Abstract
The ciliate subclass Haptoria is a diverse taxon that includes most of the free-living predators in the class Litostomatea. Phylogenetic study of this group was initially conducted using a single molecular marker small-subunit ribosomal RNA (SSU rRNA genes). Multi-gene analysis has been limited because very few other sequences were available. We performed phylogenetic analyses of Haptoria incorporating new SSU rRNA gene sequences from several debated members of the taxon, in particular, the first molecular data from Cyclotrichium. We also provided nine large-subunit ribosomal RNA (LSU rRNA) gene sequences and 10 alpha-tubulin sequences from diverse haptorians, and two possible relatives of controversial haptorians (Plagiopylea, Prostomatea). Phylogenies inferred from the different molecules showed the following: (i) Cyclotrichium and Paraspathidium were clearly separated from the haptorids and even from class Litostomatea, rejecting their high-level taxonomic assignments based on morphology. Both genera branch instead with the classes Plagiopylea, Prostomatea and Oligohymenophora. This raises the possibility that the well-known but phylogenetically problematic cyclotrichiids Mesodinium and Myrionecta may also have affinities here, rather than with litostomes; (ii) the transfer of Trachelotractus to Litostomatea is supported, especially by the analyses of SSU rRNA and LSU rRNA genes, however, Trachelotractus and Chaenea (more uncertainly) generally form the two deepest lineages within litostomes; and (iii) phylogenies of the new molecular markers are consistent with SSU rRNA gene information in recovering order Pleurostomatida as monophyletic. However, Pleurostomatida branches cladistically within order Haptorida, as does subclass Trichostomatia (on the basis of SSU rRNA phylogenies). Our results suggest that the class-level taxonomy of ciliates is still not resolved, and also that a systematic revision of litostomes is required, beginning at high taxonomic levels (taxa currently ranked as subclasses and orders).
Keywords: protist, phylogeny, ciliate, Haptoria, rRNA, tubulin
1. Introduction
Ciliates are a large group of complex unicellular organisms numbering approximately 8000 described species, currently subdivided into 11 classes [1]. One of the most frequently encountered groups is the haptorians, which are found worldwide in freshwater and marine habitats and are voracious predators of flagellates, other ciliates and even small metazoans [2,3]. Haptorians generally immobilize and kill their prey using extrusomes called toxicysts [1]. The group also includes the commonly encountered planktonic ciliates Mesodinium and Myrionecta rubra, which can harbour cryptophyte endosymbionts and/or plastids [4–6], and can form red-tide blooms in which they contribute up to 70 per cent of the local primary production.
Systematically speaking, the haptorian ciliates are generally treated as a subclass, Haptoria, within the class Litostomatea [1,7], together with subclass Trichostomatia, which lack toxicysts and which are all endosymbionts of metazoans, including fishes and humans [1]. The haptorians are characterized morphologically by telokinetal stomatogenesis, usually uniform holotrichous somatic ciliation, and orally located toxicysts [8]. The group is a diverse assemblage of loosely associated taxa, comprising over 1000 species [9]. The systematics of this group is relatively difficult to determine because few morphological and/or ontogenetic characters are available. Lynn [1] recognized three orders within this subclass—Haptorida, Pleurostomatida and Cyclotrichiida—pending molecular analyses to strongly confirm or refute these divisions. Foissner & Foissner [2] suggested six orders, namely Haptorida, Spathidiida, Pleurostomatida, Pseudoholophryida, Cyclotrichiida and Archistomatida.
Recent molecular phylogenetic analyses based on a single gene small-subunit ribosomal RNA (SSU rRNA) do not provide unambiguous support for any previously proposed taxonomy of haptorians [1,9,10]. Haptoria was not monophyletic in these analyses, with several of its branches grouping together with Trichostomatia [9,11,12]. The branching pattern within the haptorians was not well resolved, which may be due to undersampling of haptorian genera [9]. Further, the cyclotrichiids for which there are full-length SSU rRNA gene sequences—the Mesodinium/Myrionecta grouping—do not branch with other haptorians, but instead branch within a basal polytomy of the class Litostomatea [9] or more often as the sister group to all other ciliates [13]. However, since these sequences are extremely divergent, it is strongly suspected that the recovered position of the Mesodinium/Myrionecta grouping is influenced by phylogenetic analysis artefact [14].
In addition, the evolutionary positions of some key taxa of haptorian ciliates have not been resolved. Members of the genus Cyclotrichium Meunier, 1910 (figure 1q,r) are common in the marine and limnetic microzooplankton [17]. Since these organisms are fragile and highly motile, they were only superficially described in early studies, without data on infraciliature [17]. The most recent morphological study, which includes infraciliature data, follows Lynn [8] in assigning Cyclotrichium to family Didiniidae, within order Haptorida [17]. However, other recent systems assign Cyclotrichium to a different order, Cyclotrichiida because they lack an ancestral character of haptorids, the dorsal brush (DB) [2,14]. This latter assignment would imply a close relationship to Mesodinium/Myrionecta; however, the phylogenetic position of Cyclotrichium has never been studied using molecular techniques.
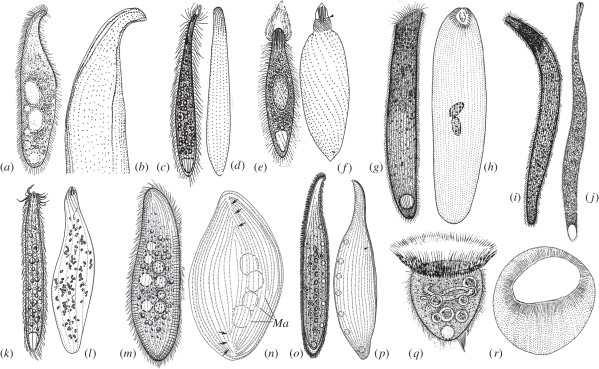
Figure 1.
Morphology and infraciliature of nine haptorian ciliates in vivo and after silver impregnation (two images (m,n) are from Pan et al. [15], others are from Lin et al. [16]). (a,b) Amphileptus marinus, (c,d) Chaenea teres, (e,f) Phialina salinarum, (g,h) Paraspathidium apofuscum, (i,j) Trachelotractus entzi, (k,l) Chaenea vorax, (m,n) Epiphyllum shenzhenense, (o,p) Loxophyllum jini, (q,r) Cyclotrichium cyclokaryon.
The genus Paraspathidium (figure 1g,h) has a haptorid-like shape and suite of morphological characters (DB, extrusomes, a slit-like, apically located cytostome, dikinetids around buccal field) [18]. It has been regarded as a gymnostome haptorid (Litostomatea) by Foissner [19]. Nonetheless, recent SSU rRNA gene phylogenies and analysis of the secondary structures of the variable region 2 (V2) and variable region 4 (V4) of this molecule support a relationship with class Plagiopylea rather than with class Litostomatea [20]. This has not been tested with other phylogenetic markers.
Trachelotractus (figure 1i,j) was traditionally considered to be a karyorelictean owing to its similarity with trachelocercids in general appearance and contractility [21,22], but was transferred to class Litostomatea, order Haptorida by Foissner [23] because of detailed similarity to typical haptorid species—for example, a peribuccal ridge with extrusomes, somatic monokinetids, oralized somatic kinetids and specialized ciliary rows curving around the pharyngeal opening [24]. However, SSU rRNA gene phylogenies including a single species of Trachelotractus recover it as a deep branch within the class Litostomatea, and not specifically related to other members of Haptorida [24]. Again it is important that this be confirmed with improved taxon sampling, and additional markers.
