Abstract
Colonies of eusocial Hymenoptera, such as ants, bees and wasps, have long been recognized as candidates for the study of genomic imprinting on the grounds of evolutionary conflicts that arise from close interactions among colony members and relatedness asymmetry owing to haplodiploidy. Although a general kinship theory of genomic imprinting predicts its occurrence under various circumstances of the colony life cycle, new theoretical approaches are required to account for the specifics of real colonies based on recent advances in molecular-level understanding of ants and honeybees. Using a multivariate quantitative genetic model, we examined the potential impact of genomic imprinting on genes that determine the carrier female's propensity to develop into the queen caste. When queen overproduction owing to the increased propensity comes at a colony-level cost, the conflict between maternally and paternally inherited genes in polyandrous (queen multiple mating) colonies favours genomic imprinting. Moreover, we show that the genomic imprinting can occur even under monandry (queen single mating), once incorporating the costs differentially experienced by new males and new queens. Our model predicts the existence of imprinted ‘genetic royal cheats’ with patriline-specific expression in polyandrous colonies, and seems consistent with the paternal effect on queen determination in monandrous Argentine ants.
Keywords: DNA methylation, genetic caste determination, intralocus sexual conflict, polyandry, social hybridogenesis, tragedy of the commons
1. Introduction
The evolution of organismal complexity is characterized by transitions that lead independent, self-replicating molecules towards being members of a higher unit of replication, such as chromosomes, sexes, eukaryotic cells, multi-cellular organisms and eusocial colonies (also known as major evolutionary transitions [1]). Once such a transition has occurred, members of the higher unit usually show extreme cohesiveness, as long as they share a common fitness interest by replicating themselves together. However, when another path of fitness acquisition becomes available for limited members of the unit, evolutionary conflict arises between these members and the others over the use of different paths. Examples include various selfish genetic elements in eukaryotes [2], male–female antagonism in sexual reproduction [3] and queen–worker conflict in eusocial insects [4].
Genomic imprinting is an epigenetic phenomenon considered to be a possible mechanism that provides a new path of fitness acquisition available only to one sex in the system of sexual reproduction (reviewed in the study of Burt & Trivers [2]). Genomic imprinting involves sex-specific differential modifications of DNA sequences during germline formation at each generation; these modifications are inherited by somatic cells of offspring and result in parent-of-origin-specific silencing or upregulation of allele expression. When fitness depends on the level of gene expression, allele-specific expression regulation provides an incentive for each allele to modify its expression differently so as to maximize its fitness. Haig's kinship theory of genomic imprinting [5,6] provides a theoretical framework that can explain genomic imprinting as a result of conflict between paternally and maternally inherited alleles over the offspring's trait. A prime example is genes involved in sibling competition for resource acquisition from their mother. When the diploid mother mates with multiple diploid males, an allele in the sibling derived from her mate (the ‘patrigene’ hereafter [7]) must compete with those in the other siblings who share the same allele with a probability less than 1/2, whereas an allele in the sibling derived from the mother (‘matrigene’) must not do so because the cost of competition falls upon those in the other siblings who share the same allele with a probability of 1/2. Once available to the focal genes, genomic imprinting is predicted to elicit specific expression of patrigenes and silencing of matrigenes. The expression patterns of several mammalian embryonic genes (e.g. Igf2) are consistent with this prediction of kinship theory [8].
In the example just given, close interaction between half-siblings and relatedness asymmetry from the standpoint of alleles underlie the evolution of genomic imprinting. A similar situation is expected in eusocial hymenopteran colonies, representing another major evolutionary transition. Eusocial Hymenoptera represents one of the best studied examples of evolutionary conflict, which arises from social interaction among family members and relatedness asymmetry owing to haplodiploidy (reviewed in the work of Ratnieks et al. [9]). As for the capability of epigenetic modification, it has recently been shown that DNA methylation occurs widely to various extents among ants, bees and wasps [10]. Moreover, emerging data from whole-genome sequences of the honeybee (Apis mellifera) and several ant species indicate that a DNA methylation toolkit is conserved across eusocial Hymenoptera [11–17]. It has long been recognized that the kinship theory of genomic imprinting might be applicable to various conflicts in eusocial Hymenoptera, including those over sex allocation, reproductive division of labour, brood rearing, policing, nest-mate discrimination and dispersal [2,6,7,18–21]. Although direct evidence is lacking, some studies suggested the occurrence of genomic imprinting in eusocial Hymenoptera by showing paternal effects on worker defensive behaviour in honeybees [22] and on queen caste determination in Argentine ants (Linepithema humile) [23].
In this study, we investigate the potential impact of genomic imprinting on the gene(s) that favours the immature carrier female developing into a queen in eusocial Hymenoptera (hereafter referred to as ‘queen gene imprinting’). Queens are generally larger than workers and consume more resources during their development. Given that resources are limited for the colony, a queen-destined female confronts costly competition with other queen-destined females for the limited resources, making the situation analogous to sibling competition for resource acquisition from the mother. Moreover, when there is a resource allocation trade-off between queen and worker production in the colony, queen overproduction should impose a cost on colony productivity owing to a decrease in the workforce. In social evolution theory, these circumstances are predisposed to the ‘tragedy of the commons’ [24] that is suffered by all siblings.
