Abstract
Cell size is an important adaptive trait that influences nearly all aspects of cellular physiology. Despite extensive characterization of the cell cycle regulatory network, the molecular mechanismscoupling growth to division, and thereby controlling cell size, have remained elusive. Recent workin yeast has reinvigorated the size control field and suggested provocative mechanisms forthe distinct functions of setting and sensing cell size. Further examination of size sensing models based on spatial gradients and molecular titration, coupled with elucidation of the pathways responsible for nutrient-modulated target size, may reveal the fundamental principles of eukaryotic cell size control.
Introduction
There is an intimate and often complex relationship between form and function in living organisms[1]. A simple and fundamental aspect of organismal form is size, which is an important determinant of cellular physiology. In unicellular organisms, adjusting the surface to volume ratio may be an adaptation to environmental conditions where surface transport is limiting. Surface-to-volume optimization islikely exhibited in the extensive fossil record of foraminifera, a group of unicellular marine protists. 60% of the variation in mean foraminifera size over the past 400 million years is accounted for by a linear model based solely on atmospheric oxygen concentration[2, 3]. Thus, cell size can be an important selective trait for survival in changing, nutrient-limited environments.
Cell size also affects internal cellular architecture. Not only are the volumes of various organelles proportional to that of the cell[4], DNA content in eukaryotic cells scales linearly with cell size over nearly a million-fold range[5-7]. This is true within as well as between species: diploid yeast cells are about twice as large as haploids [8-10].This constant ratio of DNA to cytoplasm suggests that cell size can adapt to support evolution of DNA content. Conversely, DNA content could adapt to accommodate physiologically-driven changes in cell size. Examples of the latter may be found in the yeast lineage, where a minimal genome could be viewed as an adaptation to small cellular size. With volumes as low as 10’s of femtoliters, yeast are among the smallest eukaryotes and contain relatively few genes, as well as small intergenic regions.For example, whereas metazoan cis-regulatory elements are found thousands of basepairs away from the transcription start site, yeast elements are limited to ~800 basepairs upstream [11]. Thus, evolutionary pressures on cell size may influence mechanisms of transcriptional regulation via selection to maintain the appropriate DNA-to-cytoplasm ratio.
Consistent with the physiological importance of cell size, there is much evidence suggesting that cells have evolved molecular mechanisms for bothmonitoring and controlling size[12]. Cellsof a given species typically vary little about their mean size [13], and this consistency requirestight co-regulation of cellular growth and division [14]. Single-cell studies have provided the most convincing evidence of cell size control. In these studies, variation in cell size at birth is harnessed to infer the presence or absence of cell size control: If size control is present, then cells that are smaller at birthwill grow proportionally more than larger cells in the subsequent cell division cycle[15-17].
This co-regulation of growth and division has been studied in a variety of organisms, but the yeasts Saccharomycescerevisiae and Schizosaccharomycespombe(Figure 1) have been favored model organisms for several reasons:a potentially simpler regulatory architecture, powerful systematic tools for genetic analysis, and simpler cell geometry[18]. More fundamentally, the regulation of growth and division is particularly tractable in yeast. In metazoans, both division and growth are subject to complex extrinsic signaling andmultifarious mutual regulation.In yeast, on the other hand, preventing growth arrests the cell cycle, butgrowth proceeds apace when proliferation is halted[15, 19]. Thus, yeast control size primarily by regulating division in response to growth.This requires detection of the nutrient concentrations that determine growth rate, growth rate itself, or cell size. Here, we review our current understanding of size control mechanisms in yeast, highlighting recent work and open questions.
Figure 1. Yeast models for cell size control.
S. pombe and S. cerevisiae are the preeminent model organisms for cell size control studies. Interestingly, S. cerevisiae and A. gossypii are more closely related than either is to S. pombe. Despite their divergent morphologies, these two yeasts are regulated by similar cell cycle control networks governing the DNA to cytoplasm ratio.
Size and Growth
“the form of an organism is determined by its rate of growth in various directions; hence rate of growth deserves to be studied as a necessary preliminary to the theoretical study of form” D.W. Thompson, On Growth and Form, 1942, P.79 [1].
Size is the most fundamental aspect of cellular form, and the basis of cell size control is the coupling of growth and division.The growth function f, relates a cell’s current size, V, and cell cycle phase, ϕ, to the rate of size increase, , so that . This relationship defines the requirements of cell size control. More specifically, whether cell growth is linear or exponential has been hotly debated and has important implications for size control[18]. For dV linear growth, cells increase their size at a constant rate C, so that . In this case, specifying the period of the cell cycle specifies mean cell size without requiring a direct link between size and division, because the progeny of large or small cells asymptotically approach the target size over several generations[20]. Linearly growing cells could therefore maintain an average size simply by adjusting the length of the cell cycle.On the other hand, for exponential growth, the rate of growth is proportional to cell size, so that . In this case, constant cell cycle periods allow growth to a constant multiple of birth size, so that slight fluctuations are not correctedin succeeding generations[21].Therefore, exponentially growing cells must vary cell cycle duration to damp fluctuations. This is accomplished through size-dependent cell cycle progression.
Cell size and growth may be measured by a variety of methods, summarized in Table 1. In budding yeast, an exponential model of growth is supported by both single cell analysis using time lapse microscopy and bulk experiments using radioactive labeling[17, 22, 23].Although it has recently been shown that various cell cycle arrests differentially shift growth rate [24], budding yeast adhere closely to exponential growth in an unperturbed cell cycle. The growth function for fission yeast, however, deviates from exponential (see [18] for a comprehensive review).Fission yeast growth includes two linear segments separated by a rate change point (RCP), followed by a period directly preceding division during which the cell does not increase its length. Despite this difference in growth function, both budding and fission yeasts rely on size control mechanisms to couple growth and division (Figure 2). This may occur because, even though active size controls are not required in linearly growing cells, they may providea more efficient means of controlling size than relying on agrowth rate dependent cell cycle duration.
Table 1.
Methods for measuring cell size
| method | measured quantity |
measurement principle |
advantages/ disadvantages |
references |
|---|---|---|---|---|
| flow cytometry | ||||
| resistive-pulse sensing (coulter counter) |
volume | change in resistance due to volume displacement |
fast and accurate/does not track individual cells |
[25, 152, 153] |
| fluorescent flow cytometry and fluorescence activated cell sorting (FACS) |
volume and shape |
light scatter | high-throughput, can be combined with different fluorescent labels / indirect measurement of size, does not track individual cells |
[154, 155] |
| protein content |
total fluorescence after FITC staining |
|||
| imaging | ||||
| bright field | area | volume estimate from area of focal plane |
Single cell time course capability, /difficult to accurately identify cell border |
[156] |
| fluorescent proteins | protein content |
fluorescence of constitutively expressed fluorophores |
Single cell time course capability/ indirect measurement |
[17] |
| interferometry | dry mass | phase shifts caused by biomolecules biomolecules |
Single cell time course capability, dry mass determination with subcellular resolution / complicated analysis |
[157-159] |
| microchannelresonators | buoyant mass |
Changes in mass affect resonance frequency |
Single-cell time course capability/ low throughput, specialized equipment needed |
[23, 49] |
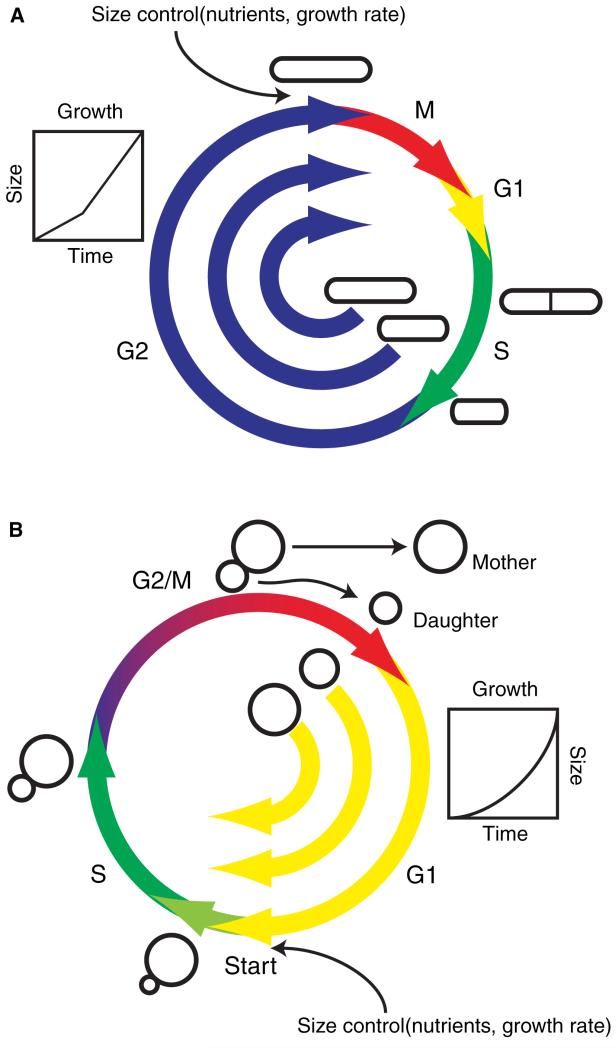
Figure 2. Size-dependent cell cycle progression in S. pombe and S. cerevisiae.