Given these important systematic uncertainties and apparent conflicts between morphology and the available SSU rRNA gene phylogenies, molecular phylogenies of haptorids with better taxon sampling and gene sampling are required. At present, there are no sequence data at all for more than 95 per cent of named haptorid species, and datasets for genes other than SSU rRNA are extremely sparse. For example, the large-subunit ribosomal RNA (LSU rRNA) gene sequence data are limited to a single full-length sequence (Spathidium) and a fragment of about 300 nucleotides from four genera [25]. No previous analysis of Litostomatea has used sequences from protein-coding genes.
In the present work, we increase the taxon sampling of these ciliates, with an emphasis on free-living Haptoria, especially phylogenetically controversial taxa. New SSU rRNA genes were sequenced from seven species, and we expanded phylogenetic analyses of the major taxa of the subclass Haptoria with new gene markers: near-full-length LSU rRNA genes and alpha-tubulin proteins, with sequences inferred from nine and 10 species, respectively. Phylogeny and morphological characters were considered together to provide a new evaluation of the phylogenetic relationships of haptorian ciliates.
2. Results
(a). Overview
In total, 29 new sequences were obtained from 14 species of ciliates representing 10 genera, predominantly from taxa traditionally and/or currently recognized as haptorians (see the electronic supplementary material, table S1). Nine species of these (from eight genera) are depicted in figure 1. For all 10 genera, this includes the first phylogenetically useful sequence data for genes other than SSU rRNA. It also includes, to our knowledge, the first molecular data of any kind from Cyclotrichium.
We ran four sets of analyses for the SSU rRNA gene data, with different taxon samples—one ‘primary’ set with 78 ciliate species and two dinoflagellates as outgroups, and three further sets that also included the divergent or partial sequences from Mesodinium, Myrionecta and/or Askenasia. We performed two sets of analyses of alpha-tubulin genes based on amino acid and nucleotide sequences, respectively, for 54 ciliate species. The single set of analyses of LSU rRNA genes included 20 species covering five classes of ciliates, and two outgroups.
(b). Small-subunit ribosomal RNA structure in Cyclotrichium
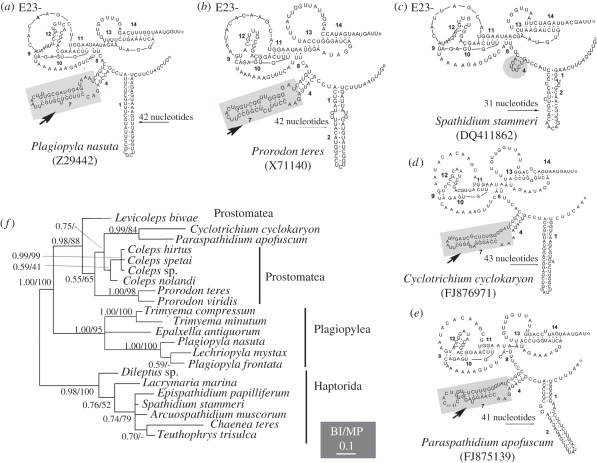
The complete Cyclotrichium cyclokaryon SSU rRNA gene is 1708 nucleotides long, which is longer than typical litostome SSU rRNA genes (approx. 1640 nucleotides). The secondary structure of V4 in Cyclotrichium does not have the deletions otherwise common to all litostome ciliates (figure 2): Litostomes, including the divergent Mesodinium and Myrionecta, have characteristic deletions in helices 23_1,2, 23_13, 23_14 and lack helix 23_7 [9,26] (figure 2c). By contrast, the total length of Helix E23-1 and 2 (or only Helix E23-1) in Cyclotrichium is markedly greater than in haptorian litostomes (43 bp versus 28–34 bp—compare figure 2c,d). Helix E23-7 is present in Cyclotrichium (arrows in figure 2) but is absent in all haptorians. However, the structure of this region in Cyclotrichium is similar to that of Paraspathidium, and of all plagiopyleans and prostomateans (figure 2a,b,e). A phylogeny of V4 regions based on combined information of the primary sequence and the secondary structure (figure 2f) shows a close relationship of Cyclotrichium and Paraspathidium (0.99 Bayesian inference (BI), 84% maximum parsimony (MP)). They cluster with Coleps spp. and form a sister group to Prorodon (0.55 BI, 65% MP) and then group with the Plagiopylea clade (1.00 BI, 100% MP). Haptorian genera formed another well-supported group (0.98 BI, 100% MP), which was clearly separated from these other taxa.
Figure 2.
Models of the secondary structure of variable region 4 (V4) of the SSU rRNA molecule, and the phylogenetic tree of 15 taxa based on V4 region primary sequences and secondary structure combined. Models of the secondary structure comprising helices E23-1,(2), 4, 7, 8, 9, 10, 11, 12, 13, 14 and two pseudoknots formed by helices E23-9 to 12, helices E23-13 and 14 are shown. The number of nucleotides in Helix E23-1 and 2 (or only E23-1) for each species is given above the long arrow. Note the Helix E23-7 in Paraspathidium–Cyclotrichium and the classes Plagiopylea and Prostomatea (arrows).
(c). Phylogenetic analyses of small-subunit ribosomal RNA genes
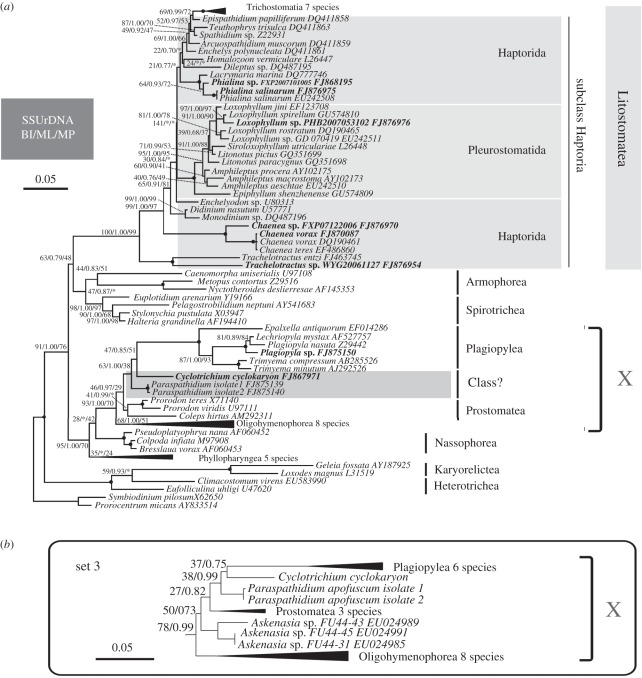
The BI, maximum likelihood (ML) and MP analyses of the primary SSU rRNA gene dataset recovered nearly identical topologies. The ML tree is shown in figure 3. Cyclotrichium and Paraspathidium do not branch with other haptorids, or even other litostomes. Instead, Cyclotrichium branches as sister to the plagiopylean clade with low support (47% ML, 0.85 BI, 51% MP). This clade then clusters with the two isolates of Paraspathidium apofuscum with moderate support values (63% ML, 1.00 BI, 38% MP). Together they form a sister group to the class Prostomatea, represented by Prorodon and Coleps, with moderate/low support (46% ML, 0.97 BI, 29% MP). Cyclotrichium, Plagiopylea (see below), Paraspathidium and Prostomatea in turn branch with Oligohymenophorea with strong support (93% ML, 1.00 BI, 70% MP), forming a group we call ‘clade X’ for simplicity (see below). These taxa group in turn with the colpodeans, nassophoreans and phyllopharyngeans with high support (95% ML, 1.00 BI, 70% MP; figure 3). The newly sequenced Plagiopyla sp. groups together with Lechriopyla mystax and Plagiopyla nasuta, forming a maximally supported clade corresponding to class Plagiopylea.