We used a formal quantitative genetic model of genomic imprinting originally developed by Mochizuki et al. [25] for the analysis of embryonic gene expression. This model allows for rigorous assessment of the feasibility of the kinship (or inclusive-fitness-based) theory of genomic imprinting. By modifying the model to incorporate haplodiploidy and eusociality, we first show that polyandry (multiple mating by the queen) leads to queen gene imprinting, in which the patrigene is expressed and the matrigene is silenced. By taking into account the differential cost of the tragedy of the commons with respect to sex of the new reproductive offspring, we identify the conditions under which queen gene imprinting with patrigene-specific expression evolves even under monandry (single mating by the queen), and those under which matrigene-specific expression evolves instead. Finally, we discuss how recent findings of the heritable components of queen caste determination in social bees and ants may relate to our model.
2. The model
A eusocial hymenopteran colony is assumed to be headed by N unrelated queens, each of which mated with P unrelated males. The mother queen produces F female and M male eggs throughout her life, and workers are assumed to be completely sterile. During her growth, a female takes the queen developmental pathway with a probability Q (0 ≤ Q ≤ 1). A colony-mean value of the probability Q represents the fraction of queens among female offspring produced in the colony. Then we consider a gene (or a set of genes) whose expression level contributes exclusively to Q in a monotonically increasing manner from the origin. The gene is assumed to be susceptible to imprinting (i.e. has an allele-dependent differential capacity to be epigenetically modified), and thus the expression of alleles can contribute to Q at a different extent depending on its parent of origin at each generation. Then the trait of the focal gene is characterized by a pair of values: the contributions when transmitted maternally (matrigene) and paternally (patrigene). Furthermore, matrigene and patrigene of the focal gene are assumed to contribute additively to Q in diploid females. Denoting the population-mean traits of matrigene and patrigene by 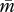 and
and 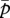 , respectively, the expected Q as a function of
, respectively, the expected Q as a function of 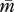 and
and 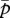 becomes
becomes
| 2.1 |
Mutations that alter the contribution of matrigenes to Q are assumed not to affect that of patrigenes, and vice versa. Denoting the mutant traits of matrigene and patrigene by m and p, respectively, the expected Q of a female that inherits a rare mutant matrigene becomes 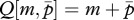 and that of one who inherits a rare mutant patrigene becomes
and that of one who inherits a rare mutant patrigene becomes 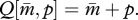 Because 0 ≤ Q ≤ 1, the ranges of m and p are constrained to fall in 0 ≤ m ≤ 1, 0 ≤ p ≤ 1 and 0 ≤ (m + p) ≤ 1 for both residents and mutants.
Because 0 ≤ Q ≤ 1, the ranges of m and p are constrained to fall in 0 ≤ m ≤ 1, 0 ≤ p ≤ 1 and 0 ≤ (m + p) ≤ 1 for both residents and mutants.
(a). Fitness functions
The increased Q may impose a cost on the productivity of the whole colony in the form of the tragedy of the commons (see §1), represented by the decreased survival of new reproductive individuals. We express the decreasing effect as a power of (1 − Q), which is a straightforward representation of a fraction of workers among female offspring. After the tragedy, the numbers of new queens and new males, respectively, from the resident colony with the population-mean traits become
| 2.2a |
and
| 2.2b |
where γ and δ are non-negative power indices that determine how the survival of the new reproductive individuals decreases by increasing Q (i.e. =0, unaffected; <1, convex; =1, linear; >1, concave). The tragedy of the commons inflicted by male offspring would be more likely to result from reduced workforce than from competition between new queens.
In the present model, fitness is defined as the multiplication rate of a gamete produced by the parental generation to the gametic phase of the offspring generation. In the haplodiploid sex determination system, an allele in a parent has the following three inheritance routes: (I) mother to daughter as matrigene, (II) mother to son as matrigene, and (III) father to daughter as patrigene, denoted by φI, φII and φIII, respectively. Thus, using formula (2.2a,b), the parent-of-origin-dependent fitness functions of a mutant allele invaded in a resident colony become
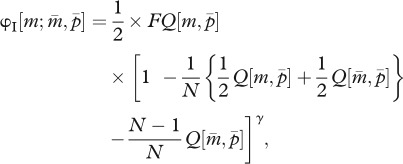 |
2.3a |
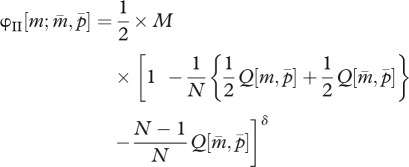 |
2.3b |
and
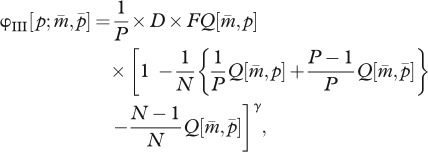 |
2.3c |
where D denotes the number of queens a male mates with. In the right-hand sides of these equations, the factors 1/2 and 1/P in the first part indicate that the fitness is evaluated at the gametic level, and the last part corresponds to the cost of the tragedy of the commons (or a power of worker fraction in the mutant-invaded colony) when one of the N mother queens harbours a mutant allele as a heterozygote (equation (2.3a,b)) and when one of the N mother queens has mated with P males, one of which had a mutant allele (equation (2.3c)).
