Abstract
Steamer ducks (Tachyeres) comprise four species, three of which are flightless. The flightless species are believed to have diverged from a flying common ancestor during the Late Pleistocene; however, their taxonomy remains contentious. Of particular interest is the previously unstudied population of flying steamer ducks in the Falkland Islands. We present the first genetic data from this insular population, and illustrate that the flying and flightless steamer ducks on the Falkland Islands are genetically indistinguishable, in contrast to their traditional classification as separate species. The three species that reside in continental South America form a genetically distinct lineage from the Falkland Island ducks. The Falkland steamer ducks diverged from their continental relatives 2.2–0.6 million years ago, coincident with a probable land bridge connecting the Falkland Islands to the mainland. The three continental species share a common ancestor approximately 15 000 years ago, possibly owing to isolation during a recent glacial advance. The continental steamer duck species are not reciprocally monophyletic, but show some amount of genetic differentiation between them. Each lineage of Tachyeres represents a different stage between flight and flightlessness. Their phylogenetic relationships suggest multiple losses of flight and/or long-term persistence of mixed-flight capability. As such, steamer ducks may provide a model system to study the evolution of flightlessness.
Keywords: flightlessness, evolution, historic DNA, Anatidae, phylogeny, bird
1. Introduction
The Pleistocene was a period of tumultuous climate change, characterized by oscillations between glacial and interglacial periods. These oscillations resulted in significant, environmentally driven changes in the amount and the distribution of habitat, forming and removing barriers to gene flow both on land and in the sea. During glacial periods, growing ice sheets separated previously connected populations and often formed impenetrable barriers to gene flow between newly created refugia. If isolation persisted for a sufficiently long time, these subpopulations would eventually become genetically distinct via genetic drift [1]. The reduction in global sea level brought about by glacial expansion also created land bridges between previously isolated land masses, creating conduits for dispersal and mixing of previously isolated populations [2]. As the temperature warmed, rising sea levels once again flooded these exposed land bridges, fragmenting populations in a way not unlike those in glacial refugia [3].
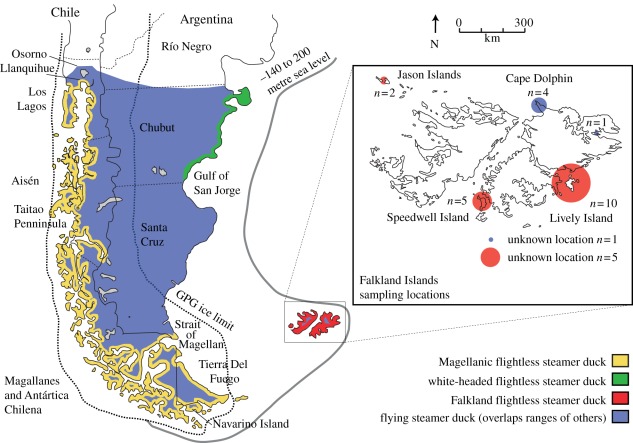
The diversification of the steamer ducks (Tachyeres) of South America (figure 1) was probably shaped by Pleistocene glaciations [6]. Two flightless species, the Magellanic flightless steamer duck (Tachyeres pteneres) and white-headed flightless steamer duck (Tachyeres leucocephalus), live on the Pacific and Atlantic coasts of southern South America, respectively, while the third flightless species, the Falkland steamer duck (Tachyeres brachypterus), is one of the only two birds endemic to the Falkland Islands (Islas Malvinas). The fourth species, the flying steamer duck (Tachyeres patachonicus), is sympatric with all three flightless species (figure 1).
Figure 1.
Steamer duck distribution. The geographical range of each species of Tachyeres, coded by colour, is based on that described by Livezey & Humphrey [4] and the International Union for Conservation of Nature (IUCN) species description. The inset provides an expanded view of the sampling locations in the Falkland Islands. The estimated maximum limit of glaciation during the great Patagonian glaciation (dashed line) follows that described by Rabassa & Coronato [5]. The expanded coastal limit (solid line) reflects sea levels 140–200 m below present.
Morphological [6] and protein electrophoretic [7] evidence has been used to suggest that the flying steamer duck is sister to the three flightless species, and that the Magellanic flightless steamer duck (T. pteneres) diverged first within the flightless clade. Based on this phylogeny, Livezey [6] hypothesized that a flying ancestral species was fragmented by Andean glaciation around 60 000 years ago, giving rise to the flying steamer duck on the eastern side of the Andes and an ancestral flightless species in a refugium along the Pacific coast. This flightless species then spread southward along the coast, becoming subdivided during the last glacial maximum (LGM) into the Magellanic flightless steamer duck on the Pacific coast, and a species ancestral to the white-headed and Falkland steamer ducks on the Atlantic coast [6]. However, one population of steamer ducks was not fully considered in this analysis: the population of flying steamer ducks that also inhabit the Falkland Islands. The origin of this population remains enigmatic, although it is presumed to have colonized the islands during the LGM. Individuals of this small population are difficult, if not impossible, to confidently differentiate from the Falkland flightless steamer duck in the field [4]. No phylogenetic or biogeographic studies have included this population, save one morphological analysis of a single incomplete specimen [8], despite general study of the evolution of the genus [6,7,9,10].
