Abstract
Cultural practices can deeply influence genetic diversity patterns. The Neolithic transitions that took place at different times and locations around the world led to major cultural and demographic changes that influenced and therefore left their marks on human genetic diversity patterns. Several studies on the European Neolithic transition suggest that mitochondrial DNA (mtDNA) and Y-chromosome data can exhibit different patterns, which could be owing to different demographic histories for females and males. Archaeological and anthropological data suggest that the transition from hunter–gatherers (HGs) to farmers' societies is probably associated with changes in social organization, particularly in post-marital residence (PMR) rules (i.e. patrilocality, matrilocality or bilocality). The movements of humans and genes associated with these rules can be seen as sex-biased short-range migrations. We developed a new individual-based simulation approach to explore the genetic consequences of 45 different scenarios, where we varied the patterns of PMR and admixture between HGs and farmers. We recorded mtDNA and Y-chromosome data and analysed their diversity patterns within and between populations, through time and space. We also collected published mtDNA and Y-chromosome data from European and Near-Eastern populations in order to identify the scenarios that would best explain them. We show that: (i) different PMR systems can lead to different patterns of genetic diversity and differentiation, (ii) asymmetries between mtDNA and Y-chromosome can be owing to different behaviours between males and females, but also to different mutations rates, and (iii) patrilocality in farmers explains the present patterns of genetic diversity better than matrilocality or bilocality. Moreover, we found that (iv) the genetic diversity of farmers change depending on the HGs PMR rules even though they are assumed to disappear more than 5000 years ago in our simulations.
Keywords: post-marital residence, migration, spatial expansion, Neolithic, Palaeolithic, admixture
1. Introduction
The Neolithic transition was one of the greatest cultural transitions in human prehistory [1–3]. The demographic and cultural changes that it triggered unquestionably changed how human genes, cultures and languages are distributed around the world today [1,4]. Although Europe is probably the most studied area, there are still major disagreements among archaeologists [1,5–7] and among geneticists [8–12] on how the transition into farming-based societies happened in this region, and on how archaeological and genetic data should be interpreted [13]. As a first approximation, the Neolithic transition has mainly been modelled by considering one or the other of the following alternative scenarios: the Cultural Diffusion model (CDM) [14] and the Demic Diffusion model (DDM) [4,15,16]. The CDM proposes that agriculture and related technologies arrived in Europe without a significant movement of farmers. It predicts that there should be no or very little genetic contribution in Europe from the Near-Eastern populations. In the DDM, the spread of Neolithic innovations was a consequence of the movement of people that either eliminated or integrated the less densely populated hunter–gatherer (HG) societies [4]. A movement of genes is thus predicted, even though its genetic consequences are much more complex than is usually acknowledged [10,17]. In the last 15 years, the CDM has gained momentum, mainly from the support of mitochondrial DNA (mtDNA) analyses [11,12], despite some criticisms which suggest that it actually supports the DDM [18]. Non-recombining region of the Y-chromosome (NRY) data were also interpreted in favour of the CDM by some authors [19], but other studies have generated opposite conclusions [10,20]. The results of both the mtDNA and NRY are thus controversial. However, if we assume that mtDNA and NRY data could indeed be interpreted in different ways, one favouring CDM and the other the DDM, respectively, this would open the possibility that the demographic histories of females and males were different [21,22]. For instance, differences in migration patterns after marriage could lead to major differences in terms of genetic diversity within and between populations when comparing mtDNA and NRY data. This is why it is very important to analyse jointly these two markers, rather than independently, as is too often done to identify possible causes for the differences obtained beyond stochasticity [23].
Archaeological and anthropological data suggest that the transition from a HG to a farming society is correlated with drastic changes in lifestyle [24] and probably also with changes in post-marital residence (PMR) systems. Moreover, the majority of today's human populations (ca 74%) are patrilocal (i.e. the woman moves to her husband's birthplace after marriage) [25,26], but HG societies appear to be more variable, with bilocality (both males and females can move after marriage, with no clear bias towards one of the sexes) and matrilocality (higher male migration rates) practices being more frequent than in other societies (i.e. farmers and pastoralists) [27]. This observation has led to the suggestion that patrilocality started to increase after the emergence of agriculture [24,27–29].
However, there has been no formal test assessing whether there was such a shift during the Neolithic transition in Europe, using genetic data. Our aim is to study the impact of different PMR systems on genetic diversity and to investigate if genetic data can give us any indication on whether such an increase in patrilocality indeed occurred after the Neolithic transition. Here, we use realistic spatial forward simulations of individuals and record their NRY and mtDNA data to explore a wide spectrum of scenarios where HG and farmer populations are allowed to be either patrilocal, bilocal or matrilocal. These post-marital rules are modelled by varying the male and female migration rates.