(b). Evolutionary dynamics
We use a multivariate quantitative genetic model that was originally developed to study sexual selection [26], and has been applied to the analyses of genomic imprinting of mammalian embryos [25,27] and social conflict [28]. The model assumes weak selection acting on the traits considered but does not require normality of the distribution of genetic values. In the following analysis, no genetic covariance is assumed to exist between m and p. Calculating per-generation changes in population-mean traits 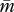 and
and 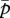 caused by selection on genetic variances of patrigenes and matrigenes, with relevant weights for the haplodiploid system (2/3 for females and 1/3 for males, which represents the sex-specific reproductive value), yields the following equations:
caused by selection on genetic variances of patrigenes and matrigenes, with relevant weights for the haplodiploid system (2/3 for females and 1/3 for males, which represents the sex-specific reproductive value), yields the following equations:
| 2.4a |
and
| 2.4b |
where the factor 1/3 corresponds to the scale of selection at the allelic level, Gm and Gp denote additive genetic variances of the traits m and p, and βm and βp are selection gradients acting on m and p. Using the fitness functions defined in equation (2.3a–c) and keeping in mind that φI and φII are independent of p and that φIII is independent of m, the selection gradients βm and βp are
| 2.5a |
and
| 2.5b |
where partial derivates are evaluated at 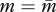 and
and 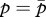 . Full details of the derivation of equations (2.4a,b) and (2.5a,b) are given by Iwasa & Pomiankowski [27], and justification of the underlying quantitative genetic model is given by Iwasa et al. [26]. Note also that the inheritance pattern of haplodiploidy is the same as that of X chromosomes analysed by Iwasa & Pomiankowski [27].
. Full details of the derivation of equations (2.4a,b) and (2.5a,b) are given by Iwasa & Pomiankowski [27], and justification of the underlying quantitative genetic model is given by Iwasa et al. [26]. Note also that the inheritance pattern of haplodiploidy is the same as that of X chromosomes analysed by Iwasa & Pomiankowski [27].
3. Results
First, we present a simplified version of our model, in which we assume that the tragedy of the commons owing to an increased Q is equally costly to new queens and new males; that is, γ = δ = c. Then, substituting equation (2.4a,b) with equation (2.5a,b) and setting 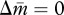 and
and 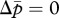 yield the following two parallel nullclines on the
yield the following two parallel nullclines on the 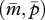 plane:
plane:
| 3.1a |
and
| 3.1b |
for 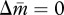 and
and 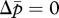 , each corresponding to the optimum for m and p, respectively. The locations of the two nullclines are determined by their
, each corresponding to the optimum for m and p, respectively. The locations of the two nullclines are determined by their 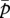 -intercepts, the right-hand sides of equation (3.1a,b).
-intercepts, the right-hand sides of equation (3.1a,b).
When c = 0, the two nullclines always overlap onto 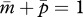 , forming a line of neutrally stable equilibrium towards which the population-mean trait
, forming a line of neutrally stable equilibrium towards which the population-mean trait 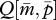 evolves from the initial state satisfying
evolves from the initial state satisfying 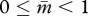 ,
, 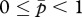 and
and 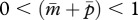 (figure 1a). Because no cost of an increased Q is assumed here,
(figure 1a). Because no cost of an increased Q is assumed here, 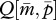 reaches 1.
reaches 1.
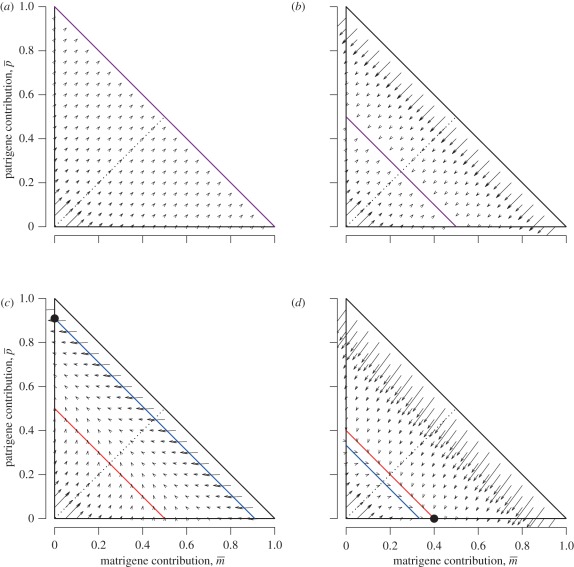
Figure 1.
Isoclines and evolutionary dynamics of queen gene imprinting. Directions of change are shown in vector fields. The equilibrium state is determined by the location of two nullclines, 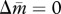 (red line) and
(red line) and 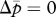 (blue line), in the triangular region. (a) In the absence of the tragedy of the commons, the two nullclines coincide and together form a line of neutrally stable equilibrium,
(blue line), in the triangular region. (a) In the absence of the tragedy of the commons, the two nullclines coincide and together form a line of neutrally stable equilibrium, 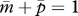 (purple line; N = 1, P = 1, γ = 0, δ = 0). (b) When new queens and new males evenly suffer the tragedy, queen gene imprinting is again a selectively neutral trait (located along the purple line) for colonies headed by monandrous queens (N = 1, P = 1, γ = 1, δ = 1). (c) Polyandry of mother queens alters the location of the nullcline
(purple line; N = 1, P = 1, γ = 0, δ = 0). (b) When new queens and new males evenly suffer the tragedy, queen gene imprinting is again a selectively neutral trait (located along the purple line) for colonies headed by monandrous queens (N = 1, P = 1, γ = 1, δ = 1). (c) Polyandry of mother queens alters the location of the nullcline 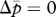 , leading to the evolution of queen gene imprinting with patrigene-specific expression (N = 1, P = 10, γ = 1, δ = 1). (d) Female-biased susceptibility to the tragedy creates a condition in which imprinting with matrigene-specific expression evolves (N = 1, P = 1, γ = 2, δ = 1). Filled circles indicate the points of evolutionary equilibrium, and dotted lines indicate m̄ = p̄. Gm = Gp = 0.01.