In contrast to traditional morphological views, a recent mitochondrial DNA analysis recovered two clades within steamer ducks: one composed of flightless Falkland steamer ducks and one Magellanic flightless steamer duck (T. pteneres), and another composed of flying steamer ducks, the white-headed steamer duck and a second individual of T. pteneres [10]. This paraphyly highlights the need for further sampling to resolve the evolutionary history of this genus. Of particular interest is the lack of monophyly of the flightless species, as it raises the possibility of more than one loss of flight within this genus and suggests that the biogeographic history of the genus should be revisited.
We present an analysis of mitochondrial and nuclear DNA sequences from more than 50 steamer ducks from the South American continent and the Falkland Islands, including the first genetic data for the flying steamer duck (T. patachonicus) from the Falkland Islands. By including more individuals from each species, we aim to resolve the phylogeny for all species of Tachyeres, test the current biogeographic scenario and create a framework for investigating the loss of flight within Tachyeres.
2. Material and methods
We obtained 23 samples of T. brachypterus, comprising 5 historic toepads collected between 1842 and 1937, 10 modern tissue samples and 8 field-collected feather and eggshell samples. We also obtained 10 modern T. patachonicus tissue samples from across Argentina and Chile and 7 historic toepad samples from the Falkland Islands. We also obtained 10 T. leucocephalus, 6 T. pteneres and 1 crested duck (Lophonetta specularioides). Sample ID, museum accession and collection information are fully detailed in electronic supplementary material, table S1.
We extracted DNA from modern samples using a Qiagen DNeasy Blood and Tissue kit. To obtain DNA from the eggshells, we removed the inner membrane from the shells and processed it as tissue. Feathers were processed by doubling the volume of all solutions prior to the wash steps and including 20 µl of 1 M dithiothreitol (DTT). Strict ancient DNA protocols were adhered to during processing of the historic toepad specimens [11]. We performed DNA extraction and PCR set-up for the historic toepads in a sterile ancient DNA laboratory at The Pennsylvania State University, which is physically isolated from all modern molecular biology research. To facilitate the digestion of the preserved tissue, we modified the Qiagen DNeasy kit extractions protocol to include (i) the addition of 20 µl of DTT during the lysis step and (ii) an extended incubation time of 48 h, during which we added an additional 20 µl of proteinase K to the sample after 24 h. We added 1 µl of Tween 20 to the final 50 µl extract to prevent the DNA from adhering to the tube walls.
We sequenced two mitochondrial (mtDNA) and four nuclear loci based on their previous utility in species-level analyses [12,13]. Details of the sequencing primers and amplification conditions are provided in electronic supplementary material, table S2. We targeted a 997 base pair (bp) fragment of mitochondrial control region (D-loop) and 738 bp fragment of ND2 in modern individuals. For the toepads, we expected the surviving DNA to be highly fragmented, so we targeted two short (less than 150 bp) overlapping fragments to obtain the first 337 bp of control region targeted in the modern samples. We cloned the PCR amplicons using a TOPO TA cloning kit (Invitrogen) following manufacturer's instructions, and used these to estimate the consensus sequence.
We targeted four nuclear loci: 303 bp of α-enolase (ENO1) intron-8, 256 bp of ornithine decarboxylase (ODC1) intron-7, 658 bp of Myoglobin (MB) intron-2 and 385 bp of chromo-helicase DNA binding protein 1 (CHD1Z). After initial tests of a subset of samples from each species, ENO1 and ODC1 were shown to be invariant. We amplified MB (either as a single fragment or as two shorter fragments targeting the four observed single nucleotide polymorphisms) and CHD1Z from all modern tissue specimens. Nuclear loci were not targeted from the toepads, as the results of the mitochondrial analyses indicated these were not sufficiently well preserved for nuclear DNA analysis. PCR conditions for all loci are described in electronic supplementary material, table S2.