2. Material and methods
(a). General framework
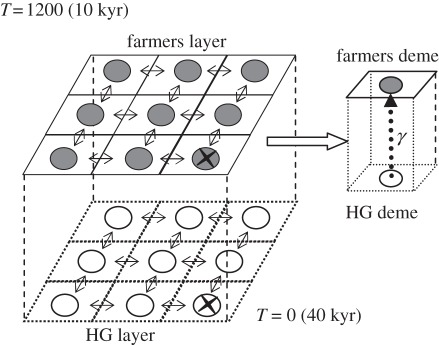
To address questions related to the changes in PMR rules during the European Neolithic transition, we developed a new forward spatial simulation approach that incorporates both geographical and demographic data, as well as several types of genetic markers. The general principle is very similar to that followed by the Splatche and Splatche2 software [30,31]. Like in those software, space is divided into ‘layers’, which are themselves subdivided into demes, as in a two-dimensional stepping-stone model (figure 1). Our framework allows to simulate different ‘layers’ (such as HG and farmers), inhabiting the same geographical space as in Currat & Excoffier [17]. Each deme can exchange migrants, at a certain rate (m), with up to four neighbours depending on its geographical location relative to the edges. Each deme is characterized by a carrying capacity (K) and a friction (F) values which can be different between layers. Density is logistically regulated within each deme (either in the HG or farmer layers), with intrinsic K and growth rate (r). Mating between layers (HG and farmers) is modelled with an admixture parameter (γ).
Figure 1.
Model of spatial expansion. Two different layers (farmers and HG) occupying the same geographical space. Demes are numbered using rows and columns, with deme 0_0 being the upper-left corner deme. The cross, in the bottom-right corner deme (deme 9_9 in the 10 × 10 lattice or deme 29_29 in the 30 × 30 lattice), indicates where the expansion starts at time T. Admixture (γ) represents gene flow between layers. In our simulations, admixture was unidirectional from the HG to the farmers layer.
However, contrary to Splatche2, our approach is not based on the coalescent. It uses a forward individual—rather than backward/coalescent gene-based simulation framework, where the demographic and genetic simulations are carried simultaneously. While computationally slower, it also has several advantages. We can: (i) model complex situations that occur in human societies (e.g. sex-biased migration) more easily, (ii) follow multi-locus genotypes within individuals, and (iii) simulate all the individuals of a deme. This last point is particularly important in studying the Neolithic transition, as one of the assumptions of the coalescent is that the effective population size is large compared with the sample size. This is unlikely to have been the case in founder HG and farmer demes, particularly if there was high variance in reproductive success.
Our approach aims at simulating in a realistic manner the movement of individuals, rather than that of genes leading to several other differences: (i) foundation events must involve at least one male and one female; (ii) the Maynard-Smith & Slatkin [32] logistic growth formula (2.1) is used and corrected to account for the fact that growth is limited by the number of reproductive females,
| 2.1 |
where Nft is the number of females at generation t; (iii) growth is not deterministic, as the number of individuals in generation t + 1 is drawn from a Poisson distribution, with mean Nt +1; (iv) the number of migrants in the different directions is also stochastically drawn from binomial distributions; and (v) sex-biased migration can be simulated using a sex ratio migration parameter given by mSR = mf/(mf + mm), where mf and mm are the female and male migration rates, respectively. Details related to K, F and admixture parameters, the growth formula, migration (including mSR parameter) and algorithm are in the electronic supplementary material, appendices A.1–A.5.
(b). Neolithic transition model
To study the properties of spatial expansions during the Neolithic transition, we simulated NRY and mtDNA data assuming a regular lattice. We assumed that: (i) there were two different layers, each corresponding to the HG and farmer layers; (ii) the first wave of expansion by HG started 40 kyr ago (1600 generations ago, assuming a generation time of 25 years [17]), corresponding to T = 0); and (iii) the second wave started 10 kyr ago (T = 1200 generations) to represent the spread of the farmers [17].
Owing to the computational cost of the simulations, our scenarios (see below) were tested with regular lattices of 100 demes (i.e. 10 by 10) per layer. The most likely scenarios were then also tested in 30 × 30 lattices (900 demes per layer). For all simulations, both the HG and farmer expansions started at the bottom-right corner deme (figure 1). Also, we decided to model the HG extinction by increasing the friction to 1 and reducing K in the HG layer at a time related to the size of the lattice used in the simulations (see the electronic supplementary material, appendix A.6 for details).
(c). Variable parameters: sex-biased migration and admixture
We were interested in determining the genetic consequences of sex-related migration patterns, in both HG and farmer societies. All combinations of bilocal, patrilocal or matrilocal societies were simulated corresponding to a total of nine scenarios (see the electronic supplementary material, table S1), using the mSR parameter. As in Currat & Excoffier [17], we assumed unidirectional admixture, from the HG to the farmer layer, in agreement with anthropological data suggesting that asymmetrical gene flow occurs when a dominant group invades a new region, as is supposed to have happened during the Bantu expansion [33] or in the colonization of Brazil by Europeans [34]. Five values of γ (0, 0.25, 0.5, 0.75 and 1) were used for each of the nine scenarios above, for a total of 45 different simulation sets carried out in the 10 × 10 lattices. For each set, 500 independent replicates were run. The results of the above simulations strongly suggested that the most probable scenarios were those with farmers patrilocality. We repeated these scenarios using a 30 × 30 lattice (i.e. on a wider geographical region), using four values for γ (0, 0.25, 0.5 and 0.75), to determine whether significantly different results would be observed with a larger lattice corresponding to a larger area. No major differences were observed in the genetic diversity within demes, in agreement with Hamilton et al. [35], who also compared lattices with different sizes in their spatial simulations (30 × 30 and 50 × 50).
