Abstract
Natural selection acts on variation that is typically assumed to be genetic in origin. But epigenetic mechanisms, which are interposed between the genome and its environment, can create diversity independently of genetic variation. Epigenetic states can respond to environmental cues, and can be heritable, thus providing a means by which environmentally responsive phenotypes might be selectable independent of genotype. Here, we have tested the possibility that environment and selection can act together to increase the penetrance of an epigenetically determined phenotype. We used isogenic Avy mice, in which the epigenetic state of the Avy allele is sensitive to dietary methyl donors. By combining methyl donor supplementation with selection for a silent Avy allele, we progressively increased the prevalence of the associated phenotype in the population over five generations. After withdrawal of the dietary supplement, the shift persisted for one generation but was lost in subsequent generations. Our data provide the first demonstration that selection for a purely epigenetic trait can result in cumulative germline effects in mammals. These results present an alternative to the paradigm that natural selection acts only on genetic variation, and suggest that epigenetic changes could underlie rapid adaptation of species in response to natural environmental fluctuations.
Keywords: epigenetic inheritance, environmental epigenetics, agouti viable yellow, DNA methylation, piRNA, adaptive evolution
1. Introduction
Epigenetic control of gene expression is fundamental to eukaryotic biology, creating somatically stable states of gene expression that underlie the diversity of cell types within a multicellular organism. Epigenetics not only creates phenotypic diversity within an individual, but also within populations, and can do so without genetic variation [1]. An epigenetic variant could thus be a novel, non-genetic substrate for natural selection [2], but this would require that the epigenetic state of the responsible locus be heritable from one generation to the next.
Epigenetic states can be meiotically heritable, resulting in transgenerational epigenetic inheritance. This can occur independently of the underlying genotype, and so the pattern of inheritance rarely follows Mendel's laws. Epigenetic inheritance is broadly distributed in nature: it is best described in plants and fungi (reviewed in [3,4]), but also occurs in species that segregate a germline, such as flies, mice and humans [5–11]. It is not known how many loci might exhibit epigenetic inheritance, and its molecular mechanism is also mysterious. There may be no one conserved carrier of intergenerational epigenetic marks across all species; candidates in mammals include cytosine methylation, chromatin proteins and small RNA molecules [4].
One of the best-studied models of epigenetic inheritance is the mouse strain agouti viable yellow (Avy). This inbred strain displays spontaneous phenotypic variation among genetically identical mice. Littermates display a characteristic spectrum of phenotypes, from yellow fur with accompanying obesity and diabetes, through various degrees of mottling to agouti fur and lean body mass (figure 1a). The basis for the variation is variable expressivity of the Avy allele, a mutant allele of the agouti coat-colour gene that arose spontaneously almost 50 years ago [12]. Avy was created by the insertion of an intracisternal A-particle (IAP) retrotransposon into an upstream non-coding exon of agouti [13,14]. The IAP is epigenetically regulated, but shows variable activity between mice and between cells in an individual mouse. When the IAP is active, transcription from a cryptic promoter in its 5′-long terminal repeat (LTR) produces an ectopic agouti transcript; constitutive agouti expression produces the obese yellow syndrome. When the IAP is epigenetically silenced, agouti is expressed in its wild-type pattern (called pseudoagouti) [15]. The activity state of Avy is inversely correlated with DNA methylation at the IAP cryptic promoter [7,16], but the factors responsible for setting its epigenetic state in each mouse are as yet unknown.
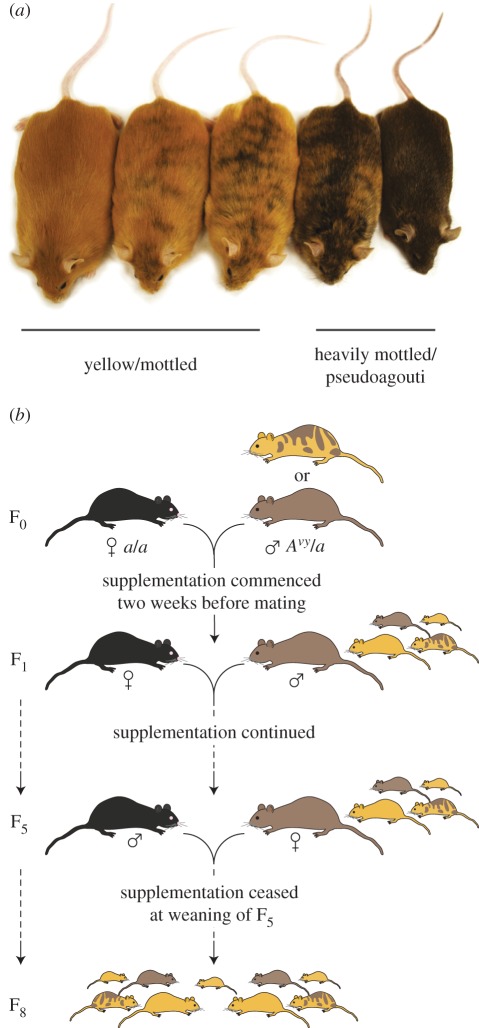
Figure 1.