Figure 3.
(a) Maximum-likelihood (ML) tree of 80 SSU rRNA gene sequences including all ciliate classes, with emphasis on the class Litostomatea. New sequences are shown in bold text. GenBank accession numbers are given after names of species. Numbers at nodes show bootstrap values from ML, Bayesian inference (BI) posterior probabilities, and bootstrap values from maximum parsimony (MP) in this order; instances in which a particular node was not recovered in the estimated BI or MP tree are designated by an asterisk (*). Nodes receiving maximal support (100% ML, 1.00 BI, 100% MP) are indicated by solid circles. (b) The subtree representing clade X from the ML tree of supplementary analysis ‘set 3’, showing the position of the Askenasia spp. clade. For clarity, only backbone support values are shown (ML bootstrap values, BI posterior probabilities). For both trees, the scale bars correspond to 0.05 expected substitutions per site. Certain groups were simplified as solid triangles to reduce the size of the figure; width and length indicates the number of sequences and the average branch length in this group.
The other new SSU rRNA gene sequences branch with or within the Litostomatea clade. The new sequence for Phialina salinarum is identical to the published sequence across the analysed sites, while the undetermined Phialina species groups with the Ph. salinarum sequences with moderate support (64% ML, 0.93 BI, 72% MP). This Phialina clade branches specifically with Lacrymaria marina, with maximal support (100% ML, 1.00 BI, 100% MP).
The new sequence from an unidentified Loxophyllum species (sampled at Qingdao, China) differs at 10 positions from another unidentified Loxophyllum species (sampled at Guangdong, China). These two sequences branch together with high support (97% ML, 1.00 BI, 97% MP) within a well-supported Loxophyllum clade (91% ML, 1.00 BI, 90% MP). Loxophyllum branches as expected in a highly nested position within a maximally supported clade that corresponds to the order Pleurostomatida (figure 3).
The new sequence from an unidentified Trachelotractus species is most similar to its congener Trachelotractus entzi, but differs at 217 nucleotide positions. It branches with T. entzi with maximal support. The new population of Chaenea vorax is identical to the published sequences from both C. vorax and Chaenea teres across all analysed sites. Trachelotractus and Chaenea occupied the two deepest positions within Litostomatea, with generally strong statistical support (99% ML, 1.00 BI, 97% MP for Trachelotractus as the deepest branch; 65% ML, 0.91 BI, 81% MP for Chaenea as the second deepest branch). This renders Haptorida (sensu lato) as a paraphyletic group, with both Pleurostomatida and Trichostomatia forming strongly supported clades that branch after the divergences of Trachelotractus and Chaenea. The optimal tree actually places Trichostomatia in a very shallow position within Haptorida, but this position, and most of the remaining backbone of the litostome tree, receives very limited support (e.g. ML bootstrap support values less than 50%).
Sets 1 and 2 of the supplementary SSU rRNA gene analyses (electronic supplementary material, figures S1–S6) included the problematic cyclotrichiids Mesodinium and Myrionecta (which have highly divergent SSU rRNA gene sequences) and Askenasia (for which only a partial sequence is available), with and without outgroups to ciliates, while set 3 included just Askenasia (and the outgroups). The overall topologies recovered in the supplementary analysis did not differ materially from those described above. In set 1, with dinoflagellates as outgroups, Mesodinium and Myrionecta form an extremely long branch that has variable positions in ML and BI trees. It branches inside Oligohymenophorea in the ML tree, while it attached at the base of the Ciliophora clade in the BI tree, but support for either placement is very weak (8% ML, 0.88 BI; electronic supplementary material, figures S1 and S2). When outgroups are excluded (set 2), Mesodinium and Myrionecta branch variably within one of the two main clades of the ciliate tree (the clade including clade X, Nassophorea and Phyllopharyngea) in ML and BI trees (52% ML, 0.92 BI; electronic supplementary material, figures S3 and S4). They branch with Nassophorea in the ML tree, and as sister group to the clade of Plagiopylea, Cyclotrichium, Paraspathidium and Prostomatea in the BI tree (support was negligible in each case). In sets 1 and 2 Askenasia forms a clade that does not branch specifically with either Mesodinium–Myrionecta or Cyclotrichium (i.e. other cyclotrichiids), however, it branches variably within clade X, either branching basally to the clade, including Plagiopylea, Paraspathidium and Cyclotrichium (ML trees and BI tree in set 1; electronic supplementary material, figures S1–S3); or as the sister group to the rest of clade X (0.77, BI tree in set 2; electronic supplementary material, figure S4). A more stable position for Askenasia is recovered in set 3, when Mesodinium and Myrionecta are excluded from the analysis (figure 3b and electronic supplementary material, figures S5 and S6). Here, Askenasia branches at the base of the clade that includes Plagiopylea, Paraspathidium and Cyclotrichium. Support for the affinity of Askenasia with the Prostomatea–Plagiopylea–Paraspathidium–Cyclotrichium clade is generally weak (50% ML, 0.73 BI in set 3) as is the support for its basal position within this clade (27% ML, 0.82 BI in set 3), however, there is stronger support for its inclusion in clade X as a whole (78% ML, 0.99 BI).
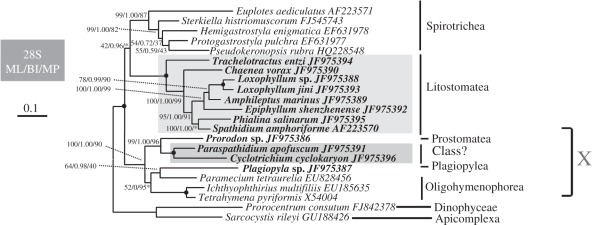
(d). Phylogenetic analyses of large-subunit ribosomal RNA genes
Taxon sampling in the LSU rRNA dataset is more limited, but overall the topology recovered is consistent with the SSU rRNA gene tree (figure 4). Paraspathidium and Cyclotrichium again do not group with litostomes, but instead branch with Prostomatea, Plagiopylea and Oligohymenophorea to form a strongly supported ‘clade X’ (100% ML, 1.00 BI, 90% MP). In contrast to the SSU rRNA gene tree, Paraspathidium and Cyclotrichium group together to a well-supported clade (100% ML, 1.00 BI, 100% MP), and this clade specifically groups with Prostomatea, represented by the new sequence from Prorodon sp., with strong support (99% ML, 1.00 BI, 96% MP). The new sequence of Plagiopyla sp., representing class Plagiopylea, forms a weakly/moderately supported clade with the oligohymenophorean Paramecium (64% ML, 0.98 BI, 40% MP); the precise position of this clade relative to the other Oligohymenophorea is unstable.
