Abstract
Elucidating the factors that shape species distributions has long been a fundamental goal in ecology and evolutionary biology. In spite of significant theoretical advancements, empirical studies of range limits have lagged behind. Specifically, little is known about how the attributes that allow species to expand their ranges and become widespread vary across phylogenies. Here, we studied the ascidian Botryllus schlosseri, a worldwide invasive species that is also characterized by marked genetic subdivision. Our study includes phylogenetic and population genetic data based on mitochondrial and nuclear genes, as well as polymorphic microsatellites for B. schlosseri colonies sampled from the southern and northern coasts of Europe and the eastern and western coasts of North America. We demonstrate that this well-known model organism comprises three highly divergent and probably reproductively isolated cryptic species (A, D and E), with two more (B and C) being suggested by data retrieved from GenBank. Among these, species A, recovered in all of the surveyed regions, is by far the most common and widespread. By contrast, species B–E, occurring mostly in sites from northern Europe, are considerably more geographically restricted. These findings, along with inferences made on transport opportunity, suggest that divergent evolutionary histories promoted differences in invasive potential between B. schlosseri sibling species, indicating that attributes that facilitate dramatic shifts in range limits can evolve more easily and frequently than previously thought. We propose environmental disturbance as a selective force that could have shaped the evolution of invasiveness in the B. schlosseri complex.
Keywords: range limit, invasiveness, biological invasions, cryptic speciation, Botryllus schlosseri, golden star tunicate
1. Introduction
A fundamental goal in ecology and evolutionary biology is to understand the factors that set the limits of species ranges [1]. Current perspectives on range limits are integrated in a wide variety of theoretical models that distinguish between the presence or absence of environmental heterogeneity and/or evolution [2]. Models without environmental gradients or evolution consider Allee effects, whereby ranges are restricted by the inability of marginal populations to grow when small [3]. Models with environmental heterogeneity but with no evolution incorporate dispersal barriers or fundamental niche limitation along ecological gradients [4]. Finally, models with environmental gradients and evolution consider the effect of gene flow and selection on adaptation to discrete habitats at a range's edge [5,6]. While considerable attention has been directed towards the theory of range limits, empirical advancements have lagged behind. A key aspect that remains to be extensively addressed is the degree to which range limits are phylogenetically conserved (i.e. do closely related lineages have similar distributional limits?).
An ideal opportunity to test the theory and patterns of range limits is offered by biological invasions. These can be viewed as ‘natural experiments’ during which species are often placed in settings outside their recent evolutionary context, where some spread extensively while others remain spatially restricted or fail to become established altogether. Early surveys across a large spectrum of taxa revealed that invasive species are often clustered within certain families like ducks (Chordata, Anatidae) or grasses (Magnoliophyta, Poaceae [7]), indicating that characteristics responsible for the invasive status of some species are highly conserved, transcending deep phylogenetic divisions. Recently, however, notable differences in the degree of spatial spread and invasion success have been reported at lower taxonomic levels between sister species [8–10], suggesting that the ability of taxa to spread and become invasive once introduced to a new environment might be a more variable attribute than previously thought.
Here, we explore these issues in the colonial ascidian Botryllus schlosseri Pallas 1766 (Chordata: Ascidiacea). Also known as the golden star tunicate, B. schlosseri is a powerful model system in the fields of immunobiology, developmental biology and evolutionary biology [11,12]. It is generally considered of European origin [13,14] (but see [15]), and was probably introduced via European shipping to the east coast of North America in the early 1800s [13]. Since then, it has spread to coastal waters of all non-polar continents, with new records of established populations being added frequently [16]. Importantly, although B. schlosseri is one of the most widespread marine invaders, it also presents the characteristics of a species likely to experience significant genetic breaks across small spatial scales and strong local adaptation. These include a sessile adult stage, a brief free-swimming larval stage [17] and gregarious settlement of kin larvae [18]. Consistent with this expectation, previous genetic surveys have documented high levels of genetic differentiation between B. schlosseri populations [16,19,20]. Despite these observations, the geographical and/or biological barriers to gene flow within this taxon remain largely unexplored. Moreover, little is known about how B. schlosseri genetic groups differ in the extent of their geographical distribution and invasion success.
We characterize the phylogenetic and population genetic structure of European and North American populations of B. schlosseri, using the mitochondrial cytochrome c oxidase subunit I (COI), the nuclear 18S rRNA and 10 polymorphic microsatellites. We aim to (i) determine the extent of genetic subdivision within this species, (ii) evaluate the degree of independence of genetic groups (i.e. do identified genetic breaks delineate unrecognized cryptic species?), and (iii) examine the spatial distribution of genetic assemblages/sibling species and determine their invasive potential. Finally, we interpret these findings in the light of evolutionary mechanisms known to enhance the ability of species to spread and become invasive.
2. Material and methods
(a). Sampling and molecular methods
Botryllus schlosseri colonies were sampled from the southern and northern coasts of Europe, and the eastern and western coasts of North America (figure 1 and table 1). DNA extractions were performed as described by Lejeusne et al. [20]. A fragment of COI was amplified using the LCO1490/HCO2198 primers [21]. Failed amplifications were repeated using the species-specific primer BsCOIR (5′-GTATTTTATTTTTAGAATTTGGTCAAG-3′) and HCO2198. For phylogenetic purposes, we used 24 additional COI sequences: 19 from GenBank [19,22] and 5 from unpublished data (A. Lacoursière-Roussel 2011, unpublished data; electronic supplementary material, appendix 2). A subset of 42 individuals from the major COI clades was analysed for the nuclear 18S rRNA gene, using the 18S1/18S4 primers [23]. Reaction chemistry and cycling parameters followed Bock et al. [24] for COI and Xu et al. [25] for 18S rRNA. Sequences were aligned in CodonCode Aligner v. 2.0.6 (CodonCode Corporation, Dedham, MA).
