stimulate maturation of the FtsZ ring and division. For simplicity in both A and B we have depicted FtsZ accumulation dictating maturation of the FtsZ ring and constriction. In the absence of data to suggest otherwise, it is equally likely that the rate-limiting step in cell division is the initiation of FtsZ ring formation. For clarity, we have also drawn the graph such that the newborn cell cultured under conditions supporting rapid growth is larger than the slow growing cell at division. In reality the sizes of these two cells likely overlap to some degree even when the difference in growth rates are at its most extreme.
Under environmental conditions that necessitate an increase in cell size, such as conditions supporting multifork replication, this model predicts the presence of an FtsZ inhibitor that is expressed and/or activated in a proportional manner in response to a specific stimulus (Figure 5B). For example, activation of a nutrient-dependent inhibitor would be proportional to the relative ease with which a particular bacterium is able to utilize a given carbon source (e.g. high activation in a carbon-rich complex medium such as LB and low activation in minimal defined medium supplemented with succinate.) Proportional activation of an inhibitor of FtsZ assembly would then lead to a proportional reduction in the pool of FtsZ available for assembly and a proportional increase in cell size (Figure 5B). This model is consistent with data indicating that even small reductions in FtsZ levels have a large impact on E. coli cell size during exponential growth under steady state conditions [114].
In this model, cell size is controlled primarily through the condition-specific reduction in the amount of FtsZ available for assembly and division. In the absence of inhibition, cells are at their smallest, default size. We favor negative regulation for several reasons. First and foremost, the impact of a transient delay in division on the cell cycle is minimal, whereas dividing earlier would impinge on preceding steps in the cell cycle including the completion of DNA replication and chromosome segregation. In addition, cells appear to be somewhat refractile to increases in the intracellular concentration of FtsZ. Increasing FtsZ levels as much as two-fold leads to only an ~10% reduction in the size of E. coli and B. subtilis cells [43]. Larger increases in FtsZ levels lead to aberrant FtsZ localization and, at concentrations ~7-fold higher than wild-type, complete division inhibition [115]. The inability to significantly reduce cell size following overexpression of FtsZ, or induce division in cells that have entered the more quiescent period of stationary phase, suggests the presence of inhibitors that are refractile to competition from excess FtsZ as well as physical constraints preventing FtsZ ring formation too early in the cell cycle. It is also possible that the inability of excess FtsZ to significantly reduce average cell size is due in part to limiting amounts of a positive factor required to promote FtsZ assembly. This view is supported by data indicating cell size is reduced by ~25% during growth in rich medium in the presence of gain-of-function mutations in the cell division protein, ftsA, which normally helps promote FtsZ assembly and is required for maintaining integrity of the cytokinetic ring [116].
Because the timing of division is tied to the availability of FtsZ for assembly into the ring, any change in cell size should only have a transient impact on the timing of division. For example, a cell that is too short for a given growth condition would delay division until it accumulates sufficient levels of FtsZ. Its normally sized daughter cells will then accumulate sufficient FtsZ within a single mass doubling period and thus be able to “divide on time”. Through its ability to correct transient aberrations in cell size, this model provides an explanation for the 40-year-old observation that E. coli cells shifted from a nutrient-poor medium to a nutrient-rich one immediately increase growth rate but delay division until they have achieved the size appropriate for the new condition [117]. We would predict that a nutrient-dependent inhibitor of FtsZ assembly is activated almost immediately upon the shift to the rich carbon source. Cells must then delay division and increase in size until they have accumulated sufficient assembly-competent FtsZ to support cytokinesis.
While other cell cycle events, most notably the initiation of DNA replication in E. coli, have been implicated in cell size control, we believe that changes in assembly, maturation, and constriction of the FtsZ ring are the primary means of coordinating cell size with cell growth in bacteria. As discussed above, although E. coli requires achievement of a specific size prior to initiating DNA replication, B. subtilis does not [2, 80]. This difference suggests that coupling replication initiation to cell growth is not a conserved means of maintaining cell size. Parsimoniously, it also seems more straightforward to control cell size by altering the timing of division, the last step in the cell cycle, than to vary the timing of initiation and risk the consequences of compensatory changes in downstream cell cycle events [25–30].