Figure 4.
ML tree estimated from the LSU rRNA sequences of 20 ciliate taxa, with myzozoan alveolates as outgroups. New sequences are shown in bold text. Numbers at nodes are the following: ML bootstrap values followed by BI posterior probability and MP bootstrap values. Nodes not present in the estimated MP tree are designated by asterisks (*). Nodes receiving maximal support (100% ML, 1.00 BI, 100% MP) are indicated by solid circles. The scale bar corresponds to 0.1 expected substitutions per site.
Class Litostomatea is otherwise monophyletic (100% ML, 1.00 BI, 100% MP). As in the SSU rRNA gene tree Trachelotractus and Chaenea branch sequentially at the base of the Litostomatea clade; statistical support is nearly maximal (100% ML, 1.00 BI, 99% MP) and high (95% ML, 1.00 BI, 91% MP), respectively. Thus, Haptorida again appears paraphyletic relative to Pleurostomatida in optimal trees (there are no LSU rRNA data for Trichostomatia). The other haptorids (Phialina and Spathidium) group together with high support in ML and BI analyses (100% ML, 1.00 BI), while in MP analyses, Spathidium and Phialina branch sequentially with the pleurostomatid clade, with weak support (56% MP, data not shown) and high support (91% MP, data not shown) for the two nodes. The four included members of order Pleurostomatida group as a single clade, with near-maximum support (100% ML, 1.00 BI, 99% MP). Inside the pleurostomatid clade, the Loxophyllum spp. clade groups with Amphileptus (78% ML, 0.99 BI, 90% MP), leaving Epiphyllum basal.
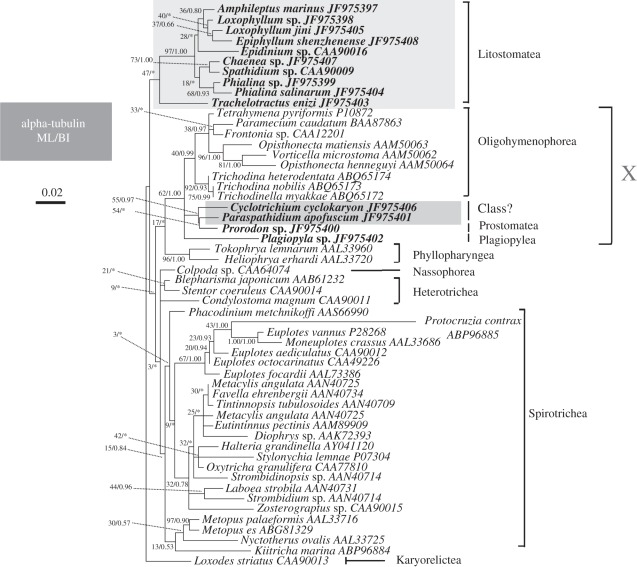
(e). Phylogenetic analyses of alpha-tubulin
The alpha-tubulin trees have more limited taxonomic sampling than the SSU rRNA trees, and are generally less well-resolved owing to the limited divergence between species. Nonetheless, we found broadly consistent phylogenetic patterns to those seen with SSU rRNA genes. The ML tree estimated for amino acid sequences is shown in figure 5. The nucleotide-level analyses based on the first two codon positions recover a similar topology to that estimated from amino acid sequences, except for the position of Trachelotractus (see below; data not shown), but statistical support is lower overall.
Figure 5.
ML tree for the 54-taxon alpha-tubulin dataset (amino acid-level analysis). Newly sequenced species are shown in bold text. Numbers at nodes are ML bootstrap values followed by BI posterior probabilities. Nodes not recovered by BI are designated by asterisks (*). The scale bar corresponds to 0.02 expected substitutions per site.
According to the amino acid analysis, C. cyclokaryon and Paraspathidium apofuscum are not closely related to other haptorids, or other Litostomatea. Instead, they show again a closer relationship to Prostomatea, Plagiopylea and Oligohymenophorea (i.e. clade X). In the ML tree, Cyclotrichium and Paraspathidium are most closely related to the prostomatean Prorodon, although this clade is not strongly supported (55% ML, 0.97 BI). These taxa then cluster with Oligohymenophorea and Plagiopylea (represented by the new Plagiopyla sequence) with limited support (62% ML, 1.00 BI).
The other litostomes, with the exception of T. entzi, form a strongly supported clade (97% ML, 1.00 BI). Trachelotractus entzi branches with this clade in the ML analyses, although this position is poorly supported (47% ML), but branches basally (i.e. in a clan with the karyorelictid Loxodes striatus) in the Bayesian analyses and in the DNA-based phylogenies. Within the litostome clade Chaenea is not recovered as deep branch, but instead shows a close relationship to Spathidium, with some support (73% ML, 1.00 BI); these two branch with Phialina spp., with almost no support. The included pleurostomatids (Loxophyllum spp., Epiphyllum shenzhenense and Amphileptus marinus) form a very weakly supported monophyletic group (36% ML, 0.80 BI). The trichostomatian Epidinium branches as sister to pleurostomatids in the ML tree, but support is negligible.
(f). Combined analyses
We also estimated phylogenies for a combined dataset of the three examined genes; the dataset included 18 ciliate genera/species, plus outgroups, with sampling of ciliates similar to the 28S rRNA analyses. The recovered topology (electronic supplementary material, figure S7) was largely similar to the 28S rRNA topology: Cyclotrichium and Paraspathidium formed a strongly supported clade, and grouped strongly with Prorodon, then with Plagiopyla and Oligohymenophorea to form a strongly supported ‘clade X’ (all ‘strongly supported’ clades receiving 98–100% ML bootstrap support). Trachelotractus and Chaenea again branched successively in the two most basal positions in the ‘true’ litostome clade, with 100% bootstrap support for both positions. Pleurostomatida, represented by Loxophyllum spp., Epiphyllum and Amphileptus, was again monophyletic, with full support.
(g). Hypothesis testing
Approximately unbiased tests were performed on each of the four datasets to test the robustness of phylogenetic associations of particular interest. A clade of Cyclotrichium and Paraspathidium with true litostomes was clearly rejected with the SSU rRNA, LSU rRNA and combined datasets (p < 0.0001 in all cases; electronic supplementary material, table S2). Trees with Chaenea basal within Litostomatea, rather than Trachelotractus, were also rejected with the SSU rRNA, LSU rRNA and combined datasets (p = 0.026 for SSU rRNA, otherwise p < 0.0001; electronic supplementary material, table S2). Neither hypothesis was rejected with the alpha-tubulin dataset alone, probably reflecting the limited information in this dataset.
3. Discussion
(a). Cyclotrichium and Paraspathidium: a separation from the class Litostomatea
Our analyses of all three examined genes examined reject a placement of both Cyclotrichium and Paraspathidium in the order Haptorida or even in the class Litostomatea. Rather, these two taxa always fall into a well-supported clade X. The predicted secondary structures of V4 regions of the SSU rRNA gene are consistent with this finding. These results, together with the earlier work using SSU rRNA data from Paraspathidium alone [10] indicate strongly that both taxa should be transferred out of the class Litostomatea. They cannot at this stage be placed in any existing order-level taxon or even class. Resolution of their higher taxonomic status should be made once the precise interrelationships between Cyclotrichium, Paraspathidium, plagiopyleans, prostomateans and oligohymenophoreans are resolved, since these varied between our analyses. Improved taxon sampling in this region of the tree for multiple genes would be valuable.