Avy mice and breeding scheme. (a) Isogenic Avy/a littermates showing the spectrum of coat-colour phenotypes: yellow, slightly mottled, half mottled, heavily mottled and pseudoagouti. (b) Schematic of methyl donor supplementation and selective breeding of the pseudoagouti phenotype. From F1 to F4, male pseudoagouti Avy/a mice were chosen for breeding; from F5 to F7, female pseudoagouti Avy/a mice were chosen. Although half of all offspring are a/a, for simplicity only Avy/a mice are shown.
While the epigenetic state of the Avy allele varies widely among isogenic mice, it is stable in somatic cells, so that the phenotype of an individual mouse will remain the same throughout its lifetime. It is also partially stable in the female germline, leading to transgenerational epigenetic inheritance, so that offspring phenotypes are biased towards that of the dam [7]. The epigenetic state of the allele is unstable in the male germline and is completely reset between generations, such that sires of all coat colours produce offspring with the same spectrum of phenotypes.
Epigenetic states can be susceptible to environmental influence, altering phenotypes within a population [17]. The epigenetic state of Avy is influenced by the addition of excess methyl donor pathway molecules to the maternal diet, which shifts the spectrum of offspring phenotypes towards pseudoagouti [14,18–20]. We have previously shown that the epigenetic effects of methyl donor supplementation on Avy are heritable [20]. This heritability provides a means by which an environmentally induced phenotype might be selected for, and rapidly pervade a population without the need for genetic change.
Here, we asked whether selection applied over multiple generations of exposure to methyl donors could fix the epigenetic state of Avy in the germline. We found that multi-generational methyl donor supplementation, coupled with selection for the pseudoagouti phenotype, steadily increased the prevalence of the pseudoagouti phenotype in the population. Our result provides the first evidence that epigenetic traits can be substrates for selection in mammals, which suggests that they may contribute to adaptive evolution.
2. Material and methods
(a). Ethics statement
All animals were handled in strict accordance with good practice as defined by the NHMRC's (Australia) Statement on Animal Experimentation and the requirements of NSW State Government legislation.
(b). Mice and diets
The mice used in this study are descended from the C57BL/6 colony maintained at Oak Ridge National Laboratories (originally obtained from the Jackson Laboratories, stock no. 000017). The Avy mutation arose spontaneously on a C3H background and the allele was backcrossed into C57BL/6. The strain, called Avy, has been maintained by brother–sister matings for over 40 years, and is thus isogenic. Mice were fed ad libitum on control (NIH-31) or supplemented (NIH-31 plus (per kilogram), 15 g choline, 15 g betaine, 7.5 g l-methionine, 150 mg zinc, 15 mg folic acid and 1.5 mg vitamin B12) diet (Specialty Feeds, WA, Australia).
(c). Breeding strategy
The breeding strategy is summarized in figure 1b. Founder breeding pairs (pseudoagouti or heavily mottled Avy/a males mated to a/a females) were supplemented from two weeks prior to mating. In generations F1–F4, male pseudoagouti Avy/a offspring were weaned onto the supplemented diet and mated with female a/a mice; supplementation was continued for the duration of breeding. We passed the Avy allele only through the sire during this part of the experiment, because the Avy allele is sensitive to methyl donors only when inherited from the sire [20]. In generation F5, female pseudoagouti Avy/a offspring were weaned onto control diet and mated with male a/a mice. Generations F6 and F7 female pseudoagouti Avy/a offspring were again selected for breeding with a/a males; mice were maintained on control diet until the end of the study. We passed the Avy allele only through the dam through this part of the experiment to maximize our chances of observing inheritance of the cumulative effect (we have previously shown that the effect of methyl donors is inherited through the dam [20], and in the absence of methyl donors, the epigenetic state of Avy is heritable only through the dam [7]). Control populations consisted of mice bred with selection for the pseudoagouti phenotype but without any methyl donor supplementation. In both supplemented and control populations, all genotypes were observed in the expected Mendelian ratios; there was no difference in litter size or breeding performance between the two groups.
(d). Phenotype scoring
Coat colour in Avy mice is tightly linked to the epigenetic state of the Avy allele. Coat colour in Avy/a offspring was assessed at mouse weaning age (21 days) by two trained, independent observers. We used a ranking system whereby coat-colour phenotypes were divided into five categories: full yellow, slightly mottled (5–30% agouti fur), half mottled (30–70% agouti fur), heavily mottled (70–95% agouti fur) or full pseudoagouti; representative phenotypes are illustrated in figure 1a. For data presentation and statistical analysis, phenotypes were divided into two groups: yellow/slightly mottled/half mottled, and heavily mottled/pseudoagouti. The five-category scores from the two observers correlated more than 82 per cent of the time, and the converted two-category scores more than 98 per cent of the time; divergent scores were randomly assigned to one or the other phenotype. For comparison of proportions of phenotypes between treatment groups, we used a chi-square test (α = 0.05).