To reduce the number of simulations required, we fixed the values of K, r and m to those from Currat & Excoffier [17], who calibrated and tested them to simulate the effect of the Neolithic expansion in Europe. For more details, see the electronic supplementary material, appendix A.7 for these and other fixed parameters used in the simulations.
(d). Summary statistics
We computed the mean expected heterozygosity (He) and mean FST [36] for NRY and mtDNA simulated data, across generations and only for demes along the diagonal. In the 10 × 10 lattices, we sampled the 10 diagonal demes from the starting deme (deme 9_9) (upper-left corner) to the last colonized, deme 0_0 (figure 1). In the pairwise FST analyses, we compared all the demes in the diagonal against the starting deme. This allowed us to study the trajectory of these statistics through time and space (in the expansion axis), for the two types of markers jointly and compare it with real data (see the electronic supplementary material, appendix A.8).
3. Results
As expected, our simulations show that both He and FST values differ for Y-linked and mtDNA markers when migration patterns differ in males and females. However, our results also demonstrate complex patterns that would have been difficult to predict without simulations. We will concentrate on the patterns observed for the farmers, since they correspond to the modern populations.
(a). General results across all scenarios
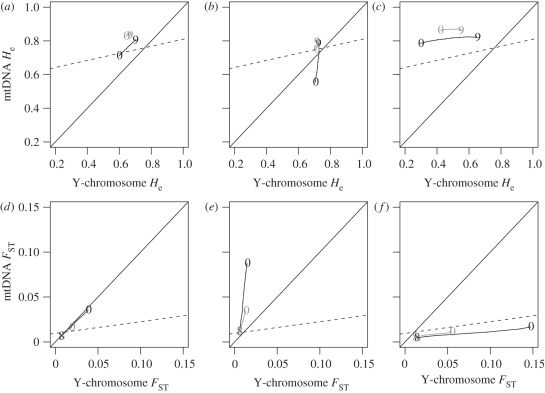
Looking at all samples obtained in a particular generation, we found that as time goes from generation 1300–1600, the set of He values becomes more concentrated in one region (figure 2a–c; see also the electronic supplementary material, figure S1). In other words, we found fewer differences on levels of genetic diversity across modern populations (generation 1600) compared with ancient populations (generation 1300).
Figure 2.
Genetic diversity and differentiation in modern populations, under no admixture. (a–c) represent average He values, whereas (d–f) represent average FST values. Only the farmer's populations were sampled, since they represent modern populations. The different columns correspond to scenarios where farmers were (a,d) bilocal, (b,e) matrilocal and (c,f) patrilocal, respectively. He and FST values were computed for the demes located in the diagonal of the 10 × 10 lattice. A line was drawn going through all demes between the plotted numbers (9 and 0) that represent the coordinates of deme 9_9 (the first deme to be colonized) and 0_0 (the last). Each colour represents a time step (black and grey are generations 1300 and 1600, respectively), for which the summary statistics were calculated (T = 1600 is the present-day generation). Each point is the mean of 500 independent simulations. The solid line represents cases where NRY and mtDNA values are equal. The dashed line is the regression obtained with the real observed data.
At the genetic differentiation level, the FST values against the starting deme (9_9) increase with distance from it, as expected. This increase can be significantly greater for the NRY compared with mtDNA data (as in figure 2f), or the opposite (as in figure 2e) depending on the scenarios (see below), but the FST values increase with distance from the starting deme. As generations go, FST values between the starting and last deme decrease with time (figures 2d–f), in agreement with the fact that He values are increasingly similar among samples. Thus, in these simulations, modern populations are genetically less differentiated than ancient ones.
(b). No admixture scenarios
In the scenarios without admixture (figure 2), the He values decrease along the axis of the expansion, with the highest values being observed in the starting deme (9_9) and the lowest in the last colonized deme (0_0). This is observed for all generations sampled. Moreover, these points typically move, as a group and across generations, from higher to lower NRY diversity and from lower to higher mtDNA diversity. In other words, present-day populations have more mtDNA diversity and slightly less NRY diversity than ancient populations, whichever the PMR pattern. Note that this is true when the set of samples from the diagonal are analysed as a group but not necessarily for each sample individually, owing to the fact that the points are also more compact, as we noted above. For instance, the starting deme loses NRY diversity (figure 2), whereas the last colonized deme actually sees its NRY diversity increase. Another very striking result was that the three scenarios (bi-, matri- and patrilocality) exhibit clearly differentiated patterns (figure 2). This can be seen in the way the points are arranged in ‘parallel lines’ through time.