A. S. pombe cells enter G2 at different sizes following S-phase. They grow in a bilinear fashion and enter mitosis upon reaching a threshold size, so that smaller cells spend more time in G2 than larger cells, as indicated. B.S. cerevisiae daughter cells are born at different sizes and grow exponentially. Smaller cells spend more time in G1 prior to Start than larger cells (as indicated), which partially compensates for initial size variation. Thus, size control, a function of nutrient conditions and growth rate, is exerted at G2-M in S. pombe and within G1 in S. cerevisiae.
Additionally, exponential and linear growth functions different have implications for the effects of cell-to-cell variability and cell size mutants on fitness. Because the instantaneous rate of growth is proportional to cell size during exponential growth, mass doubling time is independent of cell size.Therefore, size control mutations shifting cell size, but not affecting metabolic processes, are not expected to affect population doubling time in exponentially growing cells. This is the case in budding yeast, where size mutants (e.g., whi5Δ) have WT population doubling times [25]. Similarly, although fluctuations may push sizes of individual cells significantly below or above the population mean, this is not expected to affect fitness for exponentially growing cells. Consistent with this argument, budding yeast exhibit much larger size variability than fission yeast, which does not exhibit exponential growth. For haploid cells, the coefficient of variation (standard deviation / mean; CV) of S. pombe cell size at fission is ~0.06, while the CV of S. cerevisiae cell size at budding is ~0.17 [16, 17, 26]. Yet, the noise in budding yeast is still bounded, which may reflect the limits over which growth is exponential. If cells get too large, then a single genome will be unable to support exponential growth of the cytoplasm[27]. Thus, we expect the degree of noise tolerated to be related to the range over which growth is exponential.
In addition to influencing the selective pressures on size control systems, growth functions may themselves be selected for. In particular, the growth functions of fission and budding yeasts may reflect the physiological requirements of symmetric and asymmetric division patterns. Upon nutrient limitation, budding yeast will produce daughter cells less than 20% of the mother cell size [15]. This asymmetric division may select for growth functions that are efficient over a larger range in cell sizes, such as exponential growth. In turn, efficient growth over a large size rangelessensthe pressure tohaveprecisesize control. Therefore, we expect to see the degree of cell-to-cell variation tolerated in size control increase with the degree of asymmetry in division size, illustrating the potential interdependence of the growth function, division pattern, and size control.
Perfect and imperfect size thresholds
Despite different growth functions, size control in budding and fission yeasts is broadly similar. As indicated in early physiological studies,both yeasts implement continuous monitoring of a size-dependent signaland restrict cell cycle progression at specific stages in a size-dependent manner. In S. pombe, evidence for a size requirement for division came from experiments that tracked the growth and division of cells following synchronization by S-phase arrest [19]. Cells continued to lengthen during the arrest, so that they became much larger than normal cells. Upon release, the progeny of these cells returned to normal size after only a few division cycles, because each cycle took only about 60% of the normal time, preventing mass doubling before division. Cell division cycles returned to their normal durationonce the cells had regained their normal size distribution. These shortened cycles indicated that the abnormally large arrested cells had surpassed a size threshold, manifested primarily at the G2-M transition [28, 29]. Later work uncovered an additional size requirement at the G1-S transition, which is revealed by adverse growth conditions or mutations reducing cell size[30-34]. This secondary size checkpoint has been little-studied, so we do not discuss it here.
In S. cerevisiae, early single-cell studies showed that small cells spend a longer time in G1, which allows them to grow more than initially larger cells[15]. These experiments suggesteda size threshold at the G1-S transition inS. cerevisiae, rather than at the G2-M transition, as in S. pombe. Additionally, the asymmetric division pattern of budding yeast has implications for size control. Mother cells fulfilled the size requirement when they first became mothers and bud at regular intervals[35].Interestingly, even small mothers do not exhibit size control and proceed through G1 rapidly, compared to similarly-sized daughter cells [36].This difference in mother-daughter size control programs was later traced to the asymmetric distribution of the daughter-specific transcription factors Ace2 and Ash1,which regulate transcription of the G1 cyclinCLN3[37-41]. Thus, S. cerevisiae size control acts primarily on the G1-S transition in daughter cells.
Similar to the secondary size control at G1-S in fission yeast, there is evidence for additional size control during budding yeast S/G2/M. This was initially suggested by the finding that halting bud growth by depolymerizing actin induces a G2/M delay in cells with buds smaller than a threshold size[42]. This effect depends on homologues of the key regulatory proteins for mitotic entry in fission yeast (see below).A subsequent study challenged the causal role of sizeby showing that stopping bud growth through disruption of vesicular transportdoes not cause a G2/M delay [43]. This study suggested that actin depolymerization engages the morphogenesis checkpoint [44], and that the apparent size dependence is in fact due to differences in G2/M progression. Nevertheless, single-cell studies show a weak dependence of G2/M duration on size at budding (S. DiTalia, personal communication), suggesting the need for further investigation.
It is important to note that, wherever it occurs in the cell cycle, size control can be more complex than a simple threshold. Typically, cell cycle control has been viewed in terms of ‘sizers’ and ‘timers’. Sizers require that cells pass a size threshold, while timers require that cells wait a fixed amount of time independent of size[45].However, these concepts are insufficient to describe all size-dependent cell cycle progression. The degree of size control can be inferred by plotting a metric of cell growth within a given interval versus the size of the cell upon entering the interval (Figure 3A). If the slope of a curve fit to the single cell data is −1, the amount of growth in the interval will exactly compensate the difference in initial sizes. If the slope is 0, growth within the interval is uncorrelated with the entrance size and there is no size-dependent control, i.e., a timer. Imperfect size control will yield intermediate values. Haploid fission yeast cells exhibit a perfect sizer[16], whereas earlyfrog embryos exhibit a perfect timer[46]. Budding yeast G1-S control has significant but imperfect size control, as exhibited by a slope of −0.7 (Figure 3B) [17]. Thus, eukaryotic cells exhibit distinct degrees of cell size control.
Figure 3. Single-cell size control assay as applied to S. cerevisiae.
A.Size control leads to a negative correlation between cell size at birth and growth in G1. Live-cell imaging techniques allow cells to be tracked from birth to budding, which is concomitant with DNA synthesis.B. Plotting the logarithm of size at birth vs. relative growth in G1 quantifies the efficiency of G1 size control. A slope of −1 would indicate perfect size control, whereas as slope of 0 indicates a lack of size control. Wild-type S.cerevisiae exhibit imperfect but significant size control, as indicated by the −0.7 slope relating birth size to growth in G1.C. Deletion of the cell cycle control gene WHI5 greatly diminishes the efficiency of size control, as indicated by the reduced slope of −0.3. Data adapted from ref. [17].
The intermediate degree of size control exhibited by budding yeast correlates with imperfect size compensation through a single cell cycle. In other words, it will typically take more than one cycle to damp size fluctuations. Additionally, the imperfect size control means that the ‘size threshold’ for the G1-S transition is itself size-dependent and that, on average, cells that are smaller when they enter G1 will bud at smaller sizes than their larger brethren. This cannot be accounted for by a simple combination of sizer and timer modules. A full account of the molecular mechanism underlying budding yeast size control will be required to understand itscomplex size-dependence. Nevertheless, “shifting the size threshold” up or down may be interpreted as shifting the size control curve (Figure 3) right or left to generate larger or smaller cells at budding, respectively, for the same input birth size. Although common in the literature, G1 progression kinetics in budding yeast are imperfectly characterized by a single size-threshold and require a curve for accurate description.
Size-dependent cell cycle progression suggests that cells somehow sense their own size. A discussion of how cells sense their own size requires first a clarification of what we mean by size. At its most general, “size” conflates cellular mass and volume, which are often correlated. This correlation is imperfect, however, as cell density varies through the cell cycle significantly inS. pombe, and more subtly inS. cerevisiae[47-49]. Total protein or ribosome content may also represent relevant additional, correlated size metrics, whichmightbe sensed by distinct molecular mechanisms. Twosize-sensing mechanisms have emerged as favored models: a protein synthesis rate-based mechanism in budding yeast and a gradient mechanism that directly detects cell length in fission yeast.
Geometric size sensing in S. pombe
Fission yeast are rod-shaped and grow highly asymmetrically. That fission yeast maintain a constant width and increase only in length has been exploited in size control studies, as it allows measurement of a single dimension, length, to indicate three-dimensional volume [47]. Intriguingly,S. pombe cells might exploit their own well-defined geometry by using a spatial gradientto sense cell length[50, 51]. The principle issimple: a mitotic activator is localized to a region in the middle of the cell, and an inhibitor of the activator is arranged in a gradient originating from the cell poles. Thus, as the cell grows, the amount of inhibitor interacting with the activator decreases,leading to an increase inmitotic cyclin activity thatdrives mitosis.