(e). DNA methylation analysis
Genomic DNA was extracted from tail or fresh frozen whole testis by digestion with Proteinase K and extraction with phenol/chloroform. Two microgram DNA was treated with sodium bisulphite (Epitect kit, Qiagen), and 10 per cent of the sample was used in bisulphite PCR. Bisulphite allelic sequencing of the Avy promoter region (the U3 region of the 5′LTR of the inserted IAP retrotransposon) was performed as previously described [16].
(f). Small RNA sequencing and analysis
Libraries for deep sequencing were prepared from testis total RNA using the small RNA expression kit with barcoded primers (Ambion, as per the Applied Biosystems protocol). A single-emulsion PCR was used to couple the barcoded libraries to P1-coated beads as per the Applied Biosystems protocol. After emulsion PCR, templated beads were enriched in a glycerol gradient and deposited onto the surface of glass slides; sequencing was performed using 35 bp chemistry on a SOLiD machine (version 3.0). Small RNA reads were mapped to the Avy IAP 5′LTR (GenBank AF540972, verified by independent sequencing of the LTR in our laboratory) using CoronaLite allowing a maximum of two colourspace errors. The number of mapped reads from each library was normalized according to the total number of reads from that library that were mappable to the mouse genome (mm9).
3. Results
(a). Multi-generational dietary change combined with selection increases the germline stability of Avy
We exposed Avy mice to excess methyl donors for multiple generations, and coupled this with a selective breeding strategy that favoured those mice that responded to the supplement: in each generation after F0, only pseudoagouti mice were selected for further breeding (figure 1b). We chose male Avy/a mice for breeding (with congenic a/a females) as the Avy allele is sensitive to methyl donors only when inherited from the sire [20].
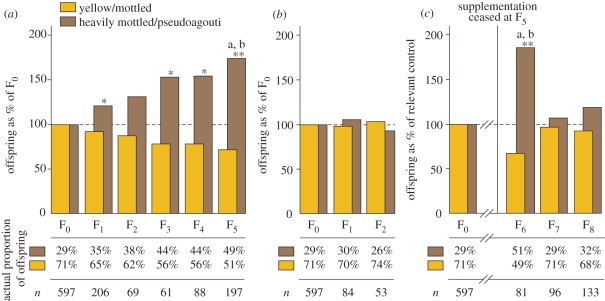
Under these conditions (environmental stimulus, phenotypic selection and genetic homogeneity), we observe that over five generations a progressively larger proportion of mice are pseudoagouti and a smaller proportion are yellow (figure 2a). Selective breeding of pseudoagouti males without supplementation does not lead to any change in the spectrum of offspring phenotypes (figure 2b); conversely, supplementation of an unselected population has previously been found to have no cumulative phenotypic effect [21,22]. Thus, a combination of diet and selection causes the silent state of Avy to become significantly more prevalent in the population.
Figure 2.
Long-term dietary methyl donor supplementation coupled with phenotypic selection increases the proportion of mice with a lean pseudoagouti phenotype. (a) Proportions of phenotypes in each generation during continual methyl donor supplementation. Proportions are expressed as a percentage of the control group (the phenotypic proportions in offspring of unsupplemented Avy/a sires). (b) Proportions of phenotypes in each generation in a population subject to selective breeding but no methyl donor supplementation. The founder population is the same as the founder population in (a). (c) Proportions of phenotypes in each generation after cessation of supplementation. Proportions are expressed as a percentage of the control group (the phenotypic proportions in offspring of unsupplemented Avy/a pseudoagouti dams). n, number of Avy/a offspring; *p < 0.05, **p < 0.005 versus control; ap < 0.05 versus F1; bp < 0.05 versus F2.
(b). The cumulative effect of dietary methyl donors is heritable for only one generation after withdrawal
To determine whether this induced germline stability is maintained when the dietary stimulus is withdrawn, at the fifth generation, we weaned mice onto an unsupplemented diet, and bred pseudoagouti mice for three further generations (figure 1b). Here, we chose Avy/a females for breeding (with a/a males) because we have previously shown that the effect of methyl donors is inherited through the dam [20] and, in the absence of methyl donors, the epigenetic state of Avy is heritable only through the dam [7]. Thus, our strategy maximized the chance of observing sustained inheritance of the cumulative effect. We observed that the phenotypic effects were maintained from F5 to F6, but in subsequent generations (F7 and F8) the phenotypes returned to baseline (figure 2c). Thus, germline stabilization of Avy appears reversible after withdrawal of the supplement, at least after exposure for a limited number of generations.