In bilocality scenarios, we predicted similar mtDNA and NRY data. In fact, the points are arranged in a direction parallel to the solid line corresponding to equal values for the x- and y-axes (i.e. for mtDNA and NRY data). Interestingly, we observed that the bilocality He values were higher for mtDNA than for NRY data (figure 2a), whereas FST values were quite similar (figure 2d). The difference in He values is probably owing to differences in the mutation rates, higher in mtDNA (see the electronic supplementary material, appendix B). Indeed, when we repeated these simulations by assuming the same mutation rate for the two markers, we found symmetrical results (not shown).
In the matrilocal scenarios, all demes had similar NRY He, hence generating values forming ‘lines’ parallel to the y-axis (figure 2b). As expected, the mtDNA FST values were higher than the NRY FST values (figure 2e), that were themselves very similar between demes (i.e. the gene flow between demes was high), generating ‘lines’ near-parallel to the y-axis. On the contrary, in scenarios with farmer patrilocality (and still no admixture), a similar behaviour is seen but inverted for the two markers (i.e. higher NRY FST), and with almost no variation along the y-axis (figure 2f). Similarly, the He values also show this behaviour (figure 2c). A particularly interesting result was that this trend was parallel to the regression obtained from the real data from modern populations (dashed line in figure 2). This was true for both FST and He values and was not observed in the other scenarios (matrilocality and bilocality).
(c). Influence of hunter–gatherer post-marital behaviour on the farmers genetic diversity (He)
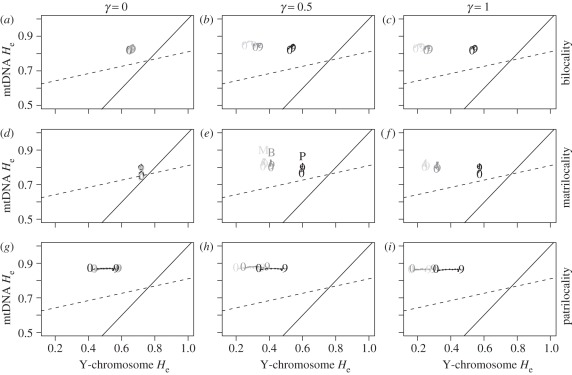
In the scenarios with admixture between HG and farmers, some significant changes are found on the level of genetic diversity (figure 3; see also the electronic supplementary material, figures S1 and S2). First, compared with the no admixture scenarios, the sets of points are shifted towards lower NRY diversity when γ increases, whereas mtDNA diversity does not change very much or shows a slight increase. Thus, in our simulations, admixture leads to a decrease in NRY diversity in all cases compared with the no admixture scenarios. Second, scenarios with patrilocality in HG populations generated fewer changes relative to the no admixture scenarios in NRY diversity compared with bilocality and matrilocality. This is true whether the farmers were patrilocal, matrilocal or bilocal and can be seen in figure 3 where the points with a P (HG patrilocality) are closer to the points of the no admixture scenarios (figure 3a,d,g) compared with the points with a B (HG bilocality) and even more with an M (HG matrilocality). In other words, in situations where HG could mate with farmers, the post-marital behaviour of HG populations clearly leads to differences in the distribution of farmers genetic diversity, in modern populations that have the same PMR system. Third, when farmers are patrilocal, a higher admixture rate (figure 3i) would tend to blur this effect and make the He pattern almost indistinguishable, whichever post-marital behaviour the HG may have had. However, the simulations that seem to better fit the trend of the observed data are the ones from patrilocality in farmers, whatever the HG's post-marital behaviour is and whatever the admixture rate is.
Figure 3.
Genetic diversity in present-day farmers, under admixture. He values that were computed for the demes located in the diagonal of the 10 × 10 lattice, between the starting deme (9_9) and the last to be colonized (0_0) represented by 9 and 0, respectively. To make the panels easier to read a line was drawn going through all demes between these two points, but the other demes identifications are not represented. Each column corresponds to one value of the admixture parameter γ (0, 0.5 and 1) and each row corresponds to one PMR system for the farmers layer (bilocality, matrilocality and patrilocality). Within each panel, the three possible scenarios for PMR system of the HG are represented. Each colour and letter represents a different residence pattern in the HG layer (colours light grey, grey and black and letters B, M and P correspond to scenarios, where HGs are bilocal, matrilocal and patrilocal, respectively). Each point is the mean of 500 independent simulations. Cases where NRY and mtDNA have the same He values would fall on the solid line. The dashed line is the regression obtained for the real observed data.
(d). Influence of hunter–gatherer post-marital behaviour on the farmers genetic differentiation (FST)
Interestingly, in a 10 × 10 lattice, the modern samples FST values seem less affected by the PMR system of the HG, than the corresponding He values (electronic supplementary material, figure S3). In particular, the analyses of only the last generation data show that the FST values were nearly identical across all HG scenarios with and without admixture. Conversely, in the generations that follow the admixture events, there were clear differences between the no admixture and admixture scenarios (electronic supplementary material, figure S4). However, in the scenarios analysed using a larger lattice (30 × 30, i.e. a larger geographical area) it was possible to separate the PMR system of the HG, on the basis of modern FST values (electronic supplementary material, figure S5).