In summary, while we are beginning to uncover the molecular mechanisms responsible for coordinating cell division with cell growth in B. subtilis and E. coli, there are still many unanswered questions. Of particular interest is FtsZ’s contribution to cell size control in E. coli and C. crescentus via its role as a regulator of longitudinal cell wall synthesis [66, 67]. In addition, we know little about cell size control in species that encode FtsZ but which do not employ binary fission as a means of reproduction, including the “giant” bacterium Epulopiscium fishelsoni [118]. Finally, the advent of high throughput sequencing technology has generated an ever expanding list of bacteria that do not encode an FtsZ homolog, including the obligate intracellular pathogen Chlamydia and its free living relative Planctomycetes [119]. How these bacteria divide, much less maintain cell size, remains an enticing question for future study.
Acknowledgments
The authors are indebted to Suckjoon Jun, Sattar Taheri-Araghi, Daniel Haeusser, and members of the Levin laboratory for helpful discussions and comments on the manuscript. We also thank all of the reviewers for their very insightful comments and suggestions. The micrograph in Figure 4B is courtesy of P. J. Buske, and we thank Isabel Erdmann and Dwight Erdmann for the photos used in Figure 3A. AC is a Lee Foundation Fellow of the McDonnell International Scholars Academy. Work in the Levin lab is supported by a Public Health Services grant (GM64671) from the NIH to PAL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Grover NB, Woldringh CL. Dimensional regulation of cell-cycle events in Escherichia coli during steady-state growth. Microbiology. 2001;147:171–181. doi: 10.1099/00221287-147-1-171. [DOI] [PubMed] [Google Scholar]
- 2.Weart RB, Lee AH, Chien AC, Haeusser DP, Hill NS, Levin PA. A metabolic sensor governing cell size in bacteria. Cell. 2007;130:335–347. doi: 10.1016/j.cell.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angert ER, Clements KD, Pace NR. The largest bacterium. Nature. 1993;362:239–241. doi: 10.1038/362239a0. [DOI] [PubMed] [Google Scholar]
- 4.Schulz HN, Jorgensen BB. Big bacteria. Annu Rev Microbiol. 2001;55:105–137. doi: 10.1146/annurev.micro.55.1.105. [DOI] [PubMed] [Google Scholar]
- 5.Razin S, Cosenza BJ. Growth phases of Mycoplasma in liquid media observed with phase-contrast microscope. J Bacteriol. 1966;91:858–869. doi: 10.1128/jb.91.2.858-869.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mendell JE, Clements KD, Choat JH, Angert ER. Extreme polyploidy in a large bacterium. Proc Natl Acad Sci USA. 2008;105:6730–6734. doi: 10.1073/pnas.0707522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Margolin W. Sculpting the bacterial cell. Curr Biol. 2009;19:R812–822. doi: 10.1016/j.cub.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young KD. Bacterial shape: two-dimensional questions and possibilities. Annu Rev Microbiol. 64:223–240. doi: 10.1146/annurev.micro.112408.134102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White CL, Gober JW. MreB: pilot or passenger of cell wall synthesis? Trends Microbiol. 2012;20:74–79. doi: 10.1016/j.tim.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 10.Donachie WD. Relationship between cell size and time of initiation of DNA replication. Nature. 1968;219:1077–1079. doi: 10.1038/2191077a0. [DOI] [PubMed] [Google Scholar]
- 11.Sharpe ME, Errington J. A fixed distance for separation of newly replicated copies of oriC in Bacillus subtilis: implications for co-ordination of chromosome segregation and cell division Mol. Microbiol. 1998;28:981–990. doi: 10.1046/j.1365-2958.1998.00857.x. [DOI] [PubMed] [Google Scholar]
- 12.Schaechter M, Maaløe O, Kjeldgaard NO. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 13.Cooper S, Helmstetter CE. Chromosome replication and the division cycle of Escherichia coli B/r. J Mol Biol. 1968;31:519–540. doi: 10.1016/0022-2836(68)90425-7. [DOI] [PubMed] [Google Scholar]