The first gene specifically identified as a size controller was Wee1, which restrains CDK activity[28] by phosphorylating Tyr 15 (Figure4A).Removal of this specific phosphate by Cdc25 is rate-limiting for mitotic entry [52-55]. The spatial size sensor model operates through the Wee1-inhibitory kinases, Cdr1 and Cdr2 [56-60], which are themselvesinhibited by Pom1, a kinase identified through its pleiotropic effects on cell morphology and growth [61]. While Pom1 is localized in a spatial gradient emanating from the cell poles [61, 62], Cdr1 and Cdr2 are localized in cortical nodes in the middle of the cell[63].As cells grow, their poles move apart, reducing medial Pom1 concentration. This leads to reduced Pom1-dependent Cdr1/2-inhibition, potentially yielding a gradient-based sizer mechanism [50, 51]. The Pom1 gradient arises due to Tea4 dependent localization to the tips followed by autophosphorylation-induced membrane dissociation [64]. In support of the gradient model, mislocalization of Pom1 lengthens cells [50, 51].
Figure 4. Models of size-dependent cell cycle regulatory networks.
Activators of cell cycle progression are colored green; inhibitors are colored red. A. In S. pombe, size control acts at the G2-M transition. The mitotic inhibitory kinase Pom1, localized to a membrane gradient originating at the cell poles, inhibits the pro-mitotic kinase Cdr2, which is localized in cortical nodes at the midcell. Cdr2, in turn, inhibits the Wee1 kinase, which inhibits CDK1. As pombe cells grow in length, the medial concentration of Pom1 decreases, allowing activation of Cdc13-CDK1 and entry into mitosis. B. In S. cerevisiae, size control acts at the G1-S transition. CDK1 in complex with the upstream G1 cyclin Cln3, whose activity is growth-dependent, activates the G1-S transcription factor SBF by direct phosphorylation and by inactivating the transcriptional inhibitor Whi5. This activates a transcriptional positive feedback loop that leads to a switch-like entry into S-phase. Importantly, Cln3 acts on promoter-bound SBF-Whi5 complexes, suggesting that Cln3 actiivty may be titrated against promoter-bound SBF.
However, the pleiotropy of Pom1 perturbations raises some doubt as to the specificity of its role in size control.Among other functions, Pom1 is required for proper positioning of the division plane[62, 65]. Furthermore, the ability of the Pom1 gradient itself to accomplish size control has been insufficiently tested. For instance, the Pom1 concentration on the membrane decays exponentially from the cell pole with a length scale of~1.5μm [66]. Since pre-mitotic S. pombe cells have an average length of more than 10 μm, this finding suggests that the medial concentration of Pom1 will drop to extremely low levels well before mitotic entry. This is particularly relevant incdc11-119mutants deficient in septation, which yield multinucleate cells containing a DNA to cytoplasm ratio similar to WT (F. Neumann personal communication). In addition, size scaling with ploidy suggests that non-spatially based mechanisms also operate in fission yeast [8].
Prior to the discovery of the Pom1 gradient mechanism, S. pombe size control studiesanticipated non-spatial mechanisms.It was suggested that accumulation of Cdc13might drive the transition[67], but most studies focused on possible Wee1 or Cdc25-mediated mechanisms. While regulation of Wee1 by Cdr2 and Cdr1kinases has been associated primarily with nutrient modulation of cell size [59, 68, 69], size sensing was more commonly ascribed to Cdc25. An early study suggested that Cdc25 (rather than Cdc13) induced mitosis at a critical size through an accumulation mechanism [70], and the finding that Cdc25 levels increase during cell cycle arrest strengthened the link between cell growth and Cdc25 synthesis[71, 72]. Finally, Cdc25 levels are disproportionately sensitive to translation initiation rate, which is mediated by uORFs and additional 5′ regulatory sequences[73]. Although these studies are far from conclusive, they suggest that a Cdc25-dependent translational sizer may act in concert with aPom1-Cdr2-Wee1 spatial sensor. Furthermore, the maintenance of a consistent, albeit greater, division length in cells with both key phosphosites mutated in Cdc2 indicates that other pathways must be involved in mitotic size control [74]. Certainly, taken together, these data suggest multiple size sensing mechanisms operating in fission yeast.
Synthesis-ratebased size sensing in S. cerevisiae
A gradient-based geometric size sensing mechanism in budding yeast cells would require the formation of linear surface gradients in these nearly spherical cells, or cytoplasmic gradients with length scales of a few microns. Although both mechanisms are feasible, as they have been observed in bacterial cells[75-77], there is strong evidence against a geometric size sensor operating in budding yeast. In particular, cell size increases linearly with ploidy over a six-fold range despite significant differences in geometrical characteristics such as cell wall curvature, surface-to-volume ratio, and eccentricity[10, 17]. It is therefore unlikely that geometric parameters other than size are responsible for initiating cell cycle progression. Furthermore, the filamentous fungusAshbyagossypii presents an interesting case in comparison to budding yeast, because it exhibits similar G1-S control, despite its distinct geometry [78](Figure 1).G1 control mutants that affect cell size in budding yeast, such as whi5 and cln3deletionmutants that make cells smaller and larger respectively, have a similar effect on the amount of cytoplasm per nucleus in A. gossypii (A. Gladfelter personal communication).This argues that the size control, as manifested in the DNA to cytoplasm ratio,is unlikely to be operating on geometric considerationsin species more closely related to budding yeast.
A mechanism that has no geometric requirement and could maintain the DNA to cytoplasm ratio for cells of any shape is a protein synthesis rate based sensor.Such a mechanism could explain the similar effects of mutations inAshbyaand buddingyeast despite gross shape differences. At its most basic, this class of models posits that a division-promoting protein is synthesized at a rate proportional to overall protein synthesis andthat the cell enters the cell cycle when that factor reaches a threshold level. Such models were proposed early on to underlie the size threshold for division in bacteria and metazoans [79-81], as well as in S. cerevisiae and S. pombe[29, 82, 83]. Consistent with this model, reducing translation rate by incubation with low doses of cycloheximide extends G1 and increases the threshold cell sizein S. cerevisiae[84-87]. Interestingly, although similar results were observed in S. pombe treated with pulses of cycloheximide[88], extended incubation of S. pombe cells in low levels of cycloheximide does not affect size at division, providing evidence against a synthesis rate mechanism in fission yeast (F. Navarro, personal communication).
Protein synthesis rate based models rely on asizer protein, whose abundanceincreases proportionally to the protein production rate.Such a sizer protein should exhibit three key properties. First, the sizer should be a dose-dependent activator of cell cycle progression; that is, it must berate limiting and not merely a necessary component for cell cycle progression. Second, its expression must be nearly constitutive during the growth period, so that it consistently indicates overall protein synthesisrate. Finally, the sizer should be unstable, so that its activity reflects current protein production rate rather than total synthesis[89-91]. The protein in S. cerevisiae that exhibits these three properties, and is thus the prime candidate for a sizer protein, is the G1 cyclin Cln3. CLN3 is a dose-dependent activator of the G1-S-transition [92-94], and unlike other cyclins, its expression oscillates only weakly through the cell cycle[40, 94-98]. Finally, Cln3 is a highly unstable protein, with a half-life of less than five minutes [92-96]. Although the properties of Cln3, when examined using bulk assays, suggest that it acts as a synthesis ratesizer, its instability has prevented its analysis in live cells. A quantitative analysis isstill required to demonstrate that, as predicted by the model, total amounts ofthe Cln3 protein correlate with size and determine precise timing of the G1-Stransition.
Although Cln3 fits the classical description of a sizer protein, there is a basic problem facing the Cln3 synthesis ratesizer model: given that the total amount of Cln3 is proportional to protein synthesis rate, which is proportional to size in exponentially growing budding yeast cells, the correlation between ribosome content and volume will render the concentration of Cln3 roughly constant. How then does the cell produce a signal whose activity increases with cell size, when the key activator remains at constant concentration? This problem is inherent to all synthesisrate sizer models and suggests that cells normalize sizerprotein abundance against a standard that does not change with cell growth.
Early formulations of this model proposed genomic DNA (or specific sites thereon, such as replication origins) as a non-growing standard [79, 83]. In budding yeast, the nucleus was proposed as a proxy for the genome, as its volume was long assumed to depend on DNA content [99, 100]. This mechanism is supported by the fact that Cln3’s primary targets are the transcription factor SBF and its inhibitor, Whi5[101, 102], and by Cln3’s active nuclear localization[103, 104]. However, the nucleus has recently been shown to grow in G1 and maintain a nearly constant nuclear to cytoplasmic volume ratio in both S. cerevisiaeand S. pombe[8, 9]. Thus, it is necessary to consider how Cln3 abundance might be compared directly to cellular DNA content. The recent finding that Cln3 is present at SBF binding sites in the promoter of a key downstream target (CLN2) suggests a model in which Cln3 activity is titrated against the fixed number of SBF binding sites (called SCBs) in the genome[105]. Evidence for this model is currently limited to the finding that transformation of yeast with a high-copy plasmid containing several SCBs increases cell size at budding in a Cln3 and Whi5-dependent manner [105]. This promising model requires further study.