4. Discussion
Altogether, our simulations allowed us to study the effect of (i) variable migration rates in males and females within the HG and farmers layers and (ii) variable admixture between layers, on the patterns of genetic diversity and differentiation in present-day and ancient populations.
(a). Main results: (i) farmers were patrilocal and (ii) different PMR systems have a different impact on human genetic patterns
Patrilocality was the most probable scenario among farmers. It was particularly obvious in the no admixture scenarios, but it was also found in the scenarios with admixture, even though not so obvious in some scenarios. This result agrees with ancient DNA (aDNA) and strontium isotope analyses that suggested patrilocality in Linear [37] and Corded [38] Ware Culture burials from Germany. Cultural phylogenetics studies also suggest that patrilocality started to increase after the advent of agriculture [29,39].
Changes in PMR systems were also found to lead to different genetic patterns in present-day populations. In particular, the farmers' He values changed significantly depending on whether the HGs were patrilocal, bilocal or matrilocal. Thus, it appears that even though HG populations disappear as far back as 5000 years before the present in our simulations, they influence present-day patterns in modern-day populations.
(b). Behaviour of summary statistics
Pairwise FST statistics were much less influenced than He by the PMR pattern of the HG populations. While FST values were different across scenarios after the start of admixture, this signal disappeared in the modern samples. However, when a larger lattice (30 × 30) is analysed (corresponding to a larger geographical area), it was possible to distinguish between the HG populations PMR systems. This is compatible with the notion that the degree of genetic differentiation between two demes depends on their geographical distance, on the migration rate between local demes and on the time since the populations started expanding. If migration rates are large and/or enough time has passed, then it may be necessary to use large lattices to avoid this homogenizing effect in FST values. This FST statistics' dependence on geographical distances implies that inferences based on local/regional sampling is valid only for the most recent history, while sampling from more distant places may be able to recover older patterns, a point that has been stressed by Wilkins & Marlowe [24].
Furthermore, the access to the genetic composition of ancient HG populations may be not only useful, but necessary to provide us with significant information on this issue. In other words, aDNA may be required to allow us to determine the post-marital behaviour of European HG populations, before and after the Neolithic transition. Currently, the number of Neolithic transition aDNA studies are slowly increasing [38,40–43] but are unfortunately limited to mtDNA. Our results suggest that obtaining NRY DNA from the same samples would be particularly important, as was done in a recent study [44].
To identify the most probable scenario, we focused on the trend observed in the statistics of both simulated and real data. Our approach was thus to some extent qualitative. To obtain a better fit, one would also need to consider the spread of populations in both the simulated and real data (not just the regression slope). In theory, simulating different scenarios should allow us to better tune migration rates, and identify the original level of diversity in both HG and farmers populations, that are compatible with the observed modern-day data.
(c). Mutation rates can generate asymmetries between NRY and mtDNA data
Although mtDNA and NRY data are often presented as symmetrical counterparts of the female and male demography, respectively, it is not necessarily that simple. The difference in mutation rates can generate an asymmetry between mtDNA and NRY He values in bilocal scenarios with no sex-related variance in reproductive success (figures 2a and 3a). This is something to keep in mind when analysing differences observed in real mtDNA and NRY data because such differences are often interpreted deterministically in terms of differences in male and female behaviours [28,45,46].
(d). Admixture decreases farmers NRY genetic diversity
We found this surprising at first, as it is usually assumed that regions where populations admix will exhibit higher levels of genetic diversity. However, the underlying assumption is that admixing populations have similar Ne. Several studies have shown that during spatial expansions the expanding population is diluted [10,47,48]. We thus believe that as admixture took place between populations with different sizes (i.e. HG having much smaller populations than the farmers), the incoming population will ‘dilute’ the farmers genetic diversity and lead to the decrease in genetic diversity. The decrease is observed even when both HG and farmers had the same pattern of PMR rules. However, this is not necessarily a general result as mtDNA genetic diversity did not always decrease. Again, the difference in mutation rates between mtDNA and NRY markers may interact in a complex way with demographic parameters leading to asymmetries in present-day data.
(e). Comparison with other sex-biased migration studies
Until now, the inference of patterns of sex-biased migration have relied mainly on the comparison and estimation of dispersal from pairwise NRY and mtDNA FST values [45,46], and by cultural phylogenetics [29,39] in modern populations.
Hamilton et al. [35] also used a spatial framework, using NRY and mtDNA data, to study matrilocal and patrilocal groups from northern Thailand. In their study, the authors applied a modified version of Splatche and were not interested in detecting shifts in PMR patterns, which were assumed to be invariant in their simulations. Instead, their aim was to compare male and female migration rates in known patrilocal and matrilocal societies that would explain present-day levels of genetic differentiation and diversity. Here, our aim was to understand how PMR (with or without shifts) interacts with admixture between different societies, to generate differences in maternally or paternally inherited markers analysed jointly.