(c). The cumulative effect of dietary methyl donors is not dependent on DNA methylation
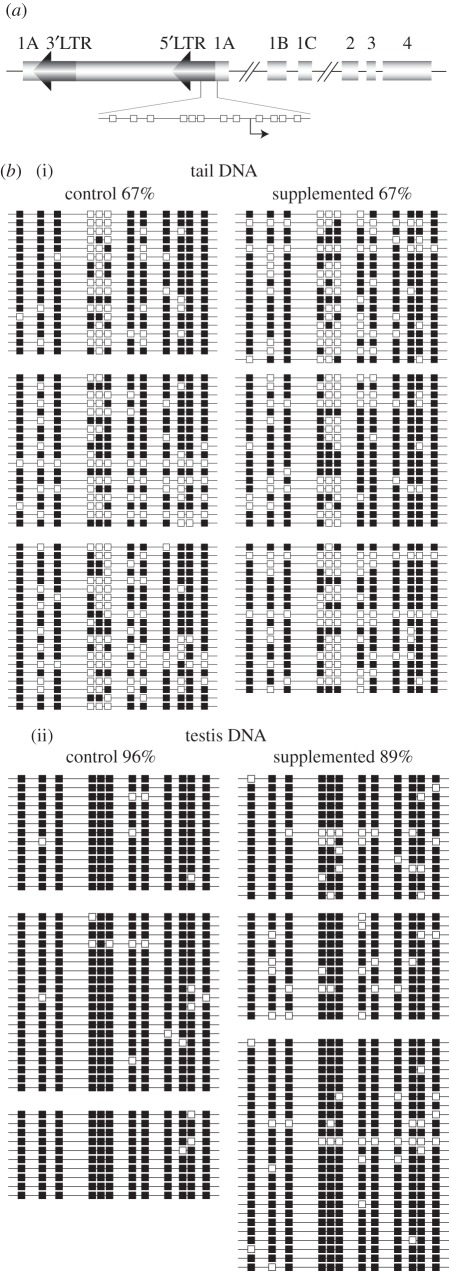
The cumulative and progressive silencing of Avy that we observe with multi-generational methyl donor exposure implies that some factor that promotes Avy silencing is accumulating in the germline over generations. This factor is either an epigenetic modification of the allele, or a molecule that interacts with it to induce an epigenetic change; cytosine methylation is an obvious candidate. We measured methylation around the Avy transcription start site (figure 3a) in unsupplemented pseudoagouti mice and in pseudoagouti mice that had been supplemented for five generations. Consistent with our previous findings [16], we find that DNA from the tails of supplemented pseudoagouti mice carries no more methylation than that from unsupplemented pseudoagouti mice (figure 3b(i)). We then examined Avy methylation in the testis to investigate whether increased DNA methylation in germ cells might be responsible for the increased stability of the Avy epigenotype. However, we found that the Avy promoter is already fully methylated in testes of control mice, and supplementation does not alter this (figure 3b(ii)).
Figure 3.
DNA methylation at the Avy promoter is unchanged by multi-generational methyl donor supplementation. (a) Schematic of the Avy locus showing the region interrogated by allelic bisulphite sequencing, which includes portions of the IAP 5′LTR and pseudoexon 1A. CpG dinucleotides are represented by boxes; the Avy cryptic promoter is marked by an arrow. (b) Bisulphite allelic sequencing profiles from tail (i) and testis (ii) of unsupplemented mice and mice supplemented for five generations. Each line represents an allele, and each box a CpG dinucleotide (white, unmethylated; black, methylated). Each block represents sequences derived from a single mouse.
(d). The Avy allele produces primary piRNAs
piRNAs are 24–30 nt RNAs that complex with members of the Piwi clade of Argonaute proteins to mediate retrotransposon silencing in the animal germline [23]. We considered the possibility that they may be the agent responsible for the cumulative germline stabilization of Avy. We used the SOLiD system (Applied Biosystems) to sequence small RNA libraries from the testes of eight pseudoagouti Avy mice (four supplemented and four unsupplemented), four unsupplemented yellow Avy mice, and an a/a mouse (a/a mice are congenic with Avy/a but do not carry the Avy allele). We obtained between 1.3 and 7.6 million mappable reads from each library (electronic supplementary material, table S1).