- 14.Løbner-Olesen A, Skarstad K, Hansen FG, von Meyenburg K, Boye E. The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell. 1989;57:881–889. doi: 10.1016/0092-8674(89)90802-7. [DOI] [PubMed] [Google Scholar]
- 15.Kaguni JM. DnaA: controlling the initiation of bacterial DNA replication and more. Annu Rev Microbiol. 2006;60:351–375. doi: 10.1146/annurev.micro.60.080805.142111. [DOI] [PubMed] [Google Scholar]
- 16.Katayama T, Ozaki S, Keyamura K, Fujimitsu K. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat Rev Microbiol. 2010;8:163–170. doi: 10.1038/nrmicro2314. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Fernandez C, Gonzalez D, Collier J. Regulation of the activity of the dual-function DnaA protein in Caulobacter crescentus. PLoS One. 6:e26028. doi: 10.1371/journal.pone.0026028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishida S, Fujimitsu K, Sekimizu K, Ohmura T, Ueda T, Katayama T. A nucleotide switch in the Escherichia coli DnaA protein initiates chromosomal replication: evidnece from a mutant DnaA protein defective in regulatory ATP hydrolysis in vitro and in vivo. J Biol Chem. 2002;277:14986–14995. doi: 10.1074/jbc.M108303200. [DOI] [PubMed] [Google Scholar]
- 19.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 20.Duderstadt KE, Mott ML, Crisona NJ, Chuang K, Yang H, Berger JM. Origin remodeling and opening in bacteria rely on distinct assembly states of the DnaA initiator. J Biol Chem. 285:28229–28239. doi: 10.1074/jbc.M110.147975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Braun RE, O’Day K, Wright A. Autoregulation of the DNA replication gene dnaA in E. coli K-12. Cell. 1985;40:159–169. doi: 10.1016/0092-8674(85)90319-8. [DOI] [PubMed] [Google Scholar]
- 22.Atlung T, Clausen ES, Hansen FG. Autoregulation of the dnaA gene of Escherichia coli K12. Mol Gen Genet. 1985;200:442–450. doi: 10.1007/BF00425729. [DOI] [PubMed] [Google Scholar]
- 23.Ogura Y, Imai Y, Ogasawara N, Moriya S. Autoregulation of the dnaA-dnaN operon and effects of DnaA protein levels on replication initiation in Bacillus subtilis. J Bacteriol. 2001;183:3833–3841. doi: 10.1128/JB.183.13.3833-3841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donachie WD, Blakely GW. Coupling the initiation of chromosome replication to cell size in Escherichia coli. Curr Opin Microbiol. 2003;6:146–150. doi: 10.1016/s1369-5274(03)00026-2. [DOI] [PubMed] [Google Scholar]
- 25.Hill NS, Kadoya R, Chattoraj DK, Levin PA. Cell size and the initiation of DNA replication in bacteria. PLoS Genetics. doi: 10.1371/journal.pgen.1002549. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atlung T, Løbner-Olesen A, Hansen FG. Overproduction of DnaA protein stimulates initiation of chromosome and minichromosome replication in Escherichia coli. Mol Gen, Genet. 1987;206:51–59. doi: 10.1007/BF00326535. [DOI] [PubMed] [Google Scholar]
- 27.Skarstad K, Løbner-Olesen A, Atlung T, von Meyenburg K, Boye E. Initiation of DNA replication in Escherichia coli after overproduction of the DnaA protein. Mol Gen Genet. 1989;218:50–56. doi: 10.1007/BF00330564. [DOI] [PubMed] [Google Scholar]
- 28.von Freiesleben U, Krekling MA, Hansen FG, Løbner-Olesen A. The eclipse period of Escherichia coli. EMBO J. 2000;19:6240–6248. doi: 10.1093/emboj/19.22.6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boye E, Stokke T, Kleckner N, Skarstad K. Coordinating DNA replication initiation with cell growth: differential roles for DnaA and SeqA proteins. Proc Natl Acad Sci USA. 1996;93:12206–12211. doi: 10.1073/pnas.93.22.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torheim NK, Boye E, Løbner-Olesen A, Stokke T, Skarstad K. The Escherichia coli SeqA protein destabilizes mutant DnaA204 protein. Mol Microbiol. 2000;37:629–638. doi: 10.1046/j.1365-2958.2000.02031.x. [DOI] [PubMed] [Google Scholar]