The fact that neither Cyclotrichium nor Paraspathidium are haptorid litostomes is broadly consistent with their rather unusual morphological characters. Cyclotrichium has a similar general appearance to core members of the haptorid family Didiniidae (i.e. Didinium and Monodinium) because it has an anterior circumoral ciliary girdle and an oval or semi-globular body shape [17]. However, Cyclotrichium is distinguished from these organisms by the densely ciliated cell surface (versus completely ‘naked’ except the ciliary girdle in Didiniidae), by the anterior proboscis, which is huge, flattened or slightly domed (versus long and conical in Didiniidae), and by the lack of DBs [8,17]. The absence of a DB is of special significance, since this feature is regarded as a synapomorphy for Litostomatea, and one of the ancestral features among the typical haptorians [27], though see below. Paraspathidium, meanwhile had been regarded as a gymnostome haptorid based on its Spathidium-like general appearance, the DB and the oralized somatic kinetids [19]. However, this taxon also has several characters found in most prostomateans, for example, a complex contractile vacuole and a dikinetidal perioral ciliature [19], which distinguishes it from typical haptorians. Since Paraspathidium is clearly not closely related to litostomes it would be interesting to re-examine the DB feature in this organism.
(b). Is Cyclotrichium related to cyclotrichiids?
Lynn [1,8] assigned Cyclotrichium to family Didiniidae because it has one girdle of ciliary kinetids, although it lacks a DB (see above). On the other hand, the absence of a DB is a character of the order Cyclotrichiida, as included in some other taxonomic systems, such as those of Foissner & Foissner [2] and Vd'acny et al. [14]. In these systems, Cyclotrichiida unites planktonic ciliates such as Askenasia, Mesodinium and Myrionecta, which have the cilia arranged in one or several girdles, but are without a DB [7,28,29]. However, in SSU rRNA gene phylogenies, Mesodinium and Myrionecta are extraordinarily divergent, and represent an extremely long branch that is not placed reliably within Ciliophora [9,13]. Unsurprisingly, we did not see any particular relationship between Mesodinium/Myrionecta and Cyclotrichium. Interestingly, however, our SSU rRNA gene phylogenies placed the partial sequences from Askenasia spp. close to Cyclotrichium and Paraspathidium, though not specifically with either. This hints at the possibility that Cyclotrichium may be related to at least some of the more familiar cyclotrichiids. If Askenasia and/or Cyclotrichium were to truly represent cyclotrichiids, the phylogenetic position of the cyclotrichiids might be closer to Oligohymenophorea, Prostomatea and Plagiopylea than to litostomes. Again, sequence data for ‘typical’ cyclotrichiids for markers other than SSU rRNA genes would be crucial.
(c). Trachelotractus and Chaenea as possible deep-branching litostomes
Trachelotractus was transferred to Litostomatea because it lacks all main infraciliary characteristics of trachelocercids and is more similar in this respect to members of the litostome order Haptorida (see §1), such as Helicoprorodon (Helicoprorodontidae) [23]. Our analyses, in which we added additional Trachelotractus sequences to the SSU rRNA dataset and considered different phylogenetic markers, further support the transfer of Trachelotractus from Karyorelictea to Litostomatea, and its deep-branching position [19,24]. Chaenea, meanwhile, was recently suggested by Vd'acny et al. [14] to be an ancestor-like form to Lacrymaria and Phialina in order Haptorida, based on a similar, but simpler morphology (e.g. no differentiation of ‘head’ region and trunk; no ‘head kineties’; only four rowed DBs). However, our analyses of several genes (except alpha tubulin), and previous phylogenies of SSU rRNA genes do not favour this scenario, with Chaenea instead forming an independent deep branch within Litostomatea [10,15]. The appropriate systematic positions of Trachelotractus and Chaenea within Litostomatea is uncertain—if the deep-branching positions recovered in most analyses are confirmed by future studies (see below), we could assume more confidently that the class Litostomatea originally evolved from a haptorid-like ancestor.
(d). New genes for phylogenetics of Litostomatea
We reported 19 new LSU rRNA genes and alpha-tubulin genes from the subclass Haptoria. In addition to the positions of problematic or potentially deep-branching haptorian litostomes discussed above, the new datasets give a broadly compatible view of litostome phylogeny to that seen with SSU rRNA genes. In particular, both LSU rRNA genes and alpha-tubulin proteins also recover Pleurostomatida as a monophyletic group. At this stage, however, the taxon sampling of markers other than SSU rRNA is still limited, and this precludes in-depth testing of several interesting hypotheses. In particular, it would be important to obtain sequences of the new markers from unsampled taxa of typical haptorids (e.g. families Dileptidae, genus Helicoprorodon), and from (additional) Trichostomatia. This would allow analyses to better resolve the evolutionary trends within litostomes. As discussed above, it would be especially important to examine the phylogenetic placement of Mesodinium, Myrionecta and Askenasia with markers other than SSU rRNA genes. This current study therefore, represents a useful foundation on which a more robust understanding of litostome phylogeny, diversity and evolution might be built.
(e). Perspectives
The current view of ciliate diversity subdivides the group into a small number of classes [1]. These classes are very much viewed as the fundamental evolutionary groups of ciliates, analogous to the division of animals into phyla. The positions of Cyclotrichium and Paraspathidium in multiple gene phylogenies illustrate that the current catalogue of ciliate classes is incomplete—very likely it will be necessary to recognize at least one class-level taxon to accommodate these organisms. This may well be an important group from a scientific standpoint—as discussed above, it might represent the true phylogenetic home of the photosynthetic Mesodinium/Myrionecta group—which are ecologically important and a fascinating evolutionary enigma [13]. Meanwhile, multiple gene phylogenies emphasize the large disparity between taxonomy and phylogeny within the true litostomes. In short, despite more than two decades of increasingly sophisticated molecular phylogenetics, the higher level phylogeny of ciliates remains substantially under-resolved. A much greater commitment to employing multiple phylogenetic markers, in parallel with improved taxon sampling, is almost certainly needed to understand the evolutionary history of this major group of eukaryotic organisms.
4. Material and methods
(a). Ciliate collection and identification
Cyclotrichium cyclokaryon, Phialina salinarum, T. entzi, Chaenea sp., E. shenzhenense and Loxophyllum jini were collected from the sandy beach of Daya Bay, Guangzhou, southern China (22°42′ N, 114°32′ E) between March 2007 and November 2009. Amphileptus marinus, Chaenea vorax, Loxophyllum sp., Paraspathidium apofuscum, Phialina sp., Plagiopyla sp. and Trachelotractus sp. were collected from sandy beaches on Jiaozhou Bay, Qingdao, China (36°08′ N, 120°43′ E) between July 2007 and November 2009. Samples were collected from the upper 0–4 cm sand layer. The specimens were investigated in vivo and impregnated with protargol following the methods of Wilbert [30] (figure 1). Species identifications of Cyclotrichium, Phialina, Trachelotractus, Paraspathidium, Loxophyllum and Chaenea were based on Long et al. [31] and Pan et al. [15]. Identification of Plagiopyla sp. was based on Lynn & Small [8]. Prorodon sp. was kindly offered by Dr Xinlu Shi (Hangzhou Normal University, China) and was collected from a freshwater puddle in the Xinjiang province of China (45°38′ N, 86°2′ S) in May 2007, and identification was following Lynn & Small [8]. Terminology and systematic classification in the present work are according to Lynn's 2008 system [1].