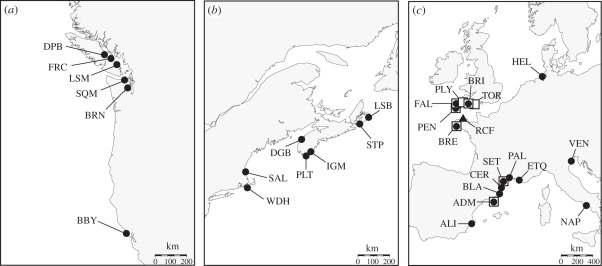
Figure 1.
Distribution of sampling locations and phylogenetic clades for B. schlosseri on the (a) west and (b) east coasts of North America, and (c) in Europe. Site IDs are defined in table 1. Filled circles, clade A; filled triangles, clade D; open squares, clade E.
Table 1.
Locations of Botryllus schlosseri sampling and genetic diversity indices for COI and microsatellite markers. N, sample size; Nh, number of haplotypes; h, haplotype diversity; π, nucleotide diversity; NA, number of alleles; NAP, number of private alleles; A, allelic richness (with rarefaction to 10 individuals); HE, mean gene diversity. For allelic richness and gene diversity calculations, only populations with N > 10 individuals were considered.
| location | ID | COI |
microsatellite |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Nh | haplotype codes | h | π | N | NA | NAP | A | HE | ||
| clade A | |||||||||||
| southern Europe | 191 | 15 | 0.511 | 0.011 | 200 | 125 | 19 | 5.2 | 0.636 | ||
| Venice, Italy | VEN | 28 | 8 | HA; HB; HO; Bs22; Bs23; Bs24; Bs25; Bs26 | 0.730 | 0.015 | 28 | 70 | 7 | 5.3 | 0.652 |
| Naples, Italy | NAP | 7 | 1 | HA | 0 | 0 | 7 | 29 | 0 | — | — |
| Estaque, France | ETQ | 11 | 2 | HA; HB | 0.509 | 0.012 | 15 | 52 | 2 | 4.9 | 0.604 |
| Palavas, France | PAL | 19 | 4 | HA; HB; Bs28; Bs30 | 0.626 | 0.014 | 19 | 59 | 2 | 5.1 | 0.599 |
| Sete, Francea | SET | 2 | 2 | HA; HB | 1.000 | 0.023 | 2 | 22 | 0 | — | — |
| Canet-en-Roussillon, France | CER | 36 | 4 | HA; HB; Bs27; Bs29 | 0.602 | 0.013 | 34 | 81 | 4 | 5.7 | 0.674 |
| Blanes, Spain | BLA | 24 | 3 | HA; HB; Bs3 | 0.540 | 0.012 | 24 | 64 | 2 | 5.3 | 0.674 |
| Arenys de Mar, Spaina | ADM | 36 | 2 | HA; HB | 0.157 | 0.004 | 38 | 67 | 2 | 5.0 | 0.609 |
| Alicante, Spain | ALI | 28 | 5 | HA; HB; Bs3; Bs5; Bs9 | 0.434 | 0.007 | 33 | 57 | 0 | 4.8 | 0.641 |
| northern Europe | 48 | 4 | 0.165 | 0.004 | 39 | 54 | 0 | 3.0 | 0.520 | ||
| Brest, Francea | BRE | 3 | 1 | HD | 0 | 0 | 3 | 22 | 0 | — | — |
| Port Pendennis, Englanda | PEN | 1 | 1 | HB | — | — | 1 | 9 | 0 | — | — |
| Falmouth, Englanda | FAL | 18 | 3 | HA; HB; HD | 0.660 | 0.014 | 18 | 39 | 0 | 3.6 | 0.506 |
| Brixham, Englanda | BRI | 7 | 1 | HD | 0 | 0 | 7 | 38 | 0 | — | — |
| Helgoland, Germany | HEL | 19 | 1 | Bs2 | 0 | 0 | 10 | 23 | 0 | 2.3 | 0.533 |
| eastern North America | 55 | 6 | 0.254 | 0.008 | 58 | 53 | 2 | 3.2 | 0.503 | ||
| Louisbourg, NS, Canada | LSB | 2 | 1 | Bs2 | 0 | 0 | 2 | 17 | 0 | — | — |
| St. Peter, NS, Canada | STP | 5 | 1 | Bs2 | 0 | 0 | 5 | 26 | 0 | — | — |
| Ingomar, NS, Canada | IGM | 8 | 2 | HO; Bs2 | 0.429 | 0.015 | 8 | 31 | 0 | — | — |
| Port La Tour, NS, Canada | PLT | 11 | 6 | HA; HO; Bs2; Bs4; Bs6; Bs7 | 0.836 | 0.027 | 12 | 34 | 0 | 3.3 | 0.526 |
| Digby, NS, Canada | DGB | 6 | 1 | Bs2 | 0 | 0 | 6 | 30 | 0 | — | — |
| Salem, MA, USA | SAL | 10 | 2 | HO; Bs2 | 0.356 | 0.012 | 11 | 26 | 0 | 2.6 | 0.429 |
| Woods Hole, MA, USA | WDH | 13 | 2 | HO; Bs2 | 0.154 | 0.005 | 14 | 38 | 2 | 3.7 | 0.555 |
| western North America | 100 | 5 | 0.323 | 0.008 | 100 | 68 | 5 | 3.3 | 0.560 | ||
| Deep Bay, BC, Canada | DPB | 9 | 1 | Bs1 | 0 | 0 | 8 | 20 | 0 | — | — |
| French Creek, BC, Canada | FRC | 23 | 1 | Bs1 | 0 | 0 | 26 | 34 | 1 | 2.9 | 0.540 |
| Ladysmith, BC, Canada | LSM | 15 | 2 | Bs1; Bs2 | 0.133 | 0.002 | 14 | 29 | 0 | 2.8 | 0.577 |
| Brinnon, WA, USA | BRN | 11 | 2 | Bs1; Bs8 | 0.436 | 0.007 | 11 | 35 | 0 | 3.5 | 0.554 |
| Sequim, WA, USA | SQM | 6 | 2 | Bs1; Bs10 | 0.600 | 0.017 | 5 | 29 | 2 | — | — |
| Bodega Bay, CA, USA | BBY | 36 | 4 | Bs1; Bs8; Bs10; Bs36 | 0.767 | 0.019 | 36 | 48 | 2 | 3.8 | 0.570 |
| clade D | |||||||||||
| northern Europe | |||||||||||
| Roscoff, France | RCF | 35 | 1 | Bs31 | 0 | 0 | 36 | — | — | — | — |
| clade E | |||||||||||
| southern Europe | 5 | 3 | 3 | ||||||||
| Sete, Francea | SET | 4 | 2 | Bs11, Bs12 | 0.667 | 0.023 | 2 | — | — | — | — |
| Arenys de Mar, Spaina | ADM | 1 | 1 | Bs21 | — | — | 1 | — | — | — | — |
| northern Europe | 128 | 9 | 0.605 | 0.005 | 118 | ||||||
| Brest, Francea | BRE | 34 | 4 | HH; Bs12; Bs18; Bs33 | 0.733 | 0.003 | 34 | — | — | — | — |
| Port Pendennis, Englanda | PEN | 17 | 2 | Bs18; Bs34 | 0.221 | 0.003 | 19 | — | — | — | — |
| Falmouth, Englanda | FAL | 25 | 2 | Bs18; Bs34 | 0.220 | 0.003 | 23 | — | — | — | — |
| Plymouth, England | PLY | 34 | 6 | Bs12; Bs18; Bs19; Bs20; Bs32; Bs34 | 0.763 | 0.007 | 25 | — | — | — | — |
| Brixham, Englanda | BRI | 14 | 5 | HH; Bs18; Bs32; Bs34; Bs35 | 0.857 | 0.007 | 14 | — | — | — | — |
| Torquay, England | TOR | 4 | 3 | HH; Bs18; Bs34 | 0.833 | 0.006 | 3 | — | — | — | — |
aLocations where different clades co-occur.