- 31.Sharpe ME, Hauser PM, Sharpe RG, Errington J. Bacillus subtilis cell cycle as studied by fluorescence microscopy: constancy of cell length at initiation of DNA replication and evidence for active nucleoid partitioning. J Bacteriol. 1998;180:547–555. doi: 10.1128/jb.180.3.547-555.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noirot-Gros MF, Dervyn E, Wu LJ, Mervelet P, Errington J, Ehrlich SD, Noirot P. An expanded view of bacterial DNA replication. Proc Natl Acad Sci USA. 2002;99:8342–8347. doi: 10.1073/pnas.122040799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noirot-Gros MF, Velten M, Yoshimura M, McGovern S, Morimoto T, Ehrlich SD, Ogasawara N, Polard P, Noirot P. Functional dissection of YabA, a negative regulator of DNA replication initiation in Bacillus subtilis. Proc Natl Acad Sci USA. 2006;103:2368–2373. doi: 10.1073/pnas.0506914103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soufo CD, Soufo HJ, Noirot-Gros MF, Steindorf A, Noirot P, Graumann PL. Cell-cycle-dependent spatial sequestration of the DnaA replication initiator protein in Bacillus subtilis. Dev Cell. 2008;15:935–941. doi: 10.1016/j.devcel.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Hayashi M, Ogura Y, Harry EJ, Ogasawara N, Moriya S. Bacillus subtilis YabA is involved in determining the timing and synchrony of replication initiation. FEMS Microbiol, Lett. 2005;247:73–79. doi: 10.1016/j.femsle.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 36.Cho E, Ogasawara N, Ishikawa S. The functional analysis of YabA, which interacts with DnaA and regulates initiation of chromosome replication in Bacillus subtils. Genes Genet Syst. 2008;83:111–125. doi: 10.1266/ggs.83.111. [DOI] [PubMed] [Google Scholar]
- 37.Goranov AI, Breier AM, Merrikh H, Grossman AD. YabA of Bacillus subtilis controls DnaA-mediated replication initiation but not the transcriptional response to replication stress. Mol Microbiol. 2009;74:454–466. doi: 10.1111/j.1365-2958.2009.06876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scholefield G, Whiting R, Errington J, Murray H. Spo0J regulates the oligomeric state of Soj to trigger its switch from an activator to an inhibitor of DNA replication initiation. Mol Microbiol. 2011;79:1089–1100. doi: 10.1111/j.1365-2958.2010.07507.x. [DOI] [PubMed] [Google Scholar]
- 39.Murray H, Errington J. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell. 2008;135:74–84. doi: 10.1016/j.cell.2008.07.044. [DOI] [PubMed] [Google Scholar]
- 40.Smits WK, Merrikh H, Bonilla CY, Grossman AD. Primosomal proteins DnaD and DnaB are recruited to chromosomal regions bound by DnaA in Bacillus subtilis. J Bacteriol. 2011;193:640–648. doi: 10.1128/JB.01253-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- 42.Erickson HP, Anderson DE, Osawa M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev. 2010;74:504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weart RB, Levin PA. Growth rate-dependent regulation of medial FtsZ ring formation. J Bacteriol. 2003;185:2826–2834. doi: 10.1128/JB.185.9.2826-2834.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirkpatrick CL, Viollier PH. Decoding Caulobacter development. FEMS Microbiol Rev. 2012;36:193–205. doi: 10.1111/j.1574-6976.2011.00309.x. [DOI] [PubMed] [Google Scholar]
- 45.de Boer PA, Crossley RE, Rothfield LI. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 46.Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem. 2007;76:539–562. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- 47.Wu LJ, Errington J. Nucleoid occlusion and bacterial cell division. Nat Rev Microbiol. 2012;10:8–12. doi: 10.1038/nrmicro2671. [DOI] [PubMed] [Google Scholar]