We aligned the resulting sequences using the Lasergene software suite and corrected the alignments by eye. For the mtDNA loci, we identified unique haplotypes using Arlequin v. 3.5.1.2 [14]. Unique alleles and allele frequencies for MB were estimated using PHASE v. 2.1.1 [15,16], following the removal of three individuals for which only two of the four single nucleotide polymorphisms were available. Five independent runs, each employing 500 iterations, a thinning interval of 2 and burn-in of 500 iterations, converged on the same set of 10 alleles and the same allele frequencies. Individuals were assigned the genotype of highest probability (all assignments were greater than 85%). For D-loop only, we calculated genetic distance (absolute and p-distance) between haplotypes using PAUP v. 4b10 [17]. We estimated a median-joining network in Network v. 4.6 [18] for each locus. D-loop sequences were trimmed to 324 bp to match the length of the historic sequences and the 5′ end of the additional sequences from Bulgarella et al. [10] from GenBank. Two individuals (BL055 and BL235) with incomplete sequences were excluded from the network. Pair-wise ΦST calculations were performed between species using analysis of molecular variance (AMOVA) [19] implemented in Arlequin.
We estimated phylogenetic trees for each locus individually as well as combined as a multi-locus dataset. We used jModeltest v. 0.1.1 [20], which implements an algorithm from Phyml [21], to identify the model of evolution that best fit the alignment data for each locus using AIC (electronic supplementary material table S3). We performed maximum-likelihood (ML) analyses using RAxML v. 7.2.8alpha [22,23] and Bayesian Markov chain Monte Carlo (MCMC) analyses using MrBayes v. 3.1.2 or 3.2 [24]. Full details are described in the electronic supplementary material.
We performed divergence time analysis for Anatidae using BEAST v. 1.6.1 [25] using only ND2 data to maximize taxonomic coverage and minimize missing data. We included one individual from each species of Tachyeres and at least one species from each genus of ducks and geese for which ND2 was available as of 1 August 2011. We calibrated the goose–duck divergence (the root of the phylogeny) using a lognormal distribution (lognormal mean = 15.9, s.d. = 0.5, offset by 20 Ma) to provide a median divergence time of 28 Ma and a 95% confidence interval spanning 42–23 Ma based on the Oligocene occurrence (Mammalian Palaeogene zones 23–24) of the fossil anserine (Cygnopterus affinis) in Belgium [26]. The Anser-Branta divergence was calibrated using a lognormal distribution (lognormal mean = 15, s.d. = 0.6, offset by 4.8 Ma) and a median age of 8 Ma to reflect the occurrence of both genera in the southern USA around 5 Ma [27,28]. We ran the MCMC chains for 50 million iterations sampling every 2000, removed the first 10 per cent of sampled states as burn-in, and visualized the results in Tracer v. 1.5 [29] to verify that parameter sampling had reached stationarity and that sufficient effective sample sizes had been reached (greater than 200) for each parameter. The maximum clade credibility (MCC) tree, summarized by treeAnnotator v. 1.6.1, was visualized in Figtree v. 1.3.1 [30].
3. Results
We successfully obtained D-loop sequence data from 53 of 57 individuals sampled, although only less than 150 bp could be obtained for BL235 and BL055. Three toepads and one field-collected non-invasive sample yielded no DNA, presumably owing to DNA degradation following environmental exposure. Only D-loop data could be obtained for four of the field-collected specimens and the 10 toepads, including six of the flying steamer ducks of the Falkland Islands. We obtained sequence data for ND2 from 36 of 39 modern specimens, CHD1Z from 34 and MB from 36 (GenBank Accession Numbers JQ408247–JQ408371; electronic supplementary material, table S1).
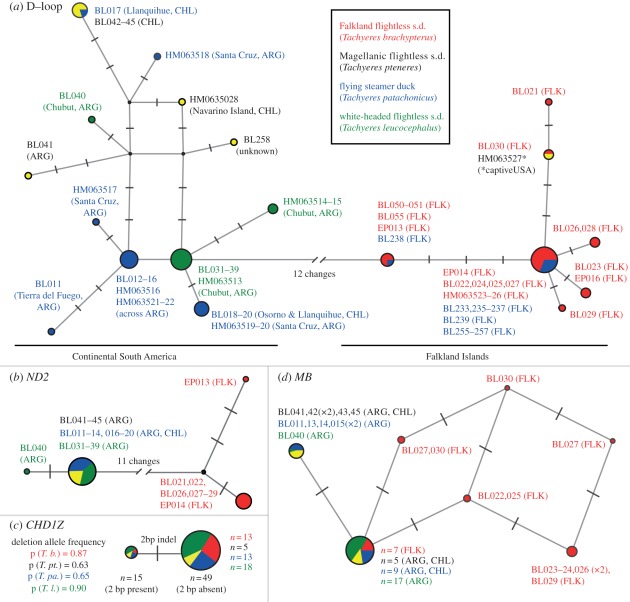
Twenty-two D-loop haplotypes were observed across the 53 new and 16 previously published Tachyeres sequences [10]. When trimmed to the length of the historic sequences for network construction, 19 distinct haplotypes remained. These data fall into two major haplogroups: one comprising only the specimens from the Falkland Islands (both flying and flightless) and one comprising the individuals from continental South America (figure 2). The only exception to this is a single T. pteneres individual (GenBank HM063527) from Bulgarella et al. [10] with a ‘Falklands’ haplotype that was obtained from a private avicultural collection in the USA. Given that individuals of T. brachypterus have been previously misidentified in American collections as T. pteneres and the former is more commonly kept in captivity [4], it is almost certainly misidentified. The possibility that this sample was misidentified was also raised by Bulgarella et al. [10].