Nuclear genetic variation was further investigated using 10 polymorphic microsatellites: BS321 [26], PB29, PB49, PB41 and PBC1 [27], and Bsm1, Bsm2, Bsm4, Bsm6 and Bsm9 [28]. Amplification conditions, fragment visualization and scoring followed Bock et al. [24]. For samples that failed to amplify, we used a modified protocol (5 cycles at 40°C annealing, and 35 cycles at 43°C annealing).
(b). Data analysis
Phylogenetic relationships were examined using neighbour-joining (NJ) analysis in MEGA v. 4 [29], and Bayesian inference (BI) in MrBayes v. 3.1 [30]. For detailed methods see electronic supplementary material, appendix 1. Relationships among COI haplotypes were also investigated using a haplotype network in TCS v. 1.21 [31]. For each population, COI haplotype and nucleotide diversities were calculated in Dnasp v. 4.50 [32]. Additionally, the COI data were used to calculate population pairwise ΦST values, with 104 permutations in Arlequin v. 3.0 [33], using the Tamura–Nei substitution model [34]. Significance levels were adjusted using sequential Bonferroni corrections [35].
Microsatellite data were checked for deviations from Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium (LD) with Genepop v. 4.0 [36]. Genetic diversity indices of allelic richness and gene diversity were calculated in FSTAT v. 2.9.3.2 [37]. Genetic relationships between multilocus genotypes were assessed using factorial correspondence analysis (FCA) in GENETIX v. 4.05 [38]. Genetic structure was investigated in Arlequin, by calculating pairwise FST values with 104 permutations and significance levels of p-values adjusted as above. To infer population structure without predefined population subdivision, we used the Bayesian clustering approach implemented in Structure v. 2.3.2 [39]. For detailed methods see electronic supplementary material, appendix 1.
3. Results
(a). Sequence variation and phylogenetic reconstructions
The COI alignment was 524 bp long, and contained 160 polymorphic sites, of which 142 were parsimony informative. Within 586 sequences, we identified 60 haplotypes: 19 new (GenBank accessions: JN248359–JN248377) and 41 reported previously ([19,20,22]; A. Lacoursière-Roussel 2011, unpublished data). The 18S rRNA alignment was 817 bp long and contained 82 polymorphic sites, of which 81 were parsimony informative. Five unique 18S rRNA sequences were identified among 42 individuals (GenBank accessions: JN248378–JN248382).
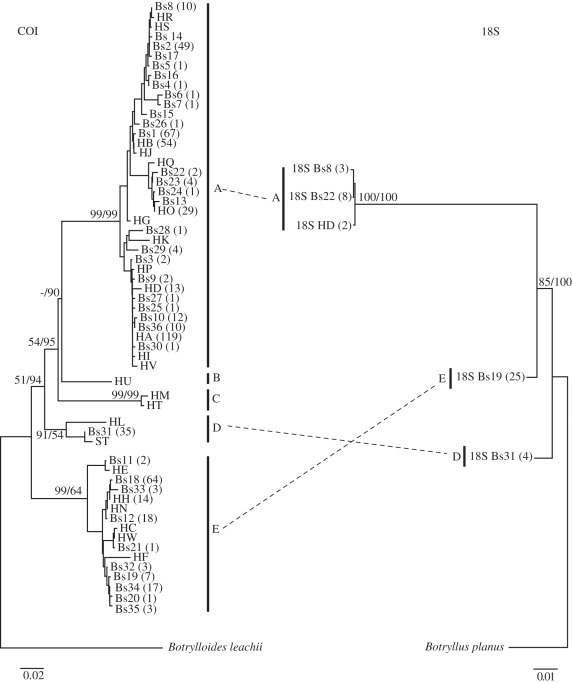
The COI phylogenetic reconstructions (identical for NJ and BI algorithms) recovered five strongly supported monophyletic clades (clades A–E; figure 2), with high interclade sequence divergences (10.8–16.5%) and comparatively lower intraclade distances (0.8–3.8%). While members of clades A, D and E were sampled in this study, clades B and C consisted of haplotypes retrieved from GenBank (figure 2; electronic supplementary material, appendix 2). The TCS network also recovered five major groups of COI haplotypes. Within each group, considerable genetic structure was detected, with haplotypes separated by up to 47 mutation steps (electronic supplementary material, appendix 3). The 18S rRNA phylogeny was congruent with that based on COI in recovering with high support the monophyly of the three lineages sampled in this study (figure 2), with high divergences (2.3–10.1%). No clear differences in colour morphs were observed between members of different phylogenetic clades.