Model-based approaches have many advantages as they allow us to identify parameters that have a significant impact on the data. However, they also rely on strong assumptions. In our study, it was necessary to make assumptions on the level and patterns of gene flow, carrying capacities and genetic make-up of the founder populations, which suggests that some of the conclusions presented here should not be taken at face value. Hence, we believe that the general trends identified are to some extent robust. For instance, our simulations were performed on a 10 × 10 lattice. But when we repeated the patrilocal scenarios on a 30 × 30 lattice, we found essentially the same results, the main difference being that the power to identify scenarios was increased in the 30 × 30 lattice.
The simulated framework introduced here owes much to the work of Currat et al. [17], but is sufficiently different to represent an interesting alternative to identify the critical assumptions that are robust and those that are not, and the type of data required to separate scenarios. Altogether, our simulations helped identify important parameters and scenarios, together with data that would be needed to study the Neolithic transition in Europe (NRY aDNA), but much work is still necessary.
There are still very few studies that have dealt with the kind of complex scenarios that involve the characterization of the expansion of two demographically different populations across the same geographical area when migration patterns and admixture levels vary, and those that exist do not deal with sex-biased migration [17,49]. Our work provides some of the first insights into the consequences of complex demographic changes that probably took place during the European Neolithic, on present-day human genetic patterns.
Acknowledgements
Part of this work was performed using HPC resources from CALMIP, Toulouse (Grant 2010-P1038). We thank B. Parreira for the stimulating comments and discussions and C. Gamba, I. Alves and anonymous reviewers for reading and constructively commenting on a previous version of the manuscript. R.R. and V.C.S. were supported by FCT grants (ref. SFRH/BD/30821/2006 and SFRH/BD/22224/2005, respectively) and P.A.B. by funding from the IGC. L.C. was funded by the FCT projects PTDC/BIA-BDE/71299/2006 and PTDC/BIA-BEC/100176/2008 and Institut Français de la Biodiversité, Programme Biodiversité des îles de l'Océan Indien (ref. CD-AOOI-07-003). This work was also partly funded by the ‘Laboratoire d'Excellence (LABEX)’ entitled TULIP (ANR-10-LABX-41).
References
- 1.Mithen S. 2007. Did farming arise from a misapplication of social intelligence? Phil. Trans. R. Soc. B 362, 705–718 10.1098/rstb.2006.2005 (doi:10.1098/rstb.2006.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis S. J. M. 2005. Why domesticate food animals? Some zoo-archaeological evidence from the Levant. J. Archaeol. Sci. 32, 1408–1416 10.1016/j.jas.2005.03.018 (doi:10.1016/j.jas.2005.03.018) [DOI] [Google Scholar]
- 3.Bellwood P. 2004. First farmers: the origins of agricultural societies. Oxford, UK: Blackwell Publishing [Google Scholar]
- 4.Ammerman A. J., Cavalli-Sforza L. L. 1984. The Neolithic transition and the genetics of populations in Europe. Princeton, NJ: Princeton University Press [Google Scholar]
- 5.Diamond J., Bellwood P. 2003. Farmers and their languages: the first expansions. Science 300, 597–603 10.1126/science.1078208 (doi:10.1126/science.1078208) [DOI] [PubMed] [Google Scholar]
- 6.Gkiasta M., Russell T., Shennan S., Steele J. 2003. Neolithic transition in Europe: the radiocarbon record revisited. Antiquity 77, 45–62 [Google Scholar]
- 7.Pinhasi R., Fort J., Ammerman A. J. 2005. Tracing the origin and spread of agriculture in Europe. PLoS Biol. 3, e410. 10.1371/journal.pbio.0030410 (doi:10.1371/journal.pbio.0030410) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbujani G., Sokal R. R., Oden N. L. 1995. Indo-European origins: a computer-simulation test of five hypotheses. Am. J. Phys. Anthropol. 96, 109–132 10.1002/ajpa.1330960202 (doi:10.1002/ajpa.1330960202) [DOI] [PubMed] [Google Scholar]
- 9.Belle E. M. S., Barbujani G. 2007. Worldwide analysis of multiple microsatellites: language diversity has a detectable influence on DNA diversity. Am. J. Phys. Anthropol. 133, 1137–1146 10.1002/ajpa.20622 (doi:10.1002/ajpa.20622) [DOI] [PubMed] [Google Scholar]