Positive feedback at the basis of the budding yeast threshold response
Althoughthe Cln3-synthesis-rate model might provide a signal whose strength increases smoothly in proportion to cell size, it does not obviously provide a threshold mechanism. A threshold in G1 was first identified by Lee Hartwell in colleagues, who defined Start as the point of commitment to the mitotic cell cycle [106]. When to exposed to mating pheromone, pre-Start cells immediately arrest their cell division cycle, whereas post-Start cells divide once more. Positive feedback loops have been shown to underlie threshold responsesin bothcell cycle and developmental transitions[107-109]. Indeed, in budding yeast, the threshold for Start is provided by a G1 cyclin positive feedback loop. The SBF inhibitor Whi5 is the rate-limiting target of Cln3 in G1, whose phosphorylation by Cln3-CDK initiates its dissociation from SBF and export from the nucleus [101, 102, 110]. Partial relief of SBF inhibition results in the transcriptional activation of the two downstream G1 cyclins, Cln1 and Cln2, completing a positive feedback loop (Figure 4b)[111-114]. Activating the Cln1/2 positive feedback loop ensures rapid export of the remainderof the nuclear Whi5 and commits the cell to division[110]. It has been proposed that ultrasensitivity in Cln3 activity is a result of cytoplasmic retention mechanisms [115, 116] or a uORF limiting translation rate [117].However, these models are inconsistent with slow and steady export of Whi5 from the nucleus in cln1Δcln2Δ cells lacking the cyclin positive feedback loop, but containing endogenous levels of Cln3[114].Importantly, progression through G1 becomes size-independent after Whi5 export[17], indicating that flipping the positive feedback-switch converts the gradually increasing size-dependent signal into a threshold response thatmarks the end of the size control program.
Increasing Cln-CDK effects S-phase entry by activating the related heterodimerictranscription factors SBF (Swi4-Swi6) and MBF (Mbp1-Swi6) to promote the coherent transcription of more than 200 genes [111, 114, 118-120]. SBF is activated both via the inhibition of Whi5 and through a Whi5-independent mechanism [101, 114]. MBF is also activated by G1-cyclin CDK activity, possibly through the Swi6 subunit shared with SBF [121]. Despite co-regulation, the transcription of the regulon through the mitotic cell cycle is temporally organized: the G1 cyclinCLN1 is the earliest activated gene (CLN2 is also relatively early) implying that G1 cyclin positive feedback is initiated prior to genome-wide changes to the transcriptional program [122]. The G1 cyclin promoters are therefore important determinants of the CDK-activity threshold required to trigger the positive feedback loop. Thus, werequire a better understanding of the molecular mechanisms that determine the susceptibility of G1-Sregulon promoters to Cln3-CDK activity in order to understand the molecular basis ofcell size regulation.
Environmental modulation of cell size
For any given environment, yeast maintain a consistent cell size by monitoring growth during the cell cycle and restricting proliferation in a size-dependent manner. However, it is well known that cell size can change dramatically as a function of extracellular conditions. Thus,the size control program is modulated according to nutrient conditions and growth rate.Typically, cells growing quickly on rich media divide at a larger size than cells growing slowly under poor nutrient conditions.
Modulation of size controlis particularly important for synthesis rate-based mechanisms.Since protein synthesis rates for slow growing cells of a given mass are lower, these cellswould be expected torequire significant increases in cell size to overcome a static threshold[90]. However, the opposite is generally the case: though there is no linear relationship between the growth rate and the mean size of the population,[25, 123]slow growth on poor nutrients tends to reduce cell size[123]. Intriguingly, a change in the available nutrients immediatelytriggers an adjustment in sizecontrol. For example, when S. cerevisiae is shifted from ethanol (slow growth) to glucose (fast growth) containing media the budding index (% cells in S/G2/M) temporarily decreases as cells rapidly adjust to a larger target size[87, 124-126]. Upon a nutrient downshift, such asa shift from glutamate to proline[30, 68, 127], or sudden glucose starvation, S. pombe immediately undergoes several rounds of division without growth phases, to attain a much smaller average cell size [128, 129].These observations have led to the conclusionthat, in addition to a size sensing mechanism, there must be a mechanism that rapidlymodulatesthe target size in response to nutrient availability[90, 130].
Several important proteins involved in metabolism and growth have also been implicated in size control. Specifically, the highly conserved kinases, target-of-rapamycin (TOR) and protein kinase A (PKA) respond to nutrient availability by sensing external cues through nutrient receptors[131], as well as internal cues such as glycolytic activity or amino acid concentrations (reviewed in [132-135]). The downstream targets of PKA and TOR include metabolic pathways and biosynthetic genes thatpromote growth. Several mutants of both the PKA and TOR pathways have been implicated in yeast size control [25][136]. As Yanagida et al[136]and Shiozaki[137]have recently provided detailed reviews on size setting in S. pombe, which highlight the importance of TOR, we will focus our discussion on summarizing recent findings in S. cerevisiae.
When screening the S. cerevisiae deletion collection for genes determining cell size, Jorgensen et al. identified SFP1 and SCH9, previously poorly characterized components of the TOR and PKA pathways [25]. Sfp1 and Sch9 activity is both modulated by nutrients andactivates ribosome production through the ribosome biosynthesis (Ribi) and ribosomal protein (RP) genes[86]. Jorgensen et al.(2004) proposed a model where ribosome biosynthesis rates set the size threshold, while current ribosome activity, i.e.,protein synthesisrate, enables passing that threshold.
Over the last decade, the pathways that connect nutrient sensing, Sfp1 and Sch9 activity, and ribosome biosynthesis have been investigated, and a more complete picture is beginning to emerge (reviewed in [134, 138]). Briefly, Sfp1 and Sch9 are both directly phosphorylated by the TOR complex 1, with crosstalk to the PKA pathway [139, 140]. Sfp1 activity is regulated by localization to the nucleus. Under unfavorable conditions such as nutrient limitation or chemical stress, Sfp1 is retained in the cytoplasm byMrs6, a component of the secretory pathway[138, 141]. In the nucleus,Sfp1 activates the transcription of the Ribi and RP genes, likely by controlling the localization of the transcription factors Fhl1 and Ifh1 [86, 142]. In turn, Sch9 activates Ribi, RP and rRNA gene expression by inactivating the transcriptional repressors Stb3, Dot6 and Tod6 [143].
Despite our increased understanding of the role of Sfp1 and Sch9 signaling in ribosome synthesis, there has been very little progress in determining how this contributes to size control. In fact, aspects of Jorgensen and Tyers’s model have been challenged by several studies: Bernstein et al.(2007) showed that disturbing the maturation of ribosomes, without inhibiting their transcription, delays Start in a Whi5-dependent manner and leads to bigger cells [144]. Another study addressed the pleiotropic effects of SFP1 deletion and showed that Sfp1 also directly targets metabolism [126]. Notably, the overexpression of Msr6, which leads to retention of Sfp1 in the cytoplasm, was shown to downregulate glycolysis[141]. Thus, Sfp1 may modulate cell size in a way that is independent of ribosome synthesis. Additionally, nutrient dependent signaling may also act on ribosome biosynthesis in a Sfp1/Sch9-independent way, e.g. possibly through transcriptional inhibitors such as Crf1 [145]. A full understanding of the roles of Sfp1 and Sch9 at Start will likely require the identification of a molecular link between these two proteins and the core cell cycle regulatory network.
Additionally, there are multiple TOR- and PKA-independent pathways in nutrient signaling that may also play a role in size control[133].For example, there is strong evidence for glucose metabolism regulating several cell cycle genes independently of PKA and TOR [146]. In this context, recent findings from Cai and Tu[147, 148] appear especially interesting: the concentration of acetyl-CoA (the endproduct of glycolysis) directly modulates the acetylation of regulatory proteins, which in turn modify histone acetylation and thereby enhance transcription of many genes required for growth and proliferation. Another interesting link between metabolism, growth and cell cycle was proposed by Futcher[149], whosuggestedthat the accumulation of storage carbohydrates could be an important determinant of Start and cell size under nutrient limited growth conditions.
Although the specific connection remains unknown, metabolic regulation must interact with cell cycle control at the G1-S transition. Studies shifting cells from poor to rich mediaindicate that size resetting is downstream of Cln3.Several studies demonstrated thatcln3 deleted cells still show nutrient modulation of size[124, 125, 150]. Baroni et al. [124] and Tokiwa et al. [150] showed that upon either glucose or cyclic adenylate-monophosphate (cAMP) addition, transcription of the G1 cyclinCLN1(and to a lesser extend CLN2) is inhibited, whereasCLN3transcription is unaffected. Flick et al [125] later mapped this inhibition to the CLN1 promoter. Inhibition of downstream G1 cyclin transcription was not only found during transient size resetting, but also played a role during steady-state growth on glucose.
Since size resetting acts downstream ofCLN3, we expect to find it acting on an element of the G1-S positive feedback loop. How is the activity of SBF modulated independently of CLN3? One likely candidate is the rate-limiting transcriptional inhibitorWhi5. While long-term steady state adjustments could simply be due to transcriptional regulation of Whi5, this would not account for immediate responses to glucose addition. We can thus speculate that modifications of Whi5 or of the Whi5-SBF interaction are responsible for rapid changes in target size. However, studies from Tyers and coworkers have shownthat size adjustment is maintained in cells lacking either or both CLN3 and WHI5[86, 102]. This implies that either a Cln3/Whi5-independent mechanism or multiple interactions set the target size. We therefore anticipate the discovery of novel transcriptional regulators that interact with SBF to fine tune the CDK requirement for passage through Start in response to nutrient conditions.