Figure 2.
Median-joining networks for each variable locus studied. Circles represent unique haplotypes, scaled by frequency and coloured by species (key inset). Each ‘tick’ represents one mutation, except between the Falkland Islands and continental clades in (a) and (b), which are separated by 12 and 11 mutations, respectively. Individuals are labelled by their DNA extraction number and source location (full specimen details in electronic supplementary material, table S1). (a,b) Mitochondrial haplotypes; (c,d) phased nuclear alleles. In some cases, the number of each allele observed is denoted by n. ARG, Argentina; CHL, Chile; FLK, Falkland Islands; s.d., steamer duck.
The uncorrected genetic distance for D-loop data between Falkland Islands individuals was 0.1–0.6 per cent and between continental individuals was 0.1–0.8 per cent, while distance between individuals originating on the continent versus the Falkland Islands was 1.3–2.5 per cent. D-loop-only analyses including an outgroup place the root of the phylogeny between the two clades, with 100 per cent maximum likelihood bootstrap percentage (MLBP) and Bayesian posterior probability (BPP) support for monophyly of the Falkland Islands clade (electronic supplementary material, figure S1).
We observed four haplotypes in ND2: two in Falkland Islands individuals and two in continental individuals (figure 2). All Falkland Islands steamer ducks differed from those from the continent by 11 fixed substitutions. We observed two alleles for CHD1Z, defined by a 2 bp indel. Both alleles are found at similar frequencies in all populations (figure 2). For the phylogenetic analyses, individuals that were heterozygous for the indel were coded as missing data for those two positions. We observed four parsimony informative sites in MB that were estimated by PHASE to correspond to 10 unique alleles (figure 2). Heterozygotes were coded as multi-state bases in phylogenetic analyses.
The combined-locus analysis strongly supports a division between the steamer ducks of the Falkland Islands (both flying and flightless) and those of continental South America (electronic supplementary material, figure S1). However, within these clades, no support was found for the monophyly of any species. All individual gene trees were compatible with the combined analysis (electronic supplementary material, figures S2–S6).
For ΦST calculation between the flying and flightless steamer ducks of the Falkland Islands, only 337 bp of D-loop were analysed, representing the complete dataset available for the flying steamer ducks from the Falkland Islands. AMOVA analyses illustrated no significant differentiation between these populations (table 1), and since no haplotypes unique to the flying steamer ducks of the Falkland Islands were recovered, these individuals were excluded from further ΦST calculations so that the entire sequence dataset could be used. Pair-wise ΦST calculations between the Falkland, white-headed, Magellanic and flying steamer ducks (of continental South America) showed significant genetic differentiation between all species at high to very high levels (table 1).
Table 1.
Pair-wise genetic differentiation (ΦST) between species calculated via AMOVA using 997 bp of mitochondrial D-loop data. Flying ducks from the Falkland Islands (FLK) are not significantly (n.s.) differentiated from Falkland steamer ducks (ΦST = 0.01706, p = 0.29) based on the short historic fragment (337 bp) and were not compared with the continental populations.
| populations | white-headed | magellanic | flying (South America) |
|---|---|---|---|
| Falkland flightless | 0.89789 (p = 0.00)** | 0.84702 (p = 0.00)** | 0.87526 (p = 0.00)** |