(b). DNA extraction, gene amplification and gene sequencing
After the identification based on several cells, one or more identical cells of each species from the same sample were isolated for DNA extraction. Genomic DNA was extracted using a REDExtract-NAmp Tissue PCR Kit (Sigma, St Louis, USA) with modifications suggested by Zhang et al. [20]. Primers used for SSU rRNA gene amplification were Euk A and Euk B [32], covering nearly the full length of the gene. Primers for partial LSU rRNA gene amplification were 28S-F2 (5′–ACSCGCTGRAYTTAAGCAT–3′) and 28S-R2: (5′–AACCTTGGAGACCTGAT–3′) [33]. The partial alpha-tubulin gene was amplified using the forward primer Tub-1 (5′–AAGGCTCTCTTGGCGTACAT–3′) and the reverse primer Tub-2 (5′–TGATGCCTTCAACACCTTCTT–3′) [34] for Paraspathidium apofuscum, C. cyclokaryon, Phialina salinarum, Phialina sp. and T. entzi, with PCR conditions following Yi et al. [35]. A different primer pair was used for Prorodon sp. and Plagiopyla sp.: Tub 371 (5′–(CUA)4 ATH CAN CCN GAY GGN CAR ATG CC) and Tub 4092 (5′ –(CAU)4 CAT NCC YTC NCC NAC RWA CCA–3′) [36]. After confirmation of the appropriate size of the amplified fragments (1.7 kb for the SSU rRNA gene, 1.9 kb for the LSU rRNA gene and 1.1 kb for the alpha-tubulin gene) on an agarose gel, each PCR product was cloned using a pUCm-T cloning vector (Sangon Company, Shanghai, China). Genes were sequenced in both directions on an ABI 3700 sequencer (Invitrogen sequencing facility, Shanghai, China), using the M13–47 and M13–48 primers. All new sequences have been deposited in the GenBank database (see the electronic supplementary material, table S1 for accession numbers).
(c). Phylogenetic analyses
(i). Analysis for small-subunit ribosomal RNA and large-subunit ribosomal RNA nucleotide sequences
The sequences of the SSU rRNA gene and LSU rRNA gene were aligned using ClustalW, as implemented in Bioedit v. 7.0.0 [37], and further modified manually using Bioedit. The datasets used for the primary phylogenetic analyses included 1495 positions for SSU rRNA and 1712 positions for LSU rRNA. Modeltest [38] and MrModeltest v. 2 [39] were used to select the best models for the ML analyses and BI. The ML trees were estimated with the PhyML v. 2.4.4 program [40] using a GTR + I + Γ model (pinvar = 0.25, α = 0.54 for SSU rRNA; pinvar = 0.11; α = 0.74 for LSU rRNA). The reliability of internal branches was assessed using non-parametric bootstrapping with 1000 replicates. BI was performed with MrBayes v. 3.1.2 [41], under a GTR + I + Γ model. Markov chain Monte Carlo (MCMC) simulations were run with two sets of four chains using the default settings, with a sampling frequency of 0.01. In each case, convergence was confirmed from the standard deviation of split frequencies (less than 0.01), and 25 per cent of generations were discarded as burn-in. A MP tree was constructed for each gene using PAUP [42]. The MP trees were found using heuristic searches with 100 random-addition sequences, and tree bisection and reconnection branch swapping. Bootstrap support was calculated from 1000 replicates.
The previously determined SSU rRNA gene sequences from cyclotrichiids—from Askenasia, Mesodinium and Myrionecta—were excluded from the primary analyses because they were only partial, or were extraordinarily divergent. To test the possible relationships between Cyclotrichium and these other cyclotrichiids, three sets of supplementary phylogenetic analyses were performed using ML and BI methods. In the first Mesodinium pulex, My. rubra and Askenasia spp. were added to the primary SSU dataset (86 species; 1436 included sites). In the second, Me. pulex, My. rubra and Askenasia were included but the dinoflagellate outgroups excluded (84 species; 1436 included sites). In set 3, the extremely long branches of Me. pulex and My. rubra were excluded, but the partial Askenasia sequences were retained, along with the outgroups (83 species; 1495 included sites). The same GTR + I + Γ models and other parameters were used as in the primary analyses.
The secondary structures of the V4 region of the SSU rRNA molecules were depicted and compared for Cyclotrichium and representative species of potentially related classes, including six plagiopyleans, seven prostomateans, seven haptorians and Paraspathidium. Information on the secondary structure of Mesodinium from Strüder-Kypke [9] is used for comparison. Default settings of the mfold website (http://mfold.bioinfo.rpi.edu/cgi-bin/rna-form1-2.3.cgi) [43] were used to produce the putative secondary structures of the V4 region. The structures were edited with RnAviz v. 2.0 [44] for aesthetic purposes under the newest eukaryotic SSU V4 model of Wuyts [26]. Phylogenetic trees based on the primary sequence and the secondary structure of the V4 region were constructed following the instructions on the MARNA website (http://biwww2.informatik.uni-freiburg.de/Software/MARNA/index.html)[45].
(ii). Analysis of alpha-tubulin proteins
The deduced amino acid translations of the alpha-tubulin gene sequences were aligned using ClustalW implemented in Bioedit v. 7.0.0, then inspected by eye and manually edited. No introns were detected in the new sequences. Three hundred and fifty-seven positions were included in the final sequence alignment. Phylogenies based on the amino acid sequences were constructed using ML, BI and MP methods. The MP tree showed very poor resolution and is not reported further. The ML tree and corresponding bootstrap support values (1000 replicates) were estimated using PhyML v. 2.4.4 [40], applying a JTT + Γ model (α = 0.64), which was selected as the best model using ProtTest v. 1.4 [46]. Amino acid alignments were also analysed in MrBayes with the amino acid model selected by the software. The MCMC simulations were run with two sets of four chains using the default settings. Chains were run for 1 × 106 generations, with a sampling frequency of 0.01, with the first 25 per cent discarded as burn-in. In addition, the first- and second-codon positions of the DNA sequences were analysed by ML using PhyML, under a GTR + I + Γ model (pinvar = 0.34; α = 0.43). Bootstrap analyses were performed on 1000 resampling replicates using identical settings.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Project no. 40976075), and the Canadian Institute for Advanced Research (CIfAR) Program in Integrated Microbial Biodiversity. Many thanks are due to Ms Feng Gao and Ms Jie Huang (Laboratory of Protozoology, OUC) for help with gene sequencing. We also want to give our appreciation to Dr Zhenzhen Yi (South China Normal University) for advice on phylogenetic analysis.