- 10.Chikhi L., Nichols R. A., Barbujani G., Beaumont M. A. 2002. Y genetic data support the Neolithic demic diffusion model. Proc. Natl Acad. Sci. USA 99, 11 008–11 013 10.1073/pnas.162158799 (doi:10.1073/pnas.162158799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richards M., et al. 2000. Tracing European founder lineages in the Near Eastern mtDNA pool. Am. J. Hum. Genet. 67, 1251–1276 10.1016/S0002-9297(07)62954-1 (doi:10.1016/S0002-9297(07)62954-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards M., Macaulay V., Torroni A., Bandelt H. 2002. In search of geographical patterns in European mitochondrial DNA. Am. J. Hum. Genet. 71, 1168–1174 10.1086/342930 (doi:10.1086/342930) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chikhi L. 2009. Update to Chikhi et al. ‘Clinal variation in the nuclear DNA of Europeans’ (1998): genetic data and storytelling–from archaeogenetics to astrologenetics? Hum. Biol. 81, 639–643 10.3378/027.081.0612 (doi:10.3378/027.081.0612) [DOI] [PubMed] [Google Scholar]
- 14.Zvelebil M., Zvelebil K. V. 1988. Agricultural transition and Indo-European dispersals. Antiquity 62, 574–583 [Google Scholar]
- 15.Itan Y., Powell A., Beaumont M. A., Burger J., Thomas M. G. 2009. The origins of lactase persistence in Europe. PLoS Comput. Biol. 5, e1000491. 10.1371/journal.pcbi.1000491 (doi:10.1371/journal.pcbi.1000491) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldman M. W., Cavalli-Sforza L. L. 1976. Cultural and biological evolutionary processes, selection for a trait under complex transmission. Theor. Popul. Biol. 9, 238–259 [DOI] [PubMed] [Google Scholar]
- 17.Currat M., Excoffier L. 2005. The effect of the Neolithic expansion on European molecular diversity. Proc. R. Soc. B 272, 679–688 10.1098/rspb.2004.2999 (doi:10.1098/rspb.2004.2999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbujani G., Bertorelle G., Chikhi L. 1998. Evidence for Paleolithic and Neolithic gene flow in Europe. Am. J. Hum. Genet. 62, 488–492 10.1086/301719 (doi:10.1086/301719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Semino O., et al. 2000. The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y-chromosome perspective. Science 290, 1155–1159 10.1126/science.290.5494.1155 (doi:10.1126/science.290.5494.1155) [DOI] [PubMed] [Google Scholar]
- 20.Balaresque P., et al. 2010. A predominantly Neolithic origin for European paternal lineages. PLoS Biol. 8, e1000285. 10.1371/journal.pbio.1000285 (doi:10.1371/journal.pbio.1000285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bentley R. A., Chikhi L., Price T. D. 2003. The Neolithic transition in Europe: comparing broad scale genetic and local scale isotopic evidence. Antiquity 77, 63–66 [Google Scholar]
- 22.Cavalli-Sforza L. L., Minch E. 1997. Paleolithic and Neolithic lineages in the European mitochondrial gene pool. Am. J. Hum. Genet. 61, 247–254 10.1016/S0002-9297(07)64303-1 (doi:10.1016/S0002-9297(07)64303-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasteiro R., Chikhi L. Submitted Female and male shared views on the Neolithic transition: clues from ancient and modern genetic data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilkins J. F., Marlowe F. W. 2006. Sex-biased migration in humans: what should we expect from genetic data? Bioessays 28, 290–300 10.1002/bies.20378 (doi:10.1002/bies.20378) [DOI] [PubMed] [Google Scholar]
- 25.Baker M., Jacobsen J. 2006. A human capital-based theory of postmarital residence rules. J. Law Econ. Organ. 23, 208–241 10.1093/jleo/ewm009 (doi:10.1093/jleo/ewm009) [DOI] [Google Scholar]
- 26.Langergraber K. E., Siedel H., Mitani J. C., Wrangham R. W., Reynolds V., Hunt K., Vigilant L. 2007. The genetic signature of sex-biased migration in patrilocal chimpanzees and humans. PLoS ONE 2, e973. 10.1371/journal.pone.0000973 (doi:10.1371/journal.pone.0000973) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marlowe F. 2004. Marital residence among foragers. Curr. Anthropol. 45, 277–283 10.1086/382256 (doi:10.1086/382256) [DOI] [Google Scholar]
- 28.Wilkins J. F. 2006. Unraveling male and female histories from human genetic data. Curr. Opin. Genet. Dev. 16, 611–617 10.1016/j.gde.2006.10.004 (doi:10.1016/j.gde.2006.10.004) [DOI] [PubMed] [Google Scholar]
- 29.Fortunato L. 2011. Reconstructing the history of residence strategies in Indo-European-speaking societies: neo-, uxori-, and virilocality. Hum. Biol. 83, 107–128 [DOI] [PubMed] [Google Scholar]
- 30.Currat M., Ray N., Excoffier L. 2004. Splatche: a program to simulate genetic diversity taking into account environmental heterogeneity. Mol. Ecol. Notes 4, 139–142 10.1046/j.1471-8286.2003.00582.x (doi:10.1046/j.1471-8286.2003.00582.x) [DOI] [Google Scholar]