Concluding remarks
The many years of work on cell size control in budding and fission yeasts have revealed the basic principles underlying the maintenance of a consistent cell size in these organisms: critical cell cycle transitionsare delayed in a size-dependent manner, while the target size is modulated in response to environmental conditions. The protein regulatory networks that effect size sensing and cell cycle control have been elucidated in both budding and fission yeasts, but understanding the mechanisms underlying nutrient modulation of target size remains a major challenge. For size sensing, the remaining challenge is to determine how the various regulatory components work together to generate a size-dependent signal. In budding yeast, a protein synthesis rate model, based on titrating sizer molecules against genomic binding sites, shows promise.In fission yeast, a gradient-based spatial sensormay play an important role. The close homology between budding yeast and metazoan G1-Sregulation, and between fission yeast and metazoan G2-Mregulation [151], suggestsconservation of the regulatory principles underlying size control. Thus, understanding size control mechanisms in yeast will help us answer the fundamental question of how eukaryotic cells sense and set their own size.
Acknowledgements
We thank F. Neumann and F. Cross for insightful comments on the manuscript. We thank A. Doncic for the images used in Figure 1. JS was funded by the BurroughsWellcome Fund, the National Institutes of Health (GM092925), and theNational Science Foundation (CAREER award #1054025). JT is supported by NIH training grant GM007276 and JE by a Swiss National Science Foundation Postdoctoral Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thompson DAW, Whyte LL. On growth and form. Cambridge University Press; Cambridge: 1942. [Google Scholar]
- 2.Payne JL, Groves JR, Jost AB, Nguyen T, Myhre S, Hill TM, Skotheim JS. Late Paleozoic fusulinoidean gigantism driven by atmospheric hyperoxia. [DOI] [PubMed]
- 3.Payne JL, Jost AB, Wang SC, Skotheim JS. A permanent shift in the evolutionary dynamics of organism size caused by the end-Permian mass extinction.
- 4.Chan Y-HM, Marshall WF. Scaling properties of cell and organelle size. Organogenesis. 2010;6:88–96. doi: 10.4161/org.6.2.11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gregory TR. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol Rev CambPhilosSoc. 2001;76:65–101. doi: 10.1017/s1464793100005595. [DOI] [PubMed] [Google Scholar]
- 6.Cavalier-Smith T. Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Ann. Bot. 2005;95:147–175. doi: 10.1093/aob/mci010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrov DA. Mutational equilibrium model of genome size evolution. TheorPopulBiol. 2002;61:531–544. doi: 10.1006/tpbi.2002.1605. [DOI] [PubMed] [Google Scholar]
- 8.Neumann FR, Nurse P. Nuclear size control in fission yeast. J Cell Biol. 2007;179:593–600. doi: 10.1083/jcb.200708054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jorgensen P, Edgington NP, Schneider BL, Rupes I, Tyers M, Futcher B. The size of the nucleus increases as yeast cells grow. MolBiol Cell. 2007;18:3523–3532. doi: 10.1091/mbc.E06-10-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mortimer RK. Radiobiological and genetic studies on a polyploid series (haploid to hexaploid) of Saccharomyces cerevisiae. Radiat. Res. 1958;9:312–326. [PubMed] [Google Scholar]
- 11.Dobi KC, Winston F. Analysis of transcriptional activation at a distance in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:5575–5586. doi: 10.1128/MCB.00459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchison JM. The Biology of the Cell Cycle. Cambridge University Press; 1971. [Google Scholar]
- 13.Adolph EF. The regulation of size as illustrated in unicellular organisms. C.C. Thomas; Springfield, IL; Baltimore, MD: 1931. [Google Scholar]
- 14.Prescott DM. Relation between cell growth and cell division. III. Changes in nuclear volume and growth rate and prevention of cell division in Amoeba proteus resulting from cytoplasmic amputations. Exp Cell Res. 1956;11:94–98. doi: 10.1016/0014-4827(56)90193-8. [DOI] [PubMed] [Google Scholar]
- 15.Johnston GC, Pringle JR, Hartwell LH. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp Cell Res. 1977;105:79–98. doi: 10.1016/0014-4827(77)90154-9. [DOI] [PubMed] [Google Scholar]
- 16.Sveiczer A, Novak B, Mitchison JM. The size control of fission yeast revisited. J Cell Sci. 1996;109(Pt 12):2947–2957. doi: 10.1242/jcs.109.12.2947. [DOI] [PubMed] [Google Scholar]
- 17.Di Talia S, Skotheim JM, Bean JM, Siggia ED, Cross FR. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature. 2007;448:947–951. doi: 10.1038/nature06072. [DOI] [PubMed] [Google Scholar]
- 18.Mitchison JM. Growth during the cell cycle. Int Rev Cytol. 2003;226:165–258. doi: 10.1016/s0074-7696(03)01004-0. [DOI] [PubMed] [Google Scholar]
- 19.Mitchison JM, Creanor J. Induction synchrony in the fission yeast. Schizosaccharomycespombe. Exp Cell Res. 1971;67:368–374. doi: 10.1016/0014-4827(71)90421-6. [DOI] [PubMed] [Google Scholar]
- 20.Conlon I, Raff M. Differences in the way a mammalian cell and yeast cells coordinate cell growth and cell-cycle progression. J. Biol. 2003;2:7. doi: 10.1186/1475-4924-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tyson JJ, Hannsgen KB. Global asymptotic stability of the size distribution in probabilistic models of the cell cycle. J Math Biol. 1985;22:61–68. doi: 10.1007/BF00276546. [DOI] [PubMed] [Google Scholar]
- 22.Elliott SG, McLaughlin CS. Rate of macromolecular synthesis through the cell cycle of the yeast Saccharomyces cerevisiae. PNAS. 1978;75:4384–4388. doi: 10.1073/pnas.75.9.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Godin M, Delgado FF, Son S, Grover WH, Bryan AK, Tzur A, Jorgensen P, Payer K, Grossman AD, Kirschner MW, et al. Using buoyant mass to measure the growth of single cells. Nat Methods. 2010;7:387–390. doi: 10.1038/nmeth.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goranov AI, Cook M, Ricicova M, Ben-Ari G, Gonzalez C, Hansen C, Tyers M, Amon A. The rate of cell growth is governed by cell cycle stage. Gene Dev. 2009;23:1408–1422. doi: 10.1101/gad.1777309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jorgensen P, Nishikawa JL, Breitkreutz B-J, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- 26.Lord PG, Wheals AE. Variability in individual cell cycles of Saccharomyces cerevisiae. J Cell Sci. 1981;50:361–376. doi: 10.1242/jcs.50.1.361. [DOI] [PubMed] [Google Scholar]
- 27.Zhurinsky J, Leonhard K, Watt S, Marguerat S, Bähler J, Nurse P. A coordinated global control over cellular transcription. CurrBiol. 2010;20:2010–2015. doi: 10.1016/j.cub.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Nurse P. Genetic control of cell size at cell division in yeast. Nature. 1975;256:547–551. doi: 10.1038/256547a0. [DOI] [PubMed] [Google Scholar]
- 29.Fantes P, Nurse P. Control of cell size at division in fission yeast by a growth-modulated size control over nuclear division. Exp Cell Res. 1977;107:377–386. doi: 10.1016/0014-4827(77)90359-7. [DOI] [PubMed] [Google Scholar]
- 30.Nurse P, Thuriaux P. Controls over the timing of DNA replication during the cell cycle of fission yeast. Exp Cell Res. 1977;107:365–375. doi: 10.1016/0014-4827(77)90358-5. [DOI] [PubMed] [Google Scholar]
- 31.Carlson CR, Grallert B, Stokke T, Boye E. Regulation of the start of DNA replication in Schizosaccharomycespombe. J Cell Sci. 1999;112(Pt 6):939–946. doi: 10.1242/jcs.112.6.939. [DOI] [PubMed] [Google Scholar]
- 32.Forsburg SL, Nurse P. Identification of a G1-type cyclin puc1+ in the fission yeast Schizosaccharomycespombe. Nature. 1991;351:245–248. doi: 10.1038/351245a0. [DOI] [PubMed] [Google Scholar]
- 33.Forsburg SL, Nurse P. Analysis of the Schizosaccharomycespombecyclin puc1: evidence for a role in cell cycle exit. J Cell Sci. 1994;107(Pt 3):601–613. [PubMed] [Google Scholar]
- 34.Martín-Castellanos C, Blanco MA, de Prada JM, Moreno S. The puc1 cyclin regulates the G1 phase of the fission yeast cell cycle in response to cell size. MolBiol Cell. 2000;11:543–554. doi: 10.1091/mbc.11.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hartwell LH, Unger MW. Unequal division in Saccharomyces cerevisiae and its implications for the control of cell division. J Cell Biol. 1977;75:422–435. doi: 10.1083/jcb.75.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lord PG, Wheals AE. Rate of cell cycle initiation of yeast cells when cell size is not a rate-determining factor. J Cell Sci. 1983;59:183–201. doi: 10.1242/jcs.59.1.183. [DOI] [PubMed] [Google Scholar]