References
- 1.Lynn D. H. 2008. The ciliated protozoa: characterization, classification and guide to the literature, 3rd edn. Dordrecht, The Netherlands: Springer [Google Scholar]
- 2.Foissner W., Foissner I. 1988. The fine structure of Fuscheria terricola Berger et al., 1983 and a proposed new classification of the subclass Haptoria Corliss, 1974 (Ciliophora, Litostomatea). Arch Protistenkd 135, 213–235 10.1016/S0003-9365(88)80070-8 (doi:10.1016/S0003-9365(88)80070-8) [DOI] [Google Scholar]
- 3.Lin X., Song W., Wilbert N., Warren A. 2005. Two new marine pleurostomatid ciliates from China, Loxophyllum jini and L. qiuianum (Ciliophora, Pleurostomatida). Acta Protozool. 44, 147–157 [Google Scholar]
- 4.Gustafson D. E., Jr, Stoecker D. K., Johnson M. D., Van Heukelem W. F., Sneider K. 2000. Cryptophyte algae are robbed of their organelles by the marine ciliate Mesodinium rubrum. Nature 405, 1049–1052 10.1038/35016570 (doi:10.1038/35016570) [DOI] [PubMed] [Google Scholar]
- 5.Hansen P. J., Fenchel T. 2006. The bloom-forming ciliate Mesodinium rubrum harbours a single permanent endosymbiont. Mar. Biol. Res. 2, 169–177 10.1080/17451000600719577 (doi:10.1080/17451000600719577) [DOI] [Google Scholar]
- 6.Johnson M. D., Oldach D., Delwiche C. F., Stoecker D. K. 2007. Retention of transcriptionally active cryptophyte nuclei by the ciliate Myrionecta rubra. Nature 445, 426–428 10.1038/nature05496 (doi:10.1038/nature05496) [DOI] [PubMed] [Google Scholar]
- 7.Foissner W., Berger H., Schaumburg J. 1999. Identification and ecology of limnetic plankton ciliates. München, Germany: Informationsberichte des Bayer, Landesamtes für Wasserwirtschaft [Google Scholar]
- 8.Lynn D. H., Small E. B. 2002. Phylum Ciliophora Doflein, 1901. In An illustrated guide to the protozoa (eds Lee J. J., Leedale G. F., Bradbury P.), pp. 371–656, 2nd edn Lawrence, KS: Society of Protozoologists [Google Scholar]
- 9.Strüder-Kypke M. C., Wright A. D., Foissner W., Chatzinotas A., Lynn D. H. 2006. Molecular phylogeny of litostome ciliates (Ciliophora, Litostomatea) with emphasis on free-living haptorian genera. Protist 157, 261–278 10.1016/j.protis.2006.03.003 (doi:10.1016/j.protis.2006.03.003) [DOI] [PubMed] [Google Scholar]
- 10.Gao S., Song W., Ma H., Clamp J., Yi Z., Al-Rasheid K., Chen Z., Lin X. 2008. Phylogeny of six genera of the subclass Haptoria (Ciliophora, Litostomatea) inferred from sequences of the gene coding for small subunit ribosomal RNA. J. Eukaryot. Microbiol. 55, 562–566 10.1111/j.1550-7408.2008.00360.x (doi:10.1111/j.1550-7408.2008.00360.x) [DOI] [PubMed] [Google Scholar]
- 11.Cameron S. L., O'Donoghue P. J. 2004. Phylogeny and biogeography of the ‘Australian’ trichostomes (Ciliophora: Litostomata). Protist 155, 215–335 10.1078/143446104774199600 (doi:10.1078/143446104774199600) [DOI] [PubMed] [Google Scholar]
- 12.Gao F., Fan X., Yi Z., Strüder-Kypke M., Song W. 2010. Phylogenetic consideration of two scuticociliate genera, Philasterides and Boveria (Protozoa, Ciliophora) based on 18S rRNA gene sequences. Parasitol. Int. 59, 549–555 10.1016/j.parint.2010.07.002 (doi:10.1016/j.parint.2010.07.002) [DOI] [PubMed] [Google Scholar]
- 13.Johnson M. D., Tengs T., Oldach D. W., Delwiche C. F., Stoecker D. K. 2004. Highly divergent SSU rRNA genes found in the marine ciliates Myrionecta rubra and Mesodinium pulex. Protist 155, 347–359 10.1078/1434461041844222 (doi:10.1078/1434461041844222) [DOI] [PubMed] [Google Scholar]
- 14.Vd'acny P., Bourland W. A., Orsi W., Epstein S. S., Foissner W. 2011. Phylogeny and classification of the Litostomatea (Protista, Ciliophora), with emphasis on free-living taxa and the 18S rRNA gene. Mol. Phylogenet. Evol. 59, 510–522 10.1016/j.ympev.2011.02.016 (doi:10.1016/j.ympev.2011.02.016) [DOI] [PubMed] [Google Scholar]
- 15.Pan H., Gao F., Li J., Lin X., Al-Farraj S. A., Al-Rasheid K. A. S. 2011. Morphology and phylogeny of two new pleurostomatid ciliates, Epiphyllum shenzhenense n. sp. and Loxophyllum spirellum n. sp. (Protozoa, Ciliophora) from a mangrove wetland, south China. J. Eukaryot. Microbiol. 57, 421–428 10.1111/j.1550-7408.2010.00492.x (doi:10.1111/j.1550-7408.2010.00492.x) [DOI] [PubMed] [Google Scholar]
- 16.Lin X., Song W., Warren A. 2009. Litostomatea. In Free-living ciliates in the Bohai and Yellow Seas, China (eds Song W., Warren A., Hu X.), pp. 19–134 Beijing, China: Science Press [Google Scholar]
- 17.Xu D., Song W., Hu X. 2005. Morphology of Cyclotrichium taniguchii sp. nov. and C. cyclokaryon with establishment of a new genus, Dicyclotrichium gen. nov. (Ciliophora: Haptorida). J. Mar. Biol. Assoc. UK 85, 787–794 10.1017/S0025315405011719 (doi:10.1017/S0025315405011719) [DOI] [Google Scholar]
- 18.Long H., Song W., Al-Rasheid K. A., Gong J. 2009. Three marine haptorid ciliates from northern China: Paraspathidium apofuscum n. sp., Trachelotractus entzi (Kahl, 1927) Foissner, 1997 and Apotrachelotractus variabialis Long, Song and Warren, 2009 (Protozoa, Ciliophora). J. Nat. Hist. 43, 1749–1761 10.1080/00222930902781038 (doi:10.1080/00222930902781038) [DOI] [Google Scholar]
- 19.Foissner W. 1997. Infraciliature and systematic position of the marine interstitial ciliates (Protozoa, Ciliophora) Lopezoterenia torpens (Kahl, 1931) nov. gen., nov. comb., Discotricha papillifera Tuffrau, 1954, and Paraspathidium fuscum (Kahl, 1928) Fjeld, 1955. Rev. Soc. Mex. Hist. Nat. 47, 41–63 [Google Scholar]
- 20.Zhang Q., Yi Z., Song W., Al-Rasheid K. A. S., Warren A. 2010. The systematic position of Paraspathidium Noland, 1937 (Ciliophora, Litostomatea?) inferred from primary SSU rRNA gene sequences and predicted secondary rRNA structure. Eur. J. Protistol. 46, 280–288 10.1016/j.ejop.2010.05.001 (doi:10.1016/j.ejop.2010.05.001) [DOI] [PubMed] [Google Scholar]
- 21.Carey P. G. 1992. Marine interstitial ciliates: an illustrated key. London, UK: Chapman and Hall [Google Scholar]