Figure 2.
Neighbour-joining (NJ) tree based on mitochondrial cytochrome c oxidase I (COI) haplotypes and nuclear 18S rRNA sequences. Numbers at phylogenetic nodes indicate NJ bootstrap support and Bayesian inference posterior probabilities. The number of samples showing a given haplotype is presented in brackets.
(b). Geographical patterns of cytochrome c oxidase subunit I diversity
Clade A was geographically widespread. It characterized most (98.5%) individuals from southern Europe and all of the ones from North America. It was also present in five sites from northern Europe (figure 1 and table 1). Clades B and C, not recovered here, were previously sampled in two European locations on the coast of Spain [19]. Clade D, also geographically restricted, was recovered in one site in the English Channel. Similarly, Clade E was mainly recovered in six populations concentrated in the English Channel. Only three individuals of clade E were sampled outside this region, in two southern European sites (figure 1 and table 1). The geographical distribution of clades was largely allopatric, with individual populations containing only one major lineage. However, in six of the sites surveyed, clades A and E were found in sympatry, with one of the two lineages generally dominating each site (table 1). These sympatric locations provided us with a unique opportunity to investigate the level of contemporary gene flow (and indirectly, reproductive isolation) between members of the two divergent clades using microsatellite data in Structure (see below).
Genetic diversity indices of haplotype diversity (h) and nucleotide diversity (π) computed for clade A populations were higher for southern European sites (h = 0.511; π = 0.011) than for those in northern Europe (h = 0.165; π = 0.004), or eastern (h = 0.254; π = 0.008) or western North America (h = 0.323; π = 0.008). However, this general trend masked substantial variation, as all surveyed regions contained monomorphic as well as highly diverse populations. Pairwise ΦST values computed for clade A populations also revealed heterogeneity in genetic structure, with instances of high and significant differentiation between sites within the surveyed regions, as well as low and non-significant differentiation between sites at macrogeographical scales (electronic supplementary material, appendix 4). For clade D, all individuals shared the same haplotype, while for clade E all populations showed high levels of genetic variation (h = 0.605; π = 0.005; table 1). With two exceptions, all pairwise ΦST values computed for clade E populations were high and significant (electronic supplementary material, appendix 5).
(c). Geographical patterns of microsatellite diversity
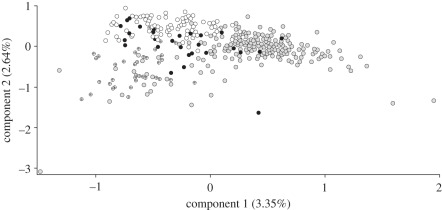
For the widely distributed clade A, we discerned 135 alleles among the 10 microsatellite markers assayed (mean: 13.5 alleles/locus). While most loci conformed to HWE expectations, 36 out of 156 cases showed significant deviations from HWE. However, no systematic deviations were observed across loci or populations (electronic supplementary material, appendix 6). Also, there was little evidence for LD, with only 6 of 720 locus pairs remaining significant after Bonferroni corrections in one of the sites surveyed. Genetic diversity indices of allelic richness (A) and gene diversity (HE) computed for clade A populations were generally higher for southern European sites (A = 5.2; HE = 0.636) than for those in northern Europe (A = 3.0; HE = 0.520), or eastern (A = 3.2; HE = 0.503) or western North America (A = 3.3; HE = 0.560; for detailed clade A microsatellite results see table 1; electronic supplementary material, appendix 6). The FCA computed using microsatellite data from clade A revealed weak clustering of genotypes based on geographical origin. Higher genetic similarities were sporadically revealed for individuals sampled between rather than within biogeographic regions (figure 3). This result was also evidenced by the low and non-significant pairwise FST values occasionally recorded between sites at macrogeographical scales (electronic supplementary material, appendix 7). For the divergent clades D and E, consistent PCR amplifications were obtained for 7 of the 10 microsatellite loci. However, the genotypic profiles of these individuals were characterized by a low number of alleles (mean: 6.3 alleles/locus) and a high incidence of homozygotes. Since these features are generally indicative of datasets with high rates of null alleles, microsatellite data for clades D and E were used only for the Structure analysis, which minimizes equilibrium requirements.
Figure 3.
Factorial correspondence analysis of clade A microsatellite data (considering 10 loci) showing genetic similarities of sampled individuals (derived from allele frequencies). Grey circles, southern Europe; black circles, northern Europe; crossed circles, eastern North America; white circles, western North America.
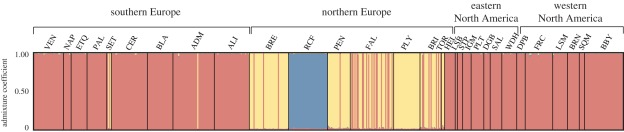
For the Structure analysis, when data from the seven microsatellite loci amplified in all samples from clades A, D and E were considered, the most likely model was the one with three clusters (electronic supplementary material, appendices 6 and 7). At K = 3, each cluster corresponded perfectly to the three major clades identified by COI and 18S rRNA (i.e. clades A, D and E; figure 4). Admixture coefficients ranged from 0.94 to 1.00, with all individuals showing 90 per cent probability intervals encompassing coefficients of 1, suggesting no contemporary gene flow between the three clusters (figure 4).
Figure 4.
Bayesian clustering of microsatellite data obtained for all samples (considering seven loci). Each individual is represented by a thin vertical line partitioned into K = 3 coloured segments (clade A, red; clade D, blue; clade E, yellow) representing the individual's estimated admixture coefficient. Sampling sites are separated by black lines. Site IDs are defined in table 1.
4. Discussion
Two important observations emerge from these results. First, our findings show that B. schlosseri, one of the most widely researched and well-known ascidians, comprises at least three previously unrecognized and probably reproductively isolated cryptic species. Second, results presented here indicate that only one B. schlosseri species is widespread globally, while its sibling species are highly geographically restricted.