- 37.Colman-Lerner A, Chin TE, Brent R. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell. 2001;107:739–750. doi: 10.1016/s0092-8674(01)00596-7. [DOI] [PubMed] [Google Scholar]
- 38.Weiss EL, Kurischko C, Zhang C, Shokat K, Drubin DG, Luca FC. The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J Cell Biol. 2002;158:885–900. doi: 10.1083/jcb.200203094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cosma MP. Daughter-specific repression of Saccharomyces cerevisiae HO: Ash1 is the commander. EMBO Reports. 2004;5:953–957. doi: 10.1038/sj.embor.7400251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Di Talia S, Wang H, Skotheim JM, Rosebrock AP, Futcher B, Cross FR. Daughter-Specific Transcription Factors Regulate Cell Size Control in Budding Yeast. PLoSBiol. 2009;7 doi: 10.1371/journal.pbio.1000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laabs TL, Markwardt DD, Slattery MG, Newcomb LL, Stillman DJ, Heideman W. ACE2 is required for daughter cell-specific G1 delay in Saccharomyces cerevisiae. PNAS. 2003;100:10275–10280. doi: 10.1073/pnas.1833999100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harvey SL, Kellogg DR. Conservation of mechanisms controlling entry into mitosis: budding yeast wee1 delays entry into mitosis and is required for cell size control. CurrBiol. 2003;13:264–275. doi: 10.1016/s0960-9822(03)00049-6. [DOI] [PubMed] [Google Scholar]
- 43.McNulty JJ, Lew DJ. Swe1p responds to cytoskeletal perturbation, not bud size, in S. cerevisiae. CurrBiol. 2005;15:2190–2198. doi: 10.1016/j.cub.2005.11.039. [DOI] [PubMed] [Google Scholar]
- 44.Howell AS, Lew DJ. Morphogenesis and the cell cycle. Genetics. 2012;190:51–77. doi: 10.1534/genetics.111.128314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Donnan L, John PC. Cell cycle control by timer and sizer in Chlamydomonas. Nature. 1983;304:630–633. doi: 10.1038/304630a0. [DOI] [PubMed] [Google Scholar]
- 46.Wang P, Hayden S, Masui Y. Transition of the blastomere cell cycle from cell size-independent to size-dependent control at the midblastula stage in Xenopuslaevis. J. Exp. Zool. 2000;287:128–144. doi: 10.1002/1097-010x(20000701)287:2<128::aid-jez3>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 47.Mitchison JM. The growth of single cells. I. Schizosaccharomycespombe. Exp Cell Res. 1957;13:244–262. doi: 10.1016/0014-4827(57)90005-8. [DOI] [PubMed] [Google Scholar]
- 48.Hartwell LH. Periodic density fluctuation during the yeast cell cycle and the selection of synchronous cultures. J Bacteriol. 1970;104:1280–1285. doi: 10.1128/jb.104.3.1280-1285.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bryan AK, Goranov A, Amon A, Manalis SR. Measurement of mass, density, and volume during the cell cycle of yeast. PNAS. 2010;107:999–1004. doi: 10.1073/pnas.0901851107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moseley JB, Mayeux A, Paoletti A, Nurse P. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature. 2009;459:857–860. doi: 10.1038/nature08074. [DOI] [PubMed] [Google Scholar]
- 51.Martin SG, Berthelot-Grosjean M. Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature. 2009;459:852–856. doi: 10.1038/nature08054. [DOI] [PubMed] [Google Scholar]
- 52.Russell P, Nurse P. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell. 1987;49:559–567. doi: 10.1016/0092-8674(87)90458-2. [DOI] [PubMed] [Google Scholar]
- 53.Gautier J, Solomon MJ, Booher RN, Bazan JF, Kirschner MW. cdc25 is a specific tyrosine phosphatase that directly activates p34cdc2. Cell. 1991;67:197–211. doi: 10.1016/0092-8674(91)90583-k. [DOI] [PubMed] [Google Scholar]
- 54.Gould KL, Nurse P. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature. 1989;342:39–45. doi: 10.1038/342039a0. [DOI] [PubMed] [Google Scholar]
- 55.Nurse P. Universal control mechanism regulating onset of M-phase. Nature. 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- 56.Coleman TRTZDWG. Negative regulation of the wee1 protein kinase by direct action of the nim1/cdr1 mitotic inducer. Cell. 1993;72:919–929. doi: 10.1016/0092-8674(93)90580-j. [DOI] [PubMed] [Google Scholar]
- 57.Parker LLWSAYPGP-WH. Phosphorylation and inactivation of the mitotic inhibitor Wee1 by the nim1/cdr1 kinase. Nature. 1993;363:736–738. doi: 10.1038/363736a0. [DOI] [PubMed] [Google Scholar]
- 58.Wu LRP. Nim1 kinase promotes mitosis by inactivating Wee1 tyrosine kinase. Nature. 1993;363:738–741. doi: 10.1038/363738a0. [DOI] [PubMed] [Google Scholar]
- 59.Breeding CSHJBMKHSMYPGGKL. The cdr2+ gene encodes a regulator of G2/M progression and cytokinesis in Schizosaccharomycespombe. MolBiol Cell. 1998;9:3399–3415. doi: 10.1091/mbc.9.12.3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanoh JRP. The protein kinase Cdr2, related to Nim1/Cdr1 mitotic inducer, regulates the onset of mitosis in fission yeast. MolBiol Cell. 1998;9:3321–3334. doi: 10.1091/mbc.9.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bähler J, Pringle JR. Pom1p, a fission yeast protein kinase that provides positional information for both polarized growth and cytokinesis. Gene Dev. 1998;12:1356–1370. doi: 10.1101/gad.12.9.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Celton-Morizur SRVSJBPA. Pom1 kinase links division plane position to cell polarity by regulating Mid1p cortical distribution. J Cell Sci. 2006;119:4710–4718. doi: 10.1242/jcs.03261. [DOI] [PubMed] [Google Scholar]
- 63.Morrell JLNCBGKL. The GIN4 family kinase, Cdr2p, acts independently of septins in fission yeast. J Cell Sci. 2004;117:5293–5302. doi: 10.1242/jcs.01409. [DOI] [PubMed] [Google Scholar]
- 64.Hachet O, Berthelot-Grosjean M, Kokkoris K, Vincenzetti V, Moosbrugger J, Martin SG. A Phosphorylation Cycle Shapes Gradients of the DYRK Family Kinase Pom1 at the Plasma Membrane. Cell. 2011;145:1116–1128. doi: 10.1016/j.cell.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 65.Padte NNMSGHMCF. The Cell-End Factor Pom1p Inhibits Mid1p in Specification of the Cell Division Plane in Fission Yeast. Current Biology. 2006;16:2480–2487. doi: 10.1016/j.cub.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 66.Saunders T, Pan K, Angel A, Guan Y, Shah J, Howard M, Chang F. Noise reduction in the intracellular pom1p gradient by a dynamic clustering mechanism. Dev Cell. 2012:164. doi: 10.1016/j.devcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sveiczer A, Tyson JJ, Novak B. A stochastic, molecular model of the fission yeast cell cycle: role of the nucleocytoplasmic ratio in cycle time regulation. Biophys. Chem. 2001;92:1–15. doi: 10.1016/s0301-4622(01)00183-1. [DOI] [PubMed] [Google Scholar]
- 68.Young PG, Fantes PA. Schizosaccharomycespombe mutants affected in their division response to starvation. J Cell Sci. 1987;88(Pt 3):295–304. doi: 10.1242/jcs.88.3.295. [DOI] [PubMed] [Google Scholar]
- 69.Belenguer P, Pelloquin L, Oustrin ML, Ducommun B. Role of the fission yeast nim 1 protein kinase in the cell cycle response to nutritional signals. Biochemical and Biophysical Research Communications. 1997;232:204–208. doi: 10.1006/bbrc.1997.6253. [DOI] [PubMed] [Google Scholar]
- 70.Moreno S, Nurse P, Russell P. Regulation of mitosis by cyclic accumulation of p80cdc25 mitotic inducer in fission yeast. Nature. 1990;344:549–552. doi: 10.1038/344549a0. [DOI] [PubMed] [Google Scholar]
- 71.Kovelman R, Russell P. Stockpiling of Cdc25 during a DNA replication checkpoint arrest in Schizosaccharomycespombe. Mol. Cell. Biol. 1996;16:86–93. doi: 10.1128/mcb.16.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rupes I, Webb BA, Mak A, Young PG. G2/M arrest caused by actin disruption is a manifestation of the cell size checkpoint in fission yeast. MolBiol Cell. 2001;12:3892–3903. doi: 10.1091/mbc.12.12.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Daga RR, Jimenez J. Translational control of the cdc25 cell cycle phosphatase: a molecular mechanism coupling mitosis to cell growth. J Cell Sci. 1999;112(Pt 18):3137–3146. doi: 10.1242/jcs.112.18.3137. [DOI] [PubMed] [Google Scholar]
- 74.Coudreuse D, Nurse P. Driving the cell cycle with a minimal CDK control network. Nature. 2010;468:1074–1079. doi: 10.1038/nature09543. [DOI] [PubMed] [Google Scholar]