, leading to the evolution of queen gene imprinting with patrigene-specific expression (N = 1, P = 10, γ = 1, δ = 1). (d) Female-biased susceptibility to the tragedy creates a condition in which imprinting with matrigene-specific expression evolves (N = 1, P = 1, γ = 2, δ = 1). Filled circles indicate the points of evolutionary equilibrium, and dotted lines indicate m̄ = p̄. Gm = Gp = 0.01.
When c > 0, both nullclines lie within the triangular region defined above (figure 1b–d). The condition of coincidence of the two nullclines is given by solving N/(N + c) = NP/(NP + c), yielding P = 1. That is, there is no selective force for queen gene imprinting when the mother queen is monandrous (figure 1b). In contrast, when the mother queen is polyandrous, even if only slightly, on average (i.e. P > 1), the nullcline 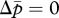 always has a larger intercept than that of
always has a larger intercept than that of 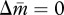 (figure 1c). This means that the optimal
(figure 1c). This means that the optimal 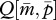 for the patrigene becomes higher than the optimum for the matrigene, leading to a conflict between the two alleles. Starting from any initial state, the population-mean trait
for the patrigene becomes higher than the optimum for the matrigene, leading to a conflict between the two alleles. Starting from any initial state, the population-mean trait 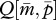 quickly evolves into the region between the two nullclines. Here,
quickly evolves into the region between the two nullclines. Here, 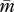 decreases according to the interest of the matrigene, whereas
decreases according to the interest of the matrigene, whereas 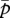 increases to satisfy the interest of the patrigene. The resulting trajectory reaches a state (black circle in figure 1c) under which the matrigene is silenced (i.e.
increases to satisfy the interest of the patrigene. The resulting trajectory reaches a state (black circle in figure 1c) under which the matrigene is silenced (i.e. 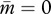 ) and the patrigene expresses solely to realize its own optimum of
) and the patrigene expresses solely to realize its own optimum of 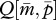 ,
,
| 3.2 |
Although the degree of polygyny with unrelated queens N affects the equilibrium value of 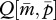 , it does not affect the evolutionary dynamics described above.
, it does not affect the evolutionary dynamics described above.
(a). Sex-specific susceptibility to the tragedy
When there is a sex difference in the susceptibility of new reproductive individuals to the tragedy of the commons (i.e. γ ≠ δ), the two nullclines 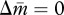 and
and 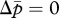 in equation (3.1a,b) are replaced with
in equation (3.1a,b) are replaced with
| 3.3a |
and
| 3.3b |
respectively. This alteration creates the opportunity for queen gene imprinting (i.e. two nullclines do not coincide) even when the mother queen is monandrous.
In a monandrous colony (P = 1), the evolutionary dynamics of queen gene imprinting depend on γ and δ, which alter the location of the two nullclines and thus the equilibrium state of 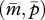 . When new males are more susceptible to the tragedy of the commons (i.e. γ < δ), the matrigene evolves to silence its expression. In contrast, when new queens are more susceptible to the tragedy (i.e. γ > δ), the nullcline
. When new males are more susceptible to the tragedy of the commons (i.e. γ < δ), the matrigene evolves to silence its expression. In contrast, when new queens are more susceptible to the tragedy (i.e. γ > δ), the nullcline 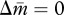 has a larger intercept than that of
has a larger intercept than that of 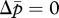 (figure 1d), leading the patrigene to evolve silencing.
(figure 1d), leading the patrigene to evolve silencing.
When a mother queen is polyandrous on average (P > 1), the evolutionary consequences become more complex. The condition of coincidence of the two nullclines is given by solving 2N/(2N + γ + δ) = NP/(NP + γ), yielding
| 3.4 |
When γ < δ, the right-hand side of equation (3.4) is always less than 1, and the matrigene is always silenced at the evolutionary equilibrium. When γ > δ, the right-hand side of equation (3.4) is larger than 1, and there arises an intermediate level of polyandry below and above which the matrigene-specific and patrigene-specific expressions evolve, respectively. Figure 2 summarizes the conditions for the evolution of queen gene imprinting.
Figure 2.
The evolution of queen gene imprinting depends on the relative cost of the tragedy of the commons (γ/δ) and degree of polyandry (P). Over a wide range of degrees of polyandry, patrigene-specific expression evolves (grey region); it can evolve even under monandry when new males are more susceptible to the tragedy (γ/δ < 1). When new queens are more susceptible (γ/δ > 1), there arises an intermediate level of polyandry (1 < P < 2), below which matrigene-specific expression evolves (black region). Queen gene imprinting is selectively neutral on the line between the two regions.