- 22.Kahl A. 1930. Urtiere oder Protozoa I: Wimpertiere oder Ciliata (Infusoria) 1. Allgemeiner Teil und Prostomata. Die Tierwelt der Deutschlands 18, 1–180 [Google Scholar]
- 23.Foissner W. 1997. Updating the Trachelocercids (Ciliophora, Karyorelictea). IV. Transfer of Trachelocerca entzi Kahl, 1927 to the Gymnostomatea as a new genus, Trachelotractus gen. n. (Helicoprorodontidae). Acta Protozool. 36, 63–74 [Google Scholar]
- 24.Gao S., Strüder-Kypke M., Al-Rasheid K., Lin X., Song W. 2010. Molecular phylogeny of three ambiguous ciliate genera: Kentrophoros, Trachelolophos and Trachelotractus (Alveolata, Ciliophora). Zool. Scr. 39, 305–313 10.1111/j.1463-6409.2010.00416.x (doi:10.1111/j.1463-6409.2010.00416.x) [DOI] [Google Scholar]
- 25.Baroin-Tourancheau A., Delgado P., Perasso R., Adoutte A. 1992. A broad molecular phylogeny of ciliates: identification of major evolutionary trends and radiations within the phylum. Proc. Natl Acad. Sci. USA 89, 9764–9768 10.1073/pnas.89.20.9764 (doi:10.1073/pnas.89.20.9764) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wuyts J., De Rijk P., Van de Peer Y., Pison G., Rousseeuw P., De Wachter R. 2000. Comparative analysis of more than 3000 sequences reveals the existence of two pseudoknots in area V4 of eukaryotic small subunit ribosomal RNA. Nucl. Acids Res. 28, 4698–4708 10.1093/nar/28.23.4698 (doi:10.1093/nar/28.23.4698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vd'ačný P., Orsi W., Foissner W. 2010. Molecular and morphological evidence for a sister group relationship of the classes Armophorea and Litostomatea (Ciliophora, Intramacronucleata, Lamellicorticata infraphyl. nov.), with an account on basal haptorid litostomateans. Eur. J. Protistol. 46, 298–309 10.1016/j.ejop.2010.07.002 (doi:10.1016/j.ejop.2010.07.002) [DOI] [PubMed] [Google Scholar]
- 28.Corliss J. O. 1979. The ciliated protozoa: characterization, classification and guide to the literature, 2nd edn. Oxford, UK: Pergamon Press [Google Scholar]
- 29.Krainer K. H., Foissner W. 1990. Revision of the genus Askenasia Blochmann, 1895, with proposal of two new species and description of Rhabdoaskenasia minima n. g., n. sp. (Ciliophora, Cyclotrichida). J. Protozool. 37, 414–427 10.1111/j.1550.7408.1990.tb01166x (doi:10.1111/j.1550.7408.1990.tb01166x) [DOI] [Google Scholar]
- 30.Wilbert N. 1975. Eine verbesserte Technik der Protargolimpragnation für Ciliaten. Mikrokosmos 64, 171–179 [Google Scholar]
- 31.Long H., Song W., Warren A. 2009. Haptorid and other lower kinetofragminophoran ciliates. In Free-living ciliates in the Bohai and Yellow Seas, China (eds Song W., Warren A., Hu X.), pp. 19–47. Beijing, China: Science Press [Google Scholar]
- 32.Medlin L., Elwood H. J., Stickel S., Sogin M. L. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71, 491–499 10.1016/0378-1119(88)90066-2 (doi:10.1016/0378-1119(88)90066-2) [DOI] [PubMed] [Google Scholar]
- 33.Moreira D., von der Heyden S., Bass D., López-Garcia P., Chao E., Cavalier-Simith T. 2007. Global eukaryote phylogeny: combined small- and large-subunit ribosomal DNA trees support monophyly of Rhizaria, Retaria and Excavata. Mol. Phylogenet. Evol. 44, 255–266 10.1016/j.ympev.2006.11.001 (doi:10.1016/j.ympev.2006.11.001) [DOI] [PubMed] [Google Scholar]
- 34.Yi Z., Song W., Stoeck T., Al-Rasheid K., Al-Khedhairy A., Gong J., Ma H., Chen Z. 2009. Phylogenetic analyses suggest that Psammomitra (Ciliophora, Urostylida) should represent an urostylid family, based on SSrRNA and alpha-tubulin gene sequence information. Zool. J. Linn. Soc. 157, 227–236 10.1111/j.1096-3642.2008.00524.x (doi:10.1111/j.1096-3642.2008.00524.x) [DOI] [Google Scholar]
- 35.Yi Z., Clamp J., Al-Rasheid K., Al-Khedhairy A., Chen Z., Song W. 2009. Evolutionary relationship and species separation of four morphologically similar stichotrichous ciliates (Protozoa, Ciliophora). Prog. Nat. Sci. 19, 581–586 10.1016/j.pnsc.2008.05.033 (doi:10.1016/j.pnsc.2008.05.033) [DOI] [Google Scholar]
- 36.Riley J. L., Katz L. A. 2001. Widespread distribution of extensive genome fragmentation in ciliates. Mol. Biol. Evol. 18, 1372–1377 10.1093/oxfordjournals.molbev.a003921 (doi:10.1093/oxfordjournals.molbev.a003921) [DOI] [PubMed] [Google Scholar]
- 37.Hall T. A. 1999. Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41, 95–98 [Google Scholar]
- 38.Posada D., Crandall K. A. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818 10.1093/bioinformatics/14.9.817 (doi:10.1093/bioinformatics/14.9.817) [DOI] [PubMed] [Google Scholar]
- 39.Nylander J. A. 2004. MrModeltest v. 2. Uppsala, Sweden: Uppsala University, Evolutionary Biology Centre [Google Scholar]
- 40.Guindon S., Gascuel O. 2003. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 10.1080/10635150390235520 (doi:10.1080/10635150390235520) [DOI] [PubMed] [Google Scholar]
- 41.Ronquist F., Huelsenbeck J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 10.1093/bioinformatics/btg180 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- 42.Swofford D. L. 2002. PAUP*, Phylogenetic analysis using parsimony (*and other methods), Version 4. Sunderland, MA: Sinauer associates [Google Scholar]
- 43.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucl. Acids Res. 31, 3406–3415 10.1093/nar/gkg595 (doi:10.1093/nar/gkg595) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rijk P. D., Wachter R. D. 1997. RNAViz, a program for the visualisation of RNA secondary structure. Nucl. Acids Res. 25, 4679–4684 10.1093/nar/25.22.4679 (doi:10.1093/nar/25.22.4679) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siebert S., Backofen R. 2005. MARNA: multiple alignment and consensus structure prediction of RNAs based on sequence structure comparisons. Bioinformatics 21, 3352–3359 10.1093/bioinformatics/bti550 (doi:10.1093/bioinformatics/bti550) [DOI] [PubMed] [Google Scholar]
- 46.Abascal F., Zardoya R., Posada D. 2005. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105 10.1093/bioinformatics/bti263 (doi:10.1093/bioinformatics/bti263) [DOI] [PubMed] [Google Scholar]