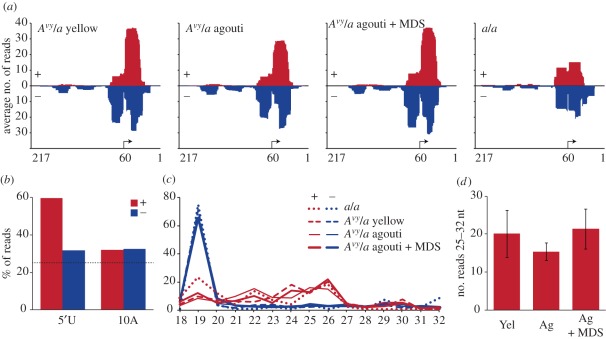
We found small RNAs that mapped to the Avy IAP 5′LTR in both the sense and antisense directions relative to Avy transcription (an average of 37 reads per million mappable reads in Avy/a mice; figure 4a). Far fewer small RNAs from the a/a mouse mapped to the Avy IAP (10 reads per million); as a/a mice do not carry the Avy allele, these RNAs presumably originated from other IAPs in the genome that have sequences very similar to that of the Avy IAP. Such reads will also be present in libraries from the Avy/a mice and it is impossible to distinguish their genomic origin, but the fact that the Avy/a mice have more than three times as many Avy-aligning reads as do congenic a/a mice indicates that most of the piRNAs are derived from the Avy allele itself. In all mice, the RNAs align almost exclusively to the U3 region of the LTR around the start point of Avy transcription. Very few small RNAs align to the R or U5 regions of the LTR, and none mapped to the adjacent pseudoexon 1A. Those RNAs that are sense with respect to the Avy transcript have a length distribution and a 5′U bias consistent with piRNAs, while those derived from the antisense strand are very short (mostly 19 nt) and have no 5′U bias (figure 4b,c).
Figure 4.
piRNAs are generated in the germline by the Avy locus but are unchanged by multigenerational methyl donor supplementation. Small RNA reads generated from the testes of a/a and Avy/a male mice were mapped to the U3 region (131–348 bp) of the 348bp Avy IAP 5′LTR; reads mapping in a sense direction relative to the Avy transcript (antisense to the LTR) are shown in red, and those mapping in an antisense direction relative to the Avy transcript (sense to the LTR) are shown in blue. (a) Alignment maps showing the numbers of reads that map across U3. The Avy cryptic promoter is indicated by an arrow. (b) Base composition of the small RNA reads. piRNAs are characterized by a 5′U; piRNAs generated by the ping–pong mechanism are characterized by a 10 nt A. The dotted line represents the proportion expected by chance. (c) Size distribution of the Avy-derived small RNA reads. (d) The average number of sense pi-sized reads generated from unsupplemented yellow (Yel) and pseudoagouti (Ag), and from fifth-generation supplemented pseudoagouti males (Ag + methyl donor supplementation (MDS)); three independently sequenced in each group. Error bars represent standard error.
Murine piRNAs are generated either as primary piRNAs, derived from the antisense strand of a retrotransposon transcript and predominantly having a 5′U, or secondary piRNAs, derived from sense transcripts by cleavage after recognition by a primary piRNA complexed with MILI (ping–pong amplification) [24]. The small RNAs that align with the Avy promoter show a strong bias for U at the 5′-terminus, but no over-representation of adenine at position 10, indicating that they are primary piRNAs derived from the Avy allele itself, and were not generated by ping–pong amplification (figure 4b). We observed no difference between yellow and pseudoagouti Avy/a mice in the abundance of these Avy-derived piRNA-like small RNAs (figure 4a,d). This is not surprising given that offspring of unsupplemented Avy/a males have the same spectrum of phenotypes regardless of the sire's phenotype [7]. But we also found no difference in the abundance of Avy-derived small RNAs in methyl donor-supplemented pseudoagouti males, making it unlikely that simple abundance of piRNAs underlies the cumulative increase in heritability driven by selection and methyl donor supplementation. While these results do not provide an explanation for the germline effects of methyl donors, they do suggest that the Avy allele is regulated in the male germline by piRNAs derived from the Avy allele itself.
4. Discussion
Our results indicate that multi-generational exposure to a dietary stimulus coupled with selection for a phenotype can progressively increase the prevalence of that phenotype in the population. The effects we observed are not genetic in origin, as all the mice in our experiment were genetically identical. The unlikely possibility that methyl donors created genetic variation that was selected is ruled out by the rapid reversion to the original spectrum of phenotypes upon withdrawal of the dietary stimulus. Thus, the effect we observed is due solely to the interaction of environment and epigenotype, demonstrating that purely epigenetic traits can provide a substrate for selection. This is completely compatible with Darwinian theory, but does not have a genetic basis. Natural selection acts on a phenotype, and an evolutionary response to selection requires that the phenotype is heritable; the Avy allele creates a heritable phenotype, which happens to be determined not by genotype, but by epigenotype.