- 48.Bernhardt TG, de Boer PA. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E. coli. Mol Cell. 2005;18:555–564. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu LJ, Errington J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell. 2004;117:915–925. doi: 10.1016/j.cell.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 50.Cho H, McManus HR, Dove SL, Bernhardt TG. Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerization antagonist. Proc Natl Acad Sci USA. 2011;108:3773–3778. doi: 10.1073/pnas.1018674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu LJ, Errington J. Nucleoid occlusion and bacterial cell division. Nat Rev Microbiol. 10:8–12. doi: 10.1038/nrmicro2671. [DOI] [PubMed] [Google Scholar]
- 52.Niki H, Yamaichi Y, Hiraga S. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev. 2000;14:212–223. [PMC free article] [PubMed] [Google Scholar]
- 53.Valens M, Penaud S, Rossignol M, Cornet F, Boccard F. Macrodomain organization of the Escherichia coli chromosome. EMBO J. 2004;23:4330–4341. doi: 10.1038/sj.emboj.7600434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu LJ, Ishikawa S, Kawai Y, Oshima T, Ogasawara N, Errington J. Noc protein binds to specific DNA sequences to coordinate cell division with chromosome segregation. EMBO J. 2009;28:1940–1952. doi: 10.1038/emboj.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu LJ, Errington J. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science. 1994;264:572–575. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- 56.McGinness T, Wake RG. Division septation in the absence of chromosome termination in Bacillus subtilis. J Mol Biol. 1979;134:251–264. doi: 10.1016/0022-2836(79)90035-4. [DOI] [PubMed] [Google Scholar]
- 57.Wu LJ, Franks AH, Wake RG. Replication through the terminus region of the Bacillus subtilis chromosome is not essential for the formation of a division septum that partitions the DNA. J Bacteriol. 1995;177:5711–5715. doi: 10.1128/jb.177.19.5711-5715.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Veiga H, Jorge AM, Pinho MG. Absence of nucleoid occlusion effector Noc impairs formation of orthogonal FtsZ rings during Staphylococcus aureus cell division. Mol Microbiol. 2011;80:1366–1380. doi: 10.1111/j.1365-2958.2011.07651.x. [DOI] [PubMed] [Google Scholar]
- 59.Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 60.Harry EJ, Rodwell J, Wake RG. Co-ordinating DNA replication with cell division in bacteria: a link between the early stages of a round of replication and mid-cell Z ring assembly. Mol Microbiol. 1999;33:33–40. doi: 10.1046/j.1365-2958.1999.01439.x. [DOI] [PubMed] [Google Scholar]
- 61.Regamey A, Harry EJ, Wake RG. Mid-cell Z ring assembly in the absence of entry into the elongation phase of the round of replication in bacteria: coordinating chromosome replication with cell division Mol. Microbiol. 2000;38:423–434. doi: 10.1046/j.1365-2958.2000.02130.x. [DOI] [PubMed] [Google Scholar]
- 62.Sun Q, Margolin W. Influence of the nucleoid on placement of FtsZ and MinE rings in Escherichia coli. J Bacteriol. 2001;183:1413–1422. doi: 10.1128/JB.183.4.1413-1422.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quardokus EM, Brun YV. DNA replication initiation is required for mid-cell positioning of FtsZ rings in Caulobacter crescentus. Mol Microbiol. 2002;45:605–616. doi: 10.1046/j.1365-2958.2002.03040.x. [DOI] [PubMed] [Google Scholar]
- 64.Harry E, Monahan L, Thompson L. Bacterial cell division: the mechanism and its precison. Int Rev Cytol. 2006;253:27–94. doi: 10.1016/S0074-7696(06)53002-5. [DOI] [PubMed] [Google Scholar]
- 65.Den Blaauwen T, Buddelmeijer N, Aarsman ME, Hameete CM, Nanninga N. Timing of FtsZ assembly in Escherichia coli. J Bacteriol. 1999;181:5167–5175. doi: 10.1128/jb.181.17.5167-5175.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol. 2007;64:938–952. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- 67.Varma A, Young KD. FtsZ collaborates with penicillin binding proteins to generate bacterial cell shape in Escherichia coli. J Bacteriol. 2004;186:6768–6774. doi: 10.1128/JB.186.20.6768-6774.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Teather RM, Collins JF, Donachie WD. Quantal behavior of a diffusable factor which initiates septum formation at potential division sites in Escherichia coli. J Bacteriol. 1974;118:407–413. doi: 10.1128/jb.118.2.407-413.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bi E, Lutkenhaus J. Interaction between the min locus and ftsZ. J Bacteriol. 1990;172:5610–5616. doi: 10.1128/jb.172.10.5610-5616.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Flardh K, Palacios P, Vicente M. Cell division genes ftsQAZ in Escherichia coli require distant cis-acting signals upstream of ddlB for full expression. Mol Microbiol. 1998;30:305–315. doi: 10.1046/j.1365-2958.1998.01064.x. [DOI] [PubMed] [Google Scholar]