4. Discussion
The present multivariate quantitative genetic model formally confirmed the applicability of the kinship theory of genomic imprinting to eusocial Hymenoptera [7,18,20,21], by explicitly showing the evolutionary process through which extreme asymmetry in matrigene and patrigene expression can evolve continuously and concurrently from the initial symmetry. The present approach also has a relative advantage in incorporating explicit colony life history and the resulting nonlinear fitness effects, which may often be confusing in inclusive fitness approaches. Our model thus extends the scope of the generalized kinship theory allowing for a single trait and can provide a rigorous basis for detailed predictions about the mode of genomic imprinting for particular traits in eusocial Hymenoptera. For simplicity, we assumed that Q is determined solely by immature females and did not analyse the correlated evolution of a workers' trait that may reduce Q (i.e. caste fate policing [29]). This treatment is equivalent to assuming the workers' effects as a fixed constant, so that the subsequent analysis of evolutionary trajectory is not affected by them.
Increasing evidence suggests that genomic imprinting may influence social traits in hymenopteran colonies [11–17,22,23] (see §1). In this study, we focused on the possibility of queen gene imprinting. Recent studies have revealed various heritable components of queen–worker caste differentiation, urging a reconsideration of the traditional view that the differentiation is controlled solely by environmental factors [30]. In the following, we discuss how queen gene imprinting relates to these recent findings and can successfully explain some of the peculiar caste determination systems.
A clear prediction of the present model is that polyandry leads to queen gene imprinting with patrigene-specific expression. The prediction itself has already been made by a previous study [7] using inclusive fitness approach: briefly, patrigene relatedness within maternal sibships (or ‘coevals’) declines with the degree of polyandry, whereas matrigene relatedness does not. Also, our initial analysis, which assumed γ = δ, corresponds to the situation in [7], where female behaviours affect both sexes of coevals evenly. Honeybees and several ant species, such as ‘AenEcDo’ army ants (sensu [31]), ‘higher’ leaf-cutting ants (Atta and Acromyrmex) and harvester ants (Pogonomyrmex), are known to show high degrees of polyandry [32]. Future studies should examine the existence of paternal genetic effects on the trait influencing queen development in these polyandrous species (for instance, by using cross-breeding experiments [22,23,33]).
The patrigene that influences queen development in polyandrous colonies under queen gene imprinting can be understood as a kind of selfish gene, because it evolves to drive the carrier female to take more colony resources at the cost of fitness of other colony members. This selfishness of patrigenes is peculiar in that its expression is realized only when transmitted paternally. Marker-based parentage analyses in honeybees and the leaf-cutting ant Acromyrmex echinator have revealed the excessive presence of certain patrilines among new queens [34–39]. One difficulty in applying a selfish-gene perspective to this phenomenon (e.g. ‘genetic royal cheats’ [39]) is that it is unclear whether and how this selfish gene can avert the tragedy of the commons, especially when transmitted maternally (i.e. at least half of the female offspring in the monogynous colony harbour the selfish allele). There is an alternative explanation that attributes this phenomenon to compatibility between matrigenes and patrigenes, which can create an apparent excess of certain patrilines in new queens, as has been demonstrated by a cross-breeding experiment in the harvester ant Pogonomyrmex rugosus [33]. Queen gene imprinting in the polyandrous colony seems to provide another explanation within the selfish-gene framework: the expression of the ‘royal cheat’ mutant trait is permitted only when transmitted paternally, because matrigenes have already evolved to be silenced. Whether and how genomic imprinting and the other possible mechanisms contribute to the observed patriline bias and the low-frequency persistence of tentative ‘royal cheat’ alleles deserves further study. Queen gene imprinting with patrigene-specific expression has further implications. For example, it might be applied to other systems of patriline-dependent genetic caste determination, such as social hybridogenesis in Pogonomyrmex harvester ants, in which the mother queen requires both within- and between-lineage matings that result in exclusive production of new queens and workers, respectively [40].
Despite the general expectation that it might be found in polyandrous species, the first evidence of a paternal effect on queen caste determination was reported from a monandrous species, the Argentine ant L. humile [23]. This species has a unicolonial social structure with multiple unrelated queens, which corresponds to N ≫ 1 and P = 1 in our model. By incorporating the ecology of eusocial hymenopteran colonies, the present model can also explain how queen gene imprinting evolves under monandry (figure 2). The sufficient condition is sex specificity in the cost of an increased Q on new reproductive offspring (i.e. γ ≠ δ in equation (2.2a,b)). This prediction corresponds to a generalized situation analysed in previous studies [7,8], where female behaviours affect differently depending on the sex of coevals. In our model, the cost is due to the tragedy of the commons (or an increased Q), an assumption that has a unique consequence in the context of eusocial hymenopteran colonies, as shown below. When new males are more susceptible to the tragedy than are new queens (γ < δ), this relative cost is inflicted only on matrigenes because haploid males inherit only matrigenes (i.e. patrigenes are unrelated to male coevals), which gives the matrigenes an incentive to contribute less to queen development and leads to their silencing even under monandry. This sex-differential susceptibility results in the trade-off in the numbers of new queens and new males, because the increased fraction of female immatures that follow the queen developmental trajectory through the increase of Q results in a decreased number of new males through the tragedy of the commons, M(1 − Q)δ, as long as the increase of Q outweighs the cost of the tragedy on the new queens themselves, (1 − Q)γ (see equation (2.2a,b)). Alternatively, the relative cost of the increased Q can also be interpreted as negative pleiotropy of the queen gene. In our formulation, this pleiotropy can arise from the expression of queen gene existing in males even if the tragedy itself is equally costly to both sexes, because equation (2.2a,b) can be in the form of (the initial number of new reproductive individuals) × (their survival): FQ × (1 − Q)γ and M(1 − Q)δ−γ × (1 − Q)γ, respectively. In both scenarios, the resulting trade-off can be viewed as a peculiar case of intralocus sexual conflict [37,41,42] caused by the queen gene. Interestingly, the observed negative correlation between the numbers of new males and new queens in controlled crosses of L. humile [23] seems consistent with this view. However, for rigorous testing of this prediction, an experimental method should be used that can evaluate the relative cost of the increased Q inflicted by males. In addition, future theoretical studies should investigate the effect of this type of sex allocation trade-off on the conflict over sex allocation, another important conflict in eusocial hymenopteran colonies [4,43,44].
In contrast, when new queens are more susceptible to the increased Q than new males (γ > δ; the extreme case δ = 0 corresponds to the analysis of Haig [18]), it is patrigenes that suffer from the relative cost, because in haplodiploids patrigenes exist solely in female offspring. In particular, patrigenes in colonies with P < 2 have higher relatedness to female coevals than their matrigene counterparts do. The subsequent evolution also depends on the degree of polyandry and on how long the focal gene stays in each sex in the long run (in haplodiploids, 2/3 of the generations in females and 1/3 in males, which represents the sex-specific reproductive value). Thus, there arises an intermediate level of polyandry, 1 < P < 2, below and above which queen gene imprinting with matrigene-specific and patrigene-specific expression evolves, respectively (figure 2).
In recent years, an increasing number of molecular resources of eusocial Hymenoptera have become available, including whole-genome sequence data of honeybees and several ant species mentioned here [12–17]. These resources provide a novel opportunity to test the theory of genomic imprinting directly at the molecular level. Among other possible alternatives, including histone modifications and trans-acting non-coding RNAs, DNA methylation is the best-studied mechanism responsible for genomic imprinting. The role of DNA methylation during queen–worker differentiation has been extensively studied in honeybees as an emerging model system, taking advantage of phenotypic distinctiveness induced by external factors such as royal jelly [45,46]. Furthermore, recent genomic and epigenomic studies have revealed that sequences coding for the DNA methyltransferase family (Dnmt1 and Dnmt3), constituting a fully functional DNA methylation toolkit, are conserved across honeybees and ants [11–17], and that genes possibly involved in queen–worker differentiation show symptoms of active methylation (i.e. reduced ratio of observed CpG dinucleotides compared with genome average) in some ants (L. humile [15], Pogonomyrmex barbatus [14]; but the underlying mechanism might be complex—see [46]). However, the current focus is mainly on de novo modification of methylation levels during development, rather than genomic imprinting during germline formation and its maintenance. Future studies should investigate whether and how the differences in methylation levels (as well as other epigenetic modifications) are influenced by genomic imprinting.
The present study provides several candidate species (polyandrous species and L. humile) that are worth exploring for the possibility of queen gene imprinting. The most definitive test is to show that the genes considered here are indeed imprinted, which will extend our understanding of epigenetics, as well as strengthen the integrity of evolutionary theory.
Acknowledgements
We thank E. Hasegawa, M. K. Hojo, K. Kobayashi, S. Nakayama, H. Ohtsuki, H. Shimoji and N. Yagi for discussions, and K. Uematsu and three anonymous reviewers for comments that significantly improved the manuscript. S.D. is supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (22-9877).
References
- 1.Maynard Smith J., Szathmáry E. 1995. The major transitions in evolution. Oxford, UK: Oxford University Press [Google Scholar]
- 2.Burt A., Trivers R. 2006. Genes in conflict. Cambridge, MA: Harvard University Press [Google Scholar]
- 3.Arnqvist G., Rowe L. 2005. Sexual conflict. Princeton, NJ: Princeton University Press [Google Scholar]
- 4.Crozier R. H., Pamilo P. 1996. Evolution of social insect colonies. Oxford, UK: Oxford University Press [Google Scholar]
- 5.Haig D., Westoby M. 1989. Parent specific gene expression and the triploid endosperm. Am. Nat. 134, 147–155 10.1086/284971 (doi:10.1086/284971) [DOI] [Google Scholar]
- 6.Haig D. 2000. The kinship theory of genomic imprinting. Ann. Rev. Ecol. Syst. 31, 9–32 10.1146/annurev.ecolsys.31.1.9 (doi:10.1146/annurev.ecolsys.31.1.9) [DOI] [Google Scholar]
- 7.Queller D. C. 2003. Theory of genomic imprinting conflict in social insects. BMC Evol. Biol. 3, 15. 10.1186/1471-2148-3-15 (doi:10.1186/1471-2148-3-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haig D., Graham C. 1991. Genomic imprinting and the strange case of the insulin-like growth factor II receptor. Cell 64, 1045–1046 10.1016/0092-8674(91)90256-X (doi:10.1016/0092-8674(91)90256-X) [DOI] [PubMed] [Google Scholar]
- 9.Ratnieks F. L. W., Foster K. R., Wenseleers T. 2006. Conflict resolution in insect societies. Ann. Rev. Entomol. 51, 581–608 10.1146/annurev.ento.51.110104.15100 (doi:10.1146/annurev.ento.51.110104.15100) [DOI] [PubMed] [Google Scholar]
- 10.Kronforst M. R., Gilley D. C., Strassmann J. E., Queller D. C. 2008. DNA methylation is widespread across social Hymenoptera. Curr. Biol. 18, R287–R288 10.1016/j.cub.2008.02.015 (doi:10.1016/j.cub.2008.02.015) [DOI] [PubMed] [Google Scholar]
- 11.Wang Y., Jorda M., Jones P. L., Maleszka R., Ling X., Robertson H. M., Mizzen C. A., Peinado M. A., Robinson G. E. 2006. Functional CpG methylation system in a social insect. Science 314, 645–647 10.1126/science.1135213 (doi:10.1126/science.1135213) [DOI] [PubMed] [Google Scholar]
- 12.Bonasio R., et al. 2010. Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science 329, 1068–1071 10.1126/science.1192428 (doi:10.1126/science.1192428) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suen G., et al. 2011. The genome sequence of the leaf-cutter ant Atta cephalotes reveals insights into its obligate symbiotic lifestyle. PLoS Genet. 7, e1002007. 10.1371/journal.pgen.1002007 (doi:10.1371/journal.pgen.1002007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith C. R., et al. 2011. Draft genome of the red harvester ant Pogonomyrmex barbatus. Proc. Natl Acad. Sci. USA 108, 5667–5672 10.1073/pnas.1007901108 (doi:10.1073/pnas.1007901108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith C. D., et al. 2011. Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile). Proc. Natl Acad. Sci. USA 108, 5673–5678 10.1073/pnas.1008617108 (doi:10.1073/pnas.1008617108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wurm Y., et al. 2011. The genome of the fire ant Solenopsis invicta. Proc. Natl Acad. Sci. USA 108, 5679–5684 10.1073/pnas.1009690108 (doi:10.1073/pnas.1009690108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nygaard S., et al. 2011. The genome of the leaf-cutting ant Acromyrmex echinatior suggests key adaptations to advanced social life and fungus farming. Genome Res. 21, 1339–1348 10.1101/gr.121392.111 (doi:10.1101/gr.121392.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haig D. 1992. Intragenomic conflict and the evolution of eusociality. J. Theor. Biol. 156, 401–403 10.1016/S0022-5193(05)80683-6 (doi:10.1016/S0022-5193(05)80683-6) [DOI] [PubMed] [Google Scholar]
- 19.Queller D. C., Strassman J. E. 2002. The many selves of social insects. Science 296, 311–313 10.1126/science.1070671 (doi:10.1126/science.1070671) [DOI] [PubMed] [Google Scholar]
- 20.Kronauer D. J. C. 2008. Genomic imprinting and kinship in the social Hymenoptera: what are the predictions? J. Theor. Biol. 254, 737–740 10.1016/j.jtbi.2008.06.019 (doi:10.1016/j.jtbi.2008.06.019) [DOI] [PubMed] [Google Scholar]
- 21.Wild G., West S. A. 2009. Genomic imprinting and sex allocation. Am. Nat. 173, E1–E14 10.1086/593305 (doi:10.1086/593305) [DOI] [PubMed] [Google Scholar]
- 22.Guzman-Novoa E., Hunt G. J., Page R. E., Uribe-Rubio J. L., Prieto-Merlos D., Becerra-Guzman F. 2005. Paternal effects on the defensive behavior of honeybees. J. Heredity 96, 376–380 10.1093/jhered/esi038 (doi:10.1093/jhered/esi038) [DOI] [PubMed] [Google Scholar]
- 23.Libbrecht R., Schwander T., Keller L. 2011. Genetic components to caste allocation in a multiple-queen ant species. Evolution 65, 2907–2915 10.1111/j.1558-5646.2011.01348.x (doi:10.1111/j.1558-5646.2011.01348.x) [DOI] [PubMed] [Google Scholar]
- 24.Hardin G. 1968. The tragedy of the commons. Science 162, 1243–1244 10.1126/science.162.3859.1243 (doi:10.1126/science.162.3859.1243) [DOI] [PubMed] [Google Scholar]
- 25.Mochizuki A., Takada Y., Iwasa Y. 1996. The evolution of genomic imprinting. Genetics 144, 1283–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwasa Y., Nee S., Pomiankowski A. 1991. The evolution of costly mate preferences. II. The handicap principle. Evolution 45, 1431–1442 [DOI] [PubMed] [Google Scholar]
- 27.Iwasa Y., Pomiankowski A. 2001. The evolution of X-linked genomic imprinting. Genetics 158, 1801–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobata S.Arms race between selfishness and policing: two-trait quantitative genetic model for caste fate conflict in eusocial Hymenoptera. Submitted. [DOI] [PubMed]
- 29.Wenseleers T., Ratnieks F. L. W., Billen J. 2003. Caste fate conflict in swarm-founding social Hymenoptera: an inclusive fitness analysis. J. Evol. Biol. 16, 647–658 10.1046/j.1420-9101.2003.00574.x (doi:10.1046/j.1420-9101.2003.00574.x) [DOI] [PubMed] [Google Scholar]
- 30.Schwander T., Lo N., Beekman M., Oldroyd B. P., Keller L. 2010. Nature versus nurture in social insect caste differentiation. Trends Ecol. Evol. 25, 275–282 10.1016/j.tree.2009.12.001 (doi:10.1016/j.tree.2009.12.001) [DOI] [PubMed] [Google Scholar]
- 31.Kronauer D. J. C. 2009. Recent advances in army ant biology (Hymenoptera: Formicidae). Myrmecol. News 12, 51–65 [Google Scholar]
- 32.Boomsma J. J., Kronauer D. J. C., Pedersen J. S. 2009. The evolution of social insect mating systems. In Organization of insect societies: from genome to sociocomplexity (eds Gadau J., Fewell J.), pp. 3–25 Cambridge, MA: Harvard University Press [Google Scholar]
- 33.Schwander T., Keller L. 2008. Genetic compatibility affects queen and worker caste determination. Science 322, 552. 10.1126/science.1162590 (doi:10.1126/science.1162590) [DOI] [PubMed] [Google Scholar]
- 34.Tilley C. A., Oldroyd B. P. 1997. Unequal subfamily proportions among honeybee queen and worker brood. Anim. Behav. 54, 1483–1490 10.1006/anbe.1997.0546 (doi:10.1006/anbe.1997.0546) [DOI] [PubMed] [Google Scholar]
- 35.Osborne K. E., Oldroyd B. P. 1999. Possible causes of reproductive dominance during emergency queen rearing by honeybees. Anim. Behav. 58, 267–272 10.1006/anbe.1999.1139 (doi:10.1006/anbe.1999.1139) [DOI] [PubMed] [Google Scholar]
- 36.Châline N., Arnold G., Papin C., Ratnieks F. L. W. 2003. Patriline differences in emergency queen rearing in the honey bee, Apis mellifera. Insect Soc. 50, 234–236 10.1007/s00040-003-0664-6 (doi:10.1007/s00040-003-0664-6) [DOI] [Google Scholar]
- 37.Moritz R., Lattorff H., Neumann P., Kraus F., Radloff S., Hepburn H. 2005. Rare royal families in honeybees, Apis mellifera. Naturwissenschaften 92, 488–491 10.1007/s00114-005-0025-6 (doi:10.1007/s00114-005-0025-6) [DOI] [PubMed] [Google Scholar]
- 38.Nanork P., Low P. A., Proft K. M., Lim J., Deowanish S., Wongsiri S., Oldroyd B. P. 2011. Actual reproductive conflict during emergency queen rearing in Apis florea. Apidologie 42, 206–210 10.1051/apido/2010052 (doi:10.1051/apido/2010052) [DOI] [Google Scholar]
- 39.Hughes W. O. H., Boomsma J. J. 2008. Genetic royal cheats in leaf-cutting ant societies. Proc. Natl Acad. Sci. USA 105, 5150–5153 10.1073/pnas.0710262105 (doi:10.1073/pnas.0710262105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Julian G. E., Fewell J. H., Gadau J., Johnson R. A., Larrabee D. 2002. Genetic determination of the queen caste in an ant hybrid zone. Proc. Natl Acad. Sci. USA 99, 8157–8160 10.1073/pnas.112222099 (doi:10.1073/pnas.112222099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rice W. R., Chippindale A. K. 2001. Intersexual ontogenetic conflict. J. Evol. Biol. 14, 685–693 10.1046/j.1420-9101.2001.00319.x (doi:10.1046/j.1420-9101.2001.00319.x) [DOI] [Google Scholar]
- 42.Bonduriansky R., Chenoweth S. F. 2009. Intralocus sexual conflict. Trends Ecol. Evol. 24, 280–288 10.1016/j.tree.2008.12.005 (doi:10.1016/j.tree.2008.12.005) [DOI] [PubMed] [Google Scholar]
- 43.Reuter M., Keller L. 2001. Sex ratio conflict and worker production in eusocial Hymenoptera. Am. Nat. 158, 166–177 10.1086/321311 (doi:10.1086/321311) [DOI] [PubMed] [Google Scholar]
- 44.Ohtsuki H., Tsuji K. 2009. Adaptive reproduction schedule as a cause of worker policing in social Hymenoptera: a dynamic game analysis. Am. Nat. 173, 747–758 10.1086/598488 (doi:10.1086/598488) [DOI] [PubMed] [Google Scholar]
- 45.Kucharski R., Maleszka J., Foret S., Maleszka R. 2008. Nutritional control of reproductive status in honeybees via DNA methylation. Science 319, 1827–1830 10.1126/science.1153069 (doi:10.1126/science.1153069) [DOI] [PubMed] [Google Scholar]
- 46.Elango N., Hunt B. G., Goodisman M. A. D., Yi S. V. 2009. DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proc. Natl Acad. Sci. USA 106, 11 206–11 211 10.1073/pnas.0900301106 (doi:10.1073/pnas.0900301106) [DOI] [PMC free article] [PubMed] [Google Scholar]