The Avy epigenotype is not normally heritable when the allele is passed through the male germline [7]; consequently, multi-generational selection alone would not be expected to have an effect on the frequency of phenotypes in the population, consistent with our observations. However, in the presence of the environmental stimulus (methyl donors) the proportions of offspring with a pseudoagouti phenotype steadily increased. This implies that in each generation, the effect of methyl donors was both inherited and augmented. Thus, methyl donors induce epigenetic inheritance of Avy through the male, allowing selection to progressively propagate the silent epigenetic state of Avy through the population.
By selecting pseudoagouti mice for breeding in each generation, we selected mice that carried a silent Avy allele and hence were potential ‘responders’ to the methyl donors. Had we instead bred the whole population without selection, any cumulative effect would necessarily have been diluted, as non-responsive mice would also have been included in the breeding population; indeed, multi-generational methyl donor supplementation without selection has no cumulative effect in laboratory-sized Avy populations [21,22].
Inheritance of the cumulative effect for one generation after withdrawal is consistent with our previous findings [20] and indicates that germ cells exposed to excess methyl donors within the developing F5 females retained a memory of the methyl donor effect, manifested in F6 mice. However, the germ cells that produced F7 mice developed in F6 embryos after methyl supplement withdrawal and displayed no memory of supplementation, indicating that the epigenetic memory was erased at some stage during the life cycle. Thus, the cumulative effect we observe is dependent on the continued presence of excess methyl donors. The observed reversion to baseline may reflect an epigenetic plasticity that could provide an advantageous ability to revert quickly to a former phenotype if environmental conditions were relatively transitory and the new phenotype no longer beneficial, something that could not be accomplished if the basis of the shift were selection for a genetic variant. It is also possible, given the selected mice were those that had potentially responded to the supplement, that the population now has an altered threshold for responding to future supplementation, so that a once-exposed lineage may respond more rapidly to renewed supplementation than a never-exposed lineage.
What is the molecular basis of the cumulative methyl donor effect, and of epigenetic inheritance itself? The increasing penetrance of the pseudoagouti phenotype in each generation indicates a gradual strengthening of epigenetic inheritance over generations, and there must be some physical basis for this effect. We found no changes in DNA methylation at the Avy promoter, nor in the abundance of Avy-derived piRNAs after five generations of supplementation. The lack of change in DNA methylation is consistent with our previous findings in the soma of supplemented mice [16] and is inconsistent with DNA methylation driving either the somatic or germline effects of methyl donors. That the abundance of piRNAs does not change with supplementation indicates that there is probably no simple relationship between the number of piRNAs and the strength of Avy inheritance. However, it may be that intergenerational differences in piRNA abundance are subtle and, given the small number of reads mapping to the Avy locus in all mice, it may be necessary to sequence much more deeply to discern them. Alternatively, given the many biological pathways and biochemical reactions involving methyl donors, there are many other potential molecular mediators of their epigenetic effects. In contrast to the genome, which is a stable molecule that is faithfully replicated and transmitted to succeeding generations, the ‘epigenome’ consists of an extremely complex assortment of proteins and chemical modifications that control the expression of information encoded by the genome. Supplementary methyl donors increase the pool of S-adenosyl methionine, which can donate methyl groups to proteins and other molecules as well as DNA [25,26]. The effects of dietary methyl donors on the Avy allele may well be indirect, mediated by changes in the behaviour of one or more proteins that influence the probability of silencing the allele in the germline or at an early stage of development.
Our data provide evidence for a non-genetic process that can drive adaptive evolution. The idea that epigenetic states might act as a substrate for adaptive evolution has been presented by several authors [2,27–29]. Our data provide probably the first direct evidence that it is possible to select for a trait with a purely epigenetic basis (certainly the first in a mammalian system), and thereby increase its frequency in a population. Our findings are reminiscent of those of Sollars et al. [30] in Drosophila Kruppel mutants, where an environmentally induced trait was selectable; that the trait could be reversed using epigenetically modifying drugs implies that it, too, had an epigenetic basis, although no responsible epiallele was identified.
Are epigenetic processes likely to contribute to adaptive evolution in nature? Although we subjected our mice to ‘artificial’ selection, a wild population of Avy mice would probably be under similar selective pressure, as the yellow obese phenotype would almost certainly be disadvantageous (yellow mice develop type II diabetes and breed poorly). The type of environment we applied in our experiment—a persistent environmental change with selective effects—is common in evolutionary history, with climate change a prominent example. Pervasive climate-associated changes in behaviour have been observed in many species [31], with some fungi exhibiting an entire additional fruiting season [32]; these changes have occurred over time frames that are consistent with an epigenetic, but not a genetic, explanation. Adaptive evolution driven by epigenetic changes may be not only more rapid, but also more flexible than that driven by genetic change. Epigenetic changes in response to an environmental stimulus can occur in multiple individuals at once, facilitating population-wide responses to environmental change with an option for reversion if the environmental stressors do not persist. But epigenetically determined phenotypes may alternatively become fixed in a population via subsequent genetic mutation of methylated cytosines [33]. It remains to be seen whether stably heritable epigenetic processes alone can drive long-term population changes that are maintained independently of the inducing stimulus and without the need for a subsequent genetic change.
Acknowledgements
All animal work was approved by the St Vincents/Garvan Animal Ethics Committee (animal research authorities no. 06/12 and no. 09/12).
This work was supported by the Australian National Health and Medical Research Council (NHMRC) and the Australian Research Council (ARC). The authors wish to thank Stephanie Hackworthy, Paul Young and Christopher Maloney for technical assistance.
References
- 1.Richards E. J. 2008. Population epigenetics. Curr. Opin. Genet. Dev. 18, 221–226 10.1016/j.gde.2008.01.014 (doi:10.1016/j.gde.2008.01.014) [DOI] [PubMed] [Google Scholar]
- 2.Jablonka E., Lamb M. J. 1989. The inheritance of acquired epigenetic variations. J. Theor. Biol. 139, 69–83 10.1016/S0022-5193(89)80058-X (doi:10.1016/S0022-5193(89)80058-X) [DOI] [PubMed] [Google Scholar]
- 3.Henderson I. R., Jacobsen S. E. 2007. Epigenetic inheritance in plants. Nature 447, 418–424 10.1038/nature05917 (doi:10.1038/nature05917) [DOI] [PubMed] [Google Scholar]
- 4.Jablonka E., Raz G. 2009. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q. Rev. Biol. 84, 131–176 10.1086/598822 (doi:10.1086/598822) [DOI] [PubMed] [Google Scholar]
- 5.Cavalli G., Paro R. 1998. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell 93, 505–518 10.1016/S0092-8674(00)81181-2 (doi:10.1016/S0092-8674(00)81181-2) [DOI] [PubMed] [Google Scholar]
- 6.Hitchins M. P., Wong J. J., Suthers G., Suter C. M., Martin D. I., Hawkins N. J., Ward R. L. 2007. Inheritance of a cancer-associated MLH1 germ-line epimutation. N. Engl. J. Med. 356, 697–705 10.1056/NEJMoa064522 (doi:10.1056/NEJMoa064522) [DOI] [PubMed] [Google Scholar]
- 7.Morgan H. D., Sutherland H. G., Martin D. I., Whitelaw E. 1999. Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 23, 314–318 10.1038/15490 (doi:10.1038/15490) [DOI] [PubMed] [Google Scholar]
- 8.Roemer I., Reik W., Dean W., Klose J. 1997. Epigenetic inheritance in the mouse. Curr. Biol. 7, 277–280 [DOI] [PubMed] [Google Scholar]
- 9.Morak M., et al. 2008. Further evidence for heritability of an epimutation in one of 12 cases with MLH1 promoter methylation in blood cells clinically displaying HNPCC. Eur. J. Hum. Genet. 16, 804–811 10.1038/ejhg.2008.25 (doi:10.1038/ejhg.2008.25) [DOI] [PubMed] [Google Scholar]
- 10.Sutherland H. G., Kearns M., Morgan H. D., Headley A. P., Morris C., Martin D. I., Whitelaw E. 2000. Reactivation of heritably silenced gene expression in mice. Mamm. Genome 11, 347–355 10.1007/s003350010066 (doi:10.1007/s003350010066) [DOI] [PubMed] [Google Scholar]
- 11.Ruden D. M., Lu X. 2008. Hsp90 affecting chromatin remodeling might explain transgenerational epigenetic inheritance in Drosophila. Curr. Genomics 9, 500–508 10.2174/138920208786241207 (doi:10.2174/138920208786241207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickies M. M. 1962. A new viable yellow mutation in the house mouse. J. Hered. 53, 84–86 [DOI] [PubMed] [Google Scholar]
- 13.Duhl D. M., Vrieling H., Miller K. A., Wolff G. L., Barsh G. S. 1994. Neomorphic agouti mutations in obese yellow mice. Nat. Genet. 8, 59–65 10.1038/ng0994-59 (doi:10.1038/ng0994-59) [DOI] [PubMed] [Google Scholar]
- 14.Waterland R. A., Jirtle R. L. 2003. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol. Cell Biol. 23, 5293–5300 10.1128/MCB.23.15.5293-5300.2003 (doi:10.1128/MCB.23.15.5293-5300.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolff G. L., Roberts D. W., Mountjoy K. G. 1999. Physiological consequences of ectopic agouti gene expression: the yellow obese mouse syndrome. Physiol. Genomics 1, 151–163 [DOI] [PubMed] [Google Scholar]
- 16.Cropley J. E., Suter C. M., Beckman K. B., Martin D. I. 2010. CpG methylation of a silent controlling element in the murine Avy allele is incomplete and unresponsive to methyl donor supplementation. PLoS ONE 5, e9055. 10.1371/journal.pone.0009055 (doi:10.1371/journal.pone.0009055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jirtle R. L., Skinner M. K. 2007. Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 8, 253–262 10.1038/nrg2045 (doi:10.1038/nrg2045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolff G. L., Kodell R. L., Moore S. R., Cooney C. A. 1998. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 12, 949–957 [PubMed] [Google Scholar]
- 19.Cooney C. A., Dave A. A., Wolff G. L. 2002. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J. Nutr. 132(8 Suppl.), 2393S–2400S [DOI] [PubMed] [Google Scholar]
- 20.Cropley J. E., Suter C. M., Beckman K. B., Martin D. I. 2006. Germ-line epigenetic modification of the murine A vy allele by nutritional supplementation. Proc. Natl Acad. Sci. USA 103, 17 308–17 312 10.1073/pnas.0607090103 (doi:10.1073/pnas.0607090103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waterland R. A., Travisano M., Tahiliani K. G. 2007. Diet-induced hypermethylation at agouti viable yellow is not inherited transgenerationally through the female. FASEB J. 21, 3380–3385 10.1096/fj.07-8229 (doi:10.1096/fj.07-8229) [DOI] [PubMed] [Google Scholar]
- 22.Cropley J. E., Suter C. M., Martin D. I. 2007. Methyl donors change the germline epigenetic state of the A(vy) allele. FASEB J. 21, 3021; author reply 3021–3022 10.1096/07-1002 (doi:10.1096/07-1002) [DOI] [PubMed] [Google Scholar]
- 23.Siomi M. C., Sato K., Pezic D., Aravin A. A. 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 12, 246–258 10.1038/nrm3089 (doi:10.1038/nrm3089) [DOI] [PubMed] [Google Scholar]
- 24.Brennecke J., Aravin A. A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G. J. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103 10.1016/j.cell.2007.01.043 (doi:10.1016/j.cell.2007.01.043) [DOI] [PubMed] [Google Scholar]
- 25.Chiang P. K., Gordon R. K., Tal J., Zeng G. C., Doctor B. P., Pardhasaradhi K., McCann P. P. 1996. S-adenosylmethionine and methylation. FASEB J. 10, 471–480 [PubMed] [Google Scholar]
- 26.Fontecave M., Atta M., Mulliez E. 2004. S-adenosylmethionine: nothing goes to waste. Trends Biochem. Sci. 29, 243–249 10.1016/j.tibs.2004.03.007 (doi:10.1016/j.tibs.2004.03.007) [DOI] [PubMed] [Google Scholar]
- 27.Guerrero-Bosagna C., Sabat P., Valladares L. 2005. Environmental signaling and evolutionary change: can exposure of pregnant mammals to environmental estrogens lead to epigenetically induced evolutionary changes in embryos? Evol. Dev. 7, 341–350 10.1111/j.1525-142X.2005.05033.x (doi:10.1111/j.1525-142X.2005.05033.x) [DOI] [PubMed] [Google Scholar]
- 28.Monk M. 1995. Epigenetic programming of differential gene expression in development and evolution. Dev. Genet. 17, 188–197 10.1002/dvg.1020170303 (doi:10.1002/dvg.1020170303) [DOI] [PubMed] [Google Scholar]
- 29.Richards E. J. 2011. Natural epigenetic variation in plant species: a view from the field. Curr. Opin. Plant Biol. 14, 204–209 10.1016/j.pbi.2011.03.009 (doi:10.1016/j.pbi.2011.03.009) [DOI] [PubMed] [Google Scholar]
- 30.Sollars V., Lu X., Xiao L., Wang X., Garfinkel M. D., Ruden D. M. 2003. Evidence for an epigenetic mechanism by which Hsp90 acts as a capacitor for morphological evolution. Nat. Genet. 33, 70–74 10.1038/ng1067 (doi:10.1038/ng1067) [DOI] [PubMed] [Google Scholar]
- 31.Parmesan C., Yohe G. 2003. A globally coherent fingerprint of climate change impacts across natural systems. Nature 421, 37–42 10.1038/nature01286 (doi:10.1038/nature01286) [DOI] [PubMed] [Google Scholar]
- 32.Gange A. C., Gange E. G., Sparks T. H., Boddy L. 2007. Rapid and recent changes in fungal fruiting patterns. Science 316, 71. 10.1126/science.1137489 (doi:10.1126/science.1137489) [DOI] [PubMed] [Google Scholar]
- 33.Feinberg A. P., Irizarry R. A. 2009. Evolution in health and medicine Sackler colloquium: stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc. Natl Acad. Sci. USA 107, 1757–1764 10.1073/pnas.0906183107 (doi:10.1073/pnas.0906183107) [DOI] [PMC free article] [PubMed] [Google Scholar]