- 71.Haeusser DP, Garza AC, Buscher AZ, Levin PA. The division inhibitor EzrA contains a seven-residue patch required for maintaining the dynamic nature of the medial FtsZ ring. J Bacteriol. 2007;189:9001–9010. doi: 10.1128/JB.01172-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Palacios P, Vicente M, Sanchez M. Dependency of Escherichia coli cell-division size, and independency of nucleoid segregation on the mode and level of ftsZ expression. Mol Microbiol. 1996;20:1093–1098. doi: 10.1111/j.1365-2958.1996.tb02549.x. [DOI] [PubMed] [Google Scholar]
- 73.Kjeldgaard NO, Maaløe O, Schaechter M. The transition between different physiological states during balanced growth of Salmonella typhimurium. J, Gen, Microbiol. 1958;19:607–616. doi: 10.1099/00221287-19-3-607. [DOI] [PubMed] [Google Scholar]
- 74.Pierucci O, Helmstetter CE, Rickert M, Weinberger M, Leonard AC. Overexpression of the dnaA gene in Escherichia coli B/r: chromosome and minichromosome replication in the presence of rifampin. J Bacteriol. 1987;169:1871–1877. doi: 10.1128/jb.169.5.1871-1877.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sargent MG. Control of cell length in Bacillus subtilis. J Bacteriol. 1975;123:7–19. doi: 10.1128/jb.123.1.7-19.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Watson SP, Clements MO, Foster SJ. Characterization of the starvation-survival response of Staphylococcus aureus. J Bacteriol. 1998;180:1750–1758. doi: 10.1128/jb.180.7.1750-1758.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fantes P, Nurse P. Control of cell size at division in fission yeast by a growth-modulated size control over nuclear division. Exp Cell Res. 1977;107:377–386. doi: 10.1016/0014-4827(77)90359-7. [DOI] [PubMed] [Google Scholar]
- 78.Trueba FJ, Woldringh CL. Changes in cell diameter during the division cycle of Escherichia coli. J Bacteriol. 1980;142:869–878. doi: 10.1128/jb.142.3.869-878.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zaritsky A, Woldringh CL, Helmstetter CE, Grover NB. Dimensional rearrangement of Escherichia coli B/r cells during a nutritional shift-down. J Gen Microbiol. 1993;139:2711–2714. doi: 10.1099/00221287-139-11-2711. [DOI] [PubMed] [Google Scholar]
- 80.Hill NS, Kadoya R, Chattoraj DK, Levin PA. Cell size and the initiation of DNA replication in bacteria. PLoS Genetics. 2012 doi: 10.1371/journal.pgen.1002549. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu M, Kleckner N. Molecular cloning and characterization of the pgm gene encoding phosphoglucomutase of Escherichia coli. J Bacteriol. 1994;176:5847–5851. doi: 10.1128/jb.176.18.5847-5851.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Allison SE, D’Elia MA, Arar S, Monteiro MA, Brown ED. Studies of the genetics, function, and kinetic mechanism of TagE, the wall teichoic acid glycosyltransferase in Bacillus subtilis 168. J Biol, Chem. 2011;286:23708–23716. doi: 10.1074/jbc.M111.241265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Glaser L, Burger MM. The Synthesis of Teichoic Acids. 3 Glucosylation of Polyglycerophosphate. J Biol Chem. 1964;239:3187–3191. [PubMed] [Google Scholar]
- 84.Debarbieux L, Bohin A, Bohin JP. Topological analysis of the membrane-bound glucosyltransferase, MdoH, required for osmoregulated periplasmic glucan synthesis in Escherichia coli. J Bacteriol. 1997;179:6692–6698. doi: 10.1128/jb.179.21.6692-6698.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Preiss J. Bacterial glycogen synthesis and its regulation. Annu Rev Microbiol. 1984;38:419–458. doi: 10.1146/annurev.mi.38.100184.002223. [DOI] [PubMed] [Google Scholar]