(a). Cryptic speciation in B. schlosseri, a model ascidian
Botryllus schlosseri was initially described over two and a half centuries ago, representing the first ascidian species ever recognized [40]. Since then, it has experienced repeated taxonomic revisions owing to extensive colour polymorphism [41]. As a result, multiple synonymies are available (Ascidiacea World Database, http://www.marinespecies.org/ascidiacea/), all of which are currently lumped under a single species designation. Our results oppose these assumptions, and demonstrate that B. schlosseri is a complex of evolutionarily distinct species, which are at least superficially morphologically indistinguishable. Both mitochondrial (COI) and nuclear (18S rRNA and microsatellite) markers consistently recovered three well-supported clades (i.e. clades A, D and E; figures 2 and 4), with two more being confirmed by COI data retrieved from GenBank (i.e. clades B and C; figure 2). The genetic divergences recorded between these lineages (10.8–16.5% COI divergence), corresponding to about 4.3–11.0 Myr of divergent evolutionary history (1.5–2.5% per Myr [42]), are significant. These levels far exceed COI divergences reported between cryptic species of other colonial tunicates, like Pseudodistoma crucigaster (2.12% [43]), Clavelina lepadiformis (5% [44]) or Pycnoclavella communis (8.55% [45]), and are comparable with those identified between widely recognized cryptic species of the solitary tunicate Ciona intestinalis (11.1–18.4% [10,46,47]). Furthermore, the complete lack of contemporary gene flow between clades, even in locations where their distribution intersects (figure 4), suggests that reproductive isolation is probably complete. Altogether, these findings indicate the need for a taxonomic re-evaluation of B. schlosseri.
Botryllus schlosseri sensu lato has been used extensively in pioneering research on the heritability of senescence and mortality, the evolutionary importance of self/non-self recognition or the genetic control of organogenesis (reviewed by Manni et al. [11] and Rosengarten & Nicotra [12]). Since accurate classification of model species is a major component for corroborating data drawn from different laboratories, the identification of cryptic speciation in B. schlosseri can be expected to impact a number of research fields. Beyond these considerations, our results highlight the importance of confirming species status via molecular data and/or crossing experiments. When such information is not available, species identifications should be interpreted with caution, even in well-known model organisms such as B. schlosseri.
(b). Contrasting distribution ranges in closely related species
Results presented here further indicate that B. schlosseri sibling species differ markedly in the extent of their geographical distribution and, probably, invasive potential. Species A is by far the most common and widespread, with a range spanning all biogeographic regions surveyed here (figure 1). Comparisons with previous molecular studies further indicate that this species is also present at additional sites in the eastern Mediterranean Sea [48], the east and west coasts of North America [49,50], South America [16], Japan, Australia [20] and New Zealand [51]. Apart from the remarkable cosmopolitan distribution, additional lines of evidence suggest that species A experiences recurrent long-distance dispersal events (most probably human-mediated), and is highly invasive. These include frequent occurrence of distantly related haplotypes within populations (table 1; electronic supplementary material, appendix 3), extensive haplotype sharing among sites within biogeographic regions (table 1) and patchy distribution of genetic variation between sites at macrogeographical scales (figure 3 and table 1; electronic supplementary material, appendices 4 and 5). By contrast, species B and C, not recovered here, have only been found in two European locations on the coast of Spain [19]. Similarly, with the exception of three individuals sampled in southern Europe, species D and E were only found in the English Channel (figure 1 and table 1). Although an important next step to confirm our findings would be to perform global sampling for B. schlosseri, particularly covering the centre of botryllid diversity in the western and southern Pacific Ocean [15], results presented here suggest that species A is much more geographically widespread than species B–E.
In colonial ascidians, because of the short-lived larval stage and sessile adult stage, the natural potential for range expansion is limited and is represented mainly by the rafting of individuals on floating debris [13]. Although this mechanism of dispersal could have contributed to intracoastal spread within the surveyed biogeographic regions, it is unlikely that it also facilitated instances of dispersal at macrogeographical scales observed here, particularly for species A. Human-mediated dispersal is a more likely candidate, as colonial ascidians are known to be problematic foulers of ship hulls and aquaculture equipment [13]. In this context, the sharp biogeographic asymmetries observed here between B. schlosseri sibling species can be explained by two scenarios that are not mutually exclusive: (i) stochastic events such as biases in human-mediated transport routes and opportunity facilitated by chance widespread introductions for B. schlosseri species A, but not for species B–E; or (ii) divergent evolutionary histories resulted in marked differences in the ability of B. schlosseri species to survive transport, establish, spread and become invasive. These scenarios are difficult to separate, but a number of clues suggest that the first one is less likely. First, biases in transport opportunity are expected to be more important in recent invaders, reflecting the stochastic nature of the invasion process. In species with long histories of anthropogenic transport like B. schlosseri, access to dispersal vectors should have eventually counterbalanced differences in spatial spread between sibling species of similar invasive potential. Second, the invasive species A co-occurs with species D and E in the English Channel, a region with extensive opportunity for transport via ship traffic. Third, species E individuals that are probably the results of recent introductions were occasionally found in southern European sites (figure 1 and table 1) [19]. Collectively, these findings oppose the possibility that lack of transport prevented the widespread occurrence of B. schlosseri species other than species A.
The alternative scenario that divergent evolutionary histories shaped differential invasive potential in B. schlosseri sibling species is intriguing. The classical perception is that attributes determining range limits are highly evolutionarily conserved [52]. This view is supported by the few available surveys reporting that closely related species have similar range sizes [52], and by the observation that invasive species are often clustered within certain families [7]. Results presented here refute this earlier assumption, and add to a growing body of literature reporting marked differences in range limits and invasion success in closely related species [8–10]. Collectively, these studies suggest that species' range limits and invasion abilities are much more evolutionarily volatile than previously thought.