| white-headed | 0.60375 (p = 0.00)** | 0.19957 (p = 0.00)** | |
| Magellanic | 0.4522 (p = 0.00)** |
**significant p-value.
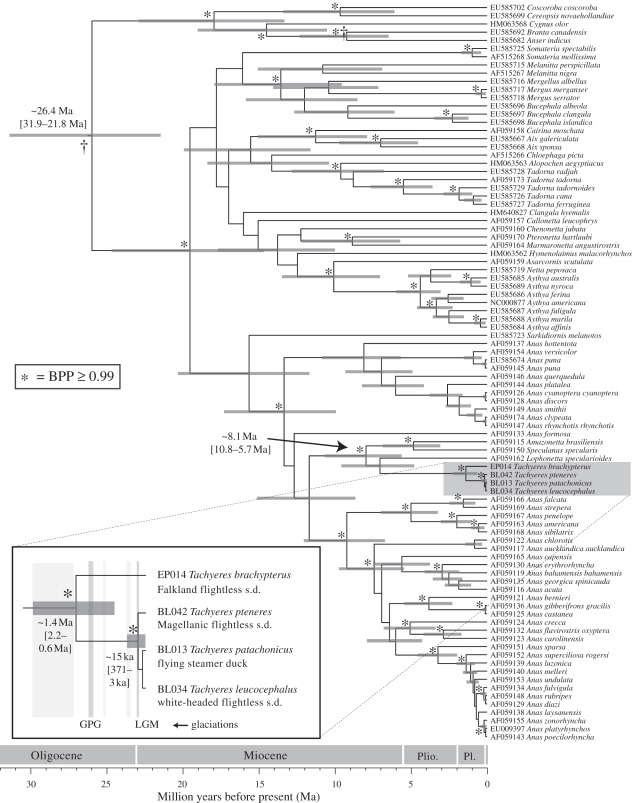
In the ND2 divergence time analysis (figure 3), geese and ducks were reciprocally monophyletic (BPP = 1.0). Tachyeres was supported as monophyletic (BPP = 1.0) with a most recent common ancestor (MRCA) living around 1.4 Ma (95% CI: 2.2–0.6 Ma). The continental steamer ducks (flying, Magellanic flightless and white-headed flightless) are supported as a single clade (BPP = 1.0), although we found no further support for resolution within the continental clade. The MRCA of this clade is estimated to around 15 ka (95% CI: 371–3 ka).
Figure 3.
Anatidae divergence times. The maximum clade credibility tree estimated in BEAST v. 1.6.1 is shown with node bars representing 95% confidence interval on divergence time. Taxon labels show GenBank accession numbers. Dagger (†) indicates fossil-calibrated nodes. The Tachyeres clade is expanded for visibility. Grey vertical bars represent glacial periods where darker bars indicate more extensive glaciations. GPG, great Patagonian glaciation; LGM, last glacial maximum; s.d., steamer duck.
4. Discussion
(a). Flying and flightless ducks on the Falkland Islands are genetically indistinguishable
Our results illustrate that the flying steamer ducks of the Falkland Islands are genetically indistinguishable from the flightless Falkland steamer ducks (figure 2 and table 1), based on the single-locus data available for the flying individuals. This clade is strongly supported (BPP = 1.0, MLBP = 96%) as genetically distinct from the three species that live in continental South America (electronic supplementary material, figure S1). Although these results are in contrast to the presently accepted taxonomy, all steamer ducks from the Falkland Islands are highly similar morphologically [4]. Thus, it seems probable that the flying and flightless ducks of the Falkland Islands comprise a single, partially flighted species. If true, the Falkland steamer duck and continental flying steamer duck may represent two of only three avian species with mixed-flight capability, the third of which, the rail Dryolimnas cuvieri, comprises distinct populations that either can or cannot fly [31].
Because only mitochondrial DNA could be obtained from flying ducks of the Falkland Islands, we cannot rule out either hybridization between flying and flightless birds or a recent mitochondrial sweep through the small flying population as the cause of the observed similarity between the flying and flightless ducks. However, flying ducks in the Falkland Islands breed almost exclusively on inland freshwater lakes. The flightless population, unable to reach these lakes, breeds along the coastline [4]. Given the consistent and exclusive pairing of flying ducks, it seems unlikely that hybridization would be sufficiently common to explain our results. These selective breeding practices may represent either reproductive isolation via different breeding habitats or possibly by positive assortative mating. This potential for reproductive isolation may actually act to encourage genetic diversification between the flying and flightless ducks, and may eventually lead to speciation between them. However, further morphological and population genetic study employing microsatellites or genome-wide single nucleotide polymorphism study from less-degraded samples will be critical in defining the extent of population structuring, if any, between flying and flightless steamer ducks of the Falkland Islands.
(b). A revised phylogeny for the South American steamer ducks
In contrast to Livezey's biogeographic hypothesis [6], in which the Falkland Islands were the last geographical region to be colonized by steamer ducks, our results place the divergence between the Falkland Islands steamer ducks and the continental steamer ducks between 2.2 and 0.6 Ma. This period includes the great Patagonian glaciation (GPG), which peaked ca 1.07 Ma, as well as several pre-GPG and one post-GPG glaciation (figure 3) [5]. At the maximum extent of the GPG, ice probably calved into the Pacific ocean, leaving little, if any, land exposed [5]. During peak glaciation, the sea level is estimated to have dropped by up to 200 m below the present level [2], exposing the sea floor between the continent and the Falkland Islands, and probably creating a land bridge between them (figure 1).
The GPG may have facilitated the divergence between Falkland Island and continental steamer ducks in two ways. First, the ancestral steamer ducks may have been divided by uninhabitable glacial conditions into two refugial populations, one restricted to the largely unglaciated [2,32] Falkland Plateau and the other to the north of the uninhabitable region. Alternatively, the entire ancestral population may have found refuge on the Falkland Plateau during the glacial period. Following deglaciation, the associated rapid rise in sea level may have formed a barrier to dispersal between the Falkland Islands and the continent, effectively subdividing the Falkland population from that in southern Argentina. We estimate the MRCA of all extant steamer ducks to have lived just prior to the GPG ca 1.4 Ma. This suggests some preference for the first scenario, in that the timing of the onset of glacial conditions corresponds more closely with that of initial subdivision, than does the timing of glacial retreat.