- 75.Chen YE, Tropini C, Jonas K, Tsokos CG, Huang KC, Laub MT. Spatial gradient of protein phosphorylation underlies replicative asymmetry in a bacterium. PNAS. 2011;108:1052–1057. doi: 10.1073/pnas.1015397108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Corbin BD, Yu X-C, Margolin W. Exploring intracellular space: function of the Min system in round-shaped Escherichia coli. EMBO J. 2002;21:1998–2008. doi: 10.1093/emboj/21.8.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang KC, Wingreen NS. Min-protein oscillations in round bacteria. Physical Biology. 2004;1:229–235. doi: 10.1088/1478-3967/1/4/005. [DOI] [PubMed] [Google Scholar]
- 78.Nair DR, D’Ausilio CA, Occhipinti P, Borsuk ME, Gladfelter AS. A conserved G regulatory circuit promotes asynchronous behavior of nuclei sharing a common cytoplasm. Cell cycle (Georgetown, Tex) 2010;9:3771–3779. doi: 10.4161/cc.9.18.12999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Donachie WD. Relationship between cell size and time of initiation of DNA replication. Nature. 1968;219:1077–1079. doi: 10.1038/2191077a0. [DOI] [PubMed] [Google Scholar]
- 80.Rossow PW, Riddle VG, Pardee AB. Synthesis of labile, serum-dependent protein in early G1 controls animal cell growth. PNAS. 1979;76:4446–4450. doi: 10.1073/pnas.76.9.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brooks RF. Continuous protein synthesis is required to maintain the probability of entry into S phase. Cell. 1977;12:311–317. doi: 10.1016/0092-8674(77)90209-4. [DOI] [PubMed] [Google Scholar]
- 82.Unger MW, Hartwell LH. Control of cell division in Saccharomyces cerevisiae by methionyl-tRNA. PNAS. 1976;73:1664–1668. doi: 10.1073/pnas.73.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fantes PA, Grant WD, Pritchard RH, Sudbery PE, Wheals AE. The regulation of cell size and the control of mitosis. J. Theor. Biol. 1975;50:213–244. doi: 10.1016/0022-5193(75)90034-x. [DOI] [PubMed] [Google Scholar]
- 84.Popolo L, Vanoni M, Alberghina L. Control of the yeast cell cycle by protein synthesis. Exp Cell Res. 1982;142:69–78. doi: 10.1016/0014-4827(82)90410-4. [DOI] [PubMed] [Google Scholar]
- 85.Moore SA. Kinetic evidence for a critical rate of protein synthesis in the Saccharomyces cerevisiae yeast cell cycle. J BiolChem. 1988;263:9674–9681. [PubMed] [Google Scholar]
- 86.Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Gene Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kief DR, Warner JR. Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol. Cell. Biol. 1981;1:1007–1015. doi: 10.1128/mcb.1.11.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Polanshek MM. Effects of heat shock and cycloheximide on growth and division of the fission yeast, Schizosaccharomycespombe. With an Appendix. Estimation of division delay for S. pombe from cell plate index curves. J Cell Sci. 1977;23:1–23. doi: 10.1242/jcs.23.1.1. [DOI] [PubMed] [Google Scholar]
- 89.Shilo B, Riddle VG, Pardee AB. Protein turnover and cell-cycle initiation in yeast. Exp Cell Res. 1979;123:221–227. doi: 10.1016/0014-4827(79)90462-2. [DOI] [PubMed] [Google Scholar]
- 90.Schneider BL, Zhang J, Markwardt J, Tokiwa G, Volpe T, Honey S, Futcher B. Growth rate and cell size modulate the synthesis of, and requirement for, G1-phase cyclins at start. Mol. Cell. Biol. 2004;24:10802–10813. doi: 10.1128/MCB.24.24.10802-10813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schneiderman MH, Dewey WC, Highfield DP. Inhibition of DNA synthesis in synchronized Chinese hamster cells treated in G1 with cycloheximide. Exp Cell Res. 1971;67:147–155. doi: 10.1016/0014-4827(71)90630-6. [DOI] [PubMed] [Google Scholar]
- 92.Nash R, Tokiwa G, Anand S, Erickson K, Futcher AB. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 1988;7:4335–4346. doi: 10.1002/j.1460-2075.1988.tb03332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cross FR. DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol. Cell. Biol. 1988;8:4675–4684. doi: 10.1128/mcb.8.11.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tyers M, Fitch I, Tokiwa G, Dahmann C, Nash R, Linskens M, Futcher B. Characterization of G1 and mitotic cyclins of budding yeast. Cold Spring Harb. Symp. Quant. Biol. 1991;56:21–32. doi: 10.1101/sqb.1991.056.01.005. [DOI] [PubMed] [Google Scholar]
- 96.Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McInerny CJ, Partridge JF, Mikesell GE, Creemer DP, Breeden LL. A novel Mcm1-dependent element in the SWI4, CLN3, CDC6, and CDC47 promoters activates M/G1-specific transcription. Gene Dev. 1997;11:1277–1288. doi: 10.1101/gad.11.10.1277. [DOI] [PubMed] [Google Scholar]
- 98.MacKay VL, Mai B, Waters L, Breeden LL. Early cell cycle box-mediated transcription of CLN3 and SWI4 contributes to the proper timing of the G1-to-S transition in budding yeast. Mol. Cell. Biol. 2001;21:4140–4148. doi: 10.1128/MCB.21.13.4140-4148.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cavalier-Smith T. Skeletal DNA and the evolution of genome size. Annu. Rev. Biophys. Bioeng. 1982;11:273–302. doi: 10.1146/annurev.bb.11.060182.001421. [DOI] [PubMed] [Google Scholar]
- 100.Futcher B. Cyclins and the wiring of the yeast cell cycle. Yeast. 1996;12:1635–1646. doi: 10.1002/(SICI)1097-0061(199612)12:16%3C1635::AID-YEA83%3E3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 101.de Bruin RAM, McDonald WH, Kalashnikova TI, Yates J, Wittenberg C. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell. 2004;117:887–898. doi: 10.1016/j.cell.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 102.Costanzo M, Nishikawa JL, Tang X, Millman JS, Schub O, Breitkreuz K, Dewar D, Rupes I, Andrews B, Tyers M. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 2004;117:899–913. doi: 10.1016/j.cell.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 103.Miller ME, Cross FR. Distinct subcellular localization patterns contribute to functional specificity of the Cln2 and Cln3 cyclins of Saccharomyces cerevisiae. Mol. Cell. Biol. 2000;20:542–555. doi: 10.1128/mcb.20.2.542-555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Edgington NP, Futcher B. Relationship between the function and the location of G1 cyclins in S. cerevisiae. J Cell Sci. 2001;114:4599–4611. doi: 10.1242/jcs.114.24.4599. [DOI] [PubMed] [Google Scholar]
- 105.Wang H, Carey LB, Cai Y, Wijnen H, Futcher B. Recruitment of Cln3 cyclin to promoters controls cell cycle entry via histone deacetylase and other targets. PLoSBiol. 2009;7:e1000189. doi: 10.1371/journal.pbio.1000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hartwell LH, Culotti J, Pringle JR, Reid BJ. Genetic control of the cell division cycle in yeast. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- 107.Ferrell JE. Tripping the switch fantastic: how a protein kinase cascade can convert graded inputs into switch-like outputs. Trends BiochemSci. 1996;21:460–466. doi: 10.1016/s0968-0004(96)20026-x. [DOI] [PubMed] [Google Scholar]
- 108.Ferrell JE, Machleder EM. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–898. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- 109.Justman QA, Serber Z, Ferrell JE, El-Samad H, Shokat KM. Tuning the activation threshold of a kinase network by nested feedback loops. Science. 2009;324:509–512. doi: 10.1126/science.1169498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Doncic A, Falleur-Fettig M, Skotheim JM. Distinct Interactions Select and Maintain a Specific Cell Fate. Mol. Cell. 2011;43:528–539. doi: 10.1016/j.molcel.2011.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nasmyth K, Dirick L. The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell. 1991;66:995–1013. doi: 10.1016/0092-8674(91)90444-4. [DOI] [PubMed] [Google Scholar]
- 112.Dirick L, Nasmyth K. Positive feedback in the activation of G1 cyclins in yeast. Nature. 1991;351:754–757. doi: 10.1038/351754a0. [DOI] [PubMed] [Google Scholar]
- 113.Cross FR, Tinkelenberg AH. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- 114.Skotheim JM, Di Talia S, Siggia ED, Cross FR. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature. 2008;454:291–296. doi: 10.1038/nature07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang H, Garí E, Vergés E, Gallego C, Aldea M. Recruitment of Cdc28 by Whi3 restricts nuclear accumulation of the G1 cyclin-Cdk complex to late G1. EMBO J. 2004;23:180–190. doi: 10.1038/sj.emboj.7600022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vergés E, Colomina N, Garí E, Gallego C, Aldea M. Cyclin Cln3 is retained at the ER and released by the J chaperone Ydj1 in late G1 to trigger cell cycle entry. Mol. Cell. 2007;26:649–662. doi: 10.1016/j.molcel.2007.04.023. [DOI] [PubMed] [Google Scholar]