An ensuing question is what prompted the evolution of differential invasion abilities in B. schlosseri species. Much of the current literature on the evolution of invasiveness stems from studies of plant taxa [53,54]. Four main types of evolutionary change are thought to contribute to plant invasiveness: hybridization, polyploidy, bottlenecks and stress-induced genome modification (reviewed by Prentis et al. [55]). In the case of B. schlosseri, environmental disturbance appears to be the most likely scenario. Recently, it has been proposed that contrasting invasion abilities can evolve between populations or sibling species subjected to different levels of disturbance in their native range [56]. Theoretical models also suggest that environmental fluctuations spanning different timescales can select for organismal flexibility and evolvability [57], both of which are favourable for invading new habitats [56]. Indeed, owing to their sessile nature, colonial ascidians such as B. schlosseri are prone to be exposed to a multitude of environmental stressors. Although the ancestral distribution range and, implicitly, historical levels of disturbance are difficult to infer for B. schlosseri because of its long and complex history of anthropogenic range expansions, the disturbance scenario provides a promising working hypothesis that remains to be tested. Ecological genomics experiments can be used in future studies to reveal the extent of ecological tolerance for different cryptic species, quantify additive genetic variance for critical invasive traits, identify genes of major phenotypic effect and ultimately clarify the evolution of invasiveness in this system.
Acknowledgements
We would like to kindly acknowledge C. W. McKindsey, A. Lacoursière-Roussel as well as J. T. Carlton, R. P. Walter, A. Zhan and students in the Cristescu and MacIsaac laboratories for stimulating discussions. We are grateful to our colleagues J. Bishop, M. Carman, P. Chevaldonné, S. Dashfield, E. Grey, A. Izquierdo-Muñoz, D. Jollivet, C. Lejeusne, L. Lévêque, M. Litvaitis, L. Manni, C. Menniti, M. Molis, J. M. Nicolas, M. Rius, P. Sordino, C. Sorte, T. W. Therriault, X. Turon, B. Vercaemer, S. Williams as well as the Roscoff Biological Station Specimen Supply Service (CNRS/FR2424) for generously providing samples. This work was supported by the NSERC Canadian Aquatic Invasive Species Network (CAISN), Fisheries and Oceans Canada, NSERC Discovery Grants to H.J.M. and M.E.C., and by an Early Researcher Award from the Ontario Ministry of Research and Innovation to M.E.C.
References
- 1.Elton C. S. 1930. Animal ecology and evolution. Oxford, UK: The Clarendon Press [Google Scholar]
- 2.Sexton J. P., McIntyre P. J., Angert A. L., Rice K. J. 2009. Evolution and ecology of species range limits. Annu. Rev. Ecol. Evol. Syst. 40, 415–436 10.1146/annurev.ecolsys.110308.120317 (doi:10.1146/annurev.ecolsys.110308.120317) [DOI] [Google Scholar]
- 3.Keitt T. H., Lewis M. A., Holt R. D. 2001. Allee effects, invasion pinning, and species' borders. Am. Nat. 157, 203–216 10.1086/318633 (doi:10.1086/318633) [DOI] [PubMed] [Google Scholar]
- 4.Pulliam H. R. 2000. On the relationship between niche and distribution. Ecol. Lett. 3, 349–361 10.1046/j.1461-0248.2000.00143.x (doi:10.1046/j.1461-0248.2000.00143.x) [DOI] [Google Scholar]
- 5.Kirkpatrick M., Barton N. H. 1997. Evolution of a species' range. Am. Nat. 150, 1–23 10.1086/286054 (doi:10.1086/286054) [DOI] [PubMed] [Google Scholar]
- 6.Ronce O., Kirkpatrick M. 2001. When sources become sinks: migrational meltdown in heterogeneous habitats. Evolution 55, 1520–1531 10.1111/j.0014-3820.2001.tb00672.x (doi:10.1111/j.0014-3820.2001.tb00672.x) [DOI] [PubMed] [Google Scholar]
- 7.McKinney M. L., Lockwood J. L. 1999. Biotic homogenization: a few winners replacing many losers in the next mass extinction. Trends Ecol. Evol. 14, 450–453 10.1016/S0169-5347(99)01679-1 (doi:10.1016/S0169-5347(99)01679-1) [DOI] [PubMed] [Google Scholar]
- 8.Neilson M. E., Stepien C. A. 2008. Evolution and phylogeography of the tubenose goby genus Proterorhinus (Gobiidae: Teleostei): evidence for new cryptic species. Biol. J. Linn. Soc. 96, 664–684 10.1111/j.1095-8312.2009.01295.x (doi:10.1111/j.1095-8312.2009.01295.x) [DOI] [Google Scholar]
- 9.Winkler G., Dodson J. J., Lee C. E. 2008. Heterogeneity within the native range: population genetic analyses of sympatric invasive and noninvasive clades of the freshwater invading copepod Eurytemora affinis. Mol. Ecol. 17, 415–430 10.1111/j.1365-294X.2007.03480.x (doi:10.1111/j.1365-294X.2007.03480.x) [DOI] [PubMed] [Google Scholar]
- 10.Zhan A., MacIsaac H. J., Cristescu M. E. 2010. Invasion genetics of the Ciona intestinalis species complex: from regional endemism to global homogeneity. Mol. Ecol. 19, 4678–4694 10.1111/j.1365-294X.2010.04837.x (doi:10.1111/j.1365-294X.2010.04837.x) [DOI] [PubMed] [Google Scholar]
- 11.Manni L., Zaniolo G., Cima F., Burighel P., Ballarin L. 2007. Botryllus schlosseri: a model ascidian for the study of asexual reproduction. Dev. Dyn. 236, 335–352 10.1002/dvdy.21037 (doi:10.1002/dvdy.21037) [DOI] [PubMed] [Google Scholar]
- 12.Rosengarten R. D., Nicotra M. L. 2011. Model systems of invertebrate allorecognition. Curr. Biol. 21, 82–92 10.1016/j.cub.2010.11.061 (doi:10.1016/j.cub.2010.11.061) [DOI] [PubMed] [Google Scholar]