Although the continental species share their MRCA with the Falkland Islands steamer ducks ca 1.4 Ma, the species on the continent did not diversify until within the last several hundred thousand years (figure 3). This very recent divergence is reflected in the absence of reciprocal monophyly (electronic supplementary material, figure S1) as lineage sorting is probably incomplete. However, unlike the flying and flightless Falkland steamer ducks, little haplotype sharing is observed among the three continental species (figure 2) and ΦST calculations indicate that the species are significantly genetically differentiated at a very high level (table 1). The mean estimated divergence ca 15.1 ka coincides with the end of a period of higher, post-glacial sea levels that lasted between 22 and 15 ka. The higher sea levels reduced the exposed coastal plain by 65 per cent, recessed the coastline by approximately 160 km, and formed many small islands along the coastline [33]. These changes could have provided the means for diversification by fragmenting existing populations or creating new habitat.
(c). Multiple losses of flight
Loss of flight in birds often accompanies isolation on islands where the energetic cost of flight outweighs the benefits [31]. Flightlessness has been observed in other island anatids, such as the Hawaiian nenes and extinct moa-nalos [34,35], and the Auckland Island and Campbell Island teals of New Zealand [36]. Although the Falkland Islands were presumably a predator-free environment at the time of steamer duck colonization, it seems unlikely that this would also be the case for those birds living on the continent. Therefore, the absence of predators does not appear to be a necessity for loss of flight in this genus [9].
Wing-loading measurements suggest that many flying steamer ducks are borderline flightless [37,38] and an estimated 25 per cent of males on the continent cannot fly, as supported by the lack of flight in these individuals when being actively pursued [37]. Flying pairs represent approximately 1 per cent of the steamer ducks in the Falkland Islands [39]. The maintenance of both flying and flightless forms (to different degrees) in two of four steamer duck species suggests that there is some benefit to the characteristics associated with flightlessness not only in insular populations, but in coastal populations as well. This could be as high as three of four species, as wing-loading measurements suggest that a small number of white-headed steamer ducks may be flight-capable [37]. Only one species of steamer duck, the Magellanic steamer duck, is entirely incapable of flight. It seems most parsimonious that the ancestral population of all steamer ducks was polymorphic for flight capability, possibly similar to the modern flying steamer duck. This polymorphism would have been retained at different levels in both the continental and Falkland Island lineages. Subsequent to their initial speciation, flight capacity would have been maintained in flying steamer ducks, but reduced or lost completely in the coastal-dwelling white-headed and Magellanic steamer ducks.
In avian species generally, flightlessness occurs more frequently in aquatic species than expected by chance [31]. In rails, the loss of flight is frequently correlated with a reduction in body size and pectoral mass, reducing their energetic requirements [40]. However, flightless ducks that use their wings for locomotion do not show a similar reduction in basal metabolism or pectoral mass [40]. Energetic cost reduction may take other forms, however, as individuals that do not or cannot fly in coastal areas may be better able to tolerate increased feather wear, avoiding energetically costly moulting, compared with inland individuals requiring flight to reach foraging grounds [38]. Other potential benefits of increased body size and reduced wing area include improved diving, defence and wing-assisted combat, thermoregulation, and locomotion via ‘steaming’ [9]. Steamer duck habitat is hospitable year-round, making migration unnecessary, and with reduced selection pressure to be volant, the benefits of large size and shorter wings may outweigh flight [9]. The relationship between resource availability and flightlessness/size on an individual level may be informative towards understanding the polymorphism of flightlessness within species and, possibly, within populations.
Given the recent MRCA of all continental steamer ducks and the even more recent emergence of two completely (or almost completely) flightless species that live in sympatry with a primarily flying species, steamer ducks may present a model system to study both the loss of flight and sympatric speciation. Within the different Tachyeres lineages, the loss of flight may be ongoing, or may represent an equilibrium point between flight and flightlessness. Partially flighted species remain an exceptional rarity in birds [31], making the South American steamer ducks an ideal resource for future evolutionary research.
Acknowledgements
We thank M. Robbins at the University of Kansas Museum of Natural History, M. Adams at the British Museum of Natural History at Tring, H. Otley and the Falkland Islands Government, and volunteers in the field who collected and donated steamer duck specimens. We also thank R. Woods and K. McCracken for helpful discussion and two anonymous reviewers for helpful comments on an earlier version. This work was supported by PSU start-up funds and by a British Ornithologists' Union small grant to B.L. Emily Pronchik helped with laboratory work and was supported by a PSU WISER undergraduate research fellowship.