- 86.Bohringer J, Fischer D, Mosler G, Hengge-Aronis R. UDP-glucose is a potential intracellular signal molecule in the control of expression of sigma S and sigma S-dependent genes in Escherichia coli. J Bacteriol. 1995;177:413–422. doi: 10.1128/jb.177.2.413-422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Price KD, Roels S, Losick R. A Bacillus subtilis gene encoding a protein similar to nucleotide sugar transferases influences cell shape and viability. J Bacteriol. 1997;179:4959–4961. doi: 10.1128/jb.179.15.4959-4961.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lazarevic V, Mauel C, Soldo B, Freymond PP, Margot P, Karamata D. Sequence analysis of the 308 degrees to 311 degrees segment of the Bacillus subtilis 168 chromosome, a region devoted to cell wall metabolism, containing non-coding grey holes which reveal chromosomal rearrangements. Microbiology. 1995;141(Pt 2):329–335. doi: 10.1099/13500872-141-2-329. [DOI] [PubMed] [Google Scholar]
- 89.Gilbert P, Byron PR, Brown MR. Application of models for envelope growth to cell length distribution data for Pseudomonas aeruginosa at various specific growth rates. Microbios. 1981;31:189–203. [PubMed] [Google Scholar]
- 90.Koppes LH, Woldringh CL, Nanninga N. Size variations and correlation of different cell cycle events in slow- growing Escherichia coli. J Bacteriol. 1978;134:423–433. doi: 10.1128/jb.134.2.423-433.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jun S, Wright A. Entropy as the driver of chromosome segregation. Nat Rev Microbiol. 2010;8:600–607. doi: 10.1038/nrmicro2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Donachie WD, Begg KJ. Cell length, nucleoid separation, and cell division of rod-shaped and spherical cells of Eschericia coli. J Bacteriol. 1989;171:4633–4639. doi: 10.1128/jb.171.9.4633-4639.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Golden JW, Yoon HS. Heterocyst development in Anabaena. Curr Opin Microbiol. 2003;6:557–563. doi: 10.1016/j.mib.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 94.Prell J, Poole P. Metabolic changes of rhizobia in legume nodules. Trends Microbiol. 2006;14:161–168. doi: 10.1016/j.tim.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 95.Willemse J, Borst JW, de Waal E, Bisseling T, van Wezel GP. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev. 2011;25:89–99. doi: 10.1101/gad.600211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schofield WB, Lim HC, Jacobs-Wagner C. Cell cycle coordination and regulation of bacterial chromosome segregation dynamics by polarly localized proteins. EMBO J. 2010;29:3068–3081. doi: 10.1038/emboj.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Judd EM, Ryan KR, Moerner WE, Shapiro L, McAdams HH. Fluorescence bleaching reveals asymmetric compartment formation prior to cell division in Caulobacter. Proc Natl Acad Sci USA. 2003;100:8235–8240. doi: 10.1073/pnas.1433105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Judd EM, Comolli LR, Chen JC, Downing KH, Moerner WE, McAdams HH. Distinct constrictive processes, separated in time and space, divide caulobacter inner and outer membranes. J Bacteriol. 2005;187:6874–6882. doi: 10.1128/JB.187.20.6874-6882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Goley ED, Iniesta AA, Shapiro L. Cell cycle regulation in Caulobacter: location, location, location. J Cell Sci. 2007;120:3501–3507. doi: 10.1242/jcs.005967. [DOI] [PubMed] [Google Scholar]
- 100.Gonin M, Quardokus EM, O’Donnol D, Maddock J, Brun YV. Regulation of stalk elongation by phosphate in Caulobacter crescentus. J Bacteriol. 2000;182:337–347. doi: 10.1128/jb.182.2.337-347.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ryter A, Schaeffer P, Ionesco H. Classification cytologique, par leur stade de blocage des mutants de sporulation de Bacillus subtilis Marburg [Cytologic classification, by their blockage stage, of sporulation mutants of Bacillus subtilis Marburg] Ann Inst Pasteur (Paris) 1966;110:305–315. [PubMed] [Google Scholar]