- 13.Carver C. E., Mallet A. L., Vercaemer B. 2006. Biological synopsis of the colonial tunicates, Botryllus schlosseri and Botrylloides violaceus. Canadian Manuscript Report of Fisheries and Aquatic Sciences 2747: v+42p
- 14.Berrill N. J. 1950. The Tunicata. With an account of the British species. London, UK: Ray Society [Google Scholar]
- 15.Carlton J. T. 2005. Setting ascidian invasions on the global stage. International Invasive Sea Squirt Conference Woods Hole Oceanographic Institution, Woods Hole, MA, April 2005; (http://www.whoi.edu/page.do?pid=11421&cid=16303&c=2) [Google Scholar]
- 16.Ben-Shlomo R., Reem E., Douek J., Rinkevich B. 2010. Population genetics of the invasive ascidian Botryllus schlosseri from South American coasts. Mar. Ecol. Prog. Ser. 412, 85–92 10.3354/meps08688 (doi:10.3354/meps08688) [DOI] [Google Scholar]
- 17.Rinkevich B., Porat R., Goren M. 1998. Ecological and life history characteristics of Botryllus schlosseri (Tunicata) populations inhabiting undersurface shallow water stones. Mar. Ecol. 19, 129–145 10.1111/j.1439-0485.1998.tb00458.x (doi:10.1111/j.1439-0485.1998.tb00458.x) [DOI] [Google Scholar]
- 18.Grosberg R. K., Quinn J. F. 1986. The genetic control and consequences of kin recognition by the larvae of a colonial marine invertebrate. Nature 322, 456–459 10.1038/322456a0 (doi:10.1038/322456a0) [DOI] [Google Scholar]
- 19.Lopez-Legentil S., Turon X., Planes S. 2006. Genetic structure of the star sea squirt, Botryllus schlosseri, introduced in southern European harbours. Mol. Ecol. 15, 3957–3967 10.1111/j.1365-294X.2006.03087.x (doi:10.1111/j.1365-294X.2006.03087.x) [DOI] [PubMed] [Google Scholar]
- 20.Lejeusne C., Bock D. G., Therriault T. W., MacIsaac H. J., Cristescu M. E. 2011. Comparative phylogeography of two colonial ascidians reveals contrasting invasion histories in North America. Biol. Inv. 13, 635–650 10.1007/s10530-010-9854-0 (doi:10.1007/s10530-010-9854-0) [DOI] [Google Scholar]
- 21.Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299 [PubMed] [Google Scholar]
- 22.Stach T., Turbeville J. M. 2002. Phylogeny of Tunicata inferred from molecular and morphological characters. Mol. Phylogenet. Evol. 25, 408–428 10.1016/S1055-7903(02)00305-6 (doi:10.1016/S1055-7903(02)00305-6) [DOI] [PubMed] [Google Scholar]
- 23.Tsagkogeorga G., et al. 2009. An updated 18S rRNA phylogeny of tunicates based on mixture and secondary structure models. BMC Evol. Biol. 9, 187. 10.1186/1471-2148-9-187 (doi:10.1186/1471-2148-9-187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bock D. G., Zhan A., Lejeusne C., MacIsaac H. J., Cristescu M. E. 2011. Looking at both sides of the invasion: patterns of colonization in the violet tunicate Botrylloides violaceus. Mol. Ecol. 20, 503–516 10.1111/j.1365-294X.2010.04971.x (doi:10.1111/j.1365-294X.2010.04971.x) [DOI] [PubMed] [Google Scholar]
- 25.Xu S., Hebert P. D. N., Kotov A. A., Cristescu M. E. 2009. The noncosmopolitanism paradigm of freshwater zooplankton: insights from the global phylogeography of the predatory cladoceran Polyphemus pediculus (Linnaeus, 1761) (Crustacea, Onychopoda). Mol. Ecol. 18, 5161–5179 10.1111/j.1365-294X.2009.04422.x (doi:10.1111/j.1365-294X.2009.04422.x) [DOI] [PubMed] [Google Scholar]
- 26.Pancer Z., Gershon Z., Rinkevich B. 1994. Direct typing of polymorphic microsatellites in the colonial ascidian Botryllus schlosseri. Biochem. Biophys. Res. Commun. 203, 646–651 10.1006/bbrc.1994.2231 (doi:10.1006/bbrc.1994.2231) [DOI] [PubMed] [Google Scholar]
- 27.Stoner D. S., Quattro J. M., Weissman I. L. 1997. Highly polymorphic microsatellite loci in the colonial ascidian Botryllus schlosseri. Mol. Mar. Biol. Biotechnol. 6, 163–171 [PubMed] [Google Scholar]
- 28.Molecular Ecology Resources Primer Development Consortium 2010. Permanent Genetic Resources added to Molecular Ecology Resources Database 1 August 2009–30 September 2009. Mol. Ecol. Res. 10, 232–236 10.1111/j.1755-0998.2009.02796.x (doi:10.1111/j.1755-0998.2009.02796.x) [DOI] [PubMed] [Google Scholar]
- 29.Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 10.1093/molbev/msm092 (doi:10.1093/molbev/msm092) [DOI] [PubMed] [Google Scholar]
- 30.Ronquist F., Huelsenbeck J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 10.1093/bioinformatics/btg180 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- 31.Clement M., Posada D., Crandall K. A. 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659 10.1046/j.1365-294x.2000.01020.x (doi:10.1046/j.1365-294x.2000.01020.x) [DOI] [PubMed] [Google Scholar]
- 32.Rozas J., Sanchez-DelBarrio J. C., Messeguer X., Rozas R. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19, 2496–2497 10.1093/bioinformatics/btg359 (doi:10.1093/bioinformatics/btg359) [DOI] [PubMed] [Google Scholar]
- 33.Excoffier L., Laval G., Schneider S. 2005. Arlequin version 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1, 47–50 [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura K., Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526 [DOI] [PubMed] [Google Scholar]
- 35.Rice W. R. 1989. Analyzing tables of statistical tests. Evolution 43, 223–225 10.2307/2409177 (doi:10.2307/2409177) [DOI] [PubMed] [Google Scholar]