- 117.Polymenis M, Schmidt EV. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Gene Dev. 1997;11:2522–2531. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Andrews BJ, Herskowitz I. Identification of a DNA binding factor involved in cell-cycle control of the yeast HO gene. Cell. 1989;57:21–29. doi: 10.1016/0092-8674(89)90168-2. [DOI] [PubMed] [Google Scholar]
- 119.Koch C, Moll T, Neuberg M, Ahorn H, Nasmyth K. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science. 1993;261:1551–1557. doi: 10.1126/science.8372350. [DOI] [PubMed] [Google Scholar]
- 120.Ferrezuelo F, Colomina N, Futcher B, Aldea M. The transcriptional network activated by Cln3 cyclin at the G1-to-S transition of the yeast cell cycle. Genome Biol. 2010;11:R67. doi: 10.1186/gb-2010-11-6-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wijnen H, Landman A, Futcher B. The G(1) cyclin Cln3 promotes cell cycle entry via the transcription factor Swi6. Mol. Cell. Biol. 2002;22:4402–4418. doi: 10.1128/MCB.22.12.4402-4418.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Eser U, Falleur-Fettig M, Johnson A, Skotheim JM. Commitment to a Cellular Transition Precedes Genome-wide Transcriptional Change. Mol. Cell. 2011;43:515–527. doi: 10.1016/j.molcel.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. MolBiol Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Baroni MD, Monti P, Alberghina L. Repression of growth-regulated G1 cyclin expression by cyclic AMP in budding yeast. Nature. 1994;371:339–342. doi: 10.1038/371339a0. [DOI] [PubMed] [Google Scholar]
- 125.Flick K, Chapman-Shimshoni D, Stuart D, Guaderrama M, Wittenberg C. Regulation of cell size by glucose is exerted via repression of the CLN1 promoter. Mol. Cell. Biol. 1998;18:2492–2501. doi: 10.1128/mcb.18.5.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cipollina C, van den Brink J, Daran-Lapujade P, Pronk JT, Porro D, de Winde JH. Saccharomyces cerevisiae SFP1: at the crossroads of central metabolism and ribosome biogenesis. Microbiology (Reading, Engl.) 2008;154:1686–1699. doi: 10.1099/mic.0.2008/017392-0. [DOI] [PubMed] [Google Scholar]
- 127.Petersen J, Nurse P. TOR signalling regulates mitotic commitment through the stress MAP kinase pathway and the Polo and Cdc2 kinases. Nat Cell Biol. 2007;9:1263–1272. doi: 10.1038/ncb1646. [DOI] [PubMed] [Google Scholar]
- 128.Costello G, Rodgers L, Beach D. Fission yeast enters the stationary phase G0 state from either mitotic G1 or G2. Curr Genet. 1986;11:119–125. [Google Scholar]
- 129.Pluskal T, Hayashi T, Saitoh S, Fujisawa A, Yanagida M. Specific biomarkers for stochastic division patterns and starvation-induced quiescence under limited glucose levels in fission yeast. FEBS J. 2011;278:1299–1315. doi: 10.1111/j.1742-4658.2011.08050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jorgensen P, Tyers M. How cells coordinate growth and division. CurrBiol. 2004;14:R1014–27. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 131.De Virgilio C, Loewith R. The TOR signalling network from yeast to man. Int. J. Biochem. Cell Biol. 2006;38:1476–1481. doi: 10.1016/j.biocel.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 132.Otsubo Y, Yamamato M. TOR signaling in fission yeast. Crit. Rev. Biochem. Mol. Biol. 2008;43:277–283. doi: 10.1080/10409230802254911. [DOI] [PubMed] [Google Scholar]
- 133.Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu. Rev. Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
- 134.Smets B, Ghillebert R, De Snijder P, Binda M, Swinnen E, De Virgilio C, Winderickx J. Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae. Curr Genet. 2010;56:1–32. doi: 10.1007/s00294-009-0287-1. [DOI] [PubMed] [Google Scholar]
- 135.Loewith R, Hall MN. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics. 2011;189:1177–1201. doi: 10.1534/genetics.111.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yanagida M, Ikai N, Shimanuki M, Sajiki K. Nutrient limitations alter cell division control and chromosome segregation through growth-related kinases and phosphatases. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2011;366:3508–3520. doi: 10.1098/rstb.2011.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shiozaki K. Nutrition-minded cell cycle. Science Signaling. 2009;2:pe74. doi: 10.1126/scisignal.296pe74. [DOI] [PubMed] [Google Scholar]
- 138.Lempiäinen H, Shore D. Growth control and ribosome biogenesis. CurrOpin Cell Biol. 2009;21:855–863. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 139.Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 140.Lempiäinen H, Uotila A, Urban J, Dohnal I, Ammerer G, Loewith R, Shore D. Sfp1 interaction with TORC1 and Mrs6 reveals feedback regulation on TOR signaling. Mol. Cell. 2009;33:704–716. doi: 10.1016/j.molcel.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 141.Singh J, Tyers M. A Rab escort protein integrates the secretion system with TOR signaling and ribosome biogenesis. Gene Dev. 2009;23:1944–1958. doi: 10.1101/gad.1804409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rudra D, Zhao Y, Warner JR. Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J. 2005;24:533–542. doi: 10.1038/sj.emboj.7600553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Huber A, French SL, Tekotte H, Yerlikaya S, Stahl M, Perepelkina MP, Tyers M, Rougemont J, Beyer AL, Loewith R. Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J. 2011;30:3052–3064. doi: 10.1038/emboj.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bernstein KA, Bleichert F, Bean JM, Cross FR, Baserga SJ. Ribosome biogenesis is sensed at the Start cell cycle checkpoint. MolBiol Cell. 2007;18:953–964. doi: 10.1091/mbc.E06-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Martin DE, Soulard A, Hall MN. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell. 2004;119:969–979. doi: 10.1016/j.cell.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 146.Newcomb LL, Diderich JA, Slattery MG, Heideman W. Glucose regulation of Saccharomyces cerevisiae cell cycle genes. Eukaryotic Cell. 2003;2:143–149. doi: 10.1128/EC.2.1.143-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cai L, Tu BP. Acetyl-CoA drives the transcriptional growth program in yeast. Cell cycle (Georgetown, Tex) 2011:10. doi: 10.4161/cc.10.18.17000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Futcher B. Metabolic cycle, cell cycle, and the finishing kick to Start. Genome Biol. 2006;7:107. doi: 10.1186/gb-2006-7-4-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tokiwa G, Tyers M, Volpe T, Futcher B. Inhibition of G1 cyclin activity by the Ras/cAMP pathway in yeast. Nature. 1994;371:342–345. doi: 10.1038/371342a0. [DOI] [PubMed] [Google Scholar]
- 151.Cross FR, Buchler NE, Skotheim JM. Evolution of networks and sequences in eukaryotic cell cycle control. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2011;366:3532–3544. doi: 10.1098/rstb.2011.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Coulter W. US Patent no 2656508 Means for counting particles suspended in a fluid. 1953
- 153.Zhang J, Schneider C, Ottmers L, Rodriguez R, Day A, Markwardt J, Schneider BL. Genomic scale mutant hunt identifies cell size homeostasis genes in S. cerevisiae. CurrBiol. 2002;12:1992–2001. doi: 10.1016/s0960-9822(02)01305-2. [DOI] [PubMed] [Google Scholar]
- 154.Herzenberg LA, Parks D, Sahaf B, Perez O, Roederer M, Herzenberg LA. The history and future of the fluorescence activated cell sorter and flow cytometry: a view from Stanford. Clin. Chem. 2002;48:1819–1827. [PubMed] [Google Scholar]
- 155.Cipollina C, Vai M, Porro D, Hatzis C. Towards understanding of the complex structure of growing yeast populations. J. Biotechnol. 2007;128:393–402. doi: 10.1016/j.jbiotec.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 156.Gordon A, Colman-Lerner A, Chin TE, Benjamin KR, Yu RC, Brent R. Single-cell quantification of molecules and rates using open-source microscope-based cytometry. Nat Methods. 2007;4:175–181. doi: 10.1038/nmeth1008. [DOI] [PubMed] [Google Scholar]
- 157.Popescu G, Park Y, Lue N, Best-Popescu C, Deflores L, Dasari RR, Feld MS, Badizadegan K. Optical imaging of cell mass and growth dynamics. Am J Physiol, Cell Physiol. 2008;295:C538–44. doi: 10.1152/ajpcell.00121.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang Z, Millet L, Mir M, Ding H, Unarunotai S, Rogers J, Gillette MU, Popescu G. Spatial light interference microscopy (SLIM) Opt Express. 2011;19:1016–1026. doi: 10.1364/OE.19.001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Barer R. Interference microscopy and mass determination. Nature. 1952;169:366–367. doi: 10.1038/169366b0. [DOI] [PubMed] [Google Scholar]