References
- 1.Hewitt G. M. 1996. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 58, 247–276 [Google Scholar]
- 2.Clapperton C. M. 1990. Quaternary glaciations in the Southern Ocean and Antarctic Peninsula area. Quat. Sci. Rev. 9, 229–252 10.1016/0277-3791(90)90020-B (doi:10.1016/0277-3791(90)90020-B) [DOI] [Google Scholar]
- 3.Vartanyan S. L., Garutt V. E., Sher A. V. 1993. Holocene dwarf mammoths from Wrangel Island in the Siberian Arctic. Nature 362, 337–340 10.1038/362337a0 (doi:10.1038/362337a0) [DOI] [PubMed] [Google Scholar]
- 4.Livezey B. C., Humphrey P. S. 1992. Taxonomy and identification of steamer-ducks (Anatidae: Tachyeres), 8th edn Lawrence, KS: The University of Kansas Museum of Natural History [Google Scholar]
- 5.Rabassa J., Coronato A. 2009. Glaciations in Patagonia and Tierra del Fuego during the Ensenadan Stage/Age (Early Pleistocene–earliest Middle Pleistocene). Quat. Int. 210, 18–36 10.1016/j.quaint.2009.06.019 (doi:10.1016/j.quaint.2009.06.019) [DOI] [Google Scholar]
- 6.Livezey B. C. 1986. Phylogeny and historical biogeography of steamer-ducks (Anatidae: Tachyeres). Syst. Zool. 35, 458–469 10.2307/2413109 (doi:10.2307/2413109) [DOI] [Google Scholar]
- 7.Corbin K. W., Livezey B. C., Humphrey P. S. 1988. Genetic differentiation among steamer-ducks (Anatidae: Tachyeres): an electrophoretic analysis. Condor 90, 773–781 10.2307/1368834 (doi:10.2307/1368834) [DOI] [Google Scholar]
- 8.Livezey B. C. 1986. Geographic variation in skeletons of flying steamer-ducks (Anatidae: Tachyeres patachonicus). J. Biogeogr. 13, 511–525 10.2307/2844815 (doi:10.2307/2844815) [DOI] [Google Scholar]
- 9.Livezey B. C., Humphrey P. S. 1986. Flightlessness in steamer-ducks (Anatidae: Tachyeres): its morphological bases and probable evolution. Evolution 40, 540–558 10.2307/2408576 (doi:10.2307/2408576) [DOI] [PubMed] [Google Scholar]
- 10.Bulgarella M., Sorenson M. D., Peters J. L., Wilson R. E., McCracken K. G. 2010. Phylogenetic relationships of Amazonetta, Speculanas, Lophonetta, and Tachyeres: four morphologically divergent duck genera endemic to South America. J. Avian Biol. 41, 186–199 10.1111/j.1600-048X.2009.04819.x (doi:10.1111/j.1600-048X.2009.04819.x) [DOI] [Google Scholar]
- 11.Wandeler P., Hoeck P. E.A., Keller L. F. 2007. Back to the future: museum specimens in population genetics. Trends Ecol. Evol. 22, 634–642 10.1016/j.tree.2007.08.017 (doi:10.1016/j.tree.2007.08.017) [DOI] [PubMed] [Google Scholar]
- 12.Peters J. L., Zhuravlev Y. N., Fefelov I., Humphries E. M., Omland K. E. 2008. Multilocus phylogeography of a holarctic duck: colonization of North America from Eurasia by gadwall (Anas strepera). Evolution 62, 1469–1483 10.1111/j.1558-5646.2008.00372.x (doi:10.1111/j.1558-5646.2008.00372.x) [DOI] [PubMed] [Google Scholar]
- 13.Irestedt M., Ohlson J. I., Zuccon D., Kallersjo M., Ericson P. G.P. 2006. Nuclear DNA from old collections of avian study skins reveals the evolutionary history of the Old World subocines (Aves, Passeriformes). Zool. Scr. 35, 567–580 10.1111/j.1463-6409.2006.00249.x (doi:10.1111/j.1463-6409.2006.00249.x) [DOI] [Google Scholar]
- 14.Excoffier L., Laval G., Schneider S. 2005. Arlequin (v. 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. 1, 47–50 [PMC free article] [PubMed] [Google Scholar]
- 15.Stephens M., Donnelly P. 2003. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 73, 1162–1169 10.1086/379378 (doi:10.1086/379378) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stephens M., Smith N. J., Donnelly P. 2001. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68, 978–989 10.1086/319501 (doi:10.1086/319501) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swofford D. L. 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods), v. 4.0. Sunderland, MA: Sinauer Associates [Google Scholar]
- 18.Bandelt H., Forster P., Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 [DOI] [PubMed] [Google Scholar]