- 31.Ray N., Currat M., Foll M., Excoffier L. 2010. Splatche2: a spatially explicit simulation framework for complex demography, genetic admixture and recombination. Bioinformatics 26, 2993–2994 [DOI] [PubMed] [Google Scholar]
- 32.Maynard-Smith J., Slatkin M. 1973. The stability of predator-prey systems. Ecology 54, 384–391 10.2307/1934346 (doi:10.2307/1934346) [DOI] [Google Scholar]
- 33.Quintana-Murci L., et al. 2008. Maternal traces of deep common ancestry and asymmetric gene flow between Pygmy hunter–gatherers and Bantu-speaking farmers. Proc. Natl Acad. Sci. USA 105, 1596–1601 10.1073/pnas.0711467105 (doi:10.1073/pnas.0711467105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salzano F. M. 2004. Interethnic variability and admixture in Latin America: social implications. Rev. Biol. Trop. 52, 405–415 [DOI] [PubMed] [Google Scholar]
- 35.Hamilton G., Stoneking M., Excoffier L. 2005. Molecular analysis reveals tighter social regulation of immigration in patrilocal populations than in matrilocal populations. Proc. Natl Acad. Sci. USA 102, 7476–7480 10.1073/pnas.0409253102 (doi:10.1073/pnas.0409253102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nei M. 1977. F-statistics and analysis of gene diversity in subdivided populations. Ann. Hum. Genet. 41, 225–233 10.1111/j.1469-1809.1977.tb01918.x (doi:10.1111/j.1469-1809.1977.tb01918.x) [DOI] [PubMed] [Google Scholar]
- 37.Bentley R. A., Price, T. D., Lüning J., Gronenborn D., Wahl J., Fullagar P. D. 2002. Human migration in early Neolithic Europe. Curr. Anthropol. 43, 799–804 [Google Scholar]
- 38.Haak W., et al. 2008. Ancient DNA, strontium isotopes, and osteological analyses shed light on social and kinship organization of the Later Stone Age. Proc. Natl Acad. Sci. USA 105, 18 226–18 231 10.1073/pnas.0807592105 (doi:10.1073/pnas.0807592105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fortunato L., Jordan F. 2010. Your place or mine? A phylogenetic comparative analysis of marital residence in Indo-European and Austronesian societies. Phil. Trans. R. Soc. B 365, 3913–3922 10.1098/rstb.2010.0017 (doi:10.1098/rstb.2010.0017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malmström H., et al. 2009. Ancient DNA reveals lack of continuity between Neolithic hunter–gatherers and contemporary Scandinavians. Curr. Biol. 19, 1758–1762 10.1016/j.cub.2009.09.017 (doi:10.1016/j.cub.2009.09.017) [DOI] [PubMed] [Google Scholar]
- 41.Haak W., et al. 2010. Ancient DNA from European Early Neolithic farmers reveals their Near Eastern affinities. PLoS Biol. 8, e1000536. 10.1371/journal.pbio.1000536 (doi:10.1371/journal.pbio.1000536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haak W., et al. 2005. Ancient DNA from the first European farmers in 7500-year-old Neolithic sites. Science 310, 1016–1018 10.1126/science.1118725 (doi:10.1126/science.1118725) [DOI] [PubMed] [Google Scholar]
- 43.Bramanti B., et al. 2009. Genetic discontinuity between local hunter–gatherers and central Europe's first farmers. Science 326, 137–140 10.1126/science.1176869 (doi:10.1126/science.1176869) [DOI] [PubMed] [Google Scholar]
- 44.Lacan M., Keyser C., Ricaut F.-X., Brucato N., Duranthon F., Guilaine J., Crubézy E., Ludes B. 2011. Ancient DNA reveals male diffusion through the Neolithic Mediterranean route. Proc. Natl Acad. Sci. USA 108, 9788–9791 10.1073/pnas.1100723108 (doi:10.1073/pnas.1100723108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilder J. A., Kingan S. B., Mobasher Z., Pilkington M. M., Hammer M. F. 2004. Global patterns of human mitochondrial DNA and Y-chromosome structure are not influenced by higher migration rates of females versus males. Nat. Genet. 36, 1122–1125 10.1038/ng1428 (doi:10.1038/ng1428) [DOI] [PubMed] [Google Scholar]
- 46.Seielstad M. T., Minch E., Cavalli-Sforza L. L. 1998. Genetic evidence for a higher female migration rate in humans. Nat. Genet. 20, 278–280 10.1038/3088 (doi:10.1038/3088) [DOI] [PubMed] [Google Scholar]
- 47.Klopfstein S., Currat M., Excoffier L. 2006. The fate of mutations surfing on the wave of a range expansion. Mol. Biol. Evol. 23, 482–490 10.1093/molbev/msj057 (doi:10.1093/molbev/msj057) [DOI] [PubMed] [Google Scholar]
- 48.Edmonds C. A., Lillie A. S., Cavalli-Sforza L. L. 2004. Mutations arising in the wave front of an expanding population. Proc. Natl Acad. Sci. USA 101, 975–979 10.1073/pnas.0308064100 (doi:10.1073/pnas.0308064100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Currat M., Excoffier L. 2004. Modern humans did not admix with Neanderthals during their range expansion into Europe. PLoS Biol. 2, e421. 10.1371/journal.pbio.0020421 (doi:10.1371/journal.pbio.0020421) [DOI] [PMC free article] [PubMed] [Google Scholar]