- 102.Piggot PJ, Hilbert DW. Sporulation of Bacillus subtilis. Curr Opin Microbiol. 2004;7:579–586. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 103.Ben-Yehuda S, Losick R. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell. 2002;109:257–266. doi: 10.1016/s0092-8674(02)00698-0. [DOI] [PubMed] [Google Scholar]
- 104.Levin PA, Losick R. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 1996;10:478–488. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- 105.Pavlendova N, Muchova K, Barak I. Chromosome segregation in Bacillus subtilis. Folia Microbiol (Praha) 2007;52:563–572. doi: 10.1007/BF02932184. [DOI] [PubMed] [Google Scholar]
- 106.Wu LJ, Errington J. Use of asymmetric cell division and spoIIIE mutants to probe chromosome orientation and organization in Bacillus subtilis. Mol Microbiol. 1998;27:777–786. doi: 10.1046/j.1365-2958.1998.00724.x. [DOI] [PubMed] [Google Scholar]
- 107.Burton BM, Marquis KA, Sullivan NL, Rapoport TA, Rudner DZ. The ATPase SpoIIIE transports DNA across fused septal membranes during sporulation in Bacillus subtilis. Cell. 2007;131:1301–1312. doi: 10.1016/j.cell.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pogliano K, Pogliano J, Becker E. Chromosome segregation in Eubacteria. Curr Opin Microbiol. 2003;6:586–593. doi: 10.1016/j.mib.2003.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zupancic ML, Tran H, Hofmeister AE. Chromosomal organization governs the timing of cell type-specific gene expression required for spore formation in Bacillus subtilis. Mol Microbiol. 2001;39:1471–1481. doi: 10.1046/j.1365-2958.2001.02331.x. [DOI] [PubMed] [Google Scholar]
- 110.Zhang L, Higgins ML, Piggot PJ. The division during bacterial sporulation is symmetrically located in Sporosarcina ureae. Mol Microbiol. 1997;25:1091–1098. doi: 10.1046/j.1365-2958.1997.5341892.x. [DOI] [PubMed] [Google Scholar]
- 111.Bi E, Lutkenhaus J. FtsZ regulates frequency of cell division in Escherichia coli. J Bacteriol. 1990;172:2765–2768. doi: 10.1128/jb.172.5.2765-2768.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Erickson HP. FtsZ, a tubulin homologue in prokaryote cell division. Trends in Cell Biology. 1997;7:362–367. doi: 10.1016/S0962-8924(97)01108-2. [DOI] [PubMed] [Google Scholar]
- 113.Vaughan S, Wickstead B, Gull K, Addinall SG. Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J Mol Evol. 2004;58:19–29. doi: 10.1007/s00239-003-2523-5. [DOI] [PubMed] [Google Scholar]
- 114.Palacios P, Vicente M, Sanchez M. Dependency of Escherichia coli cell-division size, and independency of nucleoid segregation on the mode and level of ftsZ expression. Mol Microbiol. 1996;20:1093–1098. doi: 10.1111/j.1365-2958.1996.tb02549.x. [DOI] [PubMed] [Google Scholar]
- 115.Ward JE, Jr, Lutkenhaus J. Overproduction of FtsZ induces minicell formation in E. coli. Cell. 1985;42:941–949. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]
- 116.Geissler B, Shiomi D, Margolin W. A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc Natl Acad Sci USA. 2003;100:4197–202. doi: 10.1073/pnas.0635003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cooper S. Cell division and DNA replication following a shift to a richer medium. J Mol Biol. 1969;43:1–11. doi: 10.1016/0022-2836(69)90074-6. [DOI] [PubMed] [Google Scholar]
- 118.Angert ER, Clements KD. Initiation of intracellular offspring in Epulopiscium. Mol Microbiol. 2004;51:827–835. doi: 10.1046/j.1365-2958.2003.03869.x. [DOI] [PubMed] [Google Scholar]
- 119.Erickson HP, Osawa M. Cell division without FtsZ--a variety of redundant mechanisms. Mol Microbiol. 2010;78:267–270. doi: 10.1111/j.1365-2958.2010.07321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]