- 19.Excoffier L., Smouse P. E., Quattro J. M. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes—application to human mitochondrial-DNA restriction data. Genetics 131, 479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256 10.1093/molbev/msn083 (doi:10.1093/molbev/msn083) [DOI] [PubMed] [Google Scholar]
- 21.Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 10.1080/10635150390235520 (doi:10.1080/10635150390235520) [DOI] [PubMed] [Google Scholar]
- 22.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 10.1093/bioinformatics/btl446 (doi:10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- 23.Stamatakis A., Hoover P., Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771 10.1080/10635150802429642 (doi:10.1080/10635150802429642) [DOI] [PubMed] [Google Scholar]
- 24.Ronquist F., Huelsenbeck J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 10.1093/bioinformatics/btg180 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- 25.Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howard H. 1964. Fossil Anseriformes. In The waterfowl of the world (ed. Delacour J. ), pp. 233–326 London, UK: Country Life Limited [Google Scholar]
- 27.Bickart K. J. 1990. Part I: the birds of the Late Miocene–Early Pliocene big sandy formation, Mohave County, Arizona. Recent advances in the study of Neogene fossil birds. Ornithol. Monogr. 44, 1–72 [Google Scholar]
- 28.Olson S. L., Rasumussen P. C. 2001. Miocene and Pliocene birds from the Lee Creek Mine, North Carolina. In Geology and paleontology of the Lee Creek Mine, North Carolina, III (eds Ray C. E., Bohaska D. J.). Washington, DC: Smithsonian Institution Press [Google Scholar]
- 29.Rambaut A., Drummond A. J. 2007. Tracer v. 1.4. See http://beast.bio.ed.ac.uk/Tracer [Google Scholar]
- 30.Rambaut A. 2006. Figtree v. 1.3.1. See http://tree.bio.ed.ac.uk/software/figtree [Google Scholar]
- 31.Roff D. A. 1994. The evolution of flightlessness: is history important? Evol. Ecol. 8, 639–657 10.1007/BF01237847 (doi:10.1007/BF01237847) [DOI] [Google Scholar]
- 32.Sugden D. E., Clapperton C. M. 1976. The maximum ice extent on island groups in the Scotia Sea, Antarctica. Quat. Res. 7, 268–282 10.1016/0033-5894(77)90041-2 (doi:10.1016/0033-5894(77)90041-2) [DOI] [Google Scholar]
- 33.Ponce J. F., Rabassa J., Coronato A., Borromei A. M. 2011. Palaeogeographical evolution of the Atlantic coast of Pampa and Patagonia from the last glacial maximum to the Middle Holocene. Biol. J. Linn. Soc. 103, 363–379 10.1111/j.1095-8312.2011.01653.x (doi:10.1111/j.1095-8312.2011.01653.x) [DOI] [Google Scholar]
- 34.Paxinos E. E., James H. F., Olson S. L., Sorenson M. D., Jackson J., Fleischer R. C. 2002. mtDNA from fossils reveals a radiation of Hawaiian geese recently derived from the Canada goose (Brantacanadensis). Proc. Natl Acad. Sci. USA 99, 1399–1404 10.1073/pnas.032166399 (doi:10.1073/pnas.032166399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sorenson M. D., Cooper A., Paxinos E. E., Quinn T. W., James H. F., Olson S. L., Fleischer R. C. 1999. Relationships of the extinct moa-nalos, flightless Hawaiian waterfowl, based on ancient DNA. Proc. R. Soc. Lond. B 266, 2187–2193 10.1098/rspb.1999.0907 (doi:10.1098/rspb.1999.0907) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kennedy M., Spencer H. G. 2000. Phylogeny, biogeography, and taxonomy of Australasian teals. Auk 117, 154–163 10.1642/0004-8038(2000)117[0154:PBATOA]2.0.CO;2 (doi:10.1642/0004-8038(2000)117[0154:PBATOA]2.0.CO;2) [DOI] [Google Scholar]
- 37.Humphrey P. S., Livezey B. C. 1982. Flightlessness in flying steamer-ducks. Auk 99, 368–372 [Google Scholar]
- 38.Wilson R. E., Sonsthagen S. A., Barger C. P., McCracken K. G. 2007. Asymmetric molt or feather wear in Flying Steamer Ducks (Tachyeres patachonicus) from coastal habitats in Argentina. Ornitol. Neotrop. 18, 293–300 [Google Scholar]
- 39.Woods R. W., Woods A. 2006. Birds and mammals of the Falkland Islands. Hampshire, UK: Wild Guides [Google Scholar]
- 40.McNab B. K. 1994. Energy-conservation and the evolution of flightlessness in birds. Am. Nat. 144, 628–642 10.1086/285697 (doi:10.1086/285697) [DOI] [Google Scholar]