- 36.Raymond M., Rousset F. 1995. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J. Hered. 86, 248–249 [Google Scholar]
- 37.Goudet J. 2001. FSTAT: a program to estimate and test gene diversities and fixation indices (version 2.9.3). Lausanne, Switzerland: Lausanne University [Google Scholar]
- 38.Belkhir K., Borsa P., Chikhi L., Raufaste N., Bonhomme F. 2004. GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Montpellier, France: Laboratoire Génome, Populations, Interactions, Université de Montpellier [Google Scholar]
- 39.Pritchard J. K., Stephens M., Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shenkar N., Swalla B. J. 2011. Global diversity of Ascidiacea. PLoS ONE 6, e20657. 10.1371/journal.pone.0020657 (doi:10.1371/journal.pone.0020657) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabbadin A., Graziani G. 1967. Microgeographical and ecological distribution of colour morphs of Botryllus schlosseri (Ascidiacea). Nature 213, 815–816 10.1038/213815b0 (doi:10.1038/213815b0)6031815 [DOI] [Google Scholar]
- 42.Martin A. P., Palumbi S. R. 1993. Body size, metabolic rate, generation time, and the molecular clock. Proc. Natl Acad. Sci. USA 90, 4087–4091 10.1073/pnas.90.9.4087 (doi:10.1073/pnas.90.9.4087) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tarjuelo I., Posada D., Crandall K. A., Pascual M., Turon X. 2004. Phylogeography and speciation of colour morphs in the colonial ascidian Pseudodistoma crucigaster. Mol. Ecol. 13, 3125–3136 10.1111/j.1365-294X.2004.02306.x (doi:10.1111/j.1365-294X.2004.02306.x) [DOI] [PubMed] [Google Scholar]
- 44.Tarjuelo I., Posada D., Crandall K. A., Pascual M., Turon X. 2001. Cryptic species of Clavelina (Ascidiacea) in two different habitats: harbours and rocky littoral zones in the north-western Mediterranean. Mar. Biol. 139, 455–462 10.1007/s002270100587 (doi:10.1007/s002270100587) [DOI] [Google Scholar]
- 45.Pérez-Portela R., Turon X. 2008. Cryptic divergence and strong population structure in the colonial invertebrate Pycnoclavella communis (Ascidiacea) inferred from molecular data. Zoology 111, 163–178 10.1016/j.zool.2007.06.006 (doi:10.1016/j.zool.2007.06.006) [DOI] [PubMed] [Google Scholar]
- 46.Caputi L., Andreakis N., Mastrototaro F., Cirino P., Vassillo M., Sordino P. 2007. Cryptic speciation in a model invertebrate chordate. Proc. Natl Acad. Sci. USA 104, 9364–9369 10.1073/pnas.0610158104 (doi:10.1073/pnas.0610158104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nydam M. L., Harrison R. G. 2007. Genealogical relationships within and among shallow-water Ciona species (Ascidiacea). Mar. Biol. 151, 1839–1847 10.1007/s00227-007-0617-0 (doi:10.1007/s00227-007-0617-0) [DOI] [Google Scholar]
- 48.Paz G., Douek J., Mo C., Goren M., Rinkevich B. 2003. Genetic structure of Botryllus schlosseri (Tunicata) populations from the Mediterranean coast of Israel. Mar. Ecol. Progr. Ser. 250, 153–162 10.3354/meps250153 (doi:10.3354/meps250153) [DOI] [Google Scholar]
- 49.Stoner D. S., Ben-Shlomo R., Rinkevich B., Weissman I. L. 2002. Genetic variability of Botryllys schlosseri invasions to the east and west coasts of the USA. Mar. Ecol. Prog. Ser. 243, 93–100 10.3354/meps243093 (doi:10.3354/meps243093) [DOI] [Google Scholar]
- 50.Callahan A. G., Deibel D., McKenzie C. H., Hall J. R., Rise M. L. 2010. Survey of harbours in Newfoundland for indigenous and non-indigenous ascidians and an analysis of their cytochrome c oxidase I gene sequences. Aquat. Inv. 5, 31–39 10.3391/ai.2010.5.1.5 (doi:10.3391/ai.2010.5.1.5) [DOI] [Google Scholar]
- 51.Ben-Shlomo R., Douek J., Rinkevich B. 2001. Heterozygote deficiency and chimerism in remote populations of a colonial ascidian from New Zealand. Mar. Ecol. Prog. Ser. 209, 109–117 10.3354/meps209109 (doi:10.3354/meps209109) [DOI] [Google Scholar]
- 52.Webb C. O., Ackerly D. D., McPeek M. A., Donoghue M. J. 2002. Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 33, 475–505 10.1146/annurev.ecolsys.33.010802.15044 (doi:10.1146/annurev.ecolsys.33.010802.15044) [DOI] [Google Scholar]
- 53.Ellstrand N. C., Schierenbeck K. 2000. Hybridization as a stimulus for the evolution of invasiveness in plants? Proc. Natl Acad. Sci. USA 97, 7043–7050 10.1073/pnas.97.13.7043 (doi:10.1073/pnas.97.13.7043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Colautti R. I., Eckert C. G., Barrett S. C. H. 2010. Evolutionary constraints on adaptive evolution during range expansion in an invasive plant. Proc. R. Soc. B 277, 1799–1806 10.1098/rspb.2009.2231 (doi:10.1098/rspb.2009.2231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prentis P. J., Wilson J. R. U., Dormontt E. E., Richardson D. M., Lowe A. J. 2008. Adaptive evolution in invasive species. Trends Plant Sci. 13, 288–294 10.1016/j.tplants.2008.03.004 (doi:10.1016/j.tplants.2008.03.004) [DOI] [PubMed] [Google Scholar]
- 56.Lee C. E., Gelembiuk G. W. 2008. Evolutionary origins of invasive populations. Evol. Appl. 1, 427–448 10.1111/j.1752-4571.2008.00039.x (doi:10.1111/j.1752-4571.2008.00039.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meyers L. A., Bull J. J. 2002. Fighting change with change: adaptive variation in an uncertain world. Trends Ecol. Evol. 17, 551–557 10.1016/S0169-5347(02)02633-2 (doi:10.1016/S0169-5347(02)02633-2) [DOI] [Google Scholar]