Abstract
A major challenge for social theory is to explain the importance of kin discrimination for the evolution of altruism. One way to assess the importance of kin discrimination is to test its effects on increasing relatedness within groups. The social amoeba Dictyostelium discoideum aggregates to form a fruiting body composed of dead stalk and live spores. Previous studies of a natural population showed that where D. discoideum occurs in the soil, multiple clones are often found in the same small soil samples. However, actual fruiting bodies usually contain only one clone. We here performed experiments to gauge the effect of kin-discriminatory segregation on increasing relatedness. We mixed co-occurring clones from this population using a relatedness level found in small soil samples. We found a lower proportion of uniclonal fruiting bodies and a lower level of relatedness compared with natural fruiting bodies. We found that the amount of relatedness increase attributable to kin-discriminatory segregation was small. These findings suggest a relatively minor influence of kin-discriminatory segregation on relatedness in D. discoideum. We discuss our results comparing with the results of previous studies, including those of wild clones and laboratory mutants. We ask why wild clones of D. discoideum exhibit a low degree of kin-discriminatory segregation, and what alternative factors might account for high relatedness in D. discoideum.
Keywords: population structure, recognition, altruism, Dictyostelium discoideum
1. Introduction
High genetic relatedness within groups coincided with major transitions in evolution [1]. High relatedness is thought to arise in one of two ways: by passive mechanisms of dispersal and viscosity, or by active kin discrimination [2]. The single-celled bottleneck of multi-cellular organisms [1,3] and the single individual, once-mated queen bottleneck of eusocial Hymenoptera are examples of passive mechanisms that facilitated high relatedness within groups [4,5]. Fusion compatibility systems of multi-cellular organisms that fuse somatic tissue [6,7] and colony compatibility systems of mobile eusocial insects [8,9], in contrast, are examples of behaviours that actively enforce high relatedness within groups.
Active kin discrimination may work to either maintain or generate relatedness. In organisms with unitary social development [10], relatedness is initially generated by the dispersal of one or a few individuals followed by social group formation by their descendants and behaviours that maintain colony distinctness, which can depend on kin recognition [6–9]. This form of kin discrimination is found in a number of eusocial insects [9], a number of marine invertebrates [7] and ascomycete fungi [11]. Kin discrimination also occurs in organisms that form social groups by aggregation, such as many mammals and birds [12], anuran amphibian tadpoles [13], fishes [14], sea anemones [7] and social amoebae [6,15,16]. In these organisms, kin discrimination helps generate relatedness by segregating individuals into distinct kin groups [6,7,13–16].
Mechanisms of kin discrimination generating or maintaining relatedness within groups have been studied extensively [7–9,14,16]. Manipulative experiments show that kin discrimination can segregate individuals [7,14,15,17]. Microbes provide an avenue to test the effects of kin discrimination on relatedness [15,17]. In microbes, a surprising level of cooperative behaviour could require high relatedness [18].
We studied the social amoeba Dictyostelium discoideum because data are available on natural population structure [19,20]. Studies of a natural population of D. discoideum near Mountain Lake, VA in October 2000 showed that relatedness among independently living amoebae in small soil samples (approx. 0.20 g) was 0.52 ± 0.01 on average [19]. By contrast, samples taken in October 2004–June 2005 found that relatedness in natural social groups (fruiting bodies) was 0.86–0.99 and 77–92% of fruiting bodies contained only one clone [20]. These latter samples were of fruiting bodies found primarily on deer scat [20].
Previous studies of D. discoideum showed: (i) co-occurring clones typically form chimeras [19,21], (ii) clones from different geographical regions can segregate [16,17], and (iii) variation in segregation correlates with regions of two highly polymorphic cell-adhesion genes [22,23]. In addition, a study of clones from Mountain Lake, VA showed evidence of segregation in three of three chimeric mixtures [17]. However, the overall effect of kin-discriminatory segregation on relatedness among co-occurring clones has not been assessed.
We tested the role of kin-discriminatory segregation on increasing relatedness in D. discoideum fruiting bodies. We studied pairs of clones from this Mountain Lake, VA population [19]. These clones were originally collected by plunging 6 mm diameter soda straws about 5 mm into the soil [19]. Where D. discoideum was found, on average two clones were present, with one to six clones per sample [19]. We mixed clones from the same soil samples [19] because co-occurring clones are likely to encounter each other at the microscale. We mixed clones from different soil samples because mobile animals pass spores through their guts [24], allowing clones that might otherwise inhabit different areas in the soil to encounter each other. In total, we constructed nine mixes of clones isolated from the same soil samples and nine mixes of clones isolated from different soil samples (figure 1), with three replicates per mix. We allowed slugs to migrate through soil, a substrate of natural viscosity that can be conducive for segregation of different clones [25].
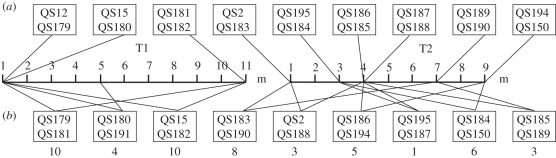
Figure 1.
Pairs of clones used in this study. Clones were taken from two transects taken near Mountain Lake Biological Station, VA [19]. Transect T1 was taken on 25 September 2000 and Transect T2 was taken on 15 October 2000 by A. Fortunato. Transect T2 was reported in a previous study of natural population structure [19]. (a) Millimetre-scale mixes were from the same soda-straw soil sample, which were separated by no more than approximately 20 mm [19]. (b) Metre-scale mixes were clones found metres apart. Numbers below sample names give the distance between clones (m). In part (b), the top clone is the clone to the left of the transect.
We examined a single round of development per replica mix because the estimates from natural fruiting bodies were based on a sampling protocol that did not allow multiple rounds of development [20]. Relatedness estimates of natural fruiting bodies were mostly based on deer scat that underwent 6 days incubation in Petri dishes [20]. Most fruiting bodies thus formed emerged after several days and were preserved in situ until the time of collection [20]. Over such a short time period in a closed container, fruiting bodies did not have time to disperse and go through an additional round of development; yet nonetheless, most fruiting bodies were uniclonal [20]. By examining the effects of a single round of development for a number of mixes (figure 1), we obtained an average value for the effects of segregation behaviour on relatedness relevant to explaining relatedness in these fruiting bodies.
Based on samples taken from a single natural population at different times, we generated a null hypothesis that a single round of development could increase relatedness from 0.50, as found in small soil samples [19], to 0.86, the lower estimate of relatedness of fruiting bodies [20]. A finding that segregation behaviour can explain this discrepancy would not mean that it does, as other factors could explain the difference between soil samples and fruiting bodies in these samples [19,20]. A finding that segregation behaviour does not explain these discrepancies, however, would suggest that it does not. Independent of our null hypothesis, our study also assesses the amount of segregation and the ability to segregate among co-occurring clones, as well as how segregation ability correlates with relatedness between clones. Studies of genetically modified clones or clones from different geographical regions suggest relatively strong effects of segregation [22,23], as well as correlations between segregation and genetic similarity [16], so we also scrutinize these hypotheses as applied to naturally co-occurring clones.
We asked three primary questions. First, we asked whether the proportion of clonal fruiting bodies and relatedness was as high as that found in natural fruiting bodies. Second, we asked what effect kin-discriminatory segregation had on relatedness. Third, we asked whether the degree of segregation correlated with relatedness between clones. Answering these questions helps explain the role of kin-discriminatory segregation in maintaining high relatedness in D. discoideum.
2. Material and methods
(a). Choice of clones
Clones were collected by plunging 6 mm diameter soda straws into soil along two 25 m transects (figure 1). We picked pairs of clones from the same 6 mm scale soil samples distinguishable using one of these three microsatellite loci: Dict13, Dict19 or Dict25 (electronic supplementary material, table S1). We used a minimum 16 bp difference between alleles to prevent overlap of stutter bands. We kept the difference as close to 16 bp as possible (electronic supplementary material, table S2). We used these same clones to construct mixes of clones from different soil samples, found metres apart (figure 1).
(b). Experiment
We collected soil from the first approximately 2.5 cm underneath leaf litter at Mountain Lake, VA. We kept the soil in Ziploc bags in a 5°C refrigerator. Before each experiment, we sieved soil through a 3 mm course wire mesh. We autoclaved and dried the soil at 37°C for 24 h. Immediately prior to the experiment, we weighed the dry soil and added sterile water to bring the moisture content to 60 per cent. This high moisture content ensured sufficient humidity for fruiting.
Prior to each experiment, we grew clones from freezer stocks and collected spores 3 days after fruiting. We counted spores with a haemocytometer and made 105 spore μl−1 spore suspensions for each clone. We mixed 150 μl of this suspension with 150 μl of a concentrated bacterial suspension (1 standard medium plate [20] of Klebsiella aerogenes in 1 ml H2O). We then mixed 125 μl of this spore/bacterial suspension of one clone with the same amount for the other clone, and deposited 200 μl (107 spores) of this chimeric suspension into a 50 ml beaker containing 35 ml of sterile buffered agar (1.98 g KH2PO4, 0.35 g Na2HPO4, 20 g agar per l ddH2O).
We allowed this suspension to dry on the agar for 3 h in a sterile laminar flow hood. We then added 8 g of soil, yielding approximately 1.5 cm depth. To maintain humidity in the beakers while also allowing some airflow, we stretched a small sheet of parafilm across the mouth of the beaker and punched three holes in the sheet. To provide directional light, we wrapped the beakers with foil around the sides and bottom. We then placed the beakers in a 25°C incubator with continuous overhead light.
After 6 days of incubation, we unwrapped the beakers. From each beaker, we haphazardly collected sori of 16 fruiting bodies at the surface of the soil. We collected the sori by touching the head of a sewing pin to each fruiting body. We placed the head of each pin in 25 μl of a 5 per cent chelex suspension in a 96-well plate. We shook the plates on a New Brunswick Scientific C1 Platform Shaker at 40 r.p.m. for 10 min to disperse the spores from the pinheads.
We repeated the experiment in whole on three different days, each time using clones raised from freezer stocks anew. We genotyped fruiting bodies using an established protocol [20] and recorded peak heights of the allele for each clone.
(c). Measurement of relatedness increase
We assessed whether there was one or two clones in a fruiting body by testing for the presence of two microsatellite peaks. For testing the presence of clones, we used microsatellite loci capable of distinguishing the two clones in a particular mix (electronic supplementary material, table S3). We estimated the proportion of clones in a mix by taking the ratio of microsatellite peak heights. We demonstrated the ability of this method to detect rare clones (electronic supplementary material text). We also showed that the amount of variation between fruiting bodies is independent of polymerase chain reaction bias (electronic supplementary material, figure S3). We calculated average relatedness of fruiting bodies within a mix as:
| 2.1 |
Here pi is the proportion of the first clone in the ith sorus (spore head), (1 − pi) the proportion of the second clone in the ith sorus and n the number of sori (D. discoideum has one sorus per fruiting body). Within the ith sorus, pi of the time a spore is the first clone and it is related to pi of the other spores by 1 and the remainder by 0, while (1 − pi) of the time a spore is the second clone and is related to (1 − pi) of the spores by 1 and the remainder by 0. A more complex calculation that took relatedness into account between clones yielded a similar result.
We decomposed R (equation (2.1)) into the starting relatedness among spores, 0.50, plus the relatedness increase caused by the increase of one clone over the other in the overall mix, Rd, plus relatedness increase caused by segregation, Rs. We calculated the component of relatedness caused by increase of one clone relative to the other as:
| 2.2 |
With no segregation behaviour, each fruiting body has the average frequency of the first clone, 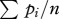 , and the average frequency of the second clone,
, and the average frequency of the second clone, 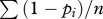 . If pi ≠ 0.5, then some relatedness increase must be caused by the domination of one clone by the other (Rd > 0).
. If pi ≠ 0.5, then some relatedness increase must be caused by the domination of one clone by the other (Rd > 0).
We calculated the component of relatedness caused by segregation within the mix as:
| 2.3 |
Here, R − 0.50 is the increase in total average relatedness and Rd the average relatedness increase caused by domination (equation (2.2)).
Rs correctly measures the amount of increase in relatedness caused by segregation, but this is an imperfect measure of the ability to segregate. The amount of segregation is constrained by the degree of clonal domination which, by increasing relatedness, reduces the amount of relatedness that segregation can increase (electronic supplementary material, figure S4). To measure the ability to segregate better, we estimated how much segregation increased relatedness out of the maximum increase possible after domination:
| 2.4 |
Using Rsp rather than Rs as a dependant variable removes variation in Rd as an explanatory effect. Rsp also puts the ability to segregate on a common scale, for all experiments and clone mixes, from 0 to 1. We discuss these calculations further in the electronic supplementary material, part (d).
We also used the arcsine square-root transformed proportions of clones within fruiting bodies as a dependant variable [16,17]. Arcsine square-root transformed proportion gives an estimate of segregation independent of the calculation of increase in relatedness. Similar to Rsp, arcsine square-root transformed proportion is independent of the mean proportion [17].
(d). Testing for an effect of relatedness between clones on the extent of segregation
We genotyped each clone for 16 variable polymorphic microsatellite loci from all six D. discoideum chromosomes (electronic supplementary material, table S1). We scored peak locations using Genotyper (Applied Biosystems) software. We calculated pairwise symmetric relatedness using Relatedness v. 5.0.8 (http://www.gsoftnet.us/GSoft.html), weighting loci equally and jackknifing across loci [26]. We used the clones in the transect as the reference population for comparing allele frequencies (electronic supplementary material, table S2).
(e). Statistical analyses
We performed all statistical analysis in JMP v. 7.0.2. To test for a correlation of relatedness between clones and the amount of segregation, we used a two-way ANCOVA model with day and soil sample (same or different) as nominal factors and relatedness as covariate. Three replicates were excluded either because no fruiting bodies were found or only one clone was present (QS2 × QS183 on day 1, QS181 × QS179 on day 2 and QS186 × QS194 on day 3). The dependant variable was either Box–Cox-transformed Rsp or Box–Cox-transformed arcsine square-root transformed proportion. Neither parameter was distributed in a manner significantly different from normal (Shapiro–Wilks W = 0.98, p = 0.51 and W = 0.99, p = 0.85, respectively; n = 51 each).
(f). Control for random variation between genotyping wells
We measured Rs in mixes of spores that did not germinate or go through a round of development. Because spores are immobile and cannot segregate, these control mixes give an estimate of variance in proportion caused by the genotyping process. We report a comparison with the 50 : 50 control mixes in figure 3. The electronic supplementary material, part (a) gives details on how these mixes were constructed.
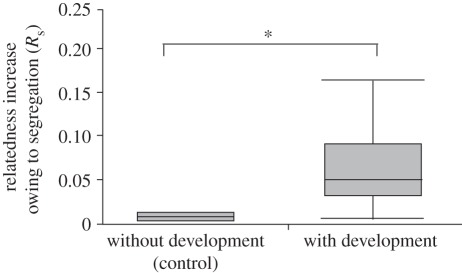
Figure 3.
Relatedness increase caused by segregation (Rs) in 50 : 50 control mixes without vegetative growth or development was significantly less than experimental mixes with vegetative growth and development (Wilcoxon rank-sum test, n = 18, Z = 5.1; *p < 0.0001).
3. Results
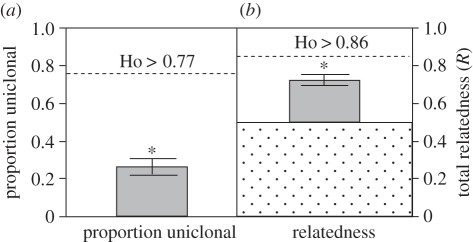
Out of 1047 fruiting bodies genotyped from three experimental days (14.6 ± 0.25 s.e. fruiting bodies genotyped per mix), 264 (25.2%) exhibited a single microsatellite peak characteristic of a uniclonal fruiting body and 783 (74.8%) exhibited two peaks characteristic of a chimeric fruiting body (D. discoideum is haploid). Averaging across the three experimental days, we found that 27.1 ± 0.05% s.e. of fruiting bodies were uniclonal and relatedness within fruiting bodies was 0.73 ± 0.02 s.e. The proportion of uniclonal fruiting bodies was significantly less than the lower 95% confidence interval (CI) estimate of 77 per cent from the natural population (one-tailed t-test, n = 18, p = 0.0001; figure 2a). The average relatedness within fruiting bodies was significantly less than the lower 95% CI estimate of 0.86 from the natural population (one-tailed t-test, n = 18, p < 0.0001; figure 2b). We used one-tailed tests because we made comparisons with the lower estimates in the natural population [20].
Figure 2.
Comparison of the proportion of uniclonal fruiting bodies and relatedness found in our study to natural fruiting bodies [20]. (a) Proportion of uniclonal fruiting bodies in the experimental mixes was significantly lower than the lowest estimate of 0.77 of uniclonal fruiting bodies found in the natural population (one-tailed t-test, n = 18, p < 0.0001). (b) Relatedness within fruiting bodies was significantly lower than the lowest estimate of 0.86 found in the natural population (one-tailed t-test, n = 18, p < 0.0001). Clones used were from soil samples [19] (figure 1), rather than scat samples [20]. Relatedness started at 0.50 among spores because of equal mixture; *p < 0.0001. See text for details.
We found a significant increase in relatedness caused by segregation (Rs) compared with the control mix. The median relatedness increase for the experimental mixes was 0.049 (quartiles 0.03, 0.09) compared with 0.001 for the control mix (quartiles 0.0004, 0.003). The median of the distribution for the experimental mixes was significantly different from the control mix (Wilcoxon rank-sum test, n = 18, Z = 5.1, p < 0.0001; figure 3). We also found a median increase of possible relatedness, Rsp, for the experimental mixes of 0.144 (quartiles 0.094, 0.26) and a median arcsine square-root transformed proportion for the experimental mixes of 0.04 rad2 (quartiles 0.10, 0.03) [17] or 147 deg2 (quartiles 329, 96) [16].
We found that 11 clones in our sample were significantly negatively related, and four clones were significantly positively related (the 95% CI for relatedness did not overlap zero; electronic supplementary material, table S3). Overall, the median relatedness among clones was −0.11 (quartiles 0.18, 0.088), which was not significantly different from zero (Wilcoxon signed-rank test, n = 18, W = −24.5, p = 0.30). We found no significant effect of the relatedness between clones, the experimental day or whether clones were from the same or different soil samples on Box–Cox-transformed Rsp or Box–Cox-transformed arcsine square-root transformed proportion (table 1).
Table 1.
Results of a two-way ANCOVA model with Box–Cox-transformed relatedness increase of possible (Rsp) or Box–Cox-transformed arcsine square-root transformed proportion as the response variable and relatedness between clones as the covariate. (Day of experiment and whether clones were from the same or different soil samples were nominal effects. Neither relatedness between clones, day of experiment or whether clones were from the same soil samples were significant predictors of Box–Cox-transformed Rsp (whole model r2 = 0.12, n = 51) or Box–Cox-transformed arcsine square-root transformed proportion (whole model r2 = 0.14, n = 51).)
| source | d.f. |
Rsp |
arcsine-transformed proportion |
||
|---|---|---|---|---|---|
| F | p | F | p | ||
| day | 2 | 0.2376 | 0.7895 | 0.4661 | 0.6304 |
| relatedness | 1 | 1.5139 | 0.2248 | 2.1802 | 0.1466 |
| soil sample | 1 | 3.6069 | 0.0638 | 3.5308 | 0.0666 |
| error | 46 | ||||
4. Discussion
Previous research on the social amoeba D. discoideum showed evidence of kin-discriminatory segregation, but the overall effects of segregation on relatedness remained unclear [16,17,22]. We asked whether kin discrimination can account for high relatedness found in natural fruiting bodies [20]. We found that kin-discriminatory segregation alone did not explain the proportion of uniclonal fruiting bodies or relatedness found in natural fruiting bodies (figure 2). Furthermore, kin-discriminatory segregation produced only a modest relatedness increase (figure 3). These findings raise questions of the adaptive basis of kin-discriminatory segregation in D. discoideum and suggest that factors other than kin-discriminatory segregation maintain high relatedness in this species.
Our findings are consistent with previous studies of wild clones. Studies of naturally co-occurring clones, including those from North Carolina [21] and Virginia [19], found that different clones usually form chimeras. However, these studies did not measure the extent of segregation within fruiting bodies. Ostrowski et al. [16] mixed laboratory strain AX4, originally isolated from North Carolina, with 14 different wild clones, and found a degree of segregation of 33–671 deg2 compared with 27–804 deg2 found here (arcsine square-root transformed proportion; electronic supplementary material, figure S5). Flowers et al. [17] found a degree of segregation of approximately 0.15 rad2 or lower, comparable with our result of 0.24 rad2 or lower here (arcsine square root-transformed proportion).
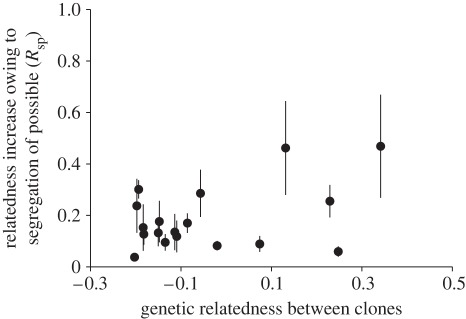
In contrast to most previous studies (but see Strassmann et al. [21]), we raised clones together from spore stage. If amoebae are like animals and learn cues from their neighbours [2,7,12,14], different clones should not segregate when raised together. Our finding of segregation (figure 3) suggests that the ability to perceive cues is innate. This is consistent with a model of kin recognition based on heterophilic cell-adhesion genes [22,23]. However, our findings are inconsistent with precise segregation based on hypervariable recognition genes [22,23]. Under such a model, unrelated clones should typically segregate into distinct, uniclonal fruiting bodies [22,23]. By contrast, we found unrelated clones usually did not segregate well (figure 2). Furthermore, we did not find a correlation between genetic relatedness and segregation ability (figure 4). Nonetheless, we did occasionally find uniclonal fruiting bodies of both clones in a mix, suggesting that perfect segregation behaviour is possible (electronic supplementary material, table S4).
Figure 4.
Relatedness increase caused by segregation of possible (Rsp) against the pairwise relatedness values between clones. Pairwise relatedness was calculated based on 16 polymorphic microsatellite loci (electronic supplementary material, table S2). Bars give s.e. of the three replicate mix experiments.
Our findings raise the question of why wild D. discoideum clones sometimes segregate, and bear variable cell-adhesion genes [22,23], if segregation behaviour is typically weak. One hypothesis is that kin-discriminatory segregation is not adaptive for this purpose. In jawed vertebrates, immunological behaviours reject allografts from non-relatives based on diverse major histocompatibility complex (MHC) genes [27]. However, it has not been conclusively established that MHC diversity evolves for rejecting tissue grafts [27], and MHC diversity is generally thought to evolve to evade parasites or promote outbreeding [28,29]. By analogy with D. discoideum, it remains possible that segregation behaviour and polymorphisms of cell-adhesion genes are maintained for other reasons. Alternatively, kin-discriminatory segregation could be adaptive. This could be the case if the costs of chimerism outweigh the benefits [7,30].
Our results draw a contrast with a previous study of Dictyostelium purpureum, which found stronger segregation [15]. Median relatedness increase caused by segregation in D. purpureum, Rs, was 0.26 (quartiles 0.07, 0.37) and median relatedness increase caused by segregation of possible Rsp, was 0.64 (quartiles 0.38, 0.82) (calculated from Mehdiabadi et al. [15]). One hypothesis to explain a lower level of segregation in D. discoideum is a stronger selective pressure for segregation in D. purpureum. In contrast to D. discoideum, D. purpureum can invest a higher proportion of cells in stalk [31]. In animals, greater investments in cooperation correlate with greater reductions of cooperative behaviour under low relatedness [12]. Less cooperation under low relatedness could produce greater costs of chimerism in D. purpureum, which would be expected to favour kin-discriminatory segregation [6,7].
What causes high relatedness in D. discoideum, if not kin-discriminatory segregation? The most obvious factor is passive, microscale population structure. The conventional explanation for passive population structure is limited dispersal, which is somewhat a misnomer because relatedness usually depends on a dispersal or global mixing stage [2]. With effective dispersal, relatedness is generated easily by mechanisms that increase local-scale population structure [2]. In D. discoideum, local-scale population structure could be enhanced by several factors. If resources are patchy and ephemeral, then the first clone to colonize a resource may expand in population and prevent other clones from colonizing the resource [32]. Alternatively, extended vegetative competition could decrease local-scale genetic diversity, as is found to occur in laboratory populations of D. discoideum [33]. Lineages might also segregate during vegetative growth as populations expand into a resource, similar to the segregation viewed in bacterial and yeast populations during population expansion [34]. Each of these mechanisms could help generate local-scale genetic population structure, which combined with spore dispersal and more global mixing, would combine to produce high relatedness [2].
In conclusion, we found that kin-discriminatory segregation in D. discoideum had only a small effect on relatedness. The amount of relatedness increase viewed in our study was not sufficient to account for the discrepancy between small soil samples and actual fruiting bodies in nature. Furthermore, the relatedness between clones did not explain variation in the degree of segregation behaviour. These findings question the adaptive value of kin-discriminatory segregation in D. discoideum and suggest that alternative factors may be important for maintaining very high relatedness in this species.
Acknowledgements
We thank members of the Strassmann and Queller laboratory for discussions and two anonymous reviewers for comments. This material is based upon work supported by the National Science Foundation under grant numbers DEB0816690 and DEB0918931 to D.C.Q. and J.E.S. O.M.G. was supported by a Wray-Todd graduate fellowship.
References
- 1.Maynard Smith J., Szathmáry E. 1995. The major transitions in evolution. Oxford, UK: W. H. Freeman [Google Scholar]
- 2.West S. A., Griffin A. S., Gardner A. 2007. Evolutionary explanations for cooperation. Curr. Biol. 17, R661–R672 10.1016/j.cub.2007.06.004 (doi:10.1016/j.cub.2007.06.004) [DOI] [PubMed] [Google Scholar]
- 3.Grosberg R. K., Strathmann R. R. 1998. One cell, two cell, red cell, blue cell: the persistence of a unicellular stage in multicellular life histories. Trends Ecol. Evol. 3, 112–116 10.1016/S0169-5347(97)01313-X (doi:10.1016/S0169-5347(97)01313-X) [DOI] [PubMed] [Google Scholar]
- 4.Hughes W. O., Oldroyd B. P., Beekman M., Ratnieks F. L. 2008. Ancestral monogamy shows kin selection is key to the evolution of eusociality. Science 320, 1213–1216 10.1126/science.1156108 (doi:10.1126/science.1156108) [DOI] [PubMed] [Google Scholar]
- 5.Boomsma J. J. 2009. Lifetime monogamy and the evolution of eusociality. Phil. Trans. R. Soc. B 364, 3191–3207 10.1098/rstb.2009.0101 (doi:10.1098/rstb.2009.0101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buss L. W. 1982. Somatic cell parasitism and the evolution of somatic tissue compatibility. Proc. Natl Acad. Sci. USA 79, 5337–5341 10.1073/pnas.79.17.5337 (doi:10.1073/pnas.79.17.5337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grosberg R. K. 1988. The evolution of allorecognition specificity in clonal invertebrates. Q. Rev. Biol. 63, 377–412 10.1086/416026 (doi:10.1086/416026) [DOI] [Google Scholar]
- 8.Crozier R. H., Dix M. W. 1979. Analysis of two genetic models for the innate components of colony odor in social Hymenoptera. Behav. Ecol. Sociobiol. 4, 217–224 10.1007/BF00297645 (doi:10.1007/BF00297645) [DOI] [Google Scholar]
- 9.Ratnieks F. L. W. 1991. The evolution of genetic odor-cue diversity in social Hymenoptera. Am. Nat. 137, 202–226 10.1086/285154 (doi:10.1086/285154) [DOI] [Google Scholar]
- 10.Queller D. C. 2000. Relatedness and the fraternal major transitions. Phil. Trans. R. Soc. B 355, 1647–1655 10.1098/rstb.2000.0727 (doi:10.1098/rstb.2000.0727) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saupe S. J. 2000. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 64, 489. 10.1128/MMBR.64.3.489-502.2000 (doi:10.1128/MMBR.64.3.489-502.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornwallis C. K., West S. A., Griffin A. S. 2009. Routes to indirect fitness in cooperatively breeding vertebrates: kin discrimination and limited dispersal. J. Evol. Biol. 22, 2445–2457 10.1111/j.1420-9101.2009.01853.x (doi:10.1111/j.1420-9101.2009.01853.x) [DOI] [PubMed] [Google Scholar]
- 13.Ward A. J. W., Hart P. J. B. 2003. The effects of kin and familiarity on interactions between fish. Fish Fish. 4, 348–358 10.1046/j.1467-2979.2003.00135.x (doi:10.1046/j.1467-2979.2003.00135.x) [DOI] [Google Scholar]
- 14.Blaustein A. R., Waldman B. 1992. Kin recognition in anuran amphibians. Anim. Behav. 44, 207–221 10.1016/0003-3472(92)90027-7 (doi:10.1016/0003-3472(92)90027-7) [DOI] [Google Scholar]
- 15.Mehdiabadi N. J., Jack C. N., Farnham T. T., Platt T. G., Kalla S. E., Shaulsky G., Queller D. C., Strassmann J. E. 2006. Social evolution: kin preference in a social microbe. Nature 442, 881–882 10.1038/442881a (doi:10.1038/442881a) [DOI] [PubMed] [Google Scholar]
- 16.Ostrowski E. A., Katoh M., Shaulsky G., Queller D. C., Strassmann J. E. 2008. Kin discrimination increases with genetic distance in a social amoeba. PLoS Biol. 6, e287. 10.1371/journal.pbio.0060287 (doi:10.1371/journal.pbio.0060287) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flowers J. M., Li S. I., Stathos A., Saxer G., Ostrowski E. A., Queller D. C., Strassmann J. E., Purugganan M. D. 2010. Variation, sex, and social cooperation: molecular population genetics of the social amoeba Dictyostelium discoideum. PLoS Genet. 6, e1001013. 10.1371/journal.pgen.1001013 (doi:10.1371/journal.pgen.1001013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foster K. R. 2010. Social behavior in microorganisms. In Social behaviour: genes, ecology and evolution (eds Szekely T., Moore A. J., Komdeur J.), pp. 331–356 Cambridge, UK: Cambridge University Press [Google Scholar]
- 19.Fortunato A., Strassmann J. E., Santorelli L., Queller D. C. 2003. Co-occurrence in nature of different clones of the social amoeba, Dictyostelium discoideum. Mol. Ecol. 12, 1031–1038 10.1046/j.1365-294X.2003.01792.x (doi:10.1046/j.1365-294X.2003.01792.x) [DOI] [PubMed] [Google Scholar]
- 20.Gilbert O. M., Foster K. R., Mehdiabadi N. J., Strassmann J. E., Queller D. C. 2007. High relatedness maintains multicellular cooperation in a social amoeba by controlling cheater mutants. Proc. Natl Acad. Sci. USA 104, 8913–8917 10.1073/pnas.0702723104 (doi:10.1073/pnas.0702723104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strassmann J. E., Zhu Y., Queller D. C. 2000. Altruism and social cheating in the social amoeba Dictyostelium discoideum. Nature 408, 965–967 10.1038/35050087 (doi:10.1038/35050087) [DOI] [PubMed] [Google Scholar]
- 22.Benabentos R., et al. 2009. Polymorphic members of the lag gene family mediate kin discrimination in Dictyostelium. Curr. Biol. 19, 567–572 10.1016/j.cub.2009.02.037 (doi:10.1016/j.cub.2009.02.037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirose S., Benabentos R., Ho H., Kuspa A., Shaulsky G. 2011. Self-recognition in social amoebae is mediated by allelic pairs of tiger genes. Science 333, 467–470 10.1126/science.1203903 (doi:10.1126/science.1203903) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevenson S. L., Landolt J. C. 1992. Vertebrates as vectors of cellular slime moulds in temperate forests. Mycol. Res. 96, 670–672 10.1016/S0953-7562(09)80495-4 (doi:10.1016/S0953-7562(09)80495-4) [DOI] [Google Scholar]
- 25.Queller D. C., Ponte E., Bozzaro S., Strassmann J. E. 2003. Single-gene greenbeard effects in the social amoeba, Dictyostelium discoideum. Science 299, 105–106 10.1126/science.1077742 (doi:10.1126/science.1077742) [DOI] [PubMed] [Google Scholar]
- 26.Queller D. C., Goodnight K. F. 1989. Estimating relatedness using genetic markers. Evolution 43, 258–275 10.2307/2409206 (doi:10.2307/2409206) [DOI] [PubMed] [Google Scholar]
- 27.Klein J. 1987. Natural history of the major histocompatibility complex. New York, NY: Wiley [Google Scholar]
- 28.Penn D. J., Potts W. K. 1999. The evolution of mating preferences and major histocompatibility complex genes. Am. Nat. 153, 145–164 10.1086/303166 (doi:10.1086/303166) [DOI] [PubMed] [Google Scholar]
- 29.Piertney S. B., Oliver M. K. 2006. The evolutionary ecology of the major histocompatibility complex. Heredity 96, 7–21 10.1038/sj.hdy.6800724 (doi:10.1038/sj.hdy.6800724) [DOI] [PubMed] [Google Scholar]
- 30.Foster K. R., Fortunato A., Strassmann J. E., Queller D. C. 2002. The costs and benefits of being a chimera. Proc. R. Soc. Lond. B 269, 2357–2362 10.1098/rspb.2002.2163 (doi:10.1098/rspb.2002.2163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raper K. B. 1984. The dictyostelids. Princeton, NJ: Princeton University Press [Google Scholar]
- 32.Gilbert O. M., Queller D. C., Strassmann J. E. 2009. Discovery of a large clonal patch of a social amoeba: implications for social evolution. Mol. Ecol. 18, 1273–1281 10.1111/j.1365-294X.2009.04108.x (doi:10.1111/j.1365-294X.2009.04108.x) [DOI] [PubMed] [Google Scholar]
- 33.Saxer G., Brock D. A., Queller D. C., Strassmann J. E. 2010. Cheating does not explain selective differences at high and low relatedness in a social amoeba. BMC Evol. Biol. 10, 5–10 10.1186/1471-2148-10-5 (doi:10.1186/1471-2148-10-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hallatschek O., Hersen P., Ramanathan S., Nelson D. R. 2007. Genetic drift at expanding frontiers promotes gene segregation. Proc. Natl Acad. Sci. USA 104, 19 926–19 930 10.1073/pnas.0710150104 (doi:10.1073/pnas.0710150104) [DOI] [PMC free article] [PubMed] [